Abstract
It is now believed that frequent jet lag or shifts of daily rhythms caused by rotating shift work can lead to deleterious health outcomes. Indeed, many serious health problems, including breast cancer, stroke, and cardiovascular disease, have been linked to an occupational history of shift work. This has heightened interest in better understanding the biological responses to jet lag and shift work, with the hope that this will pave the way to developing compounds that can help people avoid their negative health consequences. In this context, a report in this issue of the JCI takes us to a new level of understanding of the molecular control of the resetting of the multitude of internal biological clocks disrupted in a mouse model of jet lag.
Jet lag can seem like a minor discomfort, but studies are now suggesting that frequent jet lag or shifts of daily rhythms as a result of rotating shift work can lead to many deleterious health outcomes. Breast cancer, reproductive disorders, cardiovascular disease, stroke, and metabolic syndrome are all linked to an occupational history of shift work (1–3). Experiments modeling chronic jet lag have shown that it can speed up tumor growth (4) and bring on early mortality in aged mice (5), as well as lower the survival rate for hamsters with impaired heart function (6). The International Agency for Research on Cancer, a unit of the World Health Organization, has declared that night shift work is also a probable carcinogen (7). These serious health concerns heighten our interest in better understanding the biological responses to jet lag and shift work.
Why do we suffer malaise from shift work and jet lag?
The malaise from shift work and jet lag is thought to arise from the dynamic properties of a multitude of internal biological clocks responding to environmental inputs. Many of our tissues have the ability to maintain 24-hour cycles with fair accuracy (8). Because they are not precisely 24-hour clocks, but mark time for approximately (i.e., circa) a day, they are referred to as circadian clocks. They are kept aligned with the cycles of the planet in part by the regulatory influence of a small group of neurons in a region of the hypothalamus known as the suprachiasmatic nucleus (SCN; ref. 8). The SCN is a collection of many cells with molecular clocks that communicate to maintain a coupled, coherent, and rhythmic output. The SCN receives direct input from the retina and thus is informed immediately when light is detected. Light appearing at unexpected times, when the internal clocks expect to be in the darkness of night, will reset the internal rhythm of the SCN. Then the SCN cells will serve as the master pacemaker, helping all the other clocks within the organism to realign to the new light/dark cycle. The dynamics of how the SCN and this community of clocks realign to new light/dark cycles are thought to underlie jet lag.
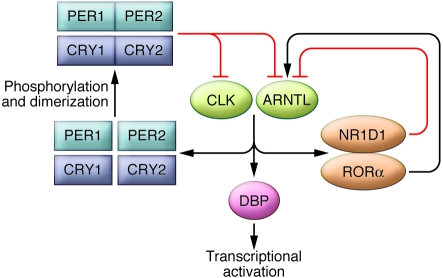
Within each SCN cell, the perception of time of day is maintained by a network of interlinked genes and proteins (9). In particular, the proteins clock (CLK) and brain and muscle aryl hydrocarbon receptor nuclear translocator–like (ARNTL; also referred to as BMAL1) act by activating transcription and driving the expression of a number of genes (Figure 1). The genes driven by CLK and ARNTL include the period genes, such as period 1 (Per1) and Per2, and the cryptochrome genes, such as cryptochrome 1 (Cry1) and Cry2. The gene products — PER1, PER2, CRY1, and CRY2, respectively — spend some time milling around in the cytoplasm of the cell, getting phosphorylated and forming dimers. Eventually, they are suited for nuclear translocation, at which time they provide a negative feedback signal inhibiting the activational drive of CLK and ARNTL that led to their own production. After some time, period/cryptochrome inhibition is released, and CLK and ARNTL are free to begin the cycle again. This is a simplified example of the molecular mechanism underlying circadian rhythms; there are several other genes playing feedback roles as well. For example, the genes encoding nuclear receptor subfamily 1, group D, member 1 (NR1D1; also known as REV-ERBα) and RAR-related orphan receptor α (RORα) mediate a feedback loop to provide rhythmic inhibitory and activating drive, respectively, to Arntl. Dbp, which encodes D-element–binding protein (DBP), is sensitive to the rhythmic transcriptional activation of CLK and ARNTL and shows a regular circadian rhythm in expression, a reflection of the intricate mechanism of the molecular clock within each cell. DBP can act as a transcriptional activator for other, downstream genes. The concurrent loops of this system are numerous, in that many of the key genes contain promoter elements sensitive to fluctuations of CLK and ARNTL, which bind E-boxes; NR1D1, which binds REV-ERBα/ROR–binding elements (RREs); and DBP, which binds D-boxes (9).
