Structural data, efficient structural comparison algorithms and appropriate datasets and filters can assist in getting an insight into time dimensionality in interaction networks; in predicting which interactions can and cannot co-exist; and in obtaining concrete predictions consistent with experiment.
Structural data, efficient structural comparison algorithms and appropriate datasets and filters can assist in getting an insight into time dimensionality in interaction networks; in predicting which interactions can and cannot co-exist; and in obtaining concrete predictions consistent with experiment.
Abstract
Inspection of protein–protein interaction maps illustrates that a hub protein can interact with a very large number of proteins, reaching tens and even hundreds. Since a single protein cannot interact with such a large number of partners at the same time, this presents a challenge: can we figure out which interactions can occur simultaneously and which are mutually excluded? Addressing this question adds a fourth dimension into interaction maps: that of time. Including the time dimension in structural networks is an immense asset; time dimensionality transforms network node-and-edge maps into cellular processes, assisting in the comprehension of cellular pathways and their regulation. While the time dimensionality can be further enhanced by linking protein complexes to time series of mRNA expression data, current robust, network experimental data are lacking. Here we outline how, using structural data, efficient structural comparison algorithms and appropriate datasets and filters can assist in getting an insight into time dimensionality in interaction networks; in predicting which interactions can and cannot co-exist; and in obtaining concrete predictions consistent with experiment. As an example, we present p53-linked processes.
The concept and introduction
In most network studies, protein–protein interactions are abstracted with a graph representation where proteins are represented as nodes and protein–protein interactions as edges. Such network representation is crucial for the comprehension of biological processes and protein function in the global sense. However, to characterize the interactions with respect to their structural and chemical properties and in particular to understand how the function is exerted, it is essential to include structural information in the networks.1–10 To date, in structural network studies, attempts have focused on elucidating the nature of the interactions and on predicting new protein interactions,11–15 affinities and kinetic constants.9 In the pioneering works of Aloy and Russell,1–4 structural information was used to interpret the molecular details of the interactions and to model the binary interactions. Aytuna et al. 11 presented a structure-based method to predict protein–protein interactions using known template interfaces; if surface regions of two proteins are structurally similar to the two sides of a template interface, the two proteins are predicted to interact. More recently, Schroeder and co-workers6 used structural templates to predict novel protein interactions specific for pancreatic cancer. Several groups have also shown how it is possible to estimate kinetic constants for the binding parameters using structural information.16–18 From a different perspective, elastic network models (i.e., eigenmode decomposition) of the protein–protein interactions can give insights into the functional organization of the proteins in the network.19
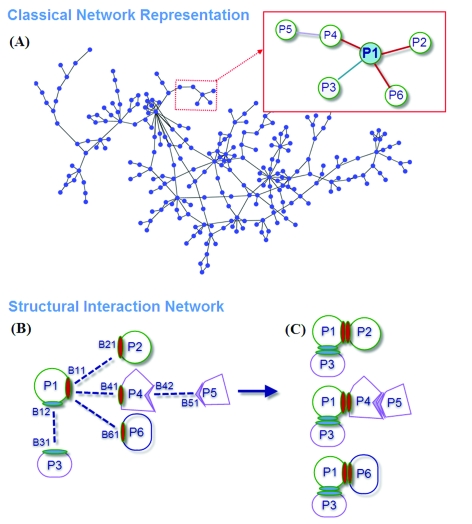
Questions to be addressed in structural networks include (i) how can a hub protein interact with many proteins with different affinities and (ii) which interactions can occur simultaneously and which are mutually excluded? Kim et al. 10 distinguished overlapping from non-overlapping interfaces in their structural interaction network to determine the interaction behavior. They grouped network hubs into singlish- and multi-interface. The former has at most two distinct binding interfaces and the interactions exclude each other whereas the latter has more than two binding interfaces with most of the interactions being simultaneously possible. This concept is illustrated in Fig. 1; the figure displays a schematic representation of a classical network and a structural interaction network. The protein labeled P1 has two different binding regions, B11 and B12: through the B12 region, the protein interacts with P3; and through the B11 region it can interact with three partners. Thus, the interactions of P1 with P2, P4 and P6 exclude each other; that is, they cannot occur at the same time. On the other hand, the interaction with P3 is simultaneously possible. In Fig. 1C, three possible interactions are displayed. This figure indicates that P1 can be used in three different complexes. This information cannot be obtained from the abstract network shown in Fig. 1A.
Fig. 1. The concept figure presenting an overview of this work.
