 Using sensitive protein sequence profile analyses we investigated the evolution of some histone chaperones and showed that Hir3p and Hpc2p have a much wider phyletic pattern than was previously known.
Using sensitive protein sequence profile analyses we investigated the evolution of some histone chaperones and showed that Hir3p and Hpc2p have a much wider phyletic pattern than was previously known.
Abstract
While histone chaperones have been intensely studied, the roles of components of the Hir–Asf1 histone chaperone complex such as Hir3p and Hpc2p are poorly understood. Using sensitive protein sequence profile analyses we investigated the evolution of these proteins and showed that Hir3p and Hpc2p have a much wider phyletic pattern than was previously known. We established the animal histone-deacetylase-complex-interacting proteins, CAIN/CABIN, to be orthologs of Hir3p. They contain a conserved core of around 30 TPR-like bi-helical repeats that are likely to form a super-helical scaffold. We identified a conserved domain, the HUN domain, in all Hpc2p homologs, including animal ubinuclein/yemanuclein and the recently discovered vertebrate cell-cycle regulator FLJ25778. The HUN domain has a characteristic pattern of conserved acidic residues based on which we predict that it is a previously unrecognized histone-tail-binding chaperone. By analyzing various high-throughput data sets, such as RNAi knock-downs, genetic and protein interaction maps and cell-cycle-specific gene expression data, we present evidence that Hpc2p homologs might be deployed in specific processes of chromatin dynamics relating to cell-cycle progression in vertebrates and schizogony in Plasmodium. Beyond the conserved HUN domain these proteins show extensive divergence patterns in different eukaryotic lineages. Hence, we propose that Hpc2p homologs are probably involved in recruitment of the ancient conserved histone-loading Hir–Asf1 complex to different lineage-specific chromatin reorganization processes.
Introduction
The unique presence of long positively charged tails in eukaryotic nucleosomal histones appears to have provided the selective pressure for evolution of a panoply of histone chaperones.1 This diverse group of proteins escorts histones right from the point of their synthesis, through their entry into the nucleus, to their repeated deposition on DNA during chromatin assembly and remodeling.1–3 The functions performed by different histone chaperones include histone storage (nucleoplasmin), shuttling of histone to the nucleus (NAP-1), and deposition of nucleosomes in conjunction with specific chromatin remodeling complexes (various nuclear histone chaperones).1,2,4–15 This latter group includes complexes with specific temporal roles, such as the CAF-1 complex required for histone deposition during replication, the FACT complex required for histone disassembly and reassembly during transcription elongation and the Hir complex that appears to be active throughout the cell cycle.1,2,10,16–21 Also included in this group are complexes specific to certain histone variants such as Swr1–Chz1 system involved in the incorporation of H2AZ and Scm3 required for cenH3 assembly.1,3,22–26 Of the characterized histone chaperones, Asf1 is required for both replication-dependent and independent nucleosome assembly via direct binding of the H3–H4 dimer.2,19,20,27 In these two distinct processes it, respectively, associates with either of two paralogous WD40 repeat proteins, Cac2p and HirA (Hir1p and Hir2p in Saccharomyces cerevisiae) to form two distinct chaperone complexes namely CAF-1 and Hir.20,28–31 Biochemical studies in S. cerevisiae have shown that the complete Hir complex comprises 4 proteins (Hir1p, Hir2p, Hir3p and Hpc2p), which also show functional interactions with a wide range of chromatin associated complexes, such as the FACT, CAF-1, PAF1 and Swi2–Snf2 complexes.19,21,32–36 Additionally, genetic studies also imply that the Hir complex might function independently of Asf1 in maintenance of kinetochore chromatin structure.32,33,36,37 Taken together the evidence suggests that the Hir complex and Asf1p are important generalized histone chaperones providing functional backup for a number of functionally specialized complexes involved in nucleosome deposition and remodeling.
