Summary
Over the past two decades, the incidence of fungal infections has dramatically increased. This is primarily due to increases in the population of immunocompromised individuals attributed to HIV/AIDS pandemic and immunosuppression therapies associated with organ transplantation, cancer, and other diseases where new immunomodulatory therapies are utilized. Significant advances have been made in understanding how fungi cause disease, but clearly much remains to be learned about the pathophysiology of these often lethal infections.
Fungal pathogens face numerous environmental challenges as they colonize and infect mammalian hosts. Regardless of a pathogen's complexity, its ability to adapt to environmental changes is critical for its survival and ability to cause disease. For example, at sites of fungal infections, the significant influx of immune effector cells and the necrosis of tissue by the invading pathogen generate hypoxic microenvironments to which both the pathogen and host cells must adapt in order to survive. However, our current knowledge of how pathogenic fungi adapt to and survive in hypoxic conditions during fungal pathogenesis is limited. Recent studies have begun to observe that the ability to adapt to various levels of hypoxia is an important component of the virulence arsenal of pathogenic fungi. In this review, we focus on known oxygen sensing mechanisms that non-pathogenic and pathogenic fungi utilize to adapt to hypoxic microenvironments and their possible relation to fungal virulence.
Keywords: Aspergillus fumigatus, Cryptococcus neoformans, Candida albicans, fungal virulence, hypoxia, sterols
Introduction/Significance of hypoxia during fungal pathogenesis
Recent advances in medical therapies, organ transplantation, HIV infections, and an increasing geriatric population have generated rising populations of immunocompromised patients. These events have all resulted in a significant increase in life-threatening human fungal infections over the last two decades (1). The limited treatment options and high mortality rates associated with these infections has consequently led to a concerted effort to better understand mechanisms of fungal pathogenesis in mammals. The general rationale behind these studies is that a better understanding of how these organisms cause disease will allow us to develop better technologies for the treatment and prevention of these often lethal infections. One increasing area of fungal pathogenesis research is related to identifying and understanding the basic metabolic pathways utilized by these fungi to survive in the harsh and highly variable mammalian host environment.
The three main fungal pathogens that cause human mycoses, Aspergillus fumigatus, Cryptococcus neoformans, and Candida albicans, are typically opportunistic pathogens. These fungi are saprophytic organisms that have evolved a unique combination of attributes to survive in their natural environments. Aspergillus fumigatus is typically found in soil and decaying organic material such as compost heaps. Cryptococcus neoformans is typically found in pigeon droppings, soil and certain trees. Unlike Aspergillus and Cryptococcus species, Candida albicans is rarely found in the soil or external environment. Instead, it is a normal inhabitant of the human microflora. Thus, C. albicans is already highly adapted to the host environment. C. albicans possess the ability to survive in disparate host environments, as illustrated by its ability to colonize diverse areas of the host (oral, vaginal, gastrointestinal areas). Coincidentally, many of these attributes, which allow fungal survival in their natural ecological niches, appear to also allow these fungi to cause disease in immunocompromised hosts. One overlooked environmental selection pressure found in natural environments of all three of the most common fungal pathogens of humans is low oxygen tension. Whether in the soil, a compost pile, pigeon guano, or the gut of a mammal, these fungi must deal with low levels of oxygen.
The focus of this review is on the increasing evidence that pathogenic fungi must adapt to rapidly changing oxygen levels during fungal infections. It is well established that oxygen levels vary throughout the mammalian body depending on numerous factors including tissue type and presence or absence of an inflammatory response. For example, oxygen levels in most mammalian tissues are found to be considerably below atmospheric levels (21%) (2-4). Even in the alveoli of healthy lungs, the most oxygen rich organ and site of infection for many fungal pathogens, the oxygen level is around 14%. By the time oxygen reaches the capillaries and diffuses into surrounding tissues its availability is much lower with levels of 2-4% reported (5, 6). In addition, it is well established that at sites of inflammation available oxygen is significantly reduced compared to surrounding tissues (7-9). Moreover, in inflamed tissues, the blood supply is often interrupted because the vessels are congested with phagocytes or the pathogen itself (10, 11). Thus, it seems highly probable that hypoxic microenvironments are generated during fungal infection.