Including the time notion in structural network is an asset, differentiating between processes. The time dimensionality can be included by linking protein complexes to time series of mRNA expression data. One of the early attempts to explain the dynamics of the interactions includes the work of Han et al.20 They classified highly connected proteins (“hub proteins”) in the yeast interactome network into date and party hubs based on their partners’ expression profiles. Party hubs, whose expression is correlated with that of their interacting partners, simultaneously bind most of their partners; on the other hand, date hubs with less correlated expression interact with their partners at different times. In their structural network, Kim et al. 10 linked the date/party hub subdivision to the number of interaction interfaces on protein surfaces. They found that multi-interface hubs correspond mostly to party hubs whereas single-interface hubs correspond to date hubs. Komurov and White21 refined the notion of dynamic modularity in the protein interaction network; they found that constitutively expressed and dynamically co-regulated proteins cluster in different parts of the protein interaction network to form static and dynamic functional modules, respectively. In another study, de Lichtenberg et al. 22 mapped microarray expression data onto yeast protein–protein interaction network. Functional linkages between proteins were inferred and analysis of the dynamics of protein complexes during the yeast cell cycle revealed previously unknown components and modules. While co-expression of a specific protein and its partners can be studied, there are no available large-scale data to infer the dynamics of protein interaction networks. The lack of clear consistent information relating to large-scale co-expression of proteins and structural data hamper the understanding of how cellular processes function in four dimensions,5 that is, considering also time.
Here, to illustrate the usefulness of structural information, we present the p53 network. p53 is a central protein playing a key role in response to a broad range of stress signals such as DNA damage and oncogene activation; as such it has a large number of binding partners in the cell. To illustrate the concept and characterize the interactions of the p53 pathway we use PRISM.11,13,15 Below, we illustrate how this concept, coupled with the PRISM strategy, allows the prediction of novel interactions for p53 and Mdm2 proteins.
Methods
In this work, we use our high performance prediction algorithm PRISM11,15 to analyze the p53 pathway. The PRISM rationale argues that if particular surface regions of any two proteins are spatially similar to the complementary partners of a known interface, in principle these two proteins can interact with each other via these regions. The prediction algorithm uses a template interface dataset and a target single protein structure dataset to predict such potential interactions between target proteins. The description of these two sets and the details of the PRISM algorithm are clarified in the following sections.
Template set
An interface can be defined as the region that links two protein chains by non-covalent interactions. The template set represents the subset of the structurally non-redundant unique interface architectures. Using the interfaces in this dataset as templates, new potentially interacting protein pairs are predicted. For this work, we used two different template sets; the first is the non-obligate template set, the other is the obligate template set. The obligate template set contains interfaces whose interactions are tight and long-lived; whereas the non-obligate set contains complexes which transiently associate and dissociate and the partner chains are stable on their own. In the non-obligate template set, there are 158 interfaces and in the obligate template set, there are 330. To generate these template sets, we considered the structurally clustered interface dataset generated by Tuncbag et al. 23 which contains 49 512 interfaces clustered into 8205 clusters. At the lowest level, these structurally distinct interface clusters contain some homologous complexes. These are next eliminated in each cluster (homology cutoff = 80%, using the ClustalW24 sequence alignment) following the procedure applied by Keskin et al. 25 For the remaining clusters, the biological relevance is addressed by removing crystal contacts using NOXclass.26 To construct an obligate template set we considered obligate interfaces: if the size of a cluster is at least two, the representative interface of that cluster joins the obligate set. A similar procedure is followed for the generation of the non-obligate template set. Since evolutionarily-related interfaces are more likely to bind similar partners, computational hotspots in the interfaces are also considered. These hotspots are obtained by the empirical formula of Hotsprint (for details, see ref. 27) which combines residue accessibility, conservation and amino acid propensity. Template interfaces are named as follows: i.e. if the PDB code of a protein (or complex) is 1axd and there is an interface between chains A and B this interface is named 1axdAB.
Target set
The target set contains proteins related to the p53 pathway; to find out if they interact directly, their surfaces are structurally compared to the template interfaces. To construct this set, all proteins known to play a role in the p53 pathway are extracted from the molecular interaction map.28 A total of 104 proteins are extracted. 47 of the 104 proteins have crystal structures. Some proteins do not have full-length structures, only those of domains or fragments. We considered all these structures in the prediction algorithm. For each, only one structure is considered; homologous structures are not added to the target set. In the structural matching with the template set, only the surface residues of the target proteins are considered. To extract surface residues, Naccess29 is applied. If the relative surface accessibility of a residue is greater than 5%, that residue is considered as surface residue.
The PRISM algorithm
PRISM predicts interfaces by spatial similarity.11,13 The PRISM prediction steps are illustrated in Fig. 2. In this example, the model template is the interface region between the homodimer of the immunoglobulin heavy chain (PDB id: 1dn2). In the PRISM nomenclature, the interface between these chains (A and B) is 1dn2AB. Each template interface is split into its two complementary partners. Using the MultiProt engine, PRISM searches whether these are structurally similar to regions on the target surfaces. MultiProt searches for spatial similarities of amino acids disregarding the order of the residues on the chain.30,31 Because template interfaces and target surfaces do not consist of contiguous chains, Multiprot is particularly appropriate for our task. For each individual alignment, MultiProt reports the 10 best substructural matches. PRISM considers the first match. To avoid a high rate of false positives, 50% of the residues of interface partners must be matched with the target surfaces. Here, we also considered evolutionary similarity (the number of identically matched target residues with template hotspots) between the target and the template; however, we did not set any cutoff for hotspot similarity. As a result, two sets are generated (T R and T L). In the first the targets are similar to the left interface partner (I L); in the second the targets are similar to the right interface partner (I R). The cross matches of these two sets give the first possible predicted protein interactions list. When the aligned target proteins are transformed onto the template interface some residues of the partner targets can collide; these candidate pairs are disallowed and removed (step 2). Corresponding target proteins of the left and right interface partners should not interpenetrate each other in their complex state. In the last step, the biological relevance of these putative protein complexes is assessed using NOXclass (see ref. 26 for NOXclass details). A NOXclass value of at least 80% moves the complexes into the final predicted complexes set. The resulting list is the putative interactions predicted by PRISM using the multi-step filtering.
