 Combining synthesis and mutagenesis, we show that a cation–π interaction between adenine of adenophostin analogues and Arg504 of IP3 receptors (IP3R) is responsible for the enhanced activity of adenophostins and can replace a phosphate–receptor interaction.
Combining synthesis and mutagenesis, we show that a cation–π interaction between adenine of adenophostin analogues and Arg504 of IP3 receptors (IP3R) is responsible for the enhanced activity of adenophostins and can replace a phosphate–receptor interaction.
Abstract
Ca2+ release by d-myo-inositol 1,4,5-trisphosphate receptors (IP3Rs) is widely considered to require the vicinal 4,5-bisphosphate motif of IP3, with P-5 and P-4 engaging the α and β domains of the binding site; using synthesis and mutagenesis we show that the adenine of synthetic glyconucleotides, through an interaction with Arg504, can replace the interaction of either P-1 or P-5 with the α-domain producing, respectively, the most potent bisphosphate agonist yet synthesised and the first agonist of IP3R without a vicinal bisphosphate motif; this will stimulate new approaches to IP3R ligand design.
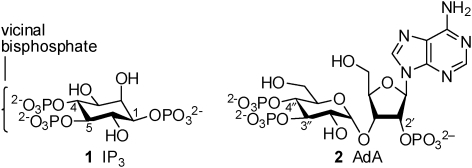
d-myo-Inositol 1,4,5-trisphosphate (IP3, 1, Fig. 1) is an intracellular messenger that evokes Ca2+ release from the intracellular stores of most animal cells by binding to IP3 receptors (IP3R), which are IP3-gated Ca2+ channels.1,2 Extensive structure–activity studies suggest that the vicinal 4,5-bisphosphate structure of IP3 is essential, while the 1-phosphate has an ancillary role that substantially increases affinity.3 For more than 20 years, this enduring conclusion has guided design of new ligands for the IP3R. Other inositol phosphate regioisomers are active through different binding modes,4 but they all have the vicinal bisphosphate structure. The fungal metabolite, adenophostin A (AdA, 2, Fig. 1),5 which is at least ten times more potent than IP3, provides additional opportunities to explore the mechanisms of IP3R activation. AdA is a glyconucleotide trisphosphate structurally related to IP3. It binds to a site that substantially overlaps the IP3-binding site,6 and structure–activity studies with synthetic analogues of AdA have established that the adenine (or another aromatic group) is essential for its enhanced affinity.6,7
Fig. 1. IP3 receptor ligands; IP3 (1) and adenophostin A (2).
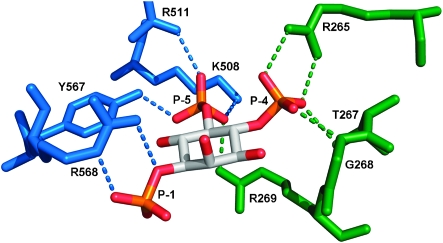
The IP3-binding core (IBC) of the IP3R forms a clam-like structure with IP3 held between two domains (α and β) linked by a flexible hinge.8 The 4-phosphate group (P-4) of IP3 interacts mainly with residues in the β-domain, while the 1-phosphate (P-1) and 5-phosphate (P-5) interact predominantly with the α-domain (Fig. 2). We therefore speculated that binding of IP3 might pull the two domains together in a manner reminiscent of glutamate binding to ionotropic glutamate receptors,9 and so cause the IBC to adopt a more constrained structure.10 Because both P-1 and P-5 interact with the α-domain, it may be possible to bridge the two domains using a ligand with only one of these groups. The fact that the bisphosphate Ins(4,5)P2 is a full agonist with low affinity11 supports this suggestion. But the inability11 of Ins(1,4)P2 to mobilise Ca2+ or bind to IP3R suggests that interaction of P-1 alone with the α-domain, where it directly contacts only Arg568 (R568, Fig. 2), is too weak to stabilise binding. Ins(1,5)P2 is also inactive12 presumably because it has insufficient interactions with the β-domain. Hence, the prevailing view is that the vicinal bisphosphate moiety of IP3 is essential for activity.
Fig. 2. Interactions of IP3 with the α (blue) and β (green) domains of the IBC of IP3R1 (PDB code 1n4k). Water molecules are omitted and only direct interactions between IP3 and residues in the IBC are shown.