Evolutionary comparisons have shown that Asf1p is highly conserved throughout eukaryotes—it is present even in organisms with highly reduced functional systems, such as microsporidians, the nucleomorph of Guillardia theta and Giardia.27,38 This suggests that Asf1 is indeed the key generalized histone chaperone that was already present in the last common ancestor of extant eukaryotes. The evolutionary history of its functional partners in generalized histone deposition, i.e. the components of the Hir complex, and the differentiation of the replication-linked Caf-1 complex are less understood. The core WD40 subunits of these complexes HirA and Cac2 are known to be widely conserved in various lineages of the eukaryotic crown group (fungi, animals and plants), with comparable functions and interactions.1 But the exact role and evolutionary pattern of the other subunits, Hir3p and Hpc2p, are unclear. We were specifically interested in understanding the origins of these less-studied Hir complex components and potentially uncovering previously uncharacterized features of their role in nucleosome assembly and deposition. Utilizing several distinct computational approaches we show that Hpc2p contains an ancient conserved domain that is likely to have a key pan-eukaryotic role in the nucleosome assembly mediated by the Hir–Asf1 complex. We also show that Hir3p is a rapidly diverging member of the complex that has possibly undergone extensive functional differentiation in the eukaryotic crown group.
Results and discussion
Phyletic and divergence patterns of components of the Hir complex suggest possible functional shifts in eukaryotic evolution
Phyletic patterns and evolutionary divergence of proteins often provide a good estimate of their functional relevance and changes in their interacting partners. They might also throw new light on their biochemical roles. To determine if components of the Hir complex showed a conservation pattern similar to Asf1p we systematically identified their orthologs across a panel of eukaryotes that included the crown group (animals, fungi, amoebozoans and plants), the Guillardia nucleomorph, chromalveolates (apicomplexans, ciliates, oomycetes and diatoms), heteroloboseans, kinetoplastids, parabasalids and diplomonads (Table S1, ESI†). The physically interacting partner of Asf1p, HirA is characterized by N- and C-terminal WD40 repeat β-propellers with an intervening linker containing a motif implicated in interactions with Asf1.1 While this configuration is conserved throughout the crown group, in saccharomycete yeasts the HirA ortholog has undergone a lineage-specific duplication spawning two genes Hir1 and Hir2, which have, respectively, diverged rapidly in their C- and N-terminal β-propellers. The primary conserved subunit of the Caf-1 complex, Cac2p is a paralog of HirA that appears to contain only an N-terminal β-propeller followed by a C-terminal Asf1p-interacting region.1,31,39 We observed that Trichomonas and Giardia contained proteins that appeared to be comparably related to both Cac2p and HirA (ESI†) suggesting that the differentiation into replication-dependent and independent chaperones had not yet occurred in the ancestral eukaryotes. All other eukaryotic lineages showed a clear differentiation of two paralogous groups (ESI†), respectively, representing HirA and Cac2 implying that the differentiation of general and temporally specific histone chaperones first emerged prior to the separation of the heterolobosean–kinetoplastid clade from other eukaryotes. However, certain lineages show the apparent loss of either the Cac2p ortholog (e.g. the Guillardia nucleomorph) or the HirA ortholog (e.g. kinetoplastids) suggesting a continued functional overlap between them. Nevertheless, the presence of at least one of the two paralogs or their common ortholog (Trichomonas and Giardia) in all eukaryotes, even the most reduced genomes, suggests that a HirA-like β-propeller provides an essential scaffold for H3–H4 assembly in conjunction with the Asf1 chaperone.