Fig. 2. Schematic representation of the PRISM algorithm.
Some proteins interact with their partners at different regions and at different time periods with different interaction strengths. PRISM supplies this information for the predicted interactions; accordingly, the binding region, time period (simultaneous or exclusive interactions) and interaction strength (whether transient or obligate) can be obtained from PRISM for the putative interactions.
Results
p53 pathway and protein interaction network
The experimental interaction data for the p53 pathway are extracted from the Molecular Interaction Map (MIM) of Kohn and his colleagues.28 In total, there are 226 interactions between 104 mammalian proteins.32 We searched for additional interactions among these 104 proteins in different interaction databases such as DIP,33 BIND34 and MINT,35 in an attempt to enrich the p53 pathway. This led to a total of 311 interactions between 104 proteins, of which 47 proteins have structural information. With these structures as input, PRISM predicts 257 interactions between 45 protein structures using the non-obligate template set; among them 52 interactions are validated using the 311 experimental interactions. PRISM further predicts 411 interactions between 47 protein structures, using the obligate template set, of which 64 interactions are available in the experimental interaction data. In Table 1, these numbers are tabulated. These predictions are between any pairs of proteins involved in the p53 pathway and survived multi-step filters. Some of the interactions are observed in both sets. In total, 76 unique interactions are verified using the experimental interaction data.
Table 1. PRISM predictions on p53 interaction network.
| Template set used | No. of protein structures | No. of putative interactions | No. of experimentally verified interactions |
| Obligate set | 45 | 257 | 52 |
| Non-obligate set | 47 | 411 | 64 |
p53 and its binding partners
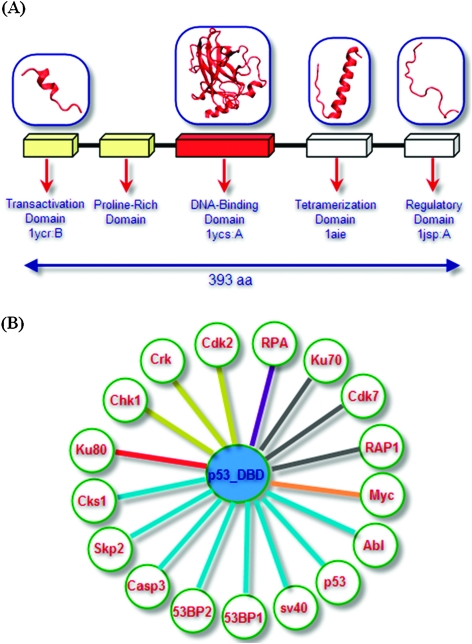
One protein can interact with different partners using the same or different surface patches. For hubs, which interact with many different proteins, this adaptation is crucial. Hub proteins can interact with their partners at different time periods through the same region (i.e., mutually exclusive interactions), or at the same time through different regions (i.e., simultaneous interactions), or both. Information relating to the time component of the interaction coupled with structural data provides an added dimensionality in protein interaction networks. As an example, let us consider p53, a central protein in the cell. p53 consists of five domains: the transactivation domain, the proline-rich domain, the DNA-binding domain, the tetramerization domain and the regulatory domain. A high resolution structure of full-length p53 is unavailable; however, structures of several individual domains are. Fig. 3A illustrates these domains with their corresponding structures. On its own, the transactivation domain is unfolded; it is folded when bound to the Mdm236 or in a membrane environment.37 The DNA-binding domain is the largest structured p53 domain with 191 residues. The tetramerization domain is structured in the p53 tetramer, forming a single helix. The regulatory domain, to which ubiquitin attaches, is unfolded.
Fig. 3. (A) The fragments of the p53 protein and the available crystal structures; (B) p53 DNA-binding domain interactions. Edges are colored according to the different binding sites; these contain both experimental and PRISM-predicted interactions.
To predict the p53 partners and their binding sites, we selected the DNA-binding domain (DBD); the helical structure of the tetramerization domain can match many helical template interfaces leading to a prediction bias. The structurally known and predicted interactions of the DBD clearly show a mechanistic multi-faced, multiple partner paradigm.
Fig. 3B illustrates known and potential PRISM-predicted p53 DBD-binding partners. In this small network, predicted interaction partners (Cdk2, Crk, Chk1, RPA, Ku70, Ku80, Abl, Casp3, RAP1A, Cdk7, Myc, Skp2, Cks1) and interaction partners known from available crystal structures of the p53 DBD complexes (53BP1, 53BP2, sv40, p53 DBD) are drawn, where edges are colored according to their binding sites; protein partners binding p53 DBD at the same binding site are depicted by similar-color edges. Fig. 4 presents the detailed picture of this small network by combining the structures of the proteins and their binding regions. The p53 DBD is represented in ribbon and colored orange. The binding regions of the p53 interacting proteins are shown in ball representation; proteins interacting through the same p53 region are depicted using the same color. For example, Chk1, Crk, and Cdk2 interact with p53 through the same region. They are colored red and their binding sites are colored yellow. Their interaction with p53 is represented by yellow edge.