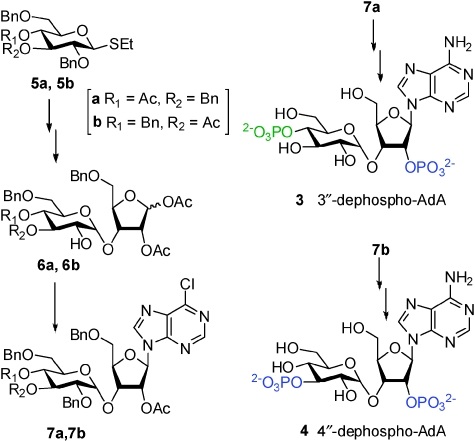
Our molecular docking experiments13 have suggested that the 2′-AMP motif of AdA interacts with the α-domain of the IBC, perhaps more strongly than P-1 of IP3. We therefore reasoned that it may be possible for 3″-dephospho-AdA (3, Scheme 1) to bridge the domains effectively and release Ca2+. Alternatively, if the 2′-AMP interacts with the β-domain, it may allow 4″-dephospho-AdA (4) to activate IP3R. We therefore developed methods to synthesise 3 and 4 (Scheme 1).‡ Briefly, thioglycosides 5a and 5b were used to glycosylate a ribofuranose acceptor leading to disaccharides 6a and 6b, which were individually subjected to Vorbrüggen condensations with silylated 6-chloropurine to give 7a and 7b, the precursors for 3 and 4, respectively. Ammonolysis of each installed the N-6 amino group and exposed the required pairs of hydroxyl groups for phosphorylation. Phosphitylation in the presence of imidazolinium triflate and in situ oxidation followed by removal of benzyl protecting groups by transfer hydrogenolysis gave 3 and 4, which were purified to homogeneity.
Scheme 1. Structures and syntheses of 3″ -and 4″-dephospho-AdA. Phosphate groups are coloured according to putative interactions with the α (blue) or β (green) domains of the IBC. Bn = benzyl.
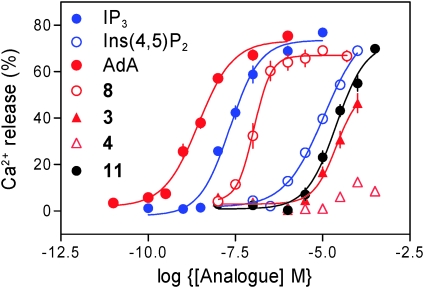
Using methods reported previously,14 we show that 4 does not evoke Ca2+ release via recombinant IP3R1, but 3 is effective, albeit at high concentrations (Fig. 3). These results support the idea that P-3″ and P-4″ of AdA mimic P-5 and P-4 of IP3, respectively, and that the 2′-AMP motif of AdA is able to bind the α-domain of the IBC more effectively than P-1 of IP3. This allows bisphosphate 3 to pull the two domains together and activate IP3R1, even though it lacks a vicinal bisphosphate. Bisphosphate 3, the AdA equivalent of Ins(1,4)P2, is the first agonist of IP3R without a vicinal bisphosphate motif.
Fig. 3. Concentration-dependent effects of ligands on Ca2+ release via IP3R1 stably expressed in DT40 cells. Results show means ± SEM; n = 8.
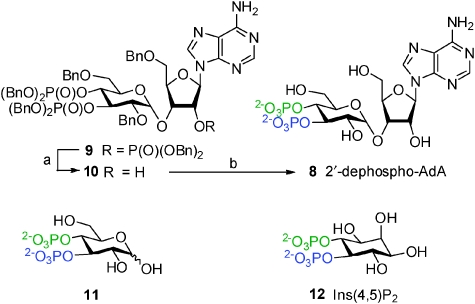
The 2′-AMP motif of AdA may allow P-2′ to bind more effectively than P-1 of IP3, or the adenosine itself may have interactions with the IBC. To distinguish between these two possible roles, we synthesised 2′-dephospho-AdA, the adenophostin equivalent of Ins(4,5)P2 (8, Scheme 2). Interestingly, treatment of a benzyl-protected version of AdA (9)15 with sodium benzoxide in BnOH led to removal of the 2′-dibenzylphosphate group giving bisphosphate 10 as the major product with minor amounts of other by-products. Use of the milder base potassium carbonate resulted in clean and highly selective conversion into 10. Deprotection and purification as before gave 8 in excellent yield. The 2′-phosphate triester in 9 is comparatively less crowded and hence more prone to nucleophilic attack than those in the 3″,4″ vicinal positions and this is likely to be the reason for this interesting high selectivity. We also synthesised the related glucose 3,4-bisphosphate (11, Scheme 2). Because 11 can be viewed as AdA without the 2′-AMP motif, any difference in the activities of 8 and 11 illustrates the role of the adenosine moiety. In functional assays, 8 was 100 times more potent than 11 (Fig. 3), confirming that the adenosine motif directly contributes to AdA binding even when it lacks the 2′-P group. Compound 8 is the first potent synthetic bisphosphate agonist of the IP3R; it is >30-fold more potent than Ins(4,5)P2 (Fig. 3, Table 1).
