Abstract
Several lines of evidence suggest that tumor cells show elevated activity of the NF-κB transcription factor, a phenomenon often resulting from constitutive activity of IκB kinase β (IKKβ). However, others have found that loss of NF-κB activity or IKKβ is tumor promoting. The role of NF-κB in tumor progression is therefore controversial and varies with tumor type. We sought to more extensively investigate the role IKKβ in melanoma tumor development by specifically disrupting Ikkb in melanocytes in an established mouse model of spontaneous melanoma, whereby HRasV12 is expressed in a melanocyte-specific, doxycycline-inducible manner in mice null for the gene encoding the tumor suppressor inhibitor cyclin-dependent kinase 4/alternative reading frame (Ink4a/Arf). Our results show that Ink4a/Arf–/– mice with melanocyte-specific deletion of Ikkb were protected from HRasV12-initiated melanoma only when p53 was expressed. This protection was accompanied by cell cycle arrest, with reduced cyclin-dependent kinase 2 (Cdk2), Cdk4, Aurora kinase A, and Aurora kinase B expression. Increased p53-mediated apoptosis was also observed, with decreased expression of the antiapoptotic proteins Bcl2 and survivin. Enhanced stabilization of p53 involved increased phosphorylation at Ser15 and reduced phosphorylation of double minute 2 (Mdm2) at Ser166. Together, our findings provide genetic and mechanistic evidence that mutant HRas initiation of tumorigenesis requires Ikkβ-mediated NF-κB activity.
Introduction
A germline mutation or deficiency in the gene that encodes inhibitor cyclin-dependent kinase 4/alternative reading frame (Ink4a/Arf), a tumor suppressor and inhibitor of the cell cycle, is frequently observed in melanoma-prone families (1, 2). Within that subgroup, the NRas gene mutation is found in 95% of primary familial melanomas (3). Interestingly, the p16Ink4a protein acts as an inhibitor of NF-κB/p65, while the Arf protein (p14Arf in human, p19Arf in mouse) activates the p53 tumor suppressor (4).
Human melanoma lesions that spontaneously arise where there is no familial genetic predisposition often exhibit both loss of the tumor suppressor INK4a/ARF and activating mutation in genes in the RAS/RAF/MAPK pathway ( http://www.sanger.ac.uk/genetics/CGP/cosmic/; Catalogue of Somatic Mutations in Cancer) (5–9). Without loss of the tumor suppressor INK4a/ARF or p53, the expression of a mutant NRAS or BRAF gene results in melanocyte senescence (10). Expression of RAS or RAF oncogenes induces the expression of inflammatory mediators, inhibitors of apoptosis, and growth factors, many of which are regulated by the transcription factor NF-κB (11–14).
There is growing evidence that tumor cells exhibit elevated NF-κB activity, often due to constitutive IκB kinase (IKK) activity (15–18). The IKK complex is mainly comprised of the catalytic subunits IKKα/1, IKKβ/2, and NF-κB essential modulator or IKKγ/3 (19, 20). Several studies, including our own, show that IKKβ is a key component in inflammation-based cancer progression (11, 12, 17, 21–24). In contrast, for some cell types, loss of NF-κB activity or IKKβ is tumor promoting (25, 26), thus adding to the confusion about the role of IKK and NF-κB in tumor progression (27, 28).
Interestingly, Aurora A kinase has been reported to regulate NF-κB by phosphorylating IκB (29). Aurora kinases are involved in the regulation of mitosis, and both Aurora A and Aurora B kinase are often amplified in tumors (30, 31). Aurora A kinase is involved in the maturation of the centromere and spindle orientation, while Aurora B kinase is required for appropriate kinetochore function during chromosome condensation and cohesion, spindle assembly, and bipolar attachment (32). Loss of either of these 2 kinases will result in cell cycle arrest, with loss of Aurora A causing arrest at the G2/M transition point and loss of Aurora B disrupting anaphase and telophase (32, 33). Inhibiting Aurora A kinase with RNAi or the inhibitor VE-465 (Merck) induces apoptosis in multiple myeloma cells, showing amplification of Aurora A kinase (34). Inhibiting Aurora A and Aurora B kinases with the inhibitor CCT129202 causes tumor cells to accumulate with a greater than 4N DNA content and undergo apoptosis, reduces double minute 2 (MDM2) levels, and induces the stability of p53 and p21. Inhibition of these aurora kinases also results in hypo-phosphorylation of RB, downregulation of thymidine kinase 1, reduced phosphorylation of histone H3, and increased cleavage of PARP (35).
When mutant HRas is expressed in melanocytes lacking Ink4a/Arf, there is tumor progression to melanoma (21, 36). Here, we show that a conditional knockout of Ikkb in Ink4a/Arf-null melanocytes expressing activated HRasV12 inhibits Aurora A kinase–mediated cell cycle progression and blocks melanoma tumor formation in FVB mice. This work demonstrates for what we believe to be the first time in a genetically modified mouse that Ikkβ is a key component in the development of melanoma. Moreover, we show that ablation of Ikkβ results in reduction in Aurora A kinase, inhibition of G2/M transition, stabilization of p53, reduction in IL-6, and increased apoptosis, demonstrating that Ikkβ contributes to tumorigenesis via mechanisms impacting cell cycle, apoptosis, and cytokine signaling. These data support the concept that IKKβ is a potential therapeutic target for melanoma.
Results
Conditional knockout of melanocyte Ikkb in the HRasV12-induced melanoma model.
The mouse model of spontaneous melanoma induced by activation of HRasV12 in mice with an underlying Ink4a/Arf-deficient background, developed by Chin et al. (36, 37), allowed us to analyze the role of the NF-κB pathway in melanoma tumorigenesis. Ikkβ has been recognized as a key mediator of the NF-κB signal transduction pathway. To generate conditional Ikkβ-deleted melanocytes in this HRasV12-induced melanoma model, we crossed FVB Ikkbflox/flox (Ikkbf/f) mice (38) with FVB TetO-Cre recombinase mice (39) to obtain the Ikkbf/f-TetO-Cre mouse strain. This strain was further interbred with FVB mice carrying TetOn-Hrasv12 on an Ink4a/Arf-null background (37) and the melanocyte tyrosinase promoter/enhancer (Tyr) controlled tetracycline reverse transcriptional activator (rtTA) (36). An outline describing this mouse model is illustrated in Figure 1A. Thus, the mice with Ikkbf/f:TetO-Cre:Tyr-rtTA:TetOn-Hrasv12:Ink4a/Arf–/– genetic background were named IkkbΔ/Δ mice, and their littermates lacking TetO-Cre were named Ikkbwt mice and used as a control group. The multiple transgenic and knockout animals were viable and developed normally into adulthood.
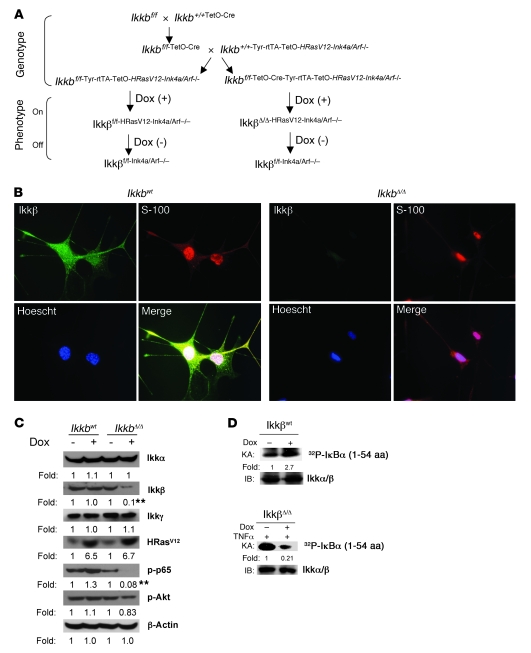
Figure 1. Generation of Ikkβ knockout mice and characterization of Ikkβ-depleted melanocytes of mice.
