Abstract
Background
Eradication of early gastric carcinoma (GC) is thought to contribute to reduction in the mortality of GC, given that most of the early GCs progress to the advanced GCs. However, early GC is alternatively considered a dormant variant of GC, and it infrequently progresses to advanced GC. The aim of this study was to clarify the extent of overlap of genetic lineages between early and advanced tubular adenocarcinomas (TUBs) of the stomach.
Methods
Immunohistochemical staining for p53 was performed using 28 surgically resected stomachs with 13 intramucosal and 15 invasive TUBs. By chromosome- and array-based comparative genomic hybridization (CGH), genomic copy number constitution was compared between the mucosal and invasive parts of the invasive TUBs and between the mucosal parts of the invasive and intramucosal TUBs, using 25 and 22 TUBs, respectively. TP53 mutation in exons 5-8 was examined in 20 TUBs.
Results
Chromosomal CGH revealed that 4q+ and 11q+ were more common in advanced and early TUBs, respectively, whereas copy number changes in 8q and 17p showed no significant differences between early and advanced TUBs. However, array CGH revealed that, of the 13 intramucosal TUBs examined, loss of MYC (MYC-) and gain of TP53 (TP53+) was detected in 9 TUBs and MYC+ and/or TP53- was detected in 3 TUBs. Of the mucosal samples of 9 invasive TUBs, 7 showed MYC-/TP53+ and none showed MYC+ and/or TP53-. Of the 9 samples from the invasive parts, 1 (from submucosal cancers) showed MYC-/TP53+ and 6 (1 from submucosal and 5 from advanced cancers) showed MYC+ and/or TP53-. The latter 6 tumours commonly showed a mutant pattern (diffuse or null) in p53 immunohistochemistry, and 4 of the 6 tumours assessable for TP53 sequence analysis revealed mutations. The overall array CGH pattern indicated that, between the mucosal and invasive parts, genetic lineage was found discontinuous in 5 advanced cancers and continuous in 3 submucosal cancers.
Conclusions
Genetic lineages often differed between early and advanced TUBs. MYC-/TP53+ and MYC + and/or TP53- may be the signatures of dormant and aggressive TUBs, respectively, in the stomach.
Background
Gastric cancer remains the second most common cause of cancer-related deaths worldwide, despite a recent decrease in its mortality in developed countries [1]. Gastric carcinomas (GCs) are classified morphologically into 2 major categories: tubular-forming type and diffuse type [2,3] and, in staging, into early cancers (involving the mucosa and the submucosa) and advanced cancers (involving the muscularis propria or deeper). Early GC is considered a curable cancer [4]; it reportedly progresses to advanced GC after varying durations as an early stage of GC [5,6]. In Japan, early GCs can be actively resected endoscopically as well as surgically [7], based on the principles of early detection and treatment. On the other hand, some pathologists maintain that the tubular-forming neoplastic lesions that are confined to the mucosa take a long time to or are unable to invade deeper tissues. Thereby, these lesions are called dysplasia [4].
Recently, a mass-screening program for neuroblastomas in Japan [8-10] was suspended because a discontinuous genetic lineage was found between the early- and the late-presenting neuroblastomas. Negative and late-presenting (≥1 year) neuroblastomas showed near-diploidy with loss of terminal 1p, whereas positive neuroblastomas in infants showed near-triploidy without 1p deletion [11,12]. On the other hand, comparative genomic hybridization (CGH)-based analysis suggested that high-grade atypical adenomatous hyperplasia in the lung is the true precursor of bronchioloalveolar carcinoma [13]. In the stomach, it is a problem to what extent genetic lineage is overlapped in early and advanced GCs.
In diffuse type GCs, we used CGH to demonstrate that chromosomal copy-number alterations in the intramucosal signet-ring cell carcinomas that showed a layered structure [14] were inherited in a fraction of poorly differentiated GCs at advanced stages [15]. The presence of a layered structure of signet ring cells in the mucosal parts of these advanced cancers also suggested that signet ring cell carcinomas may be a true precursor of poorly differentiated GCs. CGH-based lineage analysis of another subset of poorly differentiated advanced GCs with tubular components but no layered structure in the mucosal part revealed that GCs of this subset were derived from a tubular component in a tumour and were characterized by 17p- and 8q+ [16] and inactivation of the wild-type TP53 by mutation and loss of heterozygosity (LOH) [17]. In order to determine the early-stage counterpart of this type of GC, we analysed intramucosal tubular adenocarcinomas (TUBs) using CGH. These studies found that the intramucosal TUBs showed not only several chromosomal changes common to advanced GCs but also frequently showed 8q- and 17p + that were critically different from advanced and poorly differentiated GCs [18].
In the present study, copy-number changes of chromosomes and genes were examined in the mucosal and invasive parts of another series of early and advanced TUBs using array and chromosomal CGH. Based on these data, we demonstrated cases of continuous and discontinuous lineages between intramucosal and invasive parts of individual tumours and found that TP53 and MYC may be good lineage markers for gastric TUBs.
Methods
The Institutional Review Board on Medical Ethics at Shiga University of Medical Science granted approval for conducting this research on the condition that the materials used must be anonymous.
Tumour samples
This study included 28 surgically resected stomachs (Table 1) with 13 intramucosal TUBs and 15 invasive TUBs [6 submucosal and 9 advanced cancers that invaded the muscularis propria or deeper tissues (the tubular component of each tumour was assessed to be more than 30%)]. Each stomach was fixed in formalin and embedded in paraffin wax. These were selected at random from the materials diagnosed in our department from 1996 to 2008. Histological types and tumour stages were determined according to the Japanese Classification of Gastric Cancer [19] and pTNM staging, respectively. When a mucosal histological pattern was found to be heterogeneous in an individual invasive tumour, the part with the lowest grade of atypism was taken as the mucosal sample that can prevent the possibility of re-invasion of invasive cancer cells into the mucosa. In invasive cancers, DNA samples were obtained from intramucosal and invasive parts.