Scheme 2. Synthesis of 2′-dephospho-AdA and structures of glucose 3,4-bisphosphate and Ins(4,5)P2: (a) K2CO3, BnOH, 70 °C, 93%; (b) Pd(OH)2/C, cyclohexene, MeOH, H2O, 80 °C, 92%. Bn = benzyl. Phosphate groups are coloured according to putative interactions with the α (blue) or β (green) domains of the IBC.
Table 1. Ca2+ release via IP3R1. Results (means ± SEM, n = 3–8) show the EC50 values for each agonist in DT40 cells stably expressing IP3R1.
| Agonist | EC50 |
| IP3 | 35 ± 5 nM |
| Ins(4,5)P2 | 7.59 ± 2.63 μM |
| AdA | 2.9 ± 0. 9 nM |
| 8 | 240 ± 5 nM |
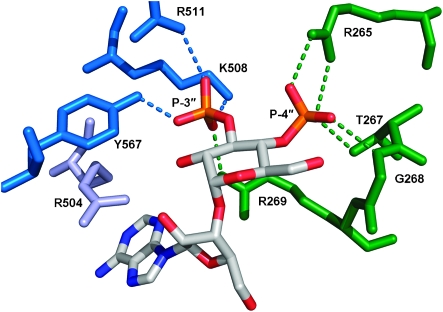
Our molecular modelling suggests a key role for arginine 504 (R504) in recognising the adenine of AdA, perhaps via a cation–π interaction.13 Using targeted mutagenesis, we established a cell line expressing only IP3R1 with R504 mutated to glutamine, to determine whether R504 selectively contributes to the activity of AdA analogues. For the mutant IP3R, the EC50 for IP3 and Ins(4,5)P2 was reduced by 57- and 8.8-fold, respectively. For AdA and 8, the reductions were 434- and 107-fold, and the response to 3 (≤3 mM) was abolished. The greater effect of this mutation on the potency of AdA and 8 is consistent with the proposed interaction between adenine and R504 (Fig. 4). Indeed, the potency of AdA and 8 at IP3R1 both depend largely on R504 because, at the mutated receptor, AdA is only equipotent with IP3, and 8 is less than 3-fold more potent than Ins(4,5)P2.
Fig. 4. Suggested interactions of 8 with the IBC of IP3R1, based on molecular docking experiments using AdA.13 The side chain of R504 is shown in pale blue.
We conclude that, while the three phosphate groups and adenine of AdA most likely make incremental contributions to IP3R binding, a vicinal bisphosphate moiety is not essential for IP3R activation. P-4 of IP3, which contacts the β-domain of the IBC, is required, but P-5, which contacts the α-domain, can (as in 3) make an interaction that can be substantially compensated by a cation–π interaction between the adenine of AdA and R504 in the α-domain. The same interaction can also serve to mitigate the loss of P-1 to provide a potent agonist (8) with only two phosphate groups. Aside from challenging the long-standing dogma that a vicinal bisphosphate is essential for agonists of IP3R, these results illustrate the potential to design less polar IP3R ligands. At their simplest, these might comprise two motifs that interact with the IBC domains linked by a suitable spacer.
Since inositol polyphosphates often bind to sites rich in Arg and Lys residues, replacing interactions with a polar phosphate by cation–π interactions should have more general applications in the chemical biology of inositol phosphate signalling and probably elsewhere.
We acknowledge the support by Programme Grants from the Wellcome Trust [082837 to AMR (Bath) and BVLP, 072084 to CWT].
Footnotes
†Dedicated to Dr Melanie N. Trusselle (1973–2008).