Indeed, we can look no further than the host response to observe that fungal pathogens are likely exposed to severely low oxygen levels during infection. Immune effector cells, such as neutrophils, function effectively in severely hypoxic microenvironments. These and other cells of the host have evolved distinct mechanisms to deal with hypoxic microenvironments generated during microbial infections. Many of these host response mechanisms are dependent upon the global transcription factor, hypoxia inducible factor (HIF) 1.
HIF 1 is a heterodimeric transcription factor that consists of one of three α-subunits (HIF-1α, HIF-2α, and HIF-3α) and one β-subunit (HIF-1β), and is the central regulator of hypoxic gene expression in mammals (reviewed in (12),(13)). Both the degradation and activity of the HIF-1α subunit are regulated by oxygen-dependent post-translational hydroxyl modifications. Under hypoxic conditions HIF-1α is not hydroxylated, leading to an accumulation of the HIF-1α subunit and expression of hypoxia-responsive genes, including those encoding many glycolytic enzymes, erythropoietin, adrenomedullin, and growth factors (14, 15).
In a recent study of Acute Respiratory Distress Syndrome (ARDS) and acute inflammatory lung injury, Thiel et al. (16) provided evidence for the importance of hypoxic microenvironments in regulation of host immune responses. ARDS patients are normally treated with a life-saving oxygen therapy, but this therapy may have a dangerous side effect in patients with uncontrolled pulmonary inflammation. Thiel et al. (16) identified a local tissue hypoxia-driven and adenosin A2A receptor (A2AR)-mediated anti-inflammatory mechanism. Their data suggest that oxygenation may lead to elimination of the A2AR-mediated lung tissue-protecting pathway and thereby further exacerbate lung injury. Taken together, the above observations and studies indicate that mammalian immune system responses to microbial infection and inflammation are critically tied to hypoxic microenvironments.
While the role of hypoxia in the immune response to fungal pathogens is relatively unknown, it follows that since immune cells of the host have evolved mechanisms to function in hypoxia, mammalian fungal pathogens like A. fumigatus, C. neoformans and C. albicans are likely exposed to hypoxic conditions during fungal pathogenesis. Indeed, during A. fumigatus infection, our laboratory has recently observed significant increases in HIF-1α activity as the fungal infection progresses and inflammation and edema increase in the lung (Grahl and Cramer, unpublished data). In addition, a recent study by Brock et al. (17) demonstrated that hypoxia likely occurs in vivo in the lung during A fumigatus infection. The authors constructed a luciferase-producing bioluminescent A. fumigatus strain, which was not attenuated in virulence in a murine model of invasive aspergillosis. Interestingly, luminescence from the lungs decreased after reaching a maximum at one day post infection, despite the high number of fungal hyphae present in histology examinations. The authors hypothesize that this phenomenon might be due to the severe tissue damage in and through the pulmonary lesions, which likely decrease the oxygen concentration in the lung tissue. Oxygen is essential for the light-producing reaction, and thus the lack of luminescence is likely attributable to the hypoxia at the site of infection (17).
Additional evidence that hypoxia may be a key component of the pathophysiology of invasive fungal infections comes from the observation that there are often significant differences in the in vivo and in vitro test results of antifungal drug efficacies. These differences have recently been postulated to be related to hypoxic conditions found in vivo as demonstrated by in vitro antifungal drug efficacy tests conducted in hypoxia (6, 18). Furthermore, recent studies have identified genes responsible for regulating fungal response to hypoxia, and some of these pathways have been observed to be essential for fungal virulence of mammals. Consequently, it seems probable that pathogenic fungi possess mechanisms to adapt to hypoxic microenvironments found in vivo during infection.