Hir3p orthologs are thus far only known from ascomycete fungi. By means of sequence profile searches with the PSI-BLAST program we were able to identify homologs of Hir3p in animals, amoebozoans, plants, chlorophyte alga, chromists and heteroloboseans (ESI†). Relative to the orthologs of the other Hir and Caf-1 complex components, such as Asf1, HirA and Cac2, these newly detected Hir3 homologs were extremely divergent in sequence (ESI†, requiring over 3 more PSI-BLAST iterations for recovery). However, this set of Hir3 homologs consistently recover each other in reciprocal searches and are present in a single copy in most eukaryotic lineages in which they are detected. This strongly suggests that they are the divergent orthologs of Hir3 in other eukaryotes. The animal ortholog of Hir3p is the calcineurin-binding protein Cabin1/Cain (e = 10–5 in iteration 3). While Cabin1/Cain has as yet not been shown to interact with other histone chaperones, it does function as a corepressor for specific transcription factors like Mef-2 by binding Sin3 and recruiting histone deacetylases.40,41 This is consistent with the repressive role of the fungal Hir complex and supports a conserved histone-associated role for the orthologs of Hir3 in other eukaryotes. Interestingly, we observed that the Hir3–Cabin1/Cain orthologs from ascomycete fungi are rapidly diverging relative to the orthologs from other crown-group eukaryotes (ESI†). This indicates a functional shift in them, perhaps related to the loss in ascomycetes of the calcium-dependent function observed in animal Cabin1/Cain proteins.40–43 The phyletic patterns of Hir3–Cabin1/Cain orthologs suggest that it probably emerged a little later in eukaryotic evolution relative to HirA–Cac2 and Asf1. It was sporadically lost in several eukaryotic lineages (e.g. alveolates, basidiomycetes, kinetoplastids and reduced genomes like microsporidians and nucleomorphs) suggesting that it is not an absolutely required component of the Hir complex. We identified a conserved core in all eukaryotic Hir3p orthologs containing about 30 tetratricopeptide repeats (TPRs) that are likely to constitute multi-rung super-helical α–α superhelix (ESI†). It has been previously observed that in certain cellular complexes TPR and β-propeller proteins cooperate to form structural scaffolds.44 Consistent with this, Hir3 might cooperate with the β-propellers of HirA to possibly constitute the scaffold of this histone chaperone complex with several distinct interaction sites.
Identification of a novel conserved domain in Hpc2 and its functional implications
Like Hir3p, Hpc2p has hitherto only been identified in ascomycete fungi. Compositional entropy analysis of the protein with the SEG program45 showed that it is a predominantly low complexity protein with a small predicted folded region at the C-terminus. To identify potential Hpc2p homologs outside of fungi we performed PSI-BLAST searches with a position specific score matrix for this region. These searches recovered as significant hits from several eukaryotes including nuclear proteins from Drosophila and mammals, respectively, yemanuclein-α and ubinuclein-1(Ubn1) and the potential mammalian cell-cycle regulator FLJ25778 (KIAA2030).46 We then prepared a hidden Markov Model of the alignment of this region culled from the PSI-BLAST searches and scanned all eukaryotic proteomes with the HMMER package to identify homologous regions. Consequently, we recovered significant hits (e < 10–4) to this region in all eukaryotic lineages with the exception of Giardia, kinetoplastids and the Guillardia nucleomorph. The relationship of the region in these newly detected eukaryotic proteins with the globular region of ascomycete Hpc2 was further confirmed by means of profile–profile comparisons with the HHpred program (p-value 10–8). We accordingly named this small conserved region (∼45–55 residues in length) the HUN (for HPC2-Ubinuclein-1) domain. The above phyletic patterns also indicated that the HUN domain was already present in the last eukaryotic common ancestor (LECA) and that the majority of eukaryotic lineages contain a homolog of yeast Hpc2p (Fig. 1). While in S. cerevisiae and related ascomycetes the HUN domain is found at the C-terminus of a largely non-globular Hpc2p, homologs from basidiomycetes, animals and plants contain the HUN domain at the N-terminus (Fig. 1). These latter proteins contain a unique all α-helical domain at their C-termini (Fig. 1, ESI†). Thus, ascomycete Hpc2 appears to have lost this domain after their divergence and undergone a drastic reorganization. Thus divergence patterns of the HUN domain proteins to certain extent parallel that of Hir3; i.e. rapid divergence in ascomycetes relative to other crown group eukaryotes. Likewise, in apicomplexan Hpc2 homologs the HUN domain is linked to a unique α-helical N-terminal domain that is not in any other lineage (Fig. 1).