(A) Ikkbf/f mice crossed with TetOn-Cre mice resulted in the Ikkbf/f-TetO-Cre strain of mice. These mice were further interbred to obtain the additional genetic background of TetOn-Hrasv12, Ink4a/Arf–/–, and melanocyte-specific Tyr-rtTA. These mice were named IkkbΔ/Δ mice, and their counterparts lacking TetOn-Cre were used as control Ikkbwt mice. On refers to tetracycline/doxycycline-inducible expression of Cre and/or Hrasv12 gene in the TetOn system. Off refers to absence of Cre and/or Hrasv12-inducible gene expression when tetracycline/doxycycline is absent. (B) E19 melanocytes isolated from Ikkbwt and IkkbΔ/Δ mice were treated with doxycycline (Dox) and stained with specific antibodies to Ikkβ (green) or S-100 (red), and nuclei were counterstained with Hoechst (blue). Original magnification, ×40. (C) Expression of the indicated proteins and their phosphorylation status were analyzed by immunoblotting, after the above cells were treated or not treated with doxycycline. Blot band densities from cultures treated with doxycycline were quantitated and normalized to the blot band density from cells of the same genetic background without doxycycline-induced HRasV12 expression. The fold change (fold) represents the value of 3 independent experiments and was statistically analyzed. **P < 0.01. (D) Cellular Ikk kinase activity (KA) was examined in vitro by monitoring phosphorylation of the GST-IκB (aa 1–54) substrate by the immunoprecipitated Ikk complex. 32P-labeled–Iκbα (32P-Iκbα) indicates phosphorylated GST-IκB (aa 1–54). Fold indicates the ratio of the immunoblotted protein band density from the doxycycline-treated culture to the without doxycycline treatment control.
To examine the efficiency of Cre/loxP-mediated deletion of Ikkb (IkkbΔ/Δ mice), melanocytes were isolated from mouse fetal skin at E19, a time when a small population of melanocytes is distributed in the skin. When the cultured melanocytes isolated from E19 embryos derived from IkkbΔ/Δ mice were treated with 1 μg/ml doxycycline, the Ikkβ subunit of the Ikk complex was reduced by over 90% in IkkbΔ/Δ melanocytes, as compared with that in melanocytes not treated with doxycycline (Figure 1, B and C). The levels of the Ikkα and Ikkγ subunits in the Ikk complex remained unchanged (Figure 1C), and the expression of HRasV12 mutant protein was induced with doxycycline treatment (Figure 1, B and C). Cre-mediated intracellular Ikkb deletion, resulting from exposure of the cells to 1 μg/ml doxycycline for 4 days, decreased NF-κB signal transduction, as shown by reduced p65 (Ser536) phosphorylation. In contrast, there was only a slight change in phospho-Akt (Ser473) (Figure 1C). To examine the potential Ikk activity induced by the expression of HRasV12 or reduced by Ikkb knockout in melanocytes, the Ikkbwt and IkkbΔ/Δ cells were treated with or without 1 μg/ml doxycycline for 4 days, and IkkbΔ/Δ cells were additionally stimulated with 20 ng/ml of TNF-α for 20 minutes prior to collection of cells. The kinase activity of the immunoprecipitated Ikk complex was determined, as described in Methods, using the glutathione-S-transferase–IκB (GST-IκB) (aa 1–54) protein as a substrate. A 2.7-fold elevation of Ikk activity was observed when the HRasV12 protein expression was induced by doxycycline in Ikkbwt melanocytes (Figure 1D). In contrast, there was a 5-fold reduction in TNF-α induction (20 ng/ml) of Ikk kinase activity when melanocytes were null for Ikkβ but expressing HRasV12 (due to doxycycline induction of TetO-Cre and Hrasv12 in melanocytes expressing Tyr-rtTA) (Figure 1D). The loss of Ikkβ activity is likely responsible for the ablation of the doxycycline-induced HRasV12-mediated signal transduction that leads to phosphorylation of p65 (Figure 1C). There is much less phosphorylation of IκBα by this immunoprecipitated complex (Figure 1D, bottom panels) than is observed when both Ikkα and Ikkβ are present in the immunoprecipitate (Figure 1D, top panels). These data are in agreement with prior studies showing that Ikkβ is more efficient at phosphorylating the GST-1–54 fragment of IκBα than Ikkα (40). This mouse model affords an efficient approach to analyze the role of Ikkβ on the NF-κB pathway involved in melanoma tumorigenesis.
Conditional deletion of Ikkb in melanocytes blocks HRas-induced melanoma tumorigenesis.
To assess the role of Ikkβ in melanoma, we used a doxycycline-inducible model, in which melanocytes with loss of the tumor suppressor Ink4a/Arf are induced to express mutant HRasV12 protein (37). Here, a cohort of 60 Ikkbwt mice at 1 month of age were treated with 1 mg/ml doxycycline in drinking water for 120 days. This resulted in 28 cases of melanoma (47% incidence), with a latency of 64 ± 20 days, whereas no skin lesions occurred in the same genetic background of Ikkbwt mice without doxycycline induction (Figure 2A). To determine the impact of Ikkb knockout on the HRas-initiated formation of melanoma, 65 IkkbΔ/Δ mice were continuously treated with doxycycline (1 mg/ml in the drinking water). The doxycycline induction of Cre-recombinase expression in melanocytes resulted in deletion of Ikkb in melanocytes. The doxycycline treatment also resulted in the targeted expression of HRasV12 in melanocytes. Moreover, when Ikkb was deleted by this Cre/LoxP-mediated gene targeting, there was a 10-fold reduction in melanoma incidence (only 4.6%; P < 0.01), with a delayed latency of 79 ± 1.7 days (Table 1). Statistical analysis showed that cumulative melanoma incidence was remarkably different between IkkbΔ/Δ and Ikkbwt mice (Figure 2A). For the few tumors that did form in the doxycycline treatment IkkbΔ/Δ group, Western blot analysis revealed that Ikkβ expression was not different than that observed in the Ikkbwt group (Figure 2B). Doxycycline induction of mutant HRas expression in FVB Ikkbwt mice (no Cre-recombinase expression) resulted in melanoma lesions located in the face (data not shown) and ear, tail, back, and anus (Figure 2, C–F). Hair was absent on the surface of all the melanoma tumors (Figure C–F). In contrast, neither melanoma, nor skin lesions were observed in the Ikkbwt or IkkbΔ/Δ mice without doxycycline in the drinking water.
Figure 2. Deletion of Ikkb blocks HRasV12-induced melanocyte transformation.
(A) The cumulative incidence of melanoma versus latency in the FVB mice following doxycycline-induced the deletion of Ikkb and/or expression of HRasV12 was analyzed according to the method described by Gray (80). Melanoma incidence among doxycyline-treated Ikkbwt mice reached 40% during 120 days compared with only 5% melanoma in IkkbΔ/Δ mice treated with doxycycline. (B) Western blot analysis of tumors from Ikkbwt and IkkbΔ/Δ mice treated with doxycycline to induce expression of mutant Ras and Cre recombinase. Tumor lysates from 3 tumors of either Ikkbwt or IkkbΔ/Δ mice were subjected to Western blot analysis and staining with antibody to murine Ikkβ and Ikkγ. (C) Spontaneous amelanotic melanoma (arrow) arising in the ear. (D) Spontaneous amelanotic melanomas (arrows) in tail and back. (E) Spontaneous amelanotic melanoma skin tumors (arrows) arising in the back skin. (F) Spontaneous amelanotic melanoma skin tumors (arrows) in the anus.
Table 1 .