Table 1.
Summary of clinical, histopathological and molecular genetic data of 25 tubular adenocarcinoma of stomach
| Case No. | Age/sex | Histological type* | Invasion** | LN** | Stage ** | CGH | p53 IHC | TP53 mutation |
|---|---|---|---|---|---|---|---|---|
| M1 | 57/M | tub1 | T1(M) | N0 | IA | c/a | + | NT |
| M2 | 72/M | tub1 | T1(M) | N0 | IA | c/a | + | NT |
| M3 | 67/F | tub1 | T1(M) | N0 | IA | c/a | - | - |
| M4 | 67/M | tub1 | T1(M) | N0 | IA | c/a | - | - |
| M5 | 74/M | tub1 | T1(M) | N0 | IA | c/a | + | - |
| M6 | 65/M | tub1 | T1(M) | N0 | IA | c/a | + | - |
| M7 | 67/F | tub1 | T1(M) | N0 | IA | c/a | + | - |
| M8 | 70/F | tub1 > pap | T1(M) | N0 | IA | c/a | - | - |
| M9 | 51/M | tub1 > tub2 | T1(M) | N0 | IA | c/a | + | - |
| M10 | 52/M | tub2 | T1(M) | N0 | IA | c/a | + | - |
| M11 | 57/F | tub2 | T1(M) | N0 | IA | NT/a | - | - |
| M12 | 89/M | tub2 | T1(M) | N0 | IA | NT/a | + | + |
| M13 | 70/M | tub1 | T1(M) | N0 | IA | NT/a | - | NA |
| S1m | 73/F | tub2 | T1(SM) | N0 | IA | c/a | + | - |
| S1i | NT/a | + | - | |||||
| S2m | 81/F | pap | T1(SM) | N1 | IB | c/a | + | - |
| S2i | NT/a | + | + | |||||
| S3m | 79/M | tub1 | T1(SM) | N0 | IA | c/a | + | + |
| S3i | NT/a | + | + | |||||
| S4m | 63/M | tub2 > tub1 | T1(SM) | N0 | IA | c/NT | + | NT |
| S5m | 75/M | pap > tub2 | T1(SM) | N1 | IB | c/NT | + | NT |
| S6m | 79/M | pap | T1(SM) | N1 | IB | c/NT | + | NT |
| A1m | 71/M | tub2 | T2(SS) | N2 | IIIA | c/a | null | - |
| A1i | NT/a | null | NA | |||||
| A2m | 73/M | tub1 > tub2 | T2(SS) | N0 | IB | c/a | + | - |
| A2i | NT/a | + | + | |||||
| A3m | 56/M | pap | T2(SS) | N1 | II | c/a | + | - |
| A3i | NT/a | + | + | |||||
| A4m | 81/M | tub2/pap | T2(SS) | N2 | IIIA | c/a | null | - |
| A4i | NT/a | null | + | |||||
| A5m | 56/M | tub1 | T2(MP) | N0 | IB | c/NT | - | NT |
| A5i | NT/NA | - | NA | |||||
| A6m | 45/F | tub2 | T2(SS) | N1 | IB | c/NT | + | NT |
| A7m | 79/F | tub2 | T2(MP) | N1 | IB | c/NT | +/- | NT |
| A8m | 69/M | tub1 | T3(SE) | N2 | IIIB | c/a | null | - |
| A8i | NT/a | null | NA | |||||
| A9m | 67/F | tub2 | T2(SS) | N1 | II | c/a | + | - |
| A9i | NT/a | + | + |
* Japanese classification of gastric carcinoma; ** pTNM classification. In the column of Case No, M = intramucosal cancer; S = cancer involving the submucosa; A = advanced cancer; m = mucosal part; i = invasive part. In the column of CGH, c = chromosomal CGH; a = array CGH. In the column of p53 immunohistochemistry (IHC); "+" indicates diffusely (>70%) positive nuclei, "-" indicates sporadically (<5%) positive nuclei; "null" indicates no immunoreactivity at all. In the column of TP53 mutation; "+" and "-" indicate presence and absence of mutation, respectively; NA = not assessable; NT = not tested.
Immunohistochemistry
Immunohistochemical staining was performed with monoclonal antibodies to p53 protein (DO-7, 1:100; Dako, Glostrup, Denmark). After antigen retrieval of tissue sections in distilled water at 121°C for 5 min, immunoreactivity was detected by an indirect biotin-streptavidin-peroxidase method using the Histofine Kit (Nichirei, Tokyo, Japan) and diaminobenzidine reaction. The sections were counterstained with haematoxylin. Slides of the negative control without the primary antibody and those of the positive control were processed in parallel.
Laser microdissection and DNA preparation
Tumour cells were obtained from 5-μm-thick tissue sections using a LMD6000 laser microdissection system (Leica Microsystems, Wetzlar, Germany). For individual tumours, cancer cells were obtained from areas >3 mm2, where cancer cells accounted for ≥90% of the total cell count. These cancer cells were digested in 200 μl of proteinase K solution at a concentration of 200 μg/ml for approximately 70 h at 37°C, followed by phenol/chloroform DNA extraction.
Whole genome amplification
Sample DNA was amplified using degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR) in 2 phases, as described previously [20], which resulted in PCR products more than 2 kb in size, suitable for nick-translation labelling for CGH.