The purpose of this review is to summarize recent advances in our understanding of mechanisms that human fungal pathogens use to adapt to hypoxic conditions and to highlight the emerging importance of this area of research in the pathogenic fungi. While studies on hypoxia adaptation in these pathogenic fungi are limited, increasing research attention to this important component of their virulence arsenal has revealed similarities and differences with each other and the model yeast S. cerevisiae and S. pombe. Importantly, we should clarify the distinction between mechanisms of hypoxia adaptation and mechanisms of hypoxic growth. It seems clear that these are two distinct biological processes requiring a distinct set of genes and mechanisms. In our literature review, we did not find detailed studies defining these two likely different processes in the fungi. It is likely that periods of adaptation are different for diverse fungi. A close examination of the methodology used in the cited studies indicated that most, if not all, studies are focused on genes allowing the fungi to adapt to hypoxia (i.e. genes expressed/required in the early phase of exposure to hypoxia (within 48 hours of a switch to hypoxia), rather than genes required for actual fungal growth in these conditions. Where possible, we have attempted to highlight these distinctions. Finally, we have divided the manuscript into sections detailing fungi with Upc2p orthologs and fungi with SREBP orthologs given the emerging importance of these pathways in oxygen sensing and hypoxia adaptation in fungi.
Fungi with the SREBP analogue Upc2p
Saccharomyces cerevisiae
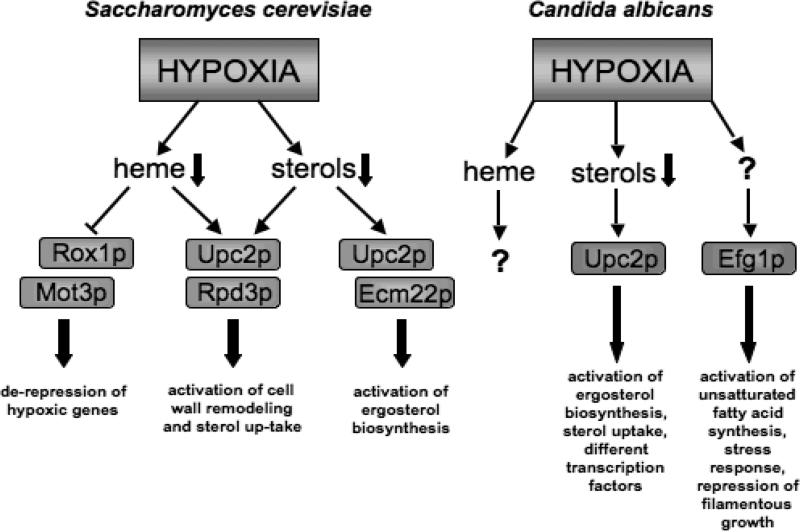
S. cerevisiae cells adapt to anaerobic conditions by inducing expression of a large number of genes, called “hypoxic genes” (19-25). The hypoxic genes encode oxygen-related functions in respiration, heme, and membrane biosynthesis that are required at higher levels when molecular oxygen is limited (20, 26). For the regulation of hypoxic genes, the cell senses oxygen availability through cellular heme levels (27, 28), and recent studies suggest that oxygen availability can also be sensed through cellular sterol levels (29).
Oxygen sensing by heme
Molecular oxygen is required as a substrate in two consecutive steps of heme biosynthesis catalyzed by the enzymes coproporphyrinogen oxidase and protoporphyrinogen IX oxidase (30). In the presence of oxygen (aerobic growth), heme accumulates, binds to the transcriptional activator Hap1p (Heme Activator Protein) and causes the formation of a Hap1p homodimer specific for DNA binding to the cis-element. Hap1p is a protein composed of a zinc-finger DNA binding domain at the N-terminus, a dimerization domain, a heme binding domain within the central region, a heme-responsive motif 7 (HRM7), and a transcriptional activation domain at the C-terminus (31-33). The heme-Hap1p-complex acts as a transcriptional activator of genes containing its recognition site (5′CGGN6CGG) (34-36), such as genes involved in respiration (reviewed in (20)).