IKKβ signature of melanoma tumorigenesis
Of note, melanocyte-specific deletion of Ikkb did not prevent formation of angiosarcoma lesions induced by loss of Ink4a/Arf (Supplemental Results, Supplemental Table 1, and Supplemental Figure 1, A–C; supplemental material available online with this article; doi: 10.1172/JCI42358DS1). Histological analysis of the melanoma lesions that arose in the doxycycline-treated Ikkbwt mice indicated that the hair follicle structure was replaced with tumor cells, suggesting melanoma arose in the melanocyte-rich hair follicle (Figure 3B). In contrast, the skin of the IkkbΔ/Δ mice was normal (Figure 3C). The spontaneous melanoma lesions were distinguished from angiosarcoma lesions by the absence of immunohistochemical (IHC) staining in the tumor proper with antibody to factor VIII, while the blood vessels were factor VIII positive (Figure 3D), and the tumor cells exhibited positive IHC staining with S-100 (Figure 3E), indicating that they were of neural crest origin. Since both melanoma cells and normal melanocytes (Figure 3F) show a positive reaction with S-100, the expression of a melanoma-specific marker, melanoma antigen recognized by T cells (Mart1), was examined to distinguish the transformed melanocytes (or melanoma cells) (Figure 3, G and H) from normal melanocytes that were negative for Mart1 (Figure 3I).
Figure 3. Distinct features of melanoma lesions.
(A) Melanoma lesions arising on the tail were photographed. (B and C) Sections of melanoma (B) and nonmelanoma skin (C) were stained with H&E. (D–I) Immunohistochemistry of factor VIII in melanoma tissue (D), S-100 in melanoma section (E) and nonmelanoma skin (F), and Mart1 in melanoma section (G and H) and nonmelanoma skin (I). Note the thickening of the epidermis over the cutaneous melanoma lesion at the arrows. Original magnification, ×20 (numerical aperture, 0.5) (B and C); ×40 (numerical aperture, 1.0) (D–I).
Knockout of Ikkb initiates apoptosis.
To analyze the mechanism by which conditional deletion of Ikkb in melanocytes resulted in blockage of HRasV12-induced melanoma formation, the E19 melanocytes from IkkbΔ/Δ and Ikkbwt mice were cultured in 1 μg/ml doxycycline, which induced HRasV12 protein expression and greatly reduced expression of Ikkβ. After 4 days of treatment with doxycycline (1 μg/ml), cells were lysed and subjected to Western blot analysis to examine the expression of apoptotic proteins. With doxycycline induction of Hrasv12 and Cre-mediated deletion of the floxed Ikkb in melanocytes, there was increased apoptosis based upon analysis of annexin V staining (Figure 4A). This was accompanied by increased expression of p53 and p21 and cleavage of caspase-3 and an attenuated expression of Bcl-2 (Figure 4B). These data suggest that loss of Ikkβ results in initiation of apoptosis through induction of p53, p21, and caspase-3, thus conferring a “braking” effect on melanoma development. To test this possibility, we first targeted p53 using lentiviral infection and selection of cells stably expressing shRNA targeting p53 in IkkbΔ/Δ melanocytes. The p53 protein was knocked down approximately 90% in melanocytes (Figure 4C), and this resulted in significant impairment of apoptosis associated with Ikkb deletion (Figure 4D). Thus, p53 expression is essential for Ikkβ-mediated apoptosis. To find the mechanism of the Ikkβ-p53 signal interaction, we examined whether Ikkβ transcriptionally regulates p53 in melanocytes. The mRNA levels of p53 were analyzed by RT-PCR (Figure 4E) and real-time RT-PCR (Supplemental Figure 3). We observed that the p53 mRNA level was not affected by Ikkβ knockout, suggesting that the accumulation of p53 protein in the Ikkβ-null cells was not due to enhanced transcription. MDM2, a p53-specific E3 ubiquitin ligase, binds p53 and regulates its turnover. The p53-MDM2 binding is negatively regulated by phosphorylation of p53 at Ser15 (41). To determine whether loss of Ikkβ might affect the phosphorylation of p53 at Ser15, lysates of Ikkbwt and IkkbΔ/Δ melanocytes, with or without doxycycline induction of mutant HRas expression, were probed with phospho-antibody specific for p53 phosphorylated on Ser15 (pSer15) in Western blot. Results show hyperphosphorylation of p53 (Ser15) in IkkbΔ/Δ cells (Figure 4F). Only low levels of p53 were present in Ikkbwt cells, and there was no detection of phospho-p53 (Ser15) in the Western blot of lysates of these cells. Moreover, expression of HRasV12 in Ikkbwt melanocytes resulted in reduced expression of p53 without detection of phospho-p53 (pSer15). Shieh et al. have demonstrated that phosphorylation of p53 at Ser15 impairs the ability of MDM2 to bind p53, contributing to the accumulation and functional activation of p53 (41). However, it remains unclear how loss of Ikkβ enhances the phosphorylation of p53 in melanocytes.
Figure 4. Deletion of Ikkb promotes apoptosis.
(A) E19 melanocytes cultured with or without doxycycline were subjected to apoptosis analysis by Annexin V. (B) Expression of the indicated proteins in melanocytes cultured with or without doxycycline-induced Ikkb deletion and/or HRasV12 expression were analyzed by immunoblotting, using specific antibodies. Immunoblot of β-actin was used as a loading control. Protein expression was quantitated and normalized to that of the same genetic background cells, without doxycycline-induced HRasV12 expression. Mean ± SD represents 3 independent experiments statistically analyzed between Ikkbwt and IkkbΔ/Δ cells with HRasV12 expression. (C) Western blot of p53 expression in IkkbΔ/Δ E19 melanocytes stably expressing lentiviral shRNA p53 or shRNA control. The expression of p53 protein in cells with p53 knockdown (9.6%) is shown in comparison with that (100%) of shRNA control–expressing cells. (D) E19 melanocytes with shRNA-mediated p53 knockdown and/or doxycycline-induced Ikkb knockout were subjected to Annexin V apoptosis analysis. shRNA-mediated p53 knockdown significantly reduced apoptosis in cells null for Ikkb (P < 0.01). (E) IkkbΔ/Δ and Ikkbwt E19 melanocytes were treated with or without doxycycline, and p53 mRNA levels were examined using RT-PCR. There was no significant change in p53 mRNA level upon deletion of Ikkb. (F) Phosphorylation of p53 (S15) and the expression of p53 protein in IkkbΔ/Δ and Ikkbwt E19 melanocyte lysates were immunoblotted with phospho-p53 (S15 antibody), and the band density is reported as the ratio between cells treated with versus without doxycycline. (G) The phosphorylation status of Mdm2 (S166) was determined by immunoblot with the β-actin–loading control. There was significant reduction of Mdm2 phosphorylation following the deletion of Ikkb (P < 0.05). *P < 0.05, **P < 0.01.
Phosphorylation of Mdm2 at serine 166 has been shown to increase its ubiquitin ligase activity and enhance p53 degradation (42). When we examined the Ser166 phosphorylation of Mdm2, using Western blot analysis of melanocytes treated or not treated with doxycycline to induce expression of HRasV12, we observed that pSer166-Mdm2 levels were greatly reduced in IkkbΔ/Δ melanocytes treated with doxycycline (Figure 4G), but doxycycline-induced expression of HRasV12 did not alter Mdm2 Ser166 phosphorylation in Ikkbwt cells. Real-time RT-PCR analysis demonstrated that the effects on p53 and Mdm2 protein expression associated with doxycycline-induced deletion of Ikkβ were not due to changes in mRNA levels. Moreover, an NF-κB binding sequence (GGGRNNYYCC) was not found in the Mdm2 promoter, upon analysis of transcriptional factor databases (UCSC Genome Bioinformatics Site, http://www.genome.ucsc.edu/; BioInformatics and Molecular Analysis Section, http://www-bimas.cit.nih.gov/molbio/proscan/). Thus, our data suggest that Ikkβ likely keeps the level of Ser166 phosphorylated Mdm2 at steady state when oncogenic mutant HRasV12 is expressed, thus contributing to p53 degradation and escape from apoptosis. By targeting Ikkβ, this steady state is perturbed, p53 is stabilized, and melanocytes exhibit increased apoptosis.