Fig. 4. Predicted partners of the p53 DNA-binding domain (left panel), with representation of some in the complexed state (right panel).
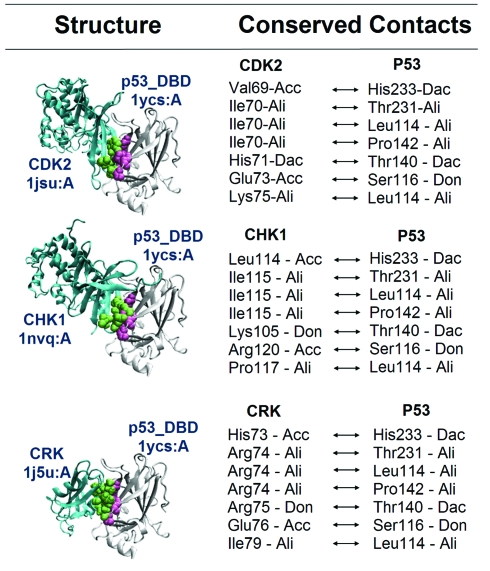
The first binding site of p53 is the region (B1) where Cdk2, Crk and Chk1 bind. These proteins are PRISM-predicted p53 interaction partners. Cdk2 and Chk1 are members of the kinase family. Crk is a proto-oncoprotein and its SH2 domain is available in the target set (1ju5:A). The phosphorylation site of p53 is located at the transactivation and regulatory domains.38–40 The catalytic site of the Cdk2 contains the residues Asp127, Lys129, Gln131, Asn132, Asp145 and Thr165. Also, the catalytic site of Chk1 contains Asp130, Asp132, Asn135 and Thr170. Here, we predict that Cdk2 and Chk1, two kinases, bind to the DBD of p53 using a region different from their catalytic sites. The predicted binding site of Cdk2 overlaps partially with the region where Cdk2 binds to CycA. The template interface for these interactions is the obligate interface formed between two oligomerization domains of the arginine repressor (1b4bAC) which binds to DNA41 with a transcription factor activity protein. Since Cdk2, Crk and Chk1 share the same binding site they are mutually exclusive and cannot interact simultaneously. When we compare their chemical contacts using MAPPIS (for details of MAPPIS see ref. 42), we observe seven structurally conserved contacts. These are illustrated in Fig. 5. Thus, even though the partners are different, their putative interactions with p53 are conserved structurally and chemically.12 NOXclass labels these interactions as biological and non-obligate, in agreement with the characteristics of mutually exclusive interactions.
Fig. 5. Conserved contacts of Cdk2, Chk1 and Crk with p53 DBD predicted with MAPPIS.
Another set of interacting proteins competing for B2, the other region of p53 DBD, are the p53 binding protein 1 (53BP1), p53 binding protein 2 (53BP2), simian virus 40 large T antigen (sv40), p53 DBD, Casp-3, Cks1, and proto-oncogene tyrosine protein kinase c-Abl. These proteins have overlapping binding sites on the DBD. The structures of the complexes of 53BP1 (1gzh:B), 53BP2 (1ycs:B), p53 DBD (2geq:A) and sv40 (2h1l:A) with the p53 DBD are available in the PDB. The other partners are PRISM-predicted interactions. The template interface of Casp3–p53 interaction is the non-obligate interface between the acetylcholine receptor and its inhibitor (2br8BG). This interaction between Casp3 and p53 DBD is predicted by the human protein interaction prediction server (PIPs),43 consistent with our results.
For two interactions to be simultaneously possible, it is insufficient that binding sites should not overlap; in addition, in the multimeric state there should not be residues overlapping between the partners. In our case here, the corresponding partners of B1 (where Cdk2, Crk, and Chk1 bind) and B2 (where 53BP1, 53BP2, sv40, p53 DBD, Casp3, Cks1, and c-Abl bind) do not interpenetrate each other in their trimeric states; consequently, simultaneous interactions of these two sets are possible. In the right column of Fig. 4, some predicted multimeric co-interacting states are shown. For example, the first complex illustrates the simultaneous interaction of Chk1, RPA and 53BP2 with p53. The next complex presents the simultaneous interactions of Cdk2, RAP1A and Casp3 with p53. However, we should note that all the possible complexes we list above are derived from static structures and the dynamics of the proteins are not considered. Small overlaps can be resolved by minor conformational changes involving side chains or some small backbone rotations.
c-Abl is a proto-oncoprotein, necessary for normal growth and development. c-Abl regulates several cell cycle control genes. The interaction between c-Abl and p53 enhances p21 transcription.44 PRISM predicts the Abl–p53 interaction and their binding regions. The template for this interaction is the interface between DBD of p53 and binding protein of p53 (53BP2) (1ycsAB). Both 53BP2 and Abl contain SH3 domain and Abl matches well the 53BP2 part of the template interface. The binding site of Abl on p53 overlaps with the binding region of 53BP1, 53BP2, sv40 etc. NOXclass classifies this interaction as biological (i.e. non-crystal).