In addition, the expression of the ROX1 (Repressor Of hypoXic genes) gene is activated by the heme-Hap1p complex and Rox1p accumulates in the cell under aerobic conditions (reviewed in (26)). The Rox1p repressor binds to its recognition site upstream of the hypoxic genes to repress their transcription (37, 38). Rox1p binds to the DNA with its HMG domain and recruits the general repression complex, Tup1/Ssn6, which binds to the Rox1p repression domain (reviewed in (26)). The repression through Rox1p varies between hypoxic genes that do not have aerobic counterparts, which are expressed at detectable levels at all oxygen concentrations but their expression is higher when oxygen decreases, like HEM13, OLE1, ERG11, and the autorepressed ROX1 itself, and hypoxic genes that have an aerobic homologue, like HMG1/HMG2, COX5A/COX5B, AAC1, AAC2/AAC3, and TIF51A/ANBI (the first gene is aerobic – last and underlined anaerobic). The anaerobic gene is then completely repressed until very low oxygen concentrations are reached (38, 39).
For some hypoxic genes, a second DNA binding protein Mot3p enhances Rox1p repression through helping recruit the Tup1p/Ssn6p complex. Two examples are the ANBI (ANaeroBically Induced) gene, encoding a subunit of eukaryotic initiation factor 5 (eIF-5a), an essential translation factor (40), and the HEM13 gene, encoding the enzyme coproporphyrinogen III oxidase, which catalyzes the rate-limiting step in heme biosynthesis (41). The hypoxic derepression of HEM13 allows the cell to continue heme biosynthesis under limited available oxygen. The strongly repressed ANBI gene has one Mot3p and two Rox1p binding sites in its promoter region, while the promoter of the partially repressed HEM13 contains one Rox1p and three Mot3p binding sites. The combination of binding sites determines the strength of repression. Multiple Mot3p binding sites plus a single Rox1p binding site are much weaker than multiple Rox1p binding sites plus a single Mot3p binding site (42). Rox1p and Mot3p both interact with Ssn6p of the general repression complex and Rox1p stabilizes Mot3p binding to DNA through interactions with Tup1p/Ssn6p (42).
Under hypoxic or anaerobic growth conditions, heme levels are reduced. Hap1p still binds to its cognate site, but in the absence of heme, Hap1p forms a biochemically distinct High-Molecular-weight Complex, HMC, which contains Hap1p and four other proteins including Hsp28p and Ydj1p. This complex represses transcription (43). Consequently, under hypoxic conditions, ROX1 and MOT3 expression is repressed resulting in the activation of hypoxic genes expression. (26).
During adaptation to anaerobic conditions, a complex program of cell wall remodeling occurs in yeast. Under anaerobic conditions, major aerobic cell wall mannoproteins, encoded by CWP1 and CWP2, are replaced by their anaerobic counterparts, encoded by the DAN/TIR genes. The DAN/TIR genes encode a group of eight cell wall mannoproteins that play a significant role in cell wall permeability (23, 44). DAN/TIR genes are regulated by heme, sterol levels, and three DNA binding transcription factors. The heme-dependent repressors Rox1p and Mot3p function synergistically to efficiently repress DAN/TIR genes under aerobic conditions (45). In addition, the sterol depletion-dependent activator Upc2p acts through a consensus site termed AR1 to induce the expression of DAN/TIR genes in anaerobic conditions (46). Sertil et al. (47) observed that the histone deacetylase and global repressor Rpd3p is required for the expression of all the DAN/TIR genes and the hypoxic gene ANBI. Moreover, the authors propose that Rpd3p is recruited to the DAN1 promoter under strict anaerobic conditions. The presence of Rpd3p at the promoter counteracts the function of the repressor Mot3p, which leads to stable binding of the activator Upc2p. Upc2p then recruits the chromatin remodeling complex Swi/Snf to reorganize chromatin, thereby facilitating the binding of the transcriptional machinery that results in the activation of gene expression (47). Upc2p, together with the transcription factor Ecm22p, is also responsible for basal and induced expression of genes encoding enzymes of ergosterol biosynthesis in yeast (ERG1, ERG2, ERG3, ERG7, ERG25, ERG26, and ERG27), and it has been implicated in the uptake of sterols under hypoxic conditions (48-52).