Melanocytes null for Ikkb undergo cell cycle arrest.
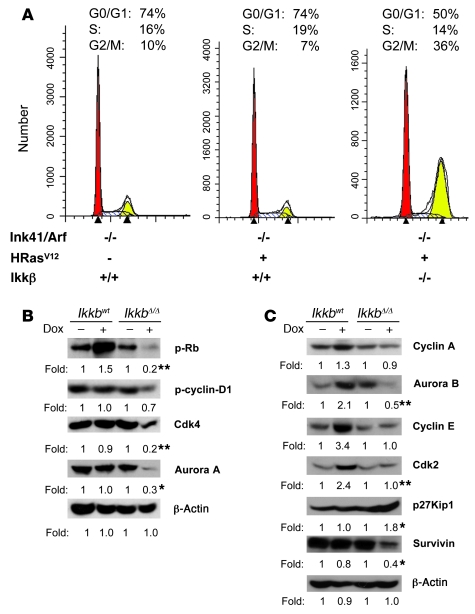
To determine the impact of Ikkb deletion on cell cycle in the melanocytes described above, cells were synchronized with a double-thymidine block for 18 hours, followed by release for 10 hours, and then cells were cultured in serum containing medium with or without 1 μg/ml doxycycline. After 4 days of doxycycline treatment, cells were serum starved for 24 hours, and the cell cycle was analyzed using FACS with CellQuest software, following staining of the fixed cells with propidium iodide. In contrast to that in Ikkbwt cells, disruption of Ikkb in IkkbΔ/Δ cells resulted in an increased percentage of cells undergoing cell cycle arrest at the G2/M phase (Figure 5A). To decipher the molecular mechanism by which IkkbΔ/Δ interrupts cell cycle progression, cell cycle checkpoint proteins were analyzed by Western immunoblot. Upon Ikkb deletion, the phosphorylation of retinoblastoma (Rb) was decreased, and expression of Cdk4, Aurora A kinase, Aurora B kinase, cyclin-dependent kinase 2 (Cdk2), and survivin were markedly reduced (Figure 5, B and C). In addition, there was an increase in p27 Kip1 protein expression (Figure 5C). Since Aurora A and Aurora B are needed for G2/M progression, as the cells are released from the arrest in S phase, the increased percentage of cells at G2/M is likely the result of the reduction of Aurora A and B kinase. It is expected that at later time points, after release from the double-thymidine block, the IkkbΔ/Δ cells would exhibit an increase in the percentage of cells in G0/G1, based upon the observed reduction in phosphorylation of Rb and reduced levels of Cdk2 and Cdk4. When the above experiment was performed on nonsynchronized cells, we observed an increase in the number of cells at the G0/G1 restriction point (Supplemental Figure 2A), indicating that the reduction of pRb, cyclin D1, Cdk4, cyclin A, and Cdk2 (Supplemental Figure 2B) that occurs with deletion of Ikkb results in a reduction or slowing of cells to transition past the G1/S restriction point. Altogether, these data show that IkkbΔ/Δ/Ink4a/Arf–/– melanocytes expressing HRasV12 exhibited enhanced apoptosis, cell cycle arrest, and significant inhibition of mutant HRas-driven melanoma formation.
Figure 5. Deletion of Ikkb interrupts cell cycle progression.
(A) Melanocytes derived from either E19 Ikkbwt or E19 IkkbΔ/Δ mice were synchronized by double-thymidine block and cell cycle was analyzed after 4 days of doxycycline induction for expression of HRasV12 and/or deletion of Ikkb. The percentage of cells in each phase of the cell cycle (G0/G1, S, G2/M) is indicated by red, white, or yellow, respectively. (B and C) Lysates from early passage E19 melanocytes, prepared by the indicated doxycycline treatment, were analyzed for expression of cell cycle checkpoint proteins by immunoblotting with antibodies as indicated. β-actin was used as a loading control. The protein expression by immunoblotting was quantified using the ImageJ program. Each value was from 3 independent experiments. The difference in the specific protein expression between Ikkbwt cells and IkkbΔ/Δ cells, upon the doxycycline-induced HRasV12 expression, was analyzed statistically. (*P < 0.05, **P < 0.01).
IL-6, as a biomarker of melanoma formation, is NF-κB dependent.
The cytokine profile in the tumor microenvironment has been shown to contribute greatly to the development of cancer. To examine the inflammatory cytokines expressed during melanoma development, we examined the expression profile of 32 cytokines in the serum of tumor-bearing and tumor-free mice. Surprisingly, the serum of mice with melanoma showed remarkable elevation in IL-6 (49-fold elevation) and a murine chemotactic cytokine, a homolog of human CXCL1 (KC) (10-fold elevation), as compared with that of melanoma-free mice of Ikkbwt and IkkbΔ/Δ genetic background (Figure 6A). IL-6 has been shown to act as tumor promoter in gastrointestinal cancers, in which it is produced largely by myeloid cells but not by the cancer cells (43). To pursue the IL-6 cellular source in this model, the tissue lysates from normal skin and melanoma and spleen, bone marrow, thymus, and serum from mice, with or without melanoma, were prepared, and IL-6 levels were determined in various organs. Surprisingly, IL-6 was highly expressed in melanoma tissue (577 ± 132 pg/g protein) and serum (200 ± 68 pg/g protein) of melanoma-bearing mice, in contrast to IL-6 in normal skin (6.2 ± 3.6 pg/g protein) or serum of tumor-free mice (6.7 ± 4.8 pg/g protein). We were unable to detect differences in spleen, bone marrow, and thymus, between mice with and without tumor (Figure 6B). These data suggest that IL-6 is more likely produced by melanoma cells than infiltrating immune cells. To confirm that IL-6 is mainly produced from melanoma cells, the melanoma cells were isolated from tumor-bearing mice, cultured in vitro, and IL-6 was detected by ELISA. Interestingly, IL-6 was significantly higher in cultured melanoma cells than in cultures of primary melanocytes (1,810 ± 290 pg/ml vs. 14 ± 2.4 pg/ml) (Figure 6B). This suggests that the melanoma tumor cells are a major source of IL-6 to the host.
Figure 6. IL-6 is a promoter of melanoma tumorigenesis.
(A) Cytokine profiles were determined in the serum of melanoma-bearing or tumor-free mice by cytokine array (n = 4). 6Ckine, chemokine (C-C motif) ligand 21; CTACK, cutaneous T cell–attracting chemokine; MCP, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; sTNFR, soluble tumor necrosis factor receptor; TARC, thymus activation regulated chemokine; TIMP, tissue inhibitors of metalloproteinase. (B) IL-6 levels in the tissues of Ikkbwt mice with or without melanoma were determined. Supernatant (Sup) IL-6 secreted by the cultured E19 melanocytes and melanoma cells derived from melanoma in Ikkbwt mice was measured by ELISA. (C) FVB mice were xenografted with melanoma cells from Ikkbwt mice. These mice were treated with doxycycline and/or BMS-345541. Tumor volume was measured, and tumor lysate was prepared for IL-6 ELISA assay. Each value represents the mean from 5 mice. BMS, BMS-345541. (D) The E19 melanocytes from Ikkbwt or IkkbΔ/Δ mice were cultured with or without doxycycline, and the supernatant IL-6 was determined by ELISA assay. (E) The cell lysates were prepared from the cultured cells above and subjected to immunoblotting for phospho-Stat3. The total Stat3 was probed as a loading control. The blots were scanned and quantified. Each value was from 3 independent experiments.