For array CGH, sample DNA was amplified using the GenomePlex Tissue Whole Genome Amplification Kit (WGA2 Kit; Sigma, St. Louis, USA) [21]. For some DNA samples that could not be sufficiently amplified, the WGA5 Kit (Sigma) was employed.
CGH (hybridization, probe DNA labelling and digital image analysis)
DOP-PCR-amplified tumour and normal DNA was labelled using fluorescein-12-dUTP and tetramethylrhodamine-5-dUTP (Roche, Mannheim, Germany), respectively, by nick translation [15]. Hybridization and image analyses were performed as described previously [22]. Gains and losses in DNA copy numbers were defined by green to red ratios >1.2 and <0.8, respectively. Chromosomes 1p32-pter, 16p, 19, 22 and Y were excluded from these analyses.
Array CGH
Oligo CGH microarray (60 K, 60-mer) (Agilent, Santa Clara, USA) was used in this study, according to the manufacturer's instructions. In brief, tumour and control DNA was non-enzymatically labelled with Cy5 and Cy3, respectively, using the Genome DNA ULS Labelling Kit (Agilent) and competitively hybridized to the microarray. Using Feature Extraction Ver.9.5.3 (Agilent), the fluorescence intensity of the tumours and controls was calculated from the hybridized array images captured using a DNA microarray scanner (Agilent). Copy-number gains and losses were defined as base 2 logarithm of the tumour signal intensity to the reference signal intensity ratio more than 0.3219 and less than -0.3219, respectively.
Mutation analysis for TP53
PCR primer sets were prepared for exons 5-8 of TP53, which hybridize to the flanking introns of each exon. See additional file 1 for primer sequences. The first PCR mixture consisted of a buffer, 200 μM dNTPs, 1.5 mM MgCl2, 0.8 μM primer, 1 ng/μl sample DNA and 0.5 U Platinum Taq (Invitrogen, California, USA) in a 25-μl final volume. After initial denaturation at 94°C for 2 min, 40 cycles of PCR were performed at 94°C for 30 sec, 54°C for 1 min and 72°C for 1 min, followed by the final extension step at 72°C for 10 min. After confirmation of PCR products by agarose gel electrophoresis, sequence PCR was performed using the BigDye Terminators v1.1 Cycle Sequencing Kit (Applied Biosystems, California, USA). The reaction mixture consisted of a buffer, 2 μl of the first PCR product, 0.15 μM primer and 0.5 μl BigDye Terminator V 1.1 in a 10-μl final volume. After initial denaturation at 96°C for 1 min, 25 cycles of PCR were performed at 96°C for 10 sec, 50°C for 5 sec and 60°C for 4 min. The forward and reverse sequences were determined using the ABI PRISM 3100 Genetic Analyser (Applied Biosystems). Mutations were detected by comparing these samples to the reference DNA sequence (GenBank accession number: HSU94788). Allelic status was determined using the ratio of mutant to wild-type peak levels in sequencing profiles.
Statistical analyses
Contingency tables were analysed by Fisher's exact test and Cochran-Armitage tests. Differences of 2-sided tests with P < 0.05 were considered to be statistically significant.
Results
Immunohistochemistry for p53
Staining patterns were considered overexpressed only when ≥70% tumour cells showed positively stained nuclei when viewed in a low-power field. Eight of the 13 intramucosal cancers, all the 6 submucosal cancers and 4 of the 9 advanced cancers showed diffusely positive nuclear staining for p53 (Table 1). A focal staining pattern was observed in case #A7. A pattern was considered negative (representing the wild-type TP53) when the presence of sparse, sporadic and weak nuclear staining was observed in <5% of the nuclei. A null staining pattern was observed in cases #A1, #A4 and #A8, suggesting a non-sense mutation of TP53 [23].
Chromosomal CGH
Twenty-five samples from 25 cases (10 mucosal, 6 submucosal and 9 advanced cancers) were used for chromosomal CGH. All these samples were obtained from intramucosal lesions that showed the lowest grade of atypism.
While 4q+ occurred in the majority (6/9 cases) of advanced TUBs, it was not detected in the 16 early TUBs (P = 0.0005). Another significantly different chromosomal change between early and advanced cancers was 11q+, which was detected in 11 of the 16 early cancers and 2 of the 9 advanced cancers (P = 0.0414). Of the 10 intramucosal TUBs examined, 3 showed loss of 8q (8q-) and/or gain of 17p (17p+), while 7 and 2 showed 8q+ and 17p-, respectively. Of the 15 mucosal samples of invasive TUBs examined, 3 and 4 showed 8q- and 17p+, respectively, while 7 and 6 showed 8q+ and 17p-, respectively.
Array CGH
Thirty-one samples from 22 cases (13 mucosal, 3 submucosal and 6 advanced cancers) were assessed using array CGH (Figure 1). DNA samples were obtained from intramucosal and/or invasive lesions.
Figure 1.
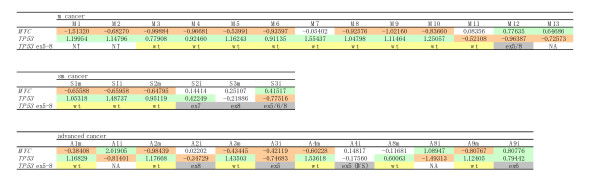
Array CGH data of MYC and TP53 and TP53 sequencing data in early and advanced tubular adenocarcinomas of stomach. Early cancers are divided into intramucosal (m) cancers and submucosal (sm) cancers. Refer to Table 1 for sample numbers. Numerals are mean test/reference ratios of array CGH. Significant losses and gains are indicated with red and green, respectively. Wild type (wt) and mutation in exon (ex) and intervening sequence (IVS) of TP53 are indicated with yellow and gray, respectively.