Ku80 is a repair protein which forms a heterodimer with Ku70; this heterodimer binds to broken DNA and repairs it; the Ku70–Ku80 heterodimer also has an important role in growth regulation.45 However, Ku70 and Ku80 also have functions independent from each other. Ku80 can move into the nucleus in its monomeric state independent from Ku70 using its own signals46 or may transiently interact with a partner. The PRISM results led us to propose that p53 DBD may be a potential partner of Ku80 in the nucleus. Deletion of Ku80 leads to an increase in p53-mediated DNA damage response.47 The predicted interaction between Ku80 (1jeq:B) and p53 is shown in the lower right portion of Fig. 4. This interaction is found to be 99.75% biologically relevant by NOXclass. The template interface for this putative interaction is the interface between the homodimer of the DCoH protein (1dchAB). The DCoH protein associates with specific DNA-binding proteins. Ku80 covers just about the entire DBD surface, blocking the interaction of other proteins. Its binding region covers the B1 binding region (where Cdk2, Crk, Chk1 bind), the B2 (where 53BP1, 53BP2, sv40, p53 DBD, Casp3, Cks1, c-Abl bind) and the B3 (where Rap1A, Ku70, Cdk7 bind). Thus, while proteins interacting through B1, B2 and B3 can interact with p53 simultaneously, none of these proteins can bind p53 when Ku80 is bound.
Replication protein-A (RPA) is a single-stranded DNA-binding protein which has several functions in the cell and contains three subunits. RPA interacts with several transcription factors including p53. The 32 kDa subunit of RPA (1z1d:A) matches the template interface formed between the homodimer of human Flt3 ligand (1eteAB).48
Mdm2 and its binding partners
Mdm2 is a negative regulator of p53. p53 promotes the transcription of Mdm2; in turn, Mdm2 binds to p53 and stimulates the ubiquitination of the p53 carboxy terminus, marking it for degradation. This negative feedback loop leads to oscillation in the levels of p53 and Mdm2 in the cell. Over-expression of Mdm2 leads to attenuation in the p53 response to stress signals. While the Mdm2–p53 interaction has been well studied, Mdm2 also has p53 independent functions49 and is a multi-interface cellular hub.
Mdm2–pRb interaction disrupts the pRb–E2F association
Like p53, mutated retinoblastoma protein (pRb) is observed in several human cancer types. Both p53 and pRb are inactivated in human tumor cells; the loss of their functions leads to tumor formation. Several viral groups target and inactivate these two tumor suppressors. Apart from the p53 dependent functions, Mdm2 physically and functionally interacts with pRb. Mdm2 negatively regulates pRb, similar to p53, and inhibits its regulatory growth function. pRb interacts with the transactivation domain of the E2F family transcription factors, which are important regulators of DNA synthesis and cell cycle progression, and blocks E2F dependent transcription. The Mdm2 interaction with pRb disrupts the pRb–E2F binding leading to an increase in the E2F-dependent transcription.50,51
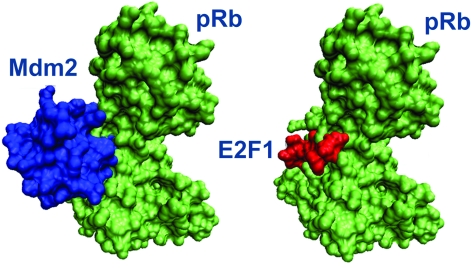
Using the interface of the homodimer of cytokine B10 (1o7zAB) as a template, PRISM predicts an Mdm2–pRb interaction. The crystal structure of the pRb–E2F1 complex is available in the PDB (1n4m; the interface is labeled as 1n4mAC).52 The pRb region interacting with E2F1 matches an Mdm2 region, suggesting that Mdm2 binds to pRb at the same region as E2F1, thus blocking its interaction with pRb (shown in Fig. 6). Consequently, the PRISM results suggest that the interactions of E2F1 and Mdm2 with pRb are mutually exclusive. Their binding sites on pRb share 16 residues, Glu533, Glu551, Glu554, His555, Ile536, Lys530, Lys537, Lys548, Lys652, Lys653, Leu649, Arg467, Arg656, Ser534, Thr645, and Val531.