To verify that Ikkβ activity is required for maintaining melanoma growth in vivo, the melanoma cells from the tumors that arose on the Ikkbf/f:Tyr-rtTA:TetO-Hrasv12:Ink4a/Arf–/– mice after doxycycline induction were xenografted into FVB strain mice (5 mice/group), and 1 mg/ml doxycycline was included in the drinking water. When tumor size reached approximate 400 mm3, the tumor-bearing mice were treated with doxycycline and/or oral administration of the Ikkβ inhibitor, BMS-345541 (75 mg/kg), twice daily for 1 week. The tumor-bearing mice without doxycycline in drinking water were used as a control group (Figure 6C, right panel, first bar). BMS-345541 (4[2′-aminoethyl] amino-1,8-dimethylimidazo[1,2-a]quinoxaline) was identified as a highly selective inhibitor of the Ikkβ (IC50 = 0.3 μm) when compared with a panel of 15 other kinases, including Stat3. Consistent with the role of BMS-345541 in inhibiting NF-κB–dependent transcription of proinflammatory cytokines both in vitro and in vivo (44), we observed that HRasV12-induced IL-6 expression in vivo (Figure 6C, right panel, second bar) was significantly suppressed when Ikkβ was inhibited by BMS-345541 (Figure 6C, right panel, third bar). The subsequent decline in intratumoral NF-κB activation with BMS-345541 treatment indicated the efficiency of Ikkβ inhibition (Supplemental Figure 4). Moreover, BMS-345541 inhibited the growth of the melanoma tumors (Figure 6C, left panel), and this was correlated with reduced IL-6 expression (r2 = 0.91).
To gain further insight into the requirement for Ikkβ in HRasV12-induced IL-6 expression in melanocytes, we established melanocyte cultures from Ikkbwt or IkkbΔ/Δ newborn mice and subjected established cultures to treatment with or without doxycycline (1 μg/ml) for 4 days, before monitoring IL-6 expression by ELISA (Figure 6D). We observed that IL-6 secretion into the culture medium was greatly diminished when Ikkb was deleted (Figure 6D, fourth bar). IL-6 is transcriptionally regulated by NF-κB (45), and IL-6 activation of the transcription factor Stat3 is very important for cancer cell proliferation (46). To examine the effect of loss of Ikkb on Stat3, we deleted Ikkb in melanocytes and measured Stat3 phosphorylation. Loss of Ikkb resulted in the reduced phosphorylation of Stat3, as compared with that in the Ikkb wild-type melanocytes (Figure 6E). Thus, the NF-κB signal transduction pathway contributes to overproduction of IL-6 in melanoma cells and thus contributes to the “cytokine storm” effect in the tumor microenvironment.
Discussion
The transcription factor NF-κB is a key regulator of the expression of genes involved in inflammation, cell cycle, apoptosis, and tumorigenesis. The contribution of NF-κB to the development of various human cancers, including melanoma, is well documented (11, 15, 22, 47). The IKK complex that activates NF-κB is hyperactivated in melanoma cells. The IKK complex is composed of 2 enzymatic units, IKKα and IKKβ, and an adaptor, IKKγ. IKKβ is required for activation of the canonical NF-κB pathway, and Ikkα is important for activation of the noncanonical NF-κB pathway (48). Although Ikkα was shown to be overexpressed in cancer cells, downregulation of IKKα by RNAi was not accompanied by significant apoptosis (49). Recently, Ikkα was described as a kinase that promotes the differentiation of keratinocytes and suppresses nonmelanoma skin cancer (49). Thus, the antiapoptotic function of NF-κB signaling is likely to be mediated by the canonical NF-κB pathway and IKKβ.
Although NF-κB is activated in melanoma, the factors leading to the constitutive NF-κB activity in melanoma cells are not entirely clear. Numerous studies, including our own, have demonstrated that expression of mutant Ras (HRasV12 or N-RasQ61) results in intrinsic activation of this pathway (50, 51). However, transgenic expression of HRasV12 alone does not efficiently induce melanoma in mouse models (21, 36). In melanocytes, mutation of either HRas or downstream BRaf alone leads to nevus formation, along with initial cell proliferation, followed by cell cycle arrest and senescence, mediated in part through expression of Ink4a-encoded proteins (10, 52). However, germline disruption of the Ink4a tumor suppressor locus may confer a familial predisposition for melanoma (53, 54). Ink4a encodes p16Ink4a, and Arf encodes p19Arf. Both Ink4a and Arf function as cell cycle and NF-κB inhibitors (55). In our murine models, mice null for the Ink4a/Arf locus develop angiosarcoma lesions in the FVB mouse strain, while in C57BL/6 mice, loss of the Ink4a/Arf locus results in development of lymphoma (36, 56). Of note, neither expression of the active Hrasv12 gene in melanocytes, nor knockout of Ink4a/Arf, alone induce melanoma (36, 56). Mouse germline manipulation of Ink4a deletion, combined with gain-of-function mutation in the HRas gene in melanocytes, results in melanocyte transformation and melanoma development (36, 37). Either activating RAS or BRAF mutations are observed in most human melanomas (3, 57, 58). Thus, this mouse melanoma model is an ideal tool to faithfully recapitulate the major genetic traits seen in human melanomas (3, 59), allowing study of the role of loss of Ink4a/Arf, Ikkβ/NF-κB, and RasV12 in melanocyte-specific tumorigenesis.
Melanocytes in mice reside mainly in the dermis of the pinna, in the epidermis of the limbs and tail, and in the hair follicles of the hairy parts of the skin (60). We observed that most melanoma lesions that formed when HRasV12 was expressed in Ink4a/Arf-null mice arose from melanocytes in the hair follicles, in which the melanoma tumor cells eventually replaced the cells of hair follicles. The expression of the melanocyte-specific marker S-100 by the tumor cells indicated that the tumor-initiating cells are most likely melanocytes in the bulb of the hair follicle or the melanocyte stem cells in the bulge region. By using Cre/LoxP-mediated deletion of the Ikkb gene in melanocytes undergoing tumorigenesis upon induction of HRasV12 expression, we have provided the first genetic demonstration to our knowledge that the absence of Ikkβ expression in melanocytes disrupts the induction of melanoma by mutant HRas in Ink4a/Arf-null melanocytes. Our data are in agreement with other observations that IKKβ is a critical component for development of certain types of tumors (61–63).
Mutation of p53 is rare in melanoma, but p53 signaling is frequently disrupted during melanoma tumor progression (41, 64–66). Melanocytes lacking p53 or expressing nonfunctional mutant p53 are more susceptible to transformation (64). In agreement with this finding, we demonstrate herein that melanocyte apoptosis associated with the deletion of Ikkb was ablated by p53 silencing. Ikkβ may play a role in the regulation of p53-mediated gene expression, through a mechanism whereby phosphorylated p65 stably associates with p53 at p53 responsive promoters, resulting in an inhibition of p53-mediated transcriptional activity (66, 67). Alternatively, Ikkβ may directly phosphorylate p53 on serines 362 and 366 to promote its turnover (68). Here, we show that Ikkb knockout in melanocytes increased p53 protein levels. However, the increase in p53 protein is not the result of an increase in p53 mRNA but is through phosphorylation of p53 at Ser15, which likely results in disruption of the p53-Mdm2 binding (41). Deletion of Ikkb also reduces the phosphorylation of Mdm2 at Ser166, which abrogates Mdm2-mediated ubiquitination and degradation of p53 (69). These data suggest that Ikkβ plays an important role in the regulation of p53 stability in melanocytes. Moreover, deletion of Ikkb leads to increased protein levels for the p53-regulated inhibitors of cell cycle progression, p21 and p27KIP, while at the same time abrogating Ras-induced Bcl-2 protein expression and activating caspase-3. Thus, melanocytes null for Ikkb may undergo cell cycle arrest and p53-mediated and caspase-3–executed apoptosis.
There are reports that KRas induction combined with loss of p53 results in nuclear localization of p65 in mouse embryonic fibroblasts (63). However, suppression of NF-κB by expression of an IκB super-repressor in KRas-transformed lung tumors resulted in apoptosis even in p53-null lung cancer lines (63). These data imply that there may be differences in the requirement for p53 in apoptosis mediated by inhibition of NF-κB, depending on the method for inhibition of NF-κB (IKK inhibition vs. inhibition of nuclear localization of p65). Alternatively, the difference in the requirement for p53 for apoptosis induction by inhibition of NF-κB, between our data and the data from Meylan et al. (63), may reflect differences in the melanoma and lung cancer models studied.