As shown in Figure 1 and Table 2, concomitant loss of MYC (MYC-) and gain of TP53 (TP53+) were detected in 9 of the 13 intramucosal TUBs, 7 of the 9 mucosal samples of invasive TUBs, 1 of the 3 invasive samples of submucosal cancers and none of the 6 invasive samples of advanced cancers. A pattern of MYC+ and/or TP53- was detected in 3 of the 13 intramucosal cancers, none of the 9 mucosal samples of invasive TUBs, 1 of the 3 invasive samples of 3 submucosal cancers and 5 of the 6 invasive samples of 6 advanced cancers.
Table 2.
Alterations of MYC and TP53 copy numbers in early and advanced tubular adenocarcinomas of stomach
| Tumour depth | Mucosa | Submucosa | MP or deeper - | |||||
|---|---|---|---|---|---|---|---|---|
| Mucosal part | SM | Mucosal part | Deep | |||||
| cCGH (10) |
aCGH (13) |
cCGH (6) |
aCGH (3) |
aCGH (3) |
cCGH (9) |
aCGH (6) |
aCGH (6) |
|
| 8q(MYC)+ | 7 | 2 (2*) | 3 | 0 | 1 (1*) | 4 | 0 | 3 (2*) |
| 8q(MYC)- | 3 | 9 (9**) | 3 | 2 (2**) | 1 (1**) | 0 | 5 (5**) | 1 (0**) |
| 17p(TP53)+ | 3 | 10(9**) | 3 | 2 (2**) | 2 (1**) | 1 | 6 (5**) | 1 (0**) |
| 17p(TP53)- | 2 | 3 (2*) | 2 | 0 | 1 (1*) | 4 | 0 | 4 (2*) |
SM, the submucosa; MP, the muscularis propria. cCGH and aCGH, chromosome- and array-based comparative genomic hybridisation; Numerals in parentheses indicate the total number of the samples examined. Bold letters indicates the results of aCGH. Asterisks in parentheses indicate concomitance of MYC+ and TP53-. Double asterisks indicate concomitance of MYC- and TP53+.
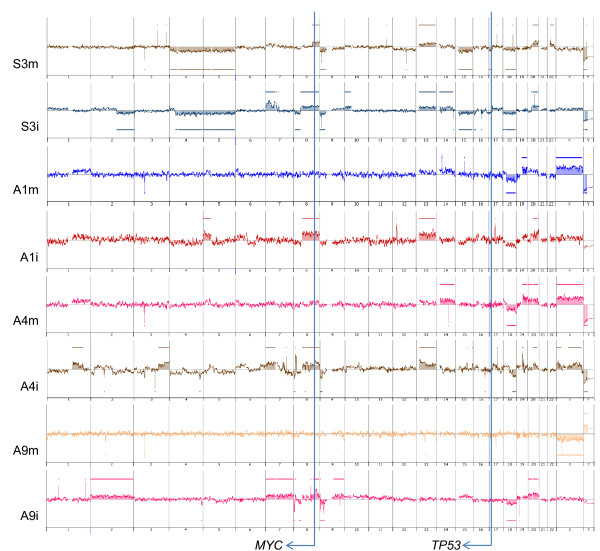
Based on the overall array CGH patterns, genetic lineages were found to be discontinuous and continuous between the mucosal and invasive parts in 5 advanced and 3 submucosal cancers, respectively. As shown in Figure 2, the sample from the mucosal part of #S3 showed 4p-, 4q-, 8q+, 9p-, 13q+, 15q-, 18q-, 20q+ and Y- , while the sample from the invasive part of #S3 inherited the same aberrations from the mucosal part, showed additional aberrations (2q-, 7p+, 8p-, 10p+, 14q+ and 20q+) and lost 4p- and 22q+. In #S1, gains of 1q, 14q, 19 and 20, and loss of 18q was common in mucosal and submucosal samples, though the gains of 1q, 14q, 19q and 20 in the submucosa were slightly less than the significant level. In #S2, a common change in both the mucosal and submucosal parts was a gain of 20p. The submucosal sample showed additional gains in 10p, 12 and 13q and losses in 3q, 4, 5, 8p, 9p, 9q, 10q, 12p, 14q, 16 and 19p. The mucosal sample showed loss of 18q, which was restored in the submucosal sample, possibly by uniparental disomy [24,25]. In #A1, #A3, #A4, #A8 and #A9, aberrations in the samples from the mucosal parts were not detected in those of the invasive parts. In #A2, the continuity of genetic lineages could not be assessed because of the absence of significant copy-number alterations in the mucosal part.
Figure 2.
Array CGH profiles. The chromosomal regions of copy number aberrations in the mucosal part of #S3 (S3m) are included basically in those in the invasive part (S3i), whereas those in the mucosal part of #A1 (A1m) are different from those of the invasive part (A1i).
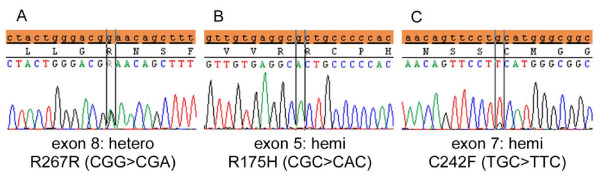
TP53 mutation analysis
Mutations were detected in 7 of the tumours that were examined for sequence analysis of exons 5-8 of TP53 (Figure 1, Table 3): 1 of the 10 intramucosal cancers, 2 of the 3 submucosal cancers and all 4 advanced cancers (only the invasive parts). In all these 7 tumours, the TP53 allele was hemizygous except in 1 intramucosal cancer (#M12) with a heterozygous pattern for an exon-8 mutation (Figure 3) and a diffuse or a null pattern for p53, as indicated by immunohistochemistry. No TP53 mutation was detected in p53- samples, as shown in Figure 4. In p53+/null samples, 2 of the 15 samples of the mucosal part showed TP53 mutation, whereas 6 of the 7 samples of invasive part showed TP53 mutation (P = 0.0023). In 1 of the submucosal cancers showing TP53+ and a hemizygous mutation (the invasive part of #S2), TP53+ may reflect uniparental polysomy [24,25].