Oxygen sensing by sterols
While heme has been thought to be the primary oxygen sensor in S. cerevisiae, recent studies suggest that sterol levels also play an important role. Upc2p and Ecm22p are functionally related to human sterol regulatory element binding protein (SREBP) with an N-terminal transcription factor domain and a C-terminal transmembrane domain. Although S. cerevisiae lacks an ortholog of SREBP, it seems that a potentially analogous oxygen-sensing mechanism exists in budding yeast regulated through Upc2p and Ecm22p. Marie et al. (53) have observed that Upc2p and Ecm22p are localized outside of the nucleus in sterol replete conditions, but in conditions of sterol depletion localization shifts toward the nucleus. The authors suggest that the N-terminal transcription factor domain is separated from the C-terminal transmembrane domain by proteolytic cleavage and enters the nucleus to activate gene expression, analogous to SREBP regulation of cholesterol biosynthesis in mammals.
Upc2p and Ecm22p both bind a sequence motif known as the sterol regulatory element (SRE) (48, 49, 54). Nearly one-third of hypoxically induced genes in S. cerevisiae contain at least one potential Upc2p/Ecm22p binding site, suggesting that these transcription factors are major players in the adaptation to hypoxia (55). The activation of target genes by Upc2p occurs in response to low sterol levels, which can be caused by blocks in ergosterol biosynthesis or by hypoxia. Davies and Rine (29) observed that both Upc2p and Ecm22p require a functional version of Hap1p for basal expression of ERG2, but when sterols are depleted Upc2p is independent of Hap1p, whereas Ecm22p still depends upon Hap1p for ERG gene activation. ERG2, ERG3, ERG10, DAN2, and DAN4 are activated by Upc2p solely in response to sterol depletion whereas DAN1 and TIR1 respond to both sterols and heme (46).
Other oxygen sensing mechanisms
An additional hypoxic regulatory pathway involving an antagonistic interaction between the Ord1p repressor and the Yap1p factor (a transcriptional activator involved in oxidative stress response) has been discovered in S. cerevisiae and regulates both TIR1 and SRP1. The hypoxic response of TIR1/SRP1 (both encode cell wall mannoproteins) depends on the absence of heme but is Rox1p-independent. Under aerobic conditions, Ord1p binds to the SRP1 promoter and expression is repressed. When conditions change to hypoxia, Yap1p also binds to the SRP1 promoter, counteracts the Ord1p effect and SRP1 is expressed (56).
Multiple pathways involved in regulating hypoxic and anoxic gene expression in yeast may exist. Studies of several other hypoxic/anaerobic genes including SUT1, encoding a putative Zn[II]2Cys6-transcription factor that facilitates the uptake and synthesis of sterols under hypoxic conditions (57), GPD2, encoding an isoenzyme of NAD-dependent glycerol 3-phosphate dehydrogenase (58), and members of the seripauperine (PAU) family, like TIR1 (59) have demonstrated Rox1p-independent hypoxic/anaerobic induction, but the mechanisms by which this occurs are not yet understood.
Another recently described possible mechanism of hypoxia signaling in yeast involves the mitochondrial respiratory chain, the cytochrome c oxidase and reactive oxygen species (60, 61). It has been shown that mitochondria from yeast, rat liver, and plants are capable of nitrite (NO2-)-dependent nitric oxide (NO) synthesis (60, 62-66). This pathway is induced when cells experience hypoxia, and furthermore, Castello et al. (60) suggest that mitochondrially produced NO functions in a signaling pathway to the nucleus by reacting with the superoxide produced by hypoxic mitochondria (67) to form peroxynitrite (ONOO-) that promotes protein tyrosine nitration of specific proteins that may be involved in a signaling pathway to the nucleus. Future research on this mechanism will likely uncover its specific role in hypoxia adaptation.
It seems clear that adaptation to hypoxia is a complex multi-faceted process regulated via the interaction of several different critical metabolic pathways in the cell. The major regulatory pathways discussed above are summarized in Figure 1 and Table 1. We now turn our attention to other fungi and discuss similarities and differences with these hypoxia adaptation mechanisms in S. cerevisiae. As many of the pathogenic fungi employ different life-styles than S. cerevisiae, it is still unclear which of these pathways involved in regulating responses to hypoxia in baker's yeast are conserved in fungi that invade mammalian hosts.