Stabilization of p53 may contribute to cell cycle arrest in IkkbΔ/Δ melanocytes. When IkkbΔ/Δ melanocytes were cell cycle synchronized in the S phase by the double-thymidine block, then released and allowed to progress, we observed downregulation of Aurora A and an increase in the percentage of cells at the G2/M phase of cell cycle. Aurora A is a centrosome kinase that plays a pivotal role in G2/M transition. However, if IkkbΔ/Δ cells were not first synchronized in S phase by the double-thymidine block, we observed an increase in the percentage of cells at the G0/G1 phase and a reduction in cells in S phase. This is likely the result of the marked reduction in expression of cyclin D1, Cdk4, cyclin E, and Cdk2 associated with loss of Ikkβ in melanocytes. These data suggest that the point in cell cycle that synchronization takes place determines whether the cells arrest at the G1 or G2/M checkpoint.
Though cyclins and Cdk genes are directly transcriptionally regulated by NF-κB, this does not appear to be the case for Aurora kinase A (Aurka). There is only 1 potential NF-κB binding element in the promoter of murine Aurka, and there are none in the human AURKA promoter. Moreover, ChIP assays performed on Ikkbwt and IkkbΔ/Δ melanocytes did not show differences in mutant HRas-induced binding to the NF-κB consensus sequence in the murine Aurka promoter (data not shown). Of interest, inhibition of Aurora A kinase by MLN8054 treatment of colon carcinoma cells released from nocodazole synchronization produces a p53-dependent G1 arrest (70). Specifically, in this melanoma study, kkb knockout, with its accompanied Aurora A downregulation, results in the reduction in progression through the G2/M phase of the cell cycle in cells synchronized by the double-thymidine block and a G1 arrest in unsynchronized cells. The stabilization of p53 is an important component of the G1 arrest. Our cumulative data suggest that deletion of Ikkb in melanocytes promotes cell cycle arrest and apoptosis, and, together, these events inhibit HRasV12-induced melanocyte transformation. IKKβ may provide a promising target for developing novel lead molecules to combat cancers.
A causal link between inflammation, NF-κB, and cancer is well recognized for several tumor types (22, 24). IL-6 production by tumor cells may be a major contributor to an autocrine and/or paracrine amplification loop (71) and constitutive activation of Stat3 signaling has been shown to be important in myeloma, melanoma (72), and Ras-induced cancer (46, 73, 74). In fact, targeted deletion of IL-6 is associated with reduced liver metastasis of lung carcinoma cells (75). Myeloid inflammatory cells also produce IL-6 and thus contribute to inflammation-associated carcinogenesis (71). We demonstrated here that elevation in the IL-6/gp130/Stat3 signaling axis in HRas-transformed mouse melanoma cells is Ikkβ dependent. Moreover, the increased expression of p53 following Ikkb knockout may also contribute to the repression of the IL6 gene promoter (76). Therefore, melanoma-derived IL-6 may provide an autocrine amplification loop at least partially via Ikkβ/IL-6/Stat3 signaling axis (Figure 7). Either genetic or small molecule (BMS-345541) targeting of Ikkb reduces IL-6 production and contributes to ablation of melanoma development or decrease of melanoma tumor burden.
Figure 7. Schematic illustration of oncogenic HRasV12-induced and Ikkβ/NF-κB–mediated melanoma tumorigenesis.
Mutation-activated HRasV12 in Ink4a/Arf–/– melanocytes triggers the Ikkβ/NF-κB signal transduction pathway, leading to inhibition of the apoptosis machinery (p53, p21, Bcl2, caspase 3 cleavage), a transcriptional burst of IL6 accompanied by activation of Stat3, cell cycle progression (with induction of Rb phosphorylation, aurora kinase, Cdk4, survivin), cell proliferation, and immortalization when p16/p19 are deleted. Deletion of Ikkb in these cells blocks cell cycle progression (reduced Rb phosphorylation, aurora kinase, Cdk4, and survivin expression), reduces proliferation, and enhances p53-mediated apoptosis. When p53 is knocked down in cells null for Ikkb, the induction of apoptosis is reversed, indicating that p53 induction (indicated by bold font) is required for the effects of loss of Ikkb on inhibition melanoma tumor growth. Therefore, deletion of Ikkb emerges as an effective target for treating melanoma tumors that express wild-type p53.
In conclusion, conditional knockout of Ikkb successfully abrogates melanoma development in Ink4a/Arf-null melanocytes expressing mutant HRasV12, which causes constitutive activation of the Ras/NF-κB/Stat3 pathway. Deletion of Ikkb results in reduced activation of Stat3, reduced IL-6 production, cell cycle arrest, and apoptosis of melanocytes (Figure 7). Thus, Ikkβ is a potential therapeutic target for clinical consideration in melanoma patients in which these pathways are involved. NF-κB inhibitors are currently in clinical trials for solid tumors (CH828; Leo Pharma), acute myeloid leukemia (AS602868; Serono International), pancreatic cancer and multiple myeloma (Curcumin; MD Anderson Cancer Center), B cell lymphoma (Seliciclib; Cyclacel Pharmaceuticals), and pancreatic cancer, lymphoma, and myeloma (RTA-402, which inhibits Stat3 and NF-κB; Raeta Pharmaceuticals, MD Anderson Cancer Center) (77). Our studies suggest that melanoma tumors exhibiting constitutive activation of the NF-κB pathway expressing wild-type p53 will benefit from treatment with IKKβ inhibitors. Identification of that specific subset of melanoma patients will be very important for future studies.
Methods
Inducible Ikkb knockout melanoma models and cell cultures.
FVB strain mice expressing TetO-Cre (39) were crossed with FVB Ikkbf/f mice (38, 78), and Ikkbf/f-TeTO-Cre mice were selected. The mice were interbred with FVB Ink4a/Arf-null mice carrying transgenic Tyr-rtTA and TetO-Hrasv12 (36, 37). Genotyping was conducted by PCR, using genomic DNA extracted from tail digestion as previously described (37–39). Animal procedures were carried out according to protocols approved by the Institutional Animal Care and Use Committee, Department of Animal Care, Vanderbilt University.
The procedure for isolation, purification, and culture of melanocytes from the skin of E19 mice was performed as previously described (56). The early-passage (passages 5–12) melanocytes were used for experiments. For conditional knockout of Ikkb in vitro, melanocytes were cultured in 254 medium containing 2× HMGS supplement (Cascade Biologics Inc.) in plates coated with collagen I (Invitrogen) and 1 μg/ml doxycycline (Sigma-Aldrich) for 96 hours. Then, cells were transferred into unsupplemented 254 medium containing 1 μg/ml doxycycline for additional 24 hours.
Immunocytochemistry and immunohistochemistry.
Immunostaining was performed with the described protocol (79), using specific antibodies against Ikkβ (MD Chemicals Inc.), S-100 (Dako North America Inc.), and Mart1, Factor VIII (Cell Signal Technology Inc.).
Western blotting, immunoprecipitation, and kinase activity assays.
Immunoblotting analysis, immunoprecipitation, and in vitro Ikk activity assays of cytoplasmic extracts from cultured primary melanocytes were performed as described previously (21), using specific antibodies against Ikkα/β (H-470) and β-Actin (both from Santa Cruz Biotechnology Inc.); HRas (Calbiochem); and Ikkβ (2C8), Ikkγ, Cdk4 (DCS156), phospho-p65 (Ser536) (93H1), phospho-Akt (Ser473) (193H12), phospho-RB (Ser780) (C84F6), phospho–Cyclin D1 (Thr286) (D29B3), phospho-Mdm2 (Ser166), phospho-p53 (Ser15), and Bcl2 (all from Cell Signal Technology Inc.).
RT-PCR.