Table 3.
Copy number changes of TP53 and MYC and p53 immunoreactivity in the samples tested for mutation analyses
| Case No. | MYC | TP53 | Mutation of TP53 exons 5-8 | Mutant allele | MDM2 | p53 IHC |
|---|---|---|---|---|---|---|
| M3 | - | + | wt | - | - | |
| M4 | - | + | wt | ns | - | |
| M5 | - | + | wt | ns | + | |
| M6 | - | + | wt | - | + | |
| M7 | ns | + | wt | - | + | |
| M8 | - | + | wt | ns | - | |
| M9 | - | + | wt | ns | + | |
| M10 | - | + | wt | - | + | |
| M11 | ns | - | wt | ns | - | |
| M12 | + | - | Exons 5/8 | hemi/hetero | ns | + |
| M13 | + | - | NA | ns | - | |
| S1m | - | + | wt | - | + | |
| S1i | - | + | wt | - | + | |
| S2m | - | + | wt | - | + | |
| S2i | ns | + | exon 7 | hemi | ns | + |
| S3m | ns | ns | exon 8 | hemi | ns | + |
| S3i | + | - | exons 5/6/8 | hemi | ns | + |
| A1m | - | + | wt | - | null | |
| A1i | + | - | NA | - | null | |
| A2m | - | + | wt | ns | + | |
| A2i | + | - | exon 8 | hemi | ns | + |
| A3m | - | + | wt | ns | + | |
| A3i | - | - | exon 5 | hemi | - | + |
| A4m | - | + | wt | - | null | |
| A4i | ns | ns | exon 5 | hemi | ns | null |
| A8m | ns | + | wt | - | null | |
| A8i | + | - | NA | ns | null | |
| A9m | - | + | wt | - | + | |
| A9i | + | + | exon 6 | hemi | ns | + |
ns = not significant; wt = wild type; NA = not assessable; hemi = hemizygous; hetero = heterozygous.
M12, exon 5: R175H (CGC > CAC), C182C (TGC > TGT), S183L (TCA > TTA); exon 8: R267R (CGG > CGA)
S2i, exon 7: C242F (TGC > TTC)
S3m, exon 8: R273S (CGT > AGT)
S3i, exon 5: A161V (GCC > GTC); exon 6: T211I (ACT > ATT); exon 8: R273S (CGT > AGT)
A2i, exon 8: R273 H (CGT > CAT)
A3i, exon 5: A161T (GCC > ACC)
A4i, exon 5: IVS5+1G > T
A9i, exon 6: Y220C (TAT > TGT)
Figure 3.
TP53 mutation patterns. A heterozygous mutation with a significant wild-type component in exon 8 (A) and a hemizygous mutation in exon 5 (B) in an intramucosal cancer (#M12). Mutations in the other tumours examined were hemizygous as shown in C (#S2).
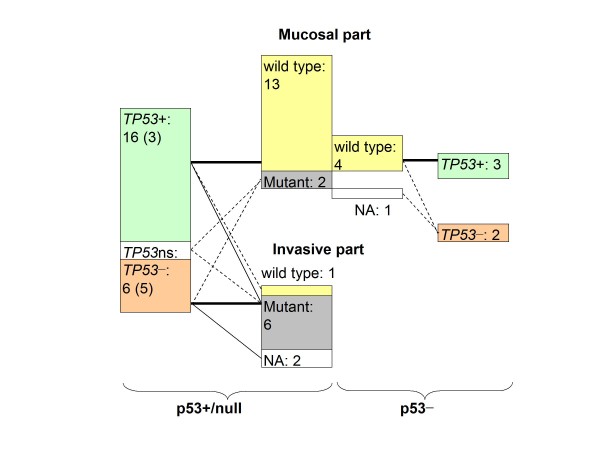
Figure 4.
Relationships among p53 immunohistochemistry, copy number of TP53 and hot spot mutation of TP53. For "p53+" and "p53-", see the legend in Table 1. "TP53 +", "TP53 -" and "TP53 n/s" indicate significant copy-number gain and loss, and no significant change, respectively. Numerals in parentheses indicate the number of samples in invasive parts. Numerals are sample numbers. Numerals in parentheses indicate the number of samples in invasive parts. NA = not assessable.
On the other hand, in the mucosal samples, the diffuse/null pattern was often associated with wild-type TP53. In order to explain this finding, the MDM2 copy number was examined using array CGH data. Of a total of 22 samples assessed for TP53 mutation analysis, MDM2 showed copy-number loss in 10 of the 14 samples with wild-type TP53 and 1 of the 8 samples with TP53 mutation (P = 0.0237).
Discussion
In intramucosal TUBs, chromosomal CGH revealed 8q- and/or 17p+ at a frequency of 3 out of 10 tumours examined; these changes were more common in a previous study that used random priming labelling [18]. In the present study, 8q+ was frequently detected both in intramucosal cancers (7/10) and in invasive cancers (7/15); however, the above-mentioned previous data rarely showed 8q+. These discrepancies may reflect differences in the labelling methods used; CGH results with nick translation labelling or random priming labelling were compared to FISH signals, and it was found that nick translation labelling not infrequently showed false-positive gains [26]. However, using the same labelling conditions, significant differences in the incidence of 4q+ and 11q+ were detected between early and advanced cancers, possibly reflecting differences in genetic lineages between them. Chromosomal CGH data of advanced cancers in this study were consistent with previous chromosomal CGH data obtained with nick translation labelling for advanced GCs (i.e. 17p- and 8q+ were quite common) [27-30].