Figure 1. An interconnected network of genes and gene products underlies circadian rhythm generation in the mammalian SCN cell.
The proteins CLK and ARNTL act by activating transcription and driving the expression of a number of genes. The products of these genes, including PER1, PER2, CRY1, and CRY2, provide a negative feedback signal to inhibit the activational drive of CLK and ARNTL. Several other genes also have feedback roles; for example, the genes encoding NR1D1 and RORα mediate a feedback loop to provide rhythmic inhibitory and activating drive, respectively, to the gene encoding ARNTL. Because Dbp is sensitive to the rhythmic transcriptional activation of CLK and ARNTL, DBP can act as a transcriptional activator for other, downstream genes.
A similar clock is present in many other cells of the body, although in some cases, the exact timekeepers are slightly altered. While the cellular clocks in the SCN can be reset by light because retinal input can increase levels of PER1 and PER2, how are clocks in cells outside of the brain reset? Levels of physical activity could be important, as could daily cycles of food intake or body temperature. Neural or humoral pathways might also be important in helping all the body clocks stay in step. One important hormonal input is corticosterone, a glucocorticoid secreted by the adrenal gland. Glucocorticoids can directly increase Per1 gene transcription (10). Glucocorticoid signaling can reset peripheral clocks (11) through what appears to be direct action on those cells; SCN cells do not express glucocorticoid receptors, but are thought to be responsive to this signal through intermediate pathways.
Jet lag varies with tissue location
A key study showed that the SCN can adjust quickly to the different light cycle of a new time zone, but the extra-SCN circadian clock in the liver can take more than a week to get in sync (12). This study used animals with a Per1-luciferase reporter transgene that allowed rhythms of cultured tissues to be measured following shifts of the light/dark cycle. Subsequent studies have shown that jet lag not only produces internal temporal disorder of the relationship between the SCN and extra-SCN clocks, but also throws the multitude of cells within the SCN into a state of temporal disarray (13–15). Cells in more ventral areas of the SCN shift more rapidly than do those in more dorsal regions (13).
A study in this issue of the JCI by Kiessling, Eichele, and Oster (16) takes us to a new level of understanding of jet lag. This study is unique in that 5 of the key genes important in generating circadian rhythms were assayed in 6 tissues of the body over multiple times and multiple days as the animal experienced jet lag. Surprisingly, the individual players in the complex network of genes and proteins constituting the molecular clock shifted to a new light/dark cycle at different rates. This result was observed both within and across tissues. For example, the period genes shifted quickly in SCN samples, but Dbp, Nr1d1, and Arntl lagged behind. Conclusions from prior work on how clocks reset using markers for period genes now must be qualified, because other genes within SCN cells apparently do not shift so rapidly, confirming an earlier report that cryptochrome genes shift more slowly than do period genes in the SCN (17). Interestingly, timing of the daily use of a running wheel shifted at a rate similar to the slowest genes in the SCN (16). The extra-SCN tissues sampled showed a variety of shifting patterns. Most different from the SCN was the pancreas, in which Nr1d1 showed the fastest resetting and Per1, Per2, Dbp, and Arntl, were slower to adjust. Thus, each tissue showed a uniquely determined pattern of jet lag, perhaps reflecting their unique input pathways (8).