Fig. 6. The Mdm2–pRb complex predicted by PRISM and the E2F1–pRb complex available in the PDB. Mdm2 associates with pRb through the same region where E2F1 interacts.
p53–Mdm2 pocket region
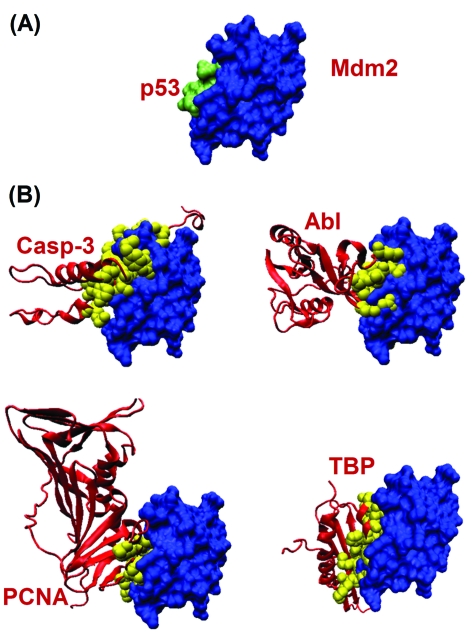
The transactivation domain of p53 interacts with the Swib domain of Mdm2. The interaction site is in a pocket region. When we focused on the PRISM-predicted putative Mdm2 interaction partners in this region, we noticed that PCNA (proliferating cell nuclear antigen), Casp3 (Caspase 3), Abl and TBP (TATA box binding protein) all bind to Mdm2, blocking its pocket region. Thus, these proteins may compete with p53. Fig. 7A illustrates the interaction between Mdm2 and the transactivation domain of p53 (PDB code: 1ycr; 1ycrAB is the PRISM labeled interface). The predicted binding sites of PCNA, Casp3, Abl and TBP are shown in Fig. 7B. This figure clearly illustrates that these putative exclusive interactions occur in the pocket region of Mdm2, just where p53 binds.
Fig. 7. Predicted partners interacting at the pocket region of Mdm2. (A) Mdm2–p53 complex taken from PDB. (B) Possible partners predicted by PRISM.
From experimental studies we know that Abl neutralizes the Mdm2-mediated degradation of p53. Abl binds to p53 and enhances its transcriptional activity, thus allowing p53 to overcome Mdm2-mediated degradation. Abl interacts with Mdm2 in vivo and in vitro. This interaction can occur in multiple Mdm2 sites.53 Here, we predict that Abl binds to Mdm2 using the Swib domain of Mdm2. This region also corresponds to the pocket region where Mdm2 binds to p53. This scenario may be a mechanism to block the Mdm2–p53 interaction decreasing Mdm2-dependent degradation of p53.
TBP is a transcription factor which binds to specific TATA box regions of DNA. TBP is predicted as a possible partner of Mdm2 protein by PRISM. The predicted binding region is illustrated in Fig. 7. This putative interaction is also available in the IntAct database.54
Mdm2–multi-interface hub protein
Mdm2 is a hub protein with multiple binding partners interacting at different binding sites. PRISM points out two distinct binding sites in the Swib domain of Mdm2. The first is detailed above. The second predicted binding site is illustrated in Fig. 8, where the left panel depicts the binding partners with their predicted binding sites. Among these, the interaction between PCAF and Mdm2 is verified in the literature. Mdm2 interacts with PCAF both in vivo and in vitro.55
Fig. 8. Predicted partners of the Swib domain of Mdm2 (left panel) and representation of some of them in the complex state (right panel).
Skp2 is also an E3 ligase like Mdm2. In several tumors, the expression levels of these two ligases are very high. However, inhibition of these ligases has more severe results in tumors.56 Mdm2 displaces Skp2 and in this way the ubiquitination of the transcription factor E2F1 is inhibited.57 The interaction between Skp2 and Mdm2 has also been validated experimentally. PRISM proposes a putative interaction between these two E3 ligases, Mdm2 and Skp2, using the template interface between the homodimer of human Flt3 ligand (1eteAB). The right panel shows some combination of interactions which can occur simultaneously; the Abl–Skp2–Mdm2, TBP–Ku80–Mdm2 and PCNA–PCAF–Mdm2 complexes are illustrated in their trimeric states. Here, none of the three proteins in each complex overlaps each other. If we find two proteins binding at different interfaces, microarray data can help in determining if they actually bind at the same time by looking at the correlation of their expression patterns. If their expression is correlated, most likely these two interactions can occur simultaneously.
Conclusions
Protein–protein interaction networks present proteins as nodes and their interactions as edges. This type of network representation is abstract; yet it is immensely useful: it provides a global picture of biological processes and protein function. At the same time, it presents a problem: some proteins have tens and even hundreds of interactions. While a node can be connected to other nodes through a large number of edges, not all edges can take place simultaneously. Since the surface area of a protein molecule is limited, a single protein cannot interact with such a large number of partners at the same time. In order to figure out which interactions can occur simultaneously and which are mutually excluded, we need to consider a fourth dimension in interaction maps, that of time. Assigning the time dimensionality in networks transforms node-and-edge maps into cellular processes, and their regulation. Here, we present a new concept integrating time into protein interaction networks using three-dimensional protein structures and interfaces. The concept is illustrated by the p53 network. p53 is a central protein, playing a key role in response to a broad range of stress signals such as DNA damage and oncogene activation; as such it has a large number of binding partners in the cell. To achieve our goal and characterize the interactions of the p53 pathway we use our PRISM server.11,13,15 PRISM is based on a simple yet well substantiated paradigm: the number of interface architectures in nature is limited. Thus, if two surface regions of two single chain proteins are similar to two sides of a crystal (or NMR) complex, they can bind. PRISM uses a template interface set to predict binding sites of two interacting proteins in a target set. As an engine, it uses an efficient structural comparison algorithm able to carry out large-scale database comparisons in impressively short time scales. Via PRISM, the structural database and the databases of interacting proteins, we are able to predict which interactions can and cannot co-exist. Here we apply this concept to the p53 and the Mdm2 hubs: for the p53 we predict four distinct binding sites on the DNA-binding domain. These sites are utilized to bind to at least 12 different proteins. Some of these interactions can occur at the same time while some others cannot. Based on this concept and strategy, we propose that Ku70, Cdk7 and RAP1A bind to p53 through the same site on p53, and hence cannot bind at the same time to p53. Additional p53 co-occurring and mutually excluded interactions are also presented. We believe that such a strategy should be immensely useful in the actual comprehension of the regulation of cellular processes beyond the common node-and-edge network picture.