Although clinically discernible early cancers are generally considered the precursors of advanced cancers [5,6], the intramucosal and invasive parts of advanced TUBs often showed discordant copy numbers of MYC and TP53 in this study: MYC- and TP53+ in 5 of the 6 samples of the mucosal parts, as commonly as in the intramucosal cancers examined, and MYC+ and/or TP53- in 5 of the 6 samples of the invasive parts. In addition, based on the overall array CGH pattern of the mucosal and invasive parts of tumours, a discontinuous genetic lineage was found in 5 of the 6 advanced cancers. Therefore, it appears that the tumour clones in the mucosal parts of the advanced cancers were not precursors of the invasive part, but minute intramucosal cancers that probably coexisted with the invasive cancers. It has been reported that multiple minute cancers are more common in TUBs than in undifferentiated-type GCs [31]. In advanced TUBs, the intramucosal precursor of an invasive component might have been lost due to ulceration in the tumour, or the precursor was not examined because it did not coincide with the part of the lowest grade of atypism, to which the examination of the mucosal part of invasive GCs was confined in the present study.
On the other hand, in submucosal cancers, a continuous genetic lineage was found in all the 3 submucosal tumours examined. It may be because the mucosal part is basically retained in early GCs and because the mucosal precursor of an invasive component showed the lowest grade of atypism and was not excluded from the present array analyses. The mucosal and invasive parts had common regions of copy number aberrations with concordant breakpoints, and most of the changes in the mucosal part were included in those of the invasive part. At the gene level, the copy numbers of MYC and TP53 were consistent between the mucosal and submucosal parts in 2 of the 3 submucosal cancers.
The TP53- and/or MYC+ pattern was detected not only in the invasive parts of 1 of the submucosal cancers and most of the advanced cancers but also in 3 of the 13 intramucosal cancers. In these tumours, p53 immunohistochemistry showed a diffuse or null (mutation) pattern (Tables 1 and 3), suggesting that the TP53- may reflect LOH and the resultant inactivation of wild-type TP53. Sequencing analyses of the mutation hot spots of TP53 have demonstrated that hemizygous mutations (i.e. mutation and LOH) were detected in the mucosal samples of 1 of the 3 intramucosal cancers and 1 submucosal cancer with a TP53- pattern, as well as the invasive parts of the 4 advanced cancers that were informative for TP53 mutation analysis. Such inactivation of wild-type TP53, which may cause further genomic instability, has been frequently reported in advanced GCs [15,17]. The MYC gene locus is also known to frequently show copy-number gains or amplifications in various advanced cancers [32]. MYC+/TP53- may thus be justified as the signature of aggressive GC.
It has been reported that LOH as well as mutations of TP53 are more frequently detected in advanced GCs than in early GCs [17,33], and that, within early GCs, TP53 mutations are more frequently detected in submucosal TUBs than in intramucosal TUBs [34,35]. This tendency was also observed in the present study, but mutation analyses indicated that hot spot mutations were not detected in the mucosal part of the tumours, except in 1 intramucosal cancer (#M12). In addition, copy-number loss of MDM2, which plays a role in p53 degradation [36], correlated with diffuse p53 overexpression in the absence of a TP53 mutation. This phenomenon is scarcely reported, but could be one of the mechanisms of p53 overexpression in early GCs.
In early cancers, there may be dormant and invasive components as well as true precursors of advanced cancers. Among the early cancers examined in the present study, an invasive part of 1 tumour (#S3) and 3 intramucosal cancers (#M11, #M12 and #M13) showed the signature of aggressive cancer (MYC+ and/or TP53-) and frequent mutations of TP53; however, in 1 submucosal cancer (#S1), the invasive parts showed the signature of dormant cancer (MYC-/TP53+). Submucosal cancers with the signature of dormant cancer may be in the submucosa for a long time, and therefore, they provide a greater opportunity for clinical detection as submucosal cancers. However, in order to eradicate advanced cancers, early cancers with the signature of aggressive cancer must be detected, such as tumours #S3 and #M11-#M13. If this hypothesis of dormant and aggressive signatures is confirmed by studies with a larger number of cases, the differentiation between the 2 signatures can be applied to gastric biopsy specimens.
Conclusions
These array CGH-based analyses have demonstrated that the copy numbers of MYC and TP53 have high discriminative power for differentiation between genetic lineages in individual gastric TUBs. This study has shown that genetic lineages are often different between early and advanced TUBs and that MYC-/TP53+ and MYC+ and/or TP53- may be signatures of dormant and aggressive TUBs, respectively, in the stomach.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NT performed most of the experiments, participated in concrete research designing and most of the data analyses and drafted the manuscript. LZQ participated in parts of experiments, in particular, chromosomal CGH. MK participated in the preparation of samples in array CGH. HT provided critical comments and suggested revisions of the manuscript. SH was a leader of this project, who conceived the study, designed the research route, and guided the experiments and the data analyses. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
TP53_primer_sequences Microsoft Excel 2003 TP53 primer sequences.
Contributor Information
Takahisa Nakayama, Email: 330601@belle.shiga-med.ac.jp.
Zhi-Qiang Ling, Email: lingzq@hotmail.com.
Ken-ichi Mukaisho, Email: mukaisho@googlemail.com.