Fig. 1. Multiple sequence alignment and domain architectures of the HUN domain. Proteins are denoted by their gene name, species abbreviations and gi. Amino acids are colored based on their functional properties and conservation (>80%) in the alignment. The predicted secondary structure is shown at the top with α-helices represented as orange cylinders and β-strands as green block arrows. Domain architectures of HUN containing protein are shown to the right. Consensus abbreviations and coloring scheme are as follows: polar (p: QNSTCHKRDE) residues shaded blue, hydrophobic (h: ACFMWYIVL), aliphatic (l: IVL) and aromatic (a: FYWH) residues colored yellow, acidic (‘-’: DE) residues shaded red, small (s: GASVTDNPC) and tiny (u: GAS) residues colored green. Species abbreviations are as follows: Acap: Ajellomyces capsulatus; Afum: Aspergillus fumigatus; Agam: Anopheles gambiae; Amel: Apis mellifera; Anid: Aspergillus nidulans; Anig: Aspergillus niger; Aory: Aspergillus oryzae; Atha: Arabidopsis thaliana; Bbov: Babesia bovis; Bfuc: Botryotinia fuckeliana; Bmal: Brugia malayi; Calb: Candida albicans; Ccin: Coprinopsis cinerea; Cgla: Candida glabrata; Cglo: Chaetomium globosum; Cneo: Cryptococcus neoformans; Cpar: Cryptosporidium parvum; Crei: Chlamydomonas reinhardtii; Ddis: Dictyostelium discoideum; Dmel: Drosophila melanogaster; Dpse: Drosophila pseudoobscura; Drer: Danio rerio; Ecun: Encephalitozoon cuniculi; Ehis: Entamoeba histolytica; Ggal: Gallus gallus; Gzea: Gibberella zeae; Hsap: Homo sapiens; Klac: Kluyveromyces lactis; Mdom: Monodelphis domestica; Mglo: Malassezia globosa; Mgri: Magnaporthe grisea; Mmus: Mus musculus; Ncra: Neurospora crassa; Nvec: Nematostella vectensis; Nvit: Nasonia vitripennis; Osat: Oryza sativa; Otau: Ostreococcus tauri; Pfal: Plasmodium falciparum; Pnod: Phaeosphaeria nodorum; Ppat: Physcomitrella patens; Ptet: Paramecium tetraurelia; Pviv: Plasmodium vivax; Scer: Saccharomyces cerevisiae; Spom: Schizosaccharomyces pombe; Spur: Strongylocentrotus purpuratus; Tadh: Trichoplax adhaerens; Tann: Theileria annulata; Tnig: Tetraodon nigroviridis; Tpar: Theileria parva; Tthe: Tetrahymena thermophila; Tvag: Trichomonas vaginalis; Umay: Ustilago maydis; Vvin: Vitis vinifera; X(Si): Xenopus (Silurana); Ylip: Yarrowia lipolytica.
The mutual information derived from the phyletic pattern of the HUN domain proteins is strongly correlated with Hir1/2 and Asf1 (>0.9) (Table S2 and Fig. S7, ESI†). These observations and the low copy number of the Hpc2 homologs point to a common conserved function for HUN domain throughout eukaryotes. The domain contains a prominent central helix followed by a region predicted to adopt an extended conformation (Fig. 1). The characteristic conserved acidic residues of the HUN domain display a periodic signature of the form DX2DX3DX2EX2–3[ED] (where X is any amino acid) and are mainly associated with the central helix. Hence, these residues are most likely to form a contiguous negatively charged exposed ridge on the protein. Crystal structures as well as mutagenesis studies have implicated the two other associated histone chaperones Asf1p and HirA in interacting with the globular head domains of the histone H3–H4 complex, but there is no evidence for these chaperones directly interacting with the non-globular tails.27,38,47–49 Based on the presence of the conserved acidic ridge we predict that the HUN domain is likely to be the histone-tail-binding subunit of the Hir complex. This prediction implies that