Acknowledgments
O.K. acknowledges a Turkish Academy of Sciences (TUBA) Distinguished Young Investigator Award. N.T. is supported by a TUBITAK fellowship. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
†This article is part of a Molecular BioSystems themed issue on Computational and Systems Biology.
References
- Aloy P., Pichaud M., Russell R. B. Curr. Opin. Struct. Biol. 2005;15(1):15–22. doi: 10.1016/j.sbi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Aloy P., Russell R. B. Proc. Natl. Acad. Sci. U. S. A. 2002;99(9):5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P., Russell R. B. Trends Biochem. Sci. 2002;27(12):633–638. doi: 10.1016/s0968-0004(02)02204-1. [DOI] [PubMed] [Google Scholar]
- Aloy P., Russell R. B. Nat. Rev. Mol. Cell Biol. 2006;7(3):188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- Bork P., Serrano L. Cell (Cambridge, Mass.) 2005;121(4):507–509. doi: 10.1016/j.cell.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Dawelbait G., Winter C., Zhang Y., Pilarsky C., Grutzmann R., Heinrich J. C., Schroeder M. Bioinformatics. 2007;23(13):i115–i124. doi: 10.1093/bioinformatics/btm188. [DOI] [PubMed] [Google Scholar]
- Gursoy A., Keskin O., Nussinov R. Biochem. Soc. Trans. 2008;36(Pt 6):1398–1403. doi: 10.1042/BST0361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O., Gursoy A., Ma B., Nussinov R. Curr. Top. Med. Chem. 2007;7(10):943–951. doi: 10.2174/156802607780906690. [DOI] [PubMed] [Google Scholar]
- Kiel C., Beltrao P., Serrano L. Annu. Rev. Biochem. 2008;77:415–441. doi: 10.1146/annurev.biochem.77.062706.133317. [DOI] [PubMed] [Google Scholar]
- Kim P. M., Lu L. J., Xia Y., Gerstein M. B. Science. 2006;314(5807):1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- Aytuna A. S., Gursoy A., Keskin O. Bioinformatics. 2005;21(12):2850–2855. doi: 10.1093/bioinformatics/bti443. [DOI] [PubMed] [Google Scholar]
- Keskin O., Nussinov R. Structure (London) 2007;15(3):341–354. doi: 10.1016/j.str.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Keskin O., Nussinov R., Gursoy A. Methods Mol. Biol. (Totowa, N. J.) 2008;484:505–521. doi: 10.1007/978-1-59745-398-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O., Tuncbag N., Gursoy A. Curr. Pharm. Biotechnol. 2008;9(2):67–76. doi: 10.2174/138920108783955191. [DOI] [PubMed] [Google Scholar]
- Ogmen U., Keskin O., Aytuna A. S., Nussinov R., Gursoy A. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C., Wohlgemuth S., Rousseau F., Schymkowitz J., Ferkinghoff-Borg J., Wittinghofer F., Serrano L. J. Mol. Biol. 2005;348(3):759–775. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- Kortemme T., Joachimiak L. A., Bullock A. N., Schuler A. D., Stoddard B. L., Baker D. Nat. Struct. Mol. Biol. 2004;11(4):371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- Selzer T., Albeck S., Schreiber G. Nat. Struct. Biol. 2000;7(7):537–541. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- Sen T. Z., Kloczkowski A., Jernigan R. L. BMC Bioinformatics. 2006;7:355. doi: 10.1186/1471-2105-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. D., Bertin N., Hao T., Goldberg D. S., Berriz G. F., Zhang L. V., Dupuy D., Walhout A. J., Cusick M. E., Roth F. P., Vidal M. Nature. 2004;430(6995):88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Komurov K., White M. Mol. Syst. Biol. 2007;3:110. doi: 10.1038/msb4100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lichtenberg U., Jensen L. J., Brunak S., Bork P. Science. 2005;307(5710):724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- Tuncbag N., Gursoy A., Guney E., Nussinov R., Keskin O. J. Mol. Biol. 2008;381(3):785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O., Tsai C. J., Wolfson H., Nussinov R. Protein Sci. 2004;13(4):1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Domingues F. S., Sommer I., Lengauer T. BMC Bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guney E., Tuncbag N., Keskin O., Gursoy A. Nucleic Acids Res. 2008;36:D662–D666. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn K. W. Mol. Biol. Cell. 1999;10(8):2703–2734. doi: 10.1091/mbc.10.8.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NACCESS [http://www.bioinf.manchester.ac.uk/naccess/].