Takanori Hattori, Email: hattori@belle.shiga-med.ac.jp.
Hiroyuki Sugihara, Email: sugihara@belle.shiga-med.ac.jp.
Acknowledgements
The authors thank Mr Katsumi Nakanishi (DNA Chip Research Inc., Yokohama, Japan) and Takafumi Fukui (FALCO biosystems Ltd., Kyoto, Japan) for valuable technical assistance and feedbacks in array CGH analysis and sequence analysis, respectively. This work was supported in part by a Grant-in Aid for Scientific Research (C-19590345) from the Japan Society of the Promotion of Science.
References
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- Carneiro F. Classification of gastric carcinomas. Curr Diagn Pathol. 1997;4:51–59. doi: 10.1016/S0968-6053(97)80008-7. [DOI] [Google Scholar]
- Jass JR. In: Oxford Textbook of Pathology. McGee JO'D, Isaacson PG, Wright NA, editor. Oxford: Oxford University Press; 1992. Tumour of the stomach; pp. 1165–1173. [Google Scholar]
- Tsukuma H, Mishima T, Oshima A. Prospective study of "early" gastric cancer. Int J Cancer. 1983;31:421–426. doi: 10.1002/ijc.2910310405. [DOI] [PubMed] [Google Scholar]
- Tsukuma H, Oshima A, Narahara H, Morii T. Natural history of early gastric cancer: a non-concurrent, long term, follow up study. Gut. 2000;47:618–621. doi: 10.1136/gut.47.5.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Ming SC, Itabashi M. In: Gastric Cancer. Nishi M, Ichikawa H, Nakajima T, Maruyama K, Tahara E, editor. Tokyo: Springer-Verlag; 1993. Pathology of early gastric cancer; pp. 66–87. [Google Scholar]
- Sawada T, Nakata T, Takasugi N, Maeda K, Hanawa Y, Shimizu K, Hirayama M, Takeda T, Mori T, Koide R, Tsunoda A, Nagahara N, Yamamoto K. Mass screening for neuroblastoma in infants in Japan. Interim report of a mass screening study group. Lancet. 1984;2(8397):271–273. doi: 10.1016/S0140-6736(84)90311-8. [DOI] [PubMed] [Google Scholar]
- Sawada T, Sugimoto T, Tanaka T, Kawakatsu H, Ishii T, Matsumura T, Horii Y. Number and cure rate of neuroblastoma cases detected by the mass screening program in Japan: future aspects. Med Pediatr Oncol. 1987;15:14–17. doi: 10.1002/mpo.2950150104. [DOI] [PubMed] [Google Scholar]
- Murphy SB, Cohn SL, Craft AW, Woods WG, Sawada T, Castleberry RP, Levy HL, Prorok PC, Hammond GD. Do children benefit from mass screening for neuroblastoma? Consensus Statement from the American Cancer Society Workshop on Neuroblastoma Screening. Lancet. 1991;337(8737):344–6. doi: 10.1016/0140-6736(91)90957-Q. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Kanda N, Maseki N, Sakurai M, Tsuchida Y, Takeda T, Okabe I, Sakurai M. Different karyotypic patterns in early and advanced stage neuroblastomas. Cancer Res. 1987;47:311–318. [PubMed] [Google Scholar]
- Kaneko Y, Kobayashi H, Watanabe N, Tomioka N, Nakagawara A. Biology of neuroblastomas that were found by mass screening at 6 months of age in Japan. Pediatr Blood Cancer. 2006;46:285–291. doi: 10.1002/pbc.20496. [DOI] [PubMed] [Google Scholar]
- Ullmann R, Bongiovanni M, Halbwedl I, Fraire AE, Cagle PT, Mori M, Papotti M, Popper HH. Is high-grade adenomatous hyperplasia an early bronchioloalveolar adenocarcinoma? J Pathol. 2003;201:371–376. doi: 10.1002/path.1460. [DOI] [PubMed] [Google Scholar]
- Sugihara H, Hattori T, Fukuda M, Fujita S. Cell proliferation and differentiation in intramucosal and advanced signet ring cell carcinomas of the human stomach. Virchows Arch A Pathol Anat Histopathol. 1987;411:117–127. doi: 10.1007/BF00712735. [DOI] [PubMed] [Google Scholar]
- Peng DF, Sugihara H, Mukaisho K, Tsubosa Y, Hattori T. Alterations of chromosomal copy number during progression of diffuse-type gastric carcinomas: metaphase- and array-based comparative genomic hybridization analyses of multiple samples from individual tumours. J Pathol. 2003;201:439–450. doi: 10.1002/path.1459. [DOI] [PubMed] [Google Scholar]
- Peng DF, Sugihara H, Mukaisho K, Ling ZQ, Hattori T. Genetic lineage of poorly differentiated gastric carcinoma with a tubular component analysed by comparative genomic hybridization. J Pathol. 2004;203:884–895. doi: 10.1002/path.1586. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Sugihara H, Ling ZQ, Peng DF, Mukaisho K, Fujiyama Y, Hattori T. How wild-type TP53 is inactivated in undifferentiated-type gastric carcinomas: analyses of intratumoral heterogeneity in deletion and mutation of TP53. Pathobiology. 2006;73:40–49. doi: 10.1159/000093090. [DOI] [PubMed] [Google Scholar]
- Kushima R, Mukaisho K, Tshukashita S, Peng D, Sugihara H, Vieth M, Stolte M, Hattori T. Molecular biological characteristics of early stomach adenocarcinomas of the completely gastric phenotype revealed by laser capture microdissection and comparative genomic hybridization. Stomach and Intestine (Tokyo) pp. 707–721.