The adrenal gland is a drag
One original insight from the study of Kiessling et al. (16) is the demonstration that a circadian oscillator remote from the SCN can impede the pacemaker action of the SCN. The clock within the adrenal gland influences the rhythm of glucocorticoid in the blood. The authors propose that this rhythm provides stabilizing feedback to the SCN and can help SCN cells resist shifts to sporadic stimuli. This was shown by the demonstration that ablation of the adrenal clock sped up adjustment of the SCN to a shifted light/dark cycle (16), confirming a prior report that ablation of the entire adrenal gland sped up adjustment of behavioral rhythms to shifted light/dark cycles (18). This demonstration of a restraining feedback from a peripheral oscillator to the SCN challenges the older, more hierarchical, understanding of the structure of the circadian system as a system with a core pacemaker and many slave oscillators (Figure 2). The present finding supports the current understanding of the circadian system as a network with feedback interactions even from the peripheral oscillators to the main pacemaker in the SCN.
Figure 2. Cellular clock genes adjust to new time zones at different rates in different body organs.
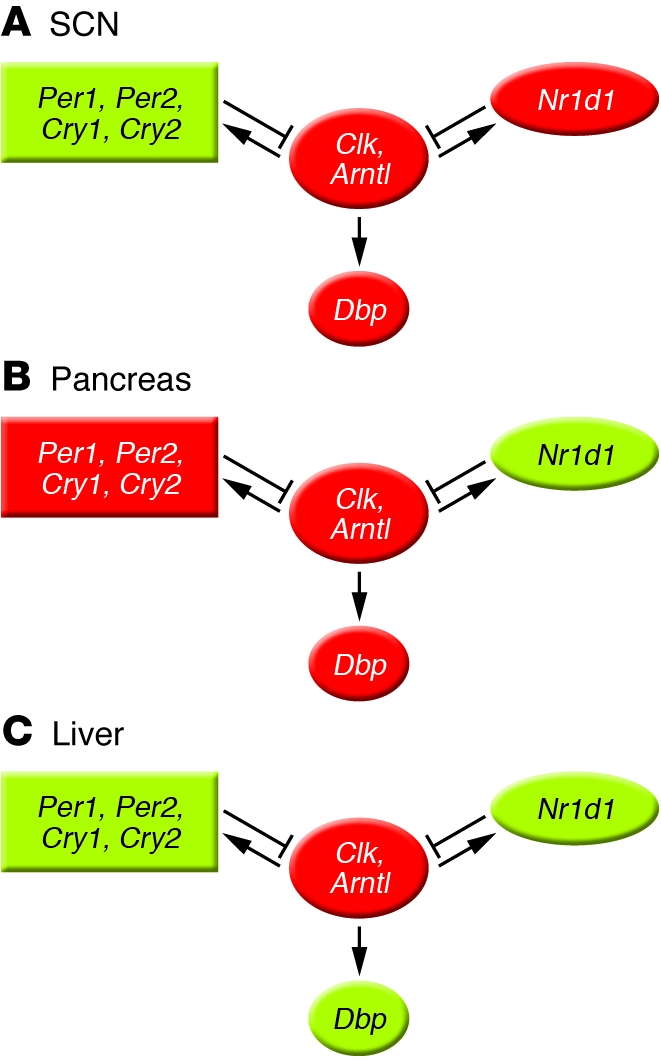
(A) In the central brain clock, the SCN, Per1 and Per2 lead the system in shifting. (B) Conversely, in the pancreas, Nr1d1 appears to take the lead. (C) The liver circadian clock shows an intermediate pattern. Red indicates genes slower to adjust to jet lag, and green denotes genes faster to respond, as indicated by Kiessling et al. (16).