the conserved HUN domain was an important innovation in LECA that was needed for deposition of the highly charged eukaryotic histones. Available experimental evidence on functions of HUN domain proteins from other eukaryotes, while not extensive, is consistent with the above proposed function. Both yem-α and Ubn1 have been shown to be strongly expressed or ubiquitous nuclear proteins (http://flybase.org and http://genome-www.stanford.edu/Human-CellCycle/Hela/search.shtml) and Ubn1 has also been shown to localize and to punctuate nuclear spots, which resembles the pattern of other histone remodeling and modifying complexes.50,51
HPC2 is critical for histone deposition in multiple contexts
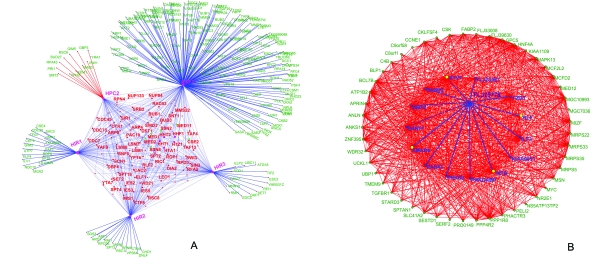
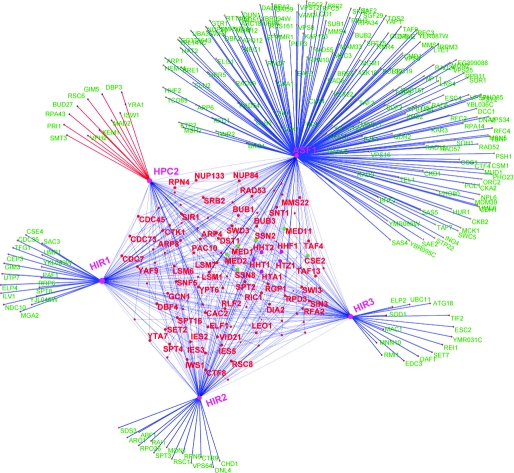
To understand the specific contexts in which the role of Hpc2p might be important, we constructed its interaction network using the available protein–protein interaction and genetic interaction data in yeast (Fig. 2A; Fig. S3, ESI†). This network showed that Hpc2p is a critical component of the Hir histone chaperone complex displaying functional interactions with genes encoding proteins belonging to various functional classes, namely: (1) chromatin remodeling complexes such as Swi–Snf, Iswi, Swr, Ino80, FACT, CAF-1, RSC and the boundary-element-associated factor Yta7; (2) histones H3, H4, H2A and H2AZ; (3) RNA polymerase subunits like RPA43 and CDC73, some basal transcription factors like TAFs and components of the CDK8, head and middle subcomplexes of the Mediator complex; (4) RNA-polymerase-elongation-associated machinery, including, TFIIS, ELF1 and prefoldin complex proteins; (5) Several DNA replication initiation and repair proteins including PCNA and primase; (6) RNA-processing machinery such as the Kem1 nuclease, PAT1 and LSM complexes associated with polyA tails and caps, and mRNA export components of the nuclear pore (Nup84); (7) kinetochore assembly and spindle checkpoint components.
Fig. 2. (A) A network representation of genetic and protein interactions of Hir complex components (Hir1, Hir2, Hir3 and Hpc2) and Asf1. Nodes and edges in the network, respectively, represent genes/proteins and interactions between them. The shared interactions between two or more of the components of the Hir complex are shown within the central pentagon module, while the unique interactions of the components (that are not shared with other components of the Hir complex) are shown outside of the pentagon module. Asf1 having functions independent of the Hir complex (e.g. the Caf-1 complex) has a large set of unique interactions. Histones H2A, H3, H4, H2AZ are shown as purple nodes within the central pentagon module and components of the mediator/CDK8 complex are shown as green nodes (Med1, Med2, Ssn2, Cse2, Ssn8 and Med11) within the module. (B) Largest dense sub-network of proteins containing HUN domain protein FLJ25778 constructed based on mutual information in their phyletic patterns. The network has 64 human proteins implicated in cell-cycle function via RNAi knockdowns, which also contains FLJ25778, are shown as nodes. These nodes are connected by an edge if a given protein pair has a high MIC (> 0.6) in their phyletic patterns (see section 4.1 in ESI† for details) and there are 908 edges in the network. The edges connecting protein FLJ25778 to other proteins in the network are shown in blue; among them transcription factors are shown as yellow nodes and the others are shown as blue nodes.