Fig. 1. Schematic of the oxygen sensing pathways in Saccharomyces cerevisiae and Candida albicans.
The proteins are defined in the text.
Table 1.
Hypoxia sensing mechanisms and pathways.
| Hypoxia sensing pathways | mammals | S. cerevisiae | C. albicans | S. pombe | C. neoformans | A. fumigatus |
|---|---|---|---|---|---|---|
| HIF pathway | a) HIF-la, HIF-2a, HIF-3a; HIF-lb | No homolog found | No homolog found | No homolog found | No homolog found | No homolog found |
| Heme level sensing pathways | No homolog found | a) Hap1p/Rox1p/Mot3p | Rfg1p* (Rox1p homolog) | No homolog found | No homolog found | No homolog found |
| No homolog found | b) Yap1p/Ord1p | Cap1px | Pap1px | No homolog found | AfYap1x | |
| Sterol sensing pathways | b) SREBP-1(a/c)*, SREBP-2*, SCAP*, INSIG-1*, INSIG-2*, S1P*, S2P* | c) Upc2py/Ecm22p | Upc2py | Sre1p, Scp1p, Ins1p* | a) Sre1p, Scp1p, Stp1p* | SrbA, InsA* |
| Tco1 hypoxia sensing pathways | no known | no known | Nik1px | no known | b) Tco1p | Bos1x |
| Regulated pathways in response to activation of hypoxia sensing pathways | regulated by a): erythropoiesis and iron metabolism (e.g. erythropoietin and transferrin); oxygen transport (e.g. adrenomedullin) glucose uptake and glycolysis (e.g. glucose transporter 1, Lactate dehydrogenase A) | regulated by a): e.g. heme biosynthesis (HEM13), cell wall remodeling (DAN/TIR genes); fatty acid synthesis (OLE1); eIF-5a factor subunit AnbIp; respiration and ATP exchange (COX5b; ACC3) | regulated by Upc2p: azole drug resistance and sterol biosynthesis (ERG11); several transcription factors (INO2, ACE2, SUT1, UPC2); corticosteroid uptatke (CBP1); | regulated by Sre1p: heme biosynthesis (HEM13, HEM14, HEM15); sphingolipid biosynthesis (e.g. SPBC887.15c); fatty acid biosynthesis (e.g. CUT6); ergosterol biosynthesis (e.g. ERG1, ERG5, ERG6, ERG11, ERG25) | regulated by a): iron uptake (e.g. SIT1 and FRE7); azol drug resistance and sterol biosynthesis (e.g. ERG3, ERG11, ERG25); crucial for virulence | regulated by SrbA: resistance to azole drugs and sterol biosynthesis (erg25, erg24, and erg3); transporters and cell wall related genes; cell polarity; crucial for virulence |
| regulated by b): lipid synthesis (cholesterol biosynthesis and uptake) | regulated by b): cell wall mannoproteins (TIR1/SRP1) | regulated by b): no impact on expression of hypoxic genes - may be post-transcriptional regulation?; required for virulence | ||||
| regulated by c): cell wall remodeling (DAN/TIR genes); ergosterol biosynthesis and sterol uptake (e.g. ERG2, ERG3, ERG10, DAN2, DAN4) |
no role in hypoxia response
role in hypoxia unknown
SREBP analog
Candida albicans
Candida albicans is an important human fungal pathogen that causes superficial skin infections as well as deep-seated infections, suggesting that its ability to switch between normoxia and hypoxia is a major determinant of its virulence (68). In C. albicans little is known about mechanisms utilized by this yeast to adapt to hypoxic microenvironments. However, our current knowledge suggests that the transcriptional response to hypoxia differs significantly between C. albicans and S. cerevisiae in important aspects. Although both are generally referred to as facultative anaerobes, genetics studies, conditions required for anaerobic growth, and genome analyses seem to suggest that these hemiascomycota yeast respond differently to changes in oxygen levels.