Developing treatments for future frequent flyers
Kiessling et al. also found that pharmacological treatments that altered levels of corticosterone could either accelerate or prolong jet lag in the mice, depending on the time of day of treatment (16). This adds to several recent observations that pharmacological treatments can speed adjustment to shifted light/dark cycles; for example, jet lag has been shown to be lessened in hamsters treated with sildenafil (19) or NAN-190, a drug that alters serotonin responses (20). Further development of such compounds might reduce the negative health consequences of jet lag and shift work. Improved methods for adjusting to new time zones could also help shorten the accompanying temporal disorder (21, 22). Whether shifting rapidly is actually better or worse for one’s health remains critical to sort out; researchers have been trying to find treatments that speed the rate of resynchronization, but we must be cautious about recommending such treatments until it is clear that these rapid shifts are not more harmful than more gradual adjustments. An important caution in interpreting the work of Kiessling et al. is that the researchers measured gene expression at the level of mRNA (16). It is important to determine the dynamics of key proteins and posttranslational modifications of proteins to more fully appreciate the relevance of these findings. Development of methods for noninvasive imaging of circadian clock dynamics in various tissues within an animal would allow studies similar to this one to become more routine and would better equip us to develop effective treatments for jet lag.
Our current understanding of jet lag incorporates disorder at many levels. Disorder can exist between the timing of the organism and the environment, within the different organs in the organism, and among the cells within a single tissue. The study in this issue demonstrates that jet lag may also include disorder among the circadian clock genes within a cell.
Acknowledgments
Tanya Leise provided helpful feedback to an earlier draft of this commentary.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(7):2265–2267. doi:10.1172/JCI43632
See the related article beginning on page 2600.
References
- 1.Knutsson A. Health disorders of shift workers. Occup Med (Lond). 2003;53(2):103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 2.Megdal SP, Kroenke CH, Laden F, Pukkala E, Schernhammer ES. Night work and breast cancer risk: a systematic review and meta-analysis. Eur J Cancer. 2005;41(13):2023–2032. doi: 10.1016/j.ejca.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Mahoney MM . Shift work, jet lag, and female reproduction [published online ahead of print March 8, 2010]. Int J Endocrinol. doi:10.1155/2010/813764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipski E, et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 2004;64(21):7879–7885. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 5.Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16(21):R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penev PD, Kolker DE, Zee PC, Turek FW. Chronic circadian desynchronization decreases the survival of animals with cardiomyopathic heart disease. . Am J Physiol. 1998;275(6 pt 2):H2334–H2337. doi: 10.1152/ajpheart.1998.275.6.H2334. [DOI] [PubMed] [Google Scholar]
- 7.Straif K, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8(12):1065–1066. doi: 10.1016/S1470-2045(07)70373-X. [DOI] [PubMed] [Google Scholar]
- 8.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 9.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto T, et al. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem. 2005;280(51):42036–42043. doi: 10.1074/jbc.M509600200. [DOI] [PubMed] [Google Scholar]
- 11.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. . Science. 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 13.Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur J Neurosci. 2009;29(1):171–180. doi: 10.1111/j.1460-9568.2008.06534.x. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25(23):5481–487. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano M, et al. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci. 2003;23(14):6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010;120(7):2600–2609. doi: 10.1172/JCI41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy AB, Field MD, Maywood ES, Hastings MH. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci. 2002;22(17):7326–7330. doi: 10.1523/JNEUROSCI.22-17-07326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sage D, et al. Influence of the corticosterone rhythm on photic entrainment of locomotor activity in rats. J Biol Rhythms. 2004;19(2):144–156. doi: 10.1177/0748730403261894. [DOI] [PubMed] [Google Scholar]
- 19.Agostino PV, Plano SA, Golombek DA. Sildenafil accelerates reentrainment of circadian rhythms after advancing light schedules. Proc Natl Acad Sci U S A. 2007;104(23):9834–9839. doi: 10.1073/pnas.0703388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler EJ, Sprouse J, Harrington ME. NAN-190 potentiates the circadian response to light and speeds re-entrainment to advanced light cycles. Neuroscience. 2008;154(4):1187–1194. doi: 10.1016/j.neuroscience.2008.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leise T, Siegelmann H. Dynamics of a multistage circadian system. J Biol Rhythms. 2006;21(4):314–323. doi: 10.1177/0748730406287281. [DOI] [PubMed] [Google Scholar]
- 22.Dean DA 2nd, Forger DB, Klerman EB. Taking the lag out of jet lag through model-based schedule design. PLoS Comput Biol. 2009;5(6):e1000418. doi: 10.1371/journal.pcbi.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]