These wide-ranging functional interactions of Hpc2p are comparable to those of Hir1p, Hir2p and Hir3p and Asf1p, suggesting that it is an integral component of this nucleosome chaperone system (Fig. 2A). In particular genetic interactions (and in some cases physical interactions) with a range of specific chromatin remodeling complexes, replication, transcription and nuclear RNA processing complexes suggest that it can function as a generalized chaperone providing a backup for several specific histone deposition systems (Fig. 2A). While the Hir–Asf1 complex has been shown to physically interact with the H3–H4 dimer, their components, including Hpc2, also showed strong epistatic interactions with H2A specifically in the context of transcription (section 3.1 and Fig. S4. ESI†). Hence, it might be of interest to investigate if the above-proposed histone chaperone function of Hpc2p might also affect assembly of H2A in certain contexts. More strikingly, Hpc2 also showed specific genetic phenotypic enhancement to the exclusion of Hir1, Hir2, Hir3 and Asf1 with Isw1 and Rsc6. These genes, respectively, encode components of the Iswi and Rsc chromatin remodeling complexes, which are important for nucleosome sliding (Fig. 2A and Fig. S3, ESI†). This raises the possibility that a histone-tail-specific chaperone function mediated by Hpc2 might be critical for this process. Independently of other histone chaperone subunits, Hpc2 also shows phenotypic suppression of synthetic lethality with specific interactions with kinetochore and spindle checkpoint genes like Mad2 and Bud27 suggesting that Hpc2’s chaperone activity might also have a specific role in organization of centromeric chromatin (Fig. 2A). Likewise, other genetic interactions unique to Hpc2 are those with the prefoldin subunit Gim5p and RNA-processing components like Kem1p, Yra1p and DBP3p. These interactions suggest that Hpc2p is potentially critical in linking histone re-deposition upon passage of the RNA polymerases to both the prefoldin complex that associates with the transcription elongation machinery52 and the RNA-processing machinery that associates with the pre-mRNA or rRNA.53
Given that the HUN domain itself is rather small, it is likely that it only mediates interactions with histones, whereas the remaining regions of these proteins are responsible for interactions with other chromatin components. In light of the wide divergence of the Hpc2 homologs beyond the HUN domain, including linkages to lineage-specific α-helical domains (Fig. 1, ESI†), it is quite possible that the interactions with chromatin components are distinct for different eukaryotic lineages. Consistent with this, vertebrate ubinuclein appears to have acquired a distinctive lineage-specific ability to bind specific transcription factors such as c-Jun.50 A computational analysis combining mutual information from phyletic patterns and RNAi knockdowns in Hela cell lines implicates the HUN domain protein FLJ25778 (but not ubinuclein) in cell-cycle progression and suggests potential functional interaction with several cell-cycle regulating transcription factors (Fig. 2B, ESI†). However, FLJ25778 mRNA levels show no significant changes throughout the cell-cycle implying that its chaperone activity or interactions could be post-translationally regulated during the course of cell-cycle progression (http://genome-www.stanford.edu/Human-CellCycle/Hela/search.shtml). Unlike yeast Hpc2 and vertebrate ubinuclein and FLJ25778, we found that the Plasmodium falciparum Hpc2 homolog is strongly elevated in conjunction with a SET domain protein (MAL6P1.131) and the RecQ/Wrn-like helicase during early schizogony and might have a specific role in chromatin reorganization or repair during this process. Thus, it is plausible that by virtue of rapid divergence and variability outside of the HUN domain Hpc2p homologs might link the ancient conserved Hir–Asf1 histone chaperone complex to distinct specific processes in different lineages.
General conclusions
Using sensitive protein sequence profile analyses we investigated the evolution of components of the Hir–Asf1 complex and showed that Hir3p and Hpc2p have a much wider phyletic pattern than was previously known. We identified the HUN domain in all Hpc2p homologs including ubinuclein and the recently discovered vertebrate cell-cycle regulator FLJ25778. The HUN domain has a characteristic pattern of acidic residues based on which we predict that it might function as a histone-tail-binding chaperone. By means of an analysis of various high-throughput data sets, such as RNAi knock-downs, genetic and protein interaction maps and cell-cycle wide gene expression, we were able to obtain evidence that Hpc2p homologs might be deployed in specific processes of chromatin dynamics relating to cell-cycle progression in vertebrates and schizogony in Plasmodium. Further, given the extensive divergence of regions of these proteins beyond the conserved HUN domain, we propose that Hpc2p homologs are important for recruitment of the ancient conserved Hir–Asf1 complex to different lineage-specific processes. We present evidence that both Hpc2 and Hir3 appear to have undergone a functional shift in ascomycete fungi implying that studying them in other model organisms might offer better clues regarding their general functions. The results and predictions presented here might help in exploring previously unexpected aspects of chromatin structure and organization.