- Japanese Gastric Cancer Association. Japanese Classification of Gastric Carcinoma - 2nd English Edition - Gastric Cancer. 1998;1:10–24. doi: 10.1007/s101209800016. [DOI] [PubMed] [Google Scholar]
- Kamitani S, Sugihara H, Shiomi H, Tani T, Hattori T. Intratumoral regional variations in copy number of the chromosomal part revealed by microdissection and combined ploidy and comparative genomic hybridization analyses in esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2002;132:30–35. doi: 10.1016/S0165-4608(01)00520-9. [DOI] [PubMed] [Google Scholar]
- Little SE, Vuononvirta R, Reis-Filho JS, Natrajan R, Iravani M, Fenwick K, Mackay A, Ashworth A, Pritchard-Jones K, Jones C. Array CGH using whole genome amplification of fresh-frozen and formalin-fixed, paraffin-embedded tumor DNA. Genomics. 2006;87:298–306. doi: 10.1016/j.ygeno.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Okada K, Sugihara H, Bamba M, Bamba T, Hattori T. Sequential numerical changes of chromosomes 7 and 18 in diffuse-type stomach cancer cell lines: combined comparative genomic hybridization, fluorescence in situ hybridization, and ploidy analyses. Cancer Genet Cytogenetl. 2000;118:99–107. doi: 10.1016/S0165-4608(99)00182-X. [DOI] [PubMed] [Google Scholar]
- Ohue M, Tomita N, Monden T, Fujita M, Fukunaga M, Takami K, Yana I, Ohnishi T, Enomoto T, Inoue M, Shimano T, Mori T. A frequent alteration of p53 gene in carcinoma in adenoma of colon. Cancer Res. 1994;54:4798–4804. [PubMed] [Google Scholar]
- Zhou X, Mok SC, Chen Z, Li Y, Wong DTW. Concurrent analysis of loss of heterozygosity (LOH) and copy number abnormality (CAN) for oral premalignancy progression using the Affymetrix 10K SNP mapping array. Hum Genet. 2004;115:327–330. doi: 10.1007/s00439-004-1163-1. [DOI] [PubMed] [Google Scholar]
- Melcher R, Al-Taie O, Kudlich T, Hartmann E, Maisch S, Steinlein C, Schmid M, Rosenwald A, Menzel T, Scheppach W, Luhrs H. SNP-Array genotyping and spectral karyotyping reveal uniparental disomy as early mutational event in MSS- and MSI-colorectal cancer cell lines. Cytogenet Genome Res. 2007;118:214–221. doi: 10.1159/000108303. [DOI] [PubMed] [Google Scholar]
- Tsubosa Y, Sugihara H, Mukaisho K, Kamitani S, Peng DF, Ling ZQ, Tani T, Hattori T. Effects of degenerate oligonucleotide-primed polymerase chain reaction amplification and labeling methods on the sensitivity and specificity of metaphase- and array-based comparat. [DOI] [PubMed]
- Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, Hagiwara A, Yamaguchi T, Takahashi T, Nakamura Y, Abe T, Inazawa J. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;24:299–305. doi: 10.1002/(SICI)1098-2264(199904)24:4<299::AID-GCC2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Guan XY, Fu SB, Xia JC, Fang Y, Sham JS, Du BD, Zhou H, Lu S, Wang BQ, Lin YZ, Liang Q, Li XM, Du B, Ning XM, Du JR, Li P, Trent JM. Recurrent chromosome changes in 62 primary gastric carcinomas detected by comparative genomic hybridization. Cancer Genet Cytogenetl. 2000;123:27–34. doi: 10.1016/S0165-4608(00)00306-X. [DOI] [PubMed] [Google Scholar]
- Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH, Noh SM. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence In situ hybridization studies. Cancer Genet Cytogenet. 2000;117:97–103. doi: 10.1016/S0165-4608(99)00152-1. [DOI] [PubMed] [Google Scholar]
- Kong G, Oga A, Park CK, Kawauchi S, Furuya T, Sasaki K. DNA sequence copy number aberrations associated with histological subtypes and DNA ploidy in gastric carcinoma. Jpn J Cancer Res. 2001;92:740–747. doi: 10.1111/j.1349-7006.2001.tb01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Itabashi M, Suzuki K, Yoshida S. Clinicopathologic study of minute and small early gastric cancer. Histogenesis of gastric cancer. Pathol Annu. 1980;15(Pt 2):1–19. [PubMed] [Google Scholar]
- Knuutila S, Björkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- Chung YJ, Choi JR, Park SW, Kim KM, Rhyu MG. Evidence for two modes of allelic loss: multifocal analysis on both early and advanced gastric carcinomas. Virchows Arch. 2001;438:31–38. doi: 10.1007/s004280000328. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Uesugi N, Jao YF, Nakamura S, Abe K, Takagane A, Terashima M. Three independent genetic profiles based on mucin expression in early differentiated-type gastric cancers--a new concept of genetic carcinogenesis of early differentiated-type adenocarcinomas. Mod Pathol. 2004;17:1223–1234. doi: 10.1038/modpathol.3800170. [DOI] [PubMed] [Google Scholar]
- Tajima Y, Yamazaki K, Makino R, Nishino N, Aoki S, Kato M, Morohara K, Kaetsu T, Kusano M. Gastric and intestinal phenotypic marker expression in early differentiated-type tumors of the stomach: clinicopathologic significance and genetic background. Clin Cancer Res. 2006;12:6469–6479. doi: 10.1158/1078-0432.CCR-06-1339. [DOI] [PubMed] [Google Scholar]
- Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TP53_primer_sequences Microsoft Excel 2003 TP53 primer sequences.