RT-PCR was performed as described previously (21). The primers used for PCR were as follows: p53 (485 bp), AGTCACAGTCGGATATCAGCC (sense) and CTCCGTCATGTGCTGTGACTT (antisense), and GAPDH (466 bp), ATGCTGGCGCTGAGTACGTC (sense) and TCAGGTCCGCCACTGACAC (antisense). The PCR products were resolved by electrophoresis in a 2% agarose gel and visualized by ethidium bromide staining.
Real-time quantitative RT-PCR.
RNA was extracted from E19 melanocytes isolated from Ikkbwt and IkkbΔ/Δ mice that had been treated or not treated with doxycycline (1 μg/ml), using QIAshredder homogenization, RNeasy Mini Kit isolation, and RNA-free DNase digestion protocols (all from Qiagen). Real-time RT-PCR was performed using Bio-Rad SYBR Green Master Mix and primers assays for Mdm2, p53, Rb1, and p21 (SABiosciences). The real-time protocol was as follows: cycle 1 (performed once), 95°C, 3 minutes; cycle 2 (performed 40 times), 95°C for 10 seconds, 60°C for 30 seconds, and 72°C for 20 seconds; cycle 3 (performed 81 times), 55°C–95°C for 30-second melt curve analysis. All experiments were performed using a Bio-Rad iCycler IQ5.
Cells with stable p53 knockdown.
For specific knockdown of p53, the stable cell line carrying p53 shRNA or control shRNA was developed by infecting E19 melanocytes with 200 μl lentiviral shRNA particles (Santa Cruz Biotechnology Inc.) in complete medium containing 10 μg/ml polybrene. Twenty-four hours after infection, stable cells expressing the shRNA were isolated via selection in the presence of 2 μg/ml puromycin. The expression of p53 protein in the stable cells, along with doxycycline induction, was evaluated by Western blot (Figure 4C). The cells that showed great reduction (over 90%) of p53 were subjected to apoptosis analysis.
Cytokine array and ELISA.
Cytokine arrays were performed using RayBio Mouse Cytokine Antibody Array G Series 2 Kit (32 cytokines) (RayBiotech Inc.), per the manufacture’s protocol. ELISA assay was performed as described previously (15).
Assessment of cell apoptosis.
Using the Annexin V FITC Apoptosis Kit (Invitrogen), expression of cell surface phosphatidylserine was evaluated as an apoptosis marker by staining, followed by flow cytometry analysis, per the manufacturer’s instructions. The TUNEL assay was performed using the FragEL DNA Fragmentation Detection Kit (Calbiochem), per the manufacturer’s protocol.
Statistics.
Results are expressed as mean ± SD from 3 independent experiments (Figure 4, A and D, and Figure 6, B–D). Prism software (GraphPad), Microsoft Excel, and correlation coefficient (r2) were used for statistical analyses. The unpaired, 2-tailed Student’s t test was used to determine the statistical difference between groups. The cumulative incidence of melanoma among groups was compared according to the method described by Gray (80). P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Catherine E. Alford (Flow Cytometry Laboratory, Nashville Veterans Affairs Medical Center) for cell cycle analysis and Melissa Downing (Vanderbilt University Immunohistochemistry Core) for immunostaining. We also thank Vivian Siegel, Hal Moses, Charles Lin, Christine Eischen, and Mark Boothby for helpful discussions. This work was supported by grants from the Department of Veterans Affairs through a VA Merit Award (to A. Richmond) and a VA Senior Research Career Scientist Award (to A. Richmond), NIH grant CA 098807 (to A. Richmond), Skin Disease Research Center grant SP30 AR 41943, Vanderbilt-Ingram Cancer Center Support grant 5P30CA068485, and an Institutional Research and Academic Career Development Award grant to Vanderbilt University School of Medicine (5K12-6M-068543).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(7):2563–2574. doi:10.1172/JCI42358.
References
- 1.Hussussian CJ, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8(1):15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein AM, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. . J Med Genet. 2007;44(2):99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskandarpour M, Hashemi J, Kanter L, Ringborg U, Platz A, Hansson J. Frequency of UV-inducible NRAS mutations in melanomas of patients with germline CDKN2A mutations. J Natl Cancer Inst. 2003;95(11):790–798. doi: 10.1093/jnci/95.11.790. [DOI] [PubMed] [Google Scholar]
- 4.Rocha S, Campbell KJ, Perkins ND. p53- and Mdm2-independent repression of NF-kappa B transactivation by the ARF tumor suppressor. Mol Cell. 2003;12(1):15–25. doi: 10.1016/S1097-2765(03)00223-5. [DOI] [PubMed] [Google Scholar]
- 5.Sini MC, et al. Molecular alterations at chromosome 9p21 in melanocytic naevi and melanoma. Br J Dermatol. 2008;158(2):243–250. doi: 10.1111/j.1365-2133.2007.08310.x. [DOI] [PubMed] [Google Scholar]
- 6.Flores JF, et al. Loss of the p16INK4a and p15INK4b genes, as well as neighboring 9p21 markers, in sporadic melanoma. Cancer Res. 1996;56(21):5023–5032. [PubMed] [Google Scholar]
- 7.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378(2):F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 8.Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22(32):5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 9.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 11.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 12.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 13.Hanson JL, Hawke NA, Kashatus D, Baldwin AS. The nuclear factor kappaB subunits RelA/p65 and c-Rel potentiate but are not required for Ras-induced cellular transformation. Cancer Res. 2004;64(20):7248–7255. doi: 10.1158/0008-5472.CAN-03-3898. [DOI] [PubMed] [Google Scholar]
- 14.Finco TS, Westwick JK, Norris JL, Beg AA, Der CJ, Baldwin AS., Jr Oncogenic Ha-Ras-induced signaling activates NF-kappaB transcriptional activity, which is required for cellular transformation. J Biol Chem. 1997;272(39):24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Richmond A. Constitutive IkappaB kinase activity correlates with nuclear factor-kappaB activation in human melanoma cells. Cancer Res. 2001;61(12):4901–4909. [PubMed] [Google Scholar]
- 16.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107(3):241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Greten FR, Karin M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett. 2004;206(2):193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Mercurio F, et al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278(5339):860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 20.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91(2):243–252. doi: 10.1016/S0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Pan WH, Clawson GA, Richmond A. Systemic targeting inhibitor of kappaB kinase inhibits melanoma tumor growth. Cancer Res. 2007;67(7):3127–3134. doi: 10.1158/0008-5472.CAN-06-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 23.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest. 2005;115(10):2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 25.Dajee M, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421(6923):639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 26.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121(7):977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Baud V, Jacque E. The alternative NF-kB activation pathway and cancer: friend or foe? [in French]. Med Sci (Paris). 2008;24(12):1083–1088. doi: 10.1051/medsci/200824121083. [DOI] [PubMed] [Google Scholar]
- 28.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8(1):33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briassouli P, Chan F, Savage K, Reis-Filho JS, Linardopoulos S. Aurora-A regulation of nuclear factor-kappaB signaling by phosphorylation of IkappaBalpha. Cancer Res. 2007;67(4):1689–1695. doi: 10.1158/0008-5472.CAN-06-2272. [DOI] [PubMed] [Google Scholar]
- 30.Chng WJ, et al. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111(3):1603–1609. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- 31.Bischoff JR, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17(11):3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Vader G, Medema RH, Lens SM. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173(6):833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans R, et al. Aurora A kinase RNAi and small molecule inhibition of Aurora kinases with VE-465 induce apoptotic death in multiple myeloma cells. Leuk Lymphoma. 2008;49(3):559–569. doi: 10.1080/10428190701824544. [DOI] [PubMed] [Google Scholar]