First, a homologue (Rfg1p) of the S. cerevisiae Rox1p has been identified in C. albicans, but Rfg1p does not play a role in the regulation of hypoxic genes in this pathogenic yeast as in S. cerevisiae (Table 1). Instead, Rfg1p is a transcriptional regulator that controls filamentous growth, and in that role, is critical for C. albicans virulence (69).
Second, S. cerevisiae genes involved in glycolysis and fermentation are not stimulated by hypoxia (25, 70), but hypoxia induces these genes and genes involved in hyphal growth in C. albicans while genes of oxidative metabolism are repressed (68). During normoxic conditions the global transcription factor Efg1p regulates the expression of genes involved in glycolysis and respiration, but it has no role in controlling the expression of respiratory genes and is not required to upregulate glycolytic gene expression in hypoxia (68, 71). Efg1p also promotes filamentation under normoxic conditions. Recently, it was observed that under hypoxic conditions Efg1p promotes the synthesis of unsaturated fatty acids, the up-regulation of genes involved in the stress response (HSP12, DDR48, CTA1) and represses filamentous growth in C. albicans. Thus, the regulatory role of Efg1p in C. albicans strongly depends on oxygen (68, 72). Transcriptional analyses observed that Efg1p is required to allow hypoxic regulation of about half of all genes that are normally regulated by hypoxia in C. albicans. In an efg1 mutant, hypoxic upregulation (e.g. CTA1) or downregulation (e.g. RIP1) of several genes is abolished, and some genes, like OLE1 encoding a fatty acid desaturase, are ineffectively expressed in hypoxia. Another major function of Efg1p is to prevent hypoxic regulation of numerous genes that are not normally up- or downregulated under hypoxia (71). Despite the fact that Efg1p is a major regulator of the hypoxic response in C. albicans, a homozygous efg1 mutant shows no severe change in virulence in comparison to the wild-type (73).
Another transcription factor in C. albicans affected by hypoxia, Ace2p, is required for filamentation in response to hypoxic conditions. Ace2p also induces fermentative growth and represses respiration, but it is possible that the effect of Ace2p on metabolism is restricted to normal oxygen conditions. This remains to be tested (74). Interestingly, an ace2 null mutant is almost avirulent in an immunocompetent mouse model, while there is only a low degree of attenuation in a neutropenic mouse model (75, 76). This may suggest that different states of the immune system may affect the development of hypoxia in vivo i.e. the lack of neutrophils in the neutropenic model minimizes the inflammatory response and hence hypoxic microenvironments encountered by the invading fungus. This remains to be examined and confirmed.
As in S. cerevisiae (23, 44, 46), the cell-wall proteome of C. albicans is sensitive to changes in environmental conditions which helps the cell to adjust to harsh environments. For example, iron deprivation and hypoxic conditions affect the expression of cell-wall protein encoding genes, such as iron acquisition and iron-uptake genes i.e. RBT5 a gene encoding a predicted GPI protein involved in iron acquisition (68, 77, 78). Numerous oxygen-dependent reactions in the cell are carried out by iron-containing enzymes (79). During hypoxic conditions, there may be competition for iron by iron-containing enzymes, which might lead to an increased expression of cell-wall protein encoding genes involved in iron-acquisition and iron-uptake (80).
Oxygen sensing in Candida albicans
In S. cerevisiae it has been observed that the cell senses oxygen availability through cellular heme and sterol levels (27-29). In C. albicans, no apparent homolog of ScHAP1 exists, but in recent studies a close ortholog of S. cerevisiae Upc2p and Ecm22p, both involved in sensing sterol depletion, has been identified. Upc2p, a transcription factor of the zinc cluster family, is an important regulator of the sterol biosynthesis and azole drug resistance in C. albicans (81, 82) (Figure 2). Hoot et al. (83) showed that transcriptional regulation of UPC2 expression occurs through Upc2p-dependent as well as a novel Upc2p-independent mechanism. Whether there is also a post-translational control mechanism as described for the mammalian sterol regulator SREBP (discussed in detail below) or as suggested for the ScUpc2p remains to be determined.