- Shatsky M., Dror O., Schneidman-Duhovny D., Nussinov R., Wolfson H. J. Nucleic Acids Res. 2004;32:W503–W507. doi: 10.1093/nar/gkh413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky M., Nussinov R., Wolfson H. J. Proteins: Struct., Funct., Genet. 2004;56(1):143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- Dartnell L., Simeonidis E., Hubank M., Tsoka S., Bogle I. D., Papageorgiou L. G. FEBS Lett. 2005;579(14):3037–3042. doi: 10.1016/j.febslet.2005.03.101. [DOI] [PubMed] [Google Scholar]
- Salwinski L., Miller C. S., Smith A. J., Pettit F. K., Bowie J. U., Eisenberg D. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G. D., Betel D., Hogue C. W. Nucleic Acids Res. 2003;31(1):248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A., Ceol A., Palazzi L. M., Nardelli G., Schneider M. V., Castagnoli L., Cesareni G. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. Science. 1996;274(5289):948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Rosal R., Pincus M. R., Brandt-Rauf P. W., Fine R. L., Michl J., Wang H. Biochemistry. 2004;43(7):1854–1861. doi: 10.1021/bi035718g. [DOI] [PubMed] [Google Scholar]
- Meek D. W., Eckhart W. Mol. Cell. Biol. 1988;8(1):461–465. doi: 10.1128/mcb.8.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A., Anderson C. W., Carroll R. B. Proc. Natl. Acad. Sci. U. S. A. 1986;83(4):897–901. doi: 10.1073/pnas.83.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Eckhart W. Proc. Natl. Acad. Sci. U. S. A. 1992;89(10):4231–4235. doi: 10.1073/pnas.89.10.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Sakanyan V., Charlier D., Glansdorff N., Van Duyne G. D. Nat. Struct. Biol. 1999;6(5):427–432. doi: 10.1038/8229. [DOI] [PubMed] [Google Scholar]
- Shulman-Peleg A., Shatsky M., Nussinov R., Wolfson H. J. Nucleic Acids Res. 2008;36:W260–W264. doi: 10.1093/nar/gkn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall M. D., Scott M. S., Barton G. J. Nucleic Acids Res. 2009;37:D651–D656. doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Wang M., Tang W., Qi T., Gu C., Hao S., Zeng X. J. Biochem. 2007;141(5):621–626. doi: 10.1093/jb/mvm068. [DOI] [PubMed] [Google Scholar]
- Walker J. R., Corpina R. A., Goldberg J. Nature. 2001;412(6847):607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Koike M., Ikuta T., Miyasaka T., Shiomi T. Oncogene. 1999;18(52):7495–7505. doi: 10.1038/sj.onc.1203247. [DOI] [PubMed] [Google Scholar]
- Holcomb V. B., Rodier F., Choi Y., Busuttil R. A., Vogel H., Vijg J., Campisi J., Hasty P. Cancer Res. 2008;68(22):9497–9502. doi: 10.1158/0008-5472.CAN-08-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides S. N., Boone T., Andrew Karplus P. Nat. Struct. Biol. 2000;7(6):486–491. doi: 10.1038/75896. [DOI] [PubMed] [Google Scholar]
- Daujat S., Neel H., Piette J. Trends Genet. 2001;17(8):459–464. doi: 10.1016/s0168-9525(01)02369-1. [DOI] [PubMed] [Google Scholar]
- Martin K., Trouche D., Hagemeier C., Sorensen T. S., La Thangue N. B., Kouzarides T. Nature. 1995;375(6533):691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- Xiao Z. X., Chen J., Levine A. J., Modjtahedi N., Xing J., Sellers W. R., Livingston D. M. Nature. 1995;375(6533):694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- Lee C., Chang J. H., Lee H. S., Cho Y. Genes Dev. 2002;16(24):3199–3212. doi: 10.1101/gad.1046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Z., Vogt Sionov R., Berger M., Zwang Y., Perets R., Van Etten R. A., Oren M., Taya Y., Haupt Y. EMBO J. 2002;21(14):3715–3727. doi: 10.1093/emboj/cdf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S., Alam-Faruque Y., Aranda B., Bancarz I., Bridge A., Derow C., Dimmer E., Feuermann M., Friedrichsen A., Huntley R., Kohler C., Khadake J., Leroy C., Liban A., Lieftink,et al. C. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Zeng S. X., Dai M. S., Yang X. J., Lu H. J. Biol. Chem. 2002;277(34):30838–30843. doi: 10.1074/jbc.M204078200. [DOI] [PubMed] [Google Scholar]
- Garber K. J. Natl. Cancer Inst. 2005;97(3):166–167. doi: 10.1093/jnci/97.3.166. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang H., Li M., Rayburn E. R., Agrawal S., Zhang R. Oncogene. 2005;24(48):7238–7247. doi: 10.1038/sj.onc.1208814. [DOI] [PubMed] [Google Scholar]