Materials and methods
Profile searches were conducted using the PSI-BLAST program with a default profile inclusion expectation (E) value threshold of 0.01.54 HMMs were constructed using hmmbuild and searches were performed using the hmmsearch from the HMMER package.55 Profile–profile comparisons were performed using the HHpred program.56 Multiple alignments were constructed using the Kalign57 program followed by manual adjustments based on PSI-BLAST results. Protein secondary structure was predicted using a multiple alignment as the input for the JPRED program.58 Then the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html) was used to cluster proteins based on their sequence similarity score densities. The genetic and protein interactions data generated by Krogan et al. and Collins et al. 59–61 were downloaded from the BIOGRID database.56 We adopted a similar strategy to identify orthologs of proteins suggested to be involved in human cell cycle as described in Madan Babu et al.;62 also see ESI† for further details. Custom written PERL scripts were used to calculate mutual information coefficients (MICs) of phyletic profiles as described in Marcotte et al. 63 and Pellegrini et al. 64 The list of 63 genomes used in the analysis is listed in ESI† Table S3. Gene expression data for intraerythrocytic developmental cycle of Plasmodium falciparum was based on Bozdech et al. 65 and PlasmoDB.66
Acknowledgments
SB, LMI and LA acknowledge the Intramural research program of National Institutes of Health, USA for funding their research.
Footnotes
References
- Eitoku M., Sato L., Senda T., Horikoshi M. Cell. Mol. Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A., Almouzni G. Biochim. Biophys. Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nat. Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Akey C. W., Luger K. Curr. Opin. Struct. Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Park Y. J., Luger K. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmen G. J., Sim R., Haushalter K. A., Ke P. C., Kadonaga J. T., Smith D. E. J. Mol. Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- Ura K., Ohsumi K. Seikagaku. 2006;78:421–425. [PubMed] [Google Scholar]
- Lejeune E., Bortfeld M., White S. A., Pidoux A. L., Ekwall K., Allshire R. C., Ladurner A. G. Curr. Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L., Svejstrup J. Q. J. Biol. Chem. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- Angelov D., Bondarenko V. A., Almagro S., Menoni H., Mongelard F., Hans F., Mietton F., Studitsky V. M., Hamiche A., Dimitrov S., Bouvet P. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., Maier-Davis B., Kornberg R. D. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P., Barbaric S., Luckenbach T., Schmid A., Schermer U. J., Blaschke D., Horz W. J. Biol. Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- Korber P., Horz W. Cell. 2004;117:5–7. doi: 10.1016/s0092-8674(04)00296-x. [DOI] [PubMed] [Google Scholar]
- Shen X., Ranallo R., Choi E., Wu C. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Moshkin Y. M., Armstrong J. A., Maeda R. K., Tamkun J. W., Verrijzer P., Kennison J. A., Karch F. Genes Dev. 2002;16:2621–2626. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M. A., Tsukiyama T., Gdula D., Wu C. Proc. Natl. Acad. Sci. U. S. A. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A., Robert F., Winston F. Mol. Cell. Biol. 2006;26:1496–1509. doi: 10.1128/MCB.26.4.1496-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Sharp J. A., Fouts E. T., Krawitz D. C., Kaufman P. D. Curr. Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., Yates, 3rd J. R., Kaufman P. D. Curr. Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Rizki G., Kaufman P. D. Genetics. 2005;171:885–899. doi: 10.1534/genetics.105.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Luk E., Vu N. D., Patteson K., Mizuguchi G., Wu W. H., Ranjan A., Backus J., Sen S., Lewis M., Bai Y., Wu C. Mol. Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M. M., Wu C. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R. E. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Iyer L. M., Wu C. Cell Cycle. 2007;6:2511–2515. doi: 10.4161/cc.6.20.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P. D., Kobayashi R., Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Kobayashi R., Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Lamour V., Lecluse Y., Desmaze C., Spector M., Bodescot M., Aurias A., Osley M. A., Lipinski M. Hum. Mol. Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- DeSilva H., Lee K., Osley M. A. Genetics. 1998;148:657–667. doi: 10.1093/genetics/148.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Ruone S., Adams M. D., Olsen A. E., Eriksson P., Yu Y., Rhoades A. R., Kaufman P. D., Stillman D. J. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Franco A. A., Osley M. A., Kaufman P. D. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz D. C., Kama T., Kaufman P. D. Mol. Cell. Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D., Nackerdien Z., Furgeson S., Eguchi S., Osley M. A. Mol. Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- Prochasson P., Florens L., Swanson S. K., Washburn M. P., Workman J. L. Genes Dev. 2005;19:2534–2539. doi: 10.1101/gad.1341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Curr. Biol. 2002;12:R256–R258. doi: 10.1016/s0960-9822(02)00786-8. [DOI] [PubMed] [Google Scholar]
- Daganzo S. M., Erzberger J. P., Lam W. M., Skordalakes E., Zhang R., Franco A. A., Brill S. J., Adams P. D., Berger J. M., Kaufman P. D. Curr. Biol. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Lorain S., Demczuk S., Lamour V., Toth S., Aurias A., Roe B. A., Lipinski M. Genome Res. 1996;6:43–50. doi: 10.1101/gr.6.1.43. [DOI] [PubMed] [Google Scholar]
- Han A., Pan F., Stroud J. C., Youn H. D., Liu J. O., Chen L. Nature. 2003;422:730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- Youn H. D., Liu J. O. Immunity. 2000;13:85–94. doi: 10.1016/s1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- Kim M. J., Jo D. G., Hong G. S., Kim B. J., Lai M., Cho D. H., Kim K. W., Bandyopadhyay A., Hong Y. M., Kim D. H., Cho C., Liu J. O., Snyder S. H., Jung Y. K. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9870–9875. doi: 10.1073/pnas.152336999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn H. D., Grozinger C. M., Liu J. O. J. Biol. Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chan D. C. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J. C. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- Kittler R., Pelletier L., Heninger A. K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., Wagner J., Surendranath V., Richter C., Bowen W., Jackson A. L., Habermann B., Hyman A. A., Buchholz F. Nat. Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Malay A. D., Umehara T., Matsubara-Malay K., Padmanabhan B., Yokoyama S. J. Biol. Chem. 2008;283:14022–14031. doi: 10.1074/jbc.M800594200. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Seol J. H., Han J. W., Youn H. D., Cho E. J. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P. D., Cohen J. L., Osley M. A. Mol. Cell. Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho S., Buisson M., Pajunen T., Ryoo Y. W., Giot J. F., Gruffat H., Sergeant A., Uitto J. J. Cell. Biol. 2000;148:1165–1176. doi: 10.1083/jcb.148.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Ahmed O., Bellon B., Capri M., Joblet C., Thomas-Delaage M. Mech. Dev. 1992;37:69–80. doi: 10.1016/0925-4773(92)90016-d. [DOI] [PubMed] [Google Scholar]
- Parusel C. T., Kritikou E. A., Hengartner M. O., Krek W., Gotta M. Development. 2006;133:621–629. doi: 10.1242/dev.02235. [DOI] [PubMed] [Google Scholar]
- Luo W., Johnson A. W., Bentley D. L. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Soding J., Biegert A., Lupas A. N. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E. L. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff J. A., Barton G. J. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O’Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. Mol. Cell. Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Madan Babu M., Teichmann S. A., Aravind L. J. Mol. Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Marcotte E. M., Pellegrini M., Ng H. L., Rice D. W., Yeates T. O., Eisenberg D. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Marcotte E. M., Thompson M. J., Eisenberg D., Yeates T. O. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z., Llinas M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl A., Brunk B., Crabtree J., Fraunholz M. J., Gajria B., Grant G. R., Ginsburg H., Gupta D., Kissinger J. C., Labo P., Li L., Mailman M. D., Pearson A. J., Roos D. S., Schug J., Stoeckert, Jr C. J., Whetzel P. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.