‡All new compounds were thoroughly characterised and exhibited satisfactory parameters using standard spectroscopic techniques. A typical example follows. Synthesis of 3′-O-(α-d-glucopyranosyl)-adenosine-3″,4″-bisphosphate (8). A suspension of bisphosphate 10 (45 mg, 0.037 mmol) and 20% Pd(OH)2 on carbon (120 mg) in a mixture of cyclohexene (1.6 mL), MeOH (3 mL) and H2O (0.22 mL) was heated at 80 °C overnight. The mixture was filtered through a membrane filter and solvents evaporated in vacuo. Purification on an AG ion-exchange column (0–100% gradient elution, with 150 mM TFA and water) afforded pure bisphosphate 8 (20 mg, 92%). 1H NMR (400 MHz, D2O): 3.74–3.90 (m, 6H, H-2″, H-5″, H-5A′, H-5B′ H-6A″, H-6B″ ), 4.10 (ddd, like a q, 1H, 9.66 Hz, 9.66 Hz, 9.66 Hz, H-4″), 4.42 (dd, 1H, 7.24 Hz, 3.86 Hz, H-4′), 4.47–4.55 (m, 2H, H-3′, H-3″ ), 4.87 (dd, like a t, 1H, 5.80 Hz, 5.31 Hz, H-2′), 5.19 (d, 1H, 3.38 Hz, H-1″), 6.19 (d, 1H, 5.80 Hz, H-1′), 8.40 (s, 1H, H-2), 8.50 (s, 1H, H-8). 13C NMR (100 MHz, D2O): 60.08 (C-6″), 60.79 (C-5′), 70.48 (C-2″, 31P coupled), 71.60 (d, 3.83 Hz, C-5″, 31P coupled), 72.83 (C-4″, 31P coupled), 73.48 (C-2′), 76.37 (C-3′), 77.71 (C-3″, 31P coupled), 83.95 (C-4′), 88.23 (C-1′), 99.02 (C-1″), 118.91 (C-5), 142.81 (C-8), 144.43 (C-2), 148.16 (C-4), 149.91 (C-6). 31P NMR (161.94 MHz, D2O with excess of TEA): 4.50, 3.62. m/z (ES+) = 590.2 [(M + H)+, 100%]; 612.1 [(M + Na)+, 100%]; m/z (ES–) 588.2 [(M – H)+, 100%]; HRMS: mass calcd for C16H26O15N5P2 [M + H]+, 590.0895; found, 590.0895.
References
- Berridge M. J. Nature. 1993;361:315. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- Foskett J. K., White C., Cheung K. H., Mak D.-O. D. Physiol. Rev. 2007;87:593. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter B. V. L., Lampe D. Angew. Chem., Int. Ed. Engl. 1995;34:1933. [Google Scholar]
- Riley A. M., Payne R., Potter B. V. L. J. Med. Chem. 1994;37:3918. doi: 10.1021/jm00049a011. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Tanzawa K., Takahashi S. J. Biol. Chem. 1994;269:369. [PubMed] [Google Scholar]
- Correa V. A., Riley A. M., Shuto S., Horne G., Nerou E. P., Marwood R. D., Potter B. V. L., Taylor C. W. Mol. Pharmacol. 2001;59:1206. doi: 10.1124/mol.59.5.1206. [DOI] [PubMed] [Google Scholar]
- Hotoda H., Murayama K., Miyamoto S., Iwata Y., Takahashi M., Kawase Y., Tanzawa K., Kaneko M. Biochemistry. 1999;38:9234. doi: 10.1021/bi990114r. [DOI] [PubMed] [Google Scholar]
- Bosanac I., Alattia J.-R., Mal T. K., Chan J., Talarico S., Tong F. K., Tong K. I., Yoshikawa F., Furuichi T., Iwai M., Michikawa T., Mikoshiba K., Ikura M. Nature. 2002;420:696. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- Taylor C. W., da Fonseca P. C. A., Morris E. P. Trends Biochem. Sci. 2004;29:210. doi: 10.1016/j.tibs.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Chan J., Whitten A. E., Jeffries C. M., Bosanac I., Mal T. K., Ito J., Porumb H., Michikawa T., Mikoshiba K., Trewhella J., Ikura M. J. Mol. Biol. 2007;373:1269. doi: 10.1016/j.jmb.2007.08.057. [DOI] [PubMed] [Google Scholar]
- Lu P. J., Gou D. M., Shieh W. R., Chen C. S. Biochemistry. 1994;33:11586. doi: 10.1021/bi00204a021. [DOI] [PubMed] [Google Scholar]
- Bello D., Aslam T., Bultynck G., Slawin A. M. Z., Roderick H. L., Bootman M. D., Conway S. J. J. Org. Chem. 2007;72:5647. doi: 10.1021/jo070611a. [DOI] [PubMed] [Google Scholar]
- Rosenberg H. J., Riley A. M., Laude A. J., Taylor C. W., Potter B. V. L. J. Med. Chem. 2003;46:4860. doi: 10.1021/jm030883f. [DOI] [PubMed] [Google Scholar]
- Laude A. J., Tovey S. C., Dedos S. G., Potter B. V. L., Lummis S. C. R., Taylor C. W. Cell Calcium. 2005;38:45. doi: 10.1016/j.ceca.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Borissow C. N., Black S. J., Paul M., Tovey S. C., Dedos S. G., Taylor C. W., Potter B. V. L. Org. Biomol. Chem. 2005;3:245. doi: 10.1039/b415229h. [DOI] [PubMed] [Google Scholar]