- 35.Chan F, et al. Mechanism of action of the Aurora kinase inhibitor CCT129202 and in vivo quantification of biological activity. Mol Cancer Ther. 2007;6(12 pt 1):3147–3157. doi: 10.1158/1535-7163.MCT-07-2156. [DOI] [PubMed] [Google Scholar]
- 36.Chin L, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11(21):2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin L, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400(6743):468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 38.Chen LW, Egan L, Li ZW, Greten FR, Kagnoff MF, Karin M. The two faces of IKK and NF-kappaB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia-reperfusion. Nat Med. 2003;9(5):575–581. doi: 10.1038/nm849. [DOI] [PubMed] [Google Scholar]
- 39.Wong MH, Saam JR, Stappenbeck TS, Rexer CH, Gordon JI. Genetic mosaic analysis based on Cre recombinase and navigated laser capture microdissection. Proc Natl Acad Sci U S A. 2000;97(23):12601–12606. doi: 10.1073/pnas.230237997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zandi E, Chen Y, Karin M. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: discrimination between free and NF-kappaB-bound substrate. Science. 1998;281(5381):1360–1363. doi: 10.1126/science.281.5381.1360. [DOI] [PubMed] [Google Scholar]
- 41.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–334. doi: 10.1016/S0092-8674(00)80416-X. [DOI] [PubMed] [Google Scholar]
- 42.Malmlof M, Roudier E, Hogberg J, Stenius U. MEK-ERK-mediated phosphorylation of Mdm2 at Ser-166 in hepatocytes. Mdm2 is activated in response to inhibited Akt signaling. J Biol Chem. 2007;282(4):2288–2296. doi: 10.1074/jbc.M604953200. [DOI] [PubMed] [Google Scholar]
- 43.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burke JR, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003;278(3):1450–1456. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 45.Miyazawa K, Mori A, Yamamoto K, Okudaira H. Transcriptional roles of CCAAT/enhancer binding protein-beta, nuclear factor-kappaB, and C-promoter binding factor 1 in interleukin (IL)-1beta-induced IL-6 synthesis by human rheumatoid fibroblast-like synoviocytes. J Biol Chem. 1998;273(13):7620–7627. doi: 10.1074/jbc.273.13.7620. [DOI] [PubMed] [Google Scholar]
- 46.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15(2):79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richmond A. Nf-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol. 2002;2(9):664–674. doi: 10.1038/nri887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 49.Descargues P, Sil AK, Karin M. IKKalpha, a critical regulator of epidermal differentiation and a suppressor of skin cancer. EMBO J. 2008;27(20):2639–2647. doi: 10.1038/emboj.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Su Y, Richmond A. Antioxidants tiron and N-acetyl-L-cysteine differentially mediate apoptosis in melanoma cells via a reactive oxygen species-independent NF-kappaB pathway. Free Radic Biol Med. 2007;42(9):1369–1380. doi: 10.1016/j.freeradbiomed.2007.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo H, et al. NF-kappa B is required for H-ras oncogene induced abnormal cell proliferation and tumorigenesis. Oncogene. 2000;19(7):841–849. doi: 10.1038/sj.onc.1203392. [DOI] [PubMed] [Google Scholar]
- 52.Mason DX, Jackson TJ, Lin AW. Molecular signature of oncogenic ras-induced senescence. Oncogene. 2004;23(57):9238–9246. doi: 10.1038/sj.onc.1208172. [DOI] [PubMed] [Google Scholar]
- 53.Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res. 2006;12(7 pt 2):2301s–2307s. doi: 10.1158/1078-0432.CCR-05-2518. [DOI] [PubMed] [Google Scholar]
- 54.Laud K, et al. Comprehensive analysis of CDKN2A (p16INK4A/p14ARF) and CDKN2B genes in 53 melanoma index cases considered to be at heightened risk of melanoma. J Med Genet. 2006;43(1):39–47. doi: 10.1136/jmg.2005.033498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolff B, Naumann M. INK4 cell cycle inhibitors direct transcriptional inactivation of NF-kappaB. Oncogene. 1999;18(16):2663–2666. doi: 10.1038/sj.onc.1202617. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, et al. Induction of melanoma in murine macrophage inflammatory protein 2 transgenic mice heterozygous for inhibitor of kinase/alternate reading frame. Cancer Res. 2001;61(22):8150–8157. [PubMed] [Google Scholar]
- 57.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 58.Patton EE, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15(3):249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 59.Platz A, Ringborg U, Grafstrom E, Hoog A, Lagerlof B. Immunohistochemical analysis of the N-ras p21 and the p53 proteins in naevi, primary tumours and metastases of human cutaneous malignant melanoma: increased immunopositivity in hereditary melanoma. Melanoma Res. 1995;5(2):101–106. doi: 10.1097/00008390-199504000-00005. [DOI] [PubMed] [Google Scholar]
- 60.Steingrimsson E, Copeland NG, Jenkins NA. Melanocyte stem cell maintenance and hair graying. Cell. 2005;121(1):9–12. doi: 10.1016/j.cell.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 61.Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118(6):671–674. doi: 10.1016/j.cell.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh S, Karin M. Missing pieces in the NF–kappaB puzzle. Cell. 2002;109(suppl): S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 63.Meylan E, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol. 2001;21(6):2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katayama H, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36(1):55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 66.Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1(5):493–503. doi: 10.1016/S1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 67.Jeong SJ, Radonovich M, Brady JN, Pise-Masison CA. HTLV-I Tax induces a novel interaction between p65/RelA and p53 that results in inhibition of p53 transcriptional activity. Blood. 2004;104(5):1490–1497. doi: 10.1182/blood-2003-12-4174. [DOI] [PubMed] [Google Scholar]
- 68.Xia Y, Padre RC, De Mendoza TH, Bottero V, Tergaonkar VB, Verma IM. Phosphorylation of p53 by IkappaB kinase 2 promotes its degradation by beta-TrCP. Proc Natl Acad Sci U S A. 2009;106(8):2629–2634. doi: 10.1073/pnas.0812256106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pise-Masison CA, et al. Inactivation of p53 by human T-cell lymphotropic virus type 1 Tax requires activation of the NF-kappaB pathway and is dependent on p53 phosphorylation. Mol Cell Biol. 2000;20(10):3377–3386. doi: 10.1128/mcb.20.10.3377-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaestner P, Stolz A, Bastians H. Determinants for the efficiency of anticancer drugs targeting either Aurora-A or Aurora-B kinases in human colon carcinoma cells. Mol Cancer Ther. 2009;8(7):2046–2056. doi: 10.1158/1535-7163.MCT-09-0323. [DOI] [PubMed] [Google Scholar]
- 71.Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18(1):19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Catlett-Falcone R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10(1):105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 73.Ancrile B, Lim KH, Counter CM. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 2007;21(14):1714–1719. doi: 10.1101/gad.1549407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15(4):283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maeda S, et al. Ikappa B kinasebeta/nuclear factor–kappaB activation controls the development of liver metastasis by way of interleukin–6 expression. Hepatology. 2009;50(6):1851–1860. doi: 10.1002/hep.23199. [DOI] [PubMed] [Google Scholar]
- 76.Santhanam U, Ray A, Sehgal PB. Repression of the interleukin 6 gene promoter by p53 and the retinoblastoma susceptibility gene product. Proc Natl Acad Sci U S A. 1991;88(17):7605–7609. doi: 10.1073/pnas.88.17.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sethi G, Tergaonkar V. Potential pharmacological control of the NF-kappaB pathway. Trends Pharmacol Sci. 2009;30(6):313–321. doi: 10.1016/j.tips.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 78.Pasparakis M, et al. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature. 2002;417(6891):861–866. doi: 10.1038/nature00820. [DOI] [PubMed] [Google Scholar]
- 79.Yang J, Zaja-Milatovic S, Thu YM, Lee F, Smykla R, Richmond A. Molecular determinants of melanoma malignancy: selecting targets for improved efficacy of chemotherapy. Mol Cancer Ther. 2009;8(3):636–647. doi: 10.1158/1535-7163.MCT-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of competing risk. Ann Stat. 1988;16(3):1141–1154. doi: 10.1214/aos/1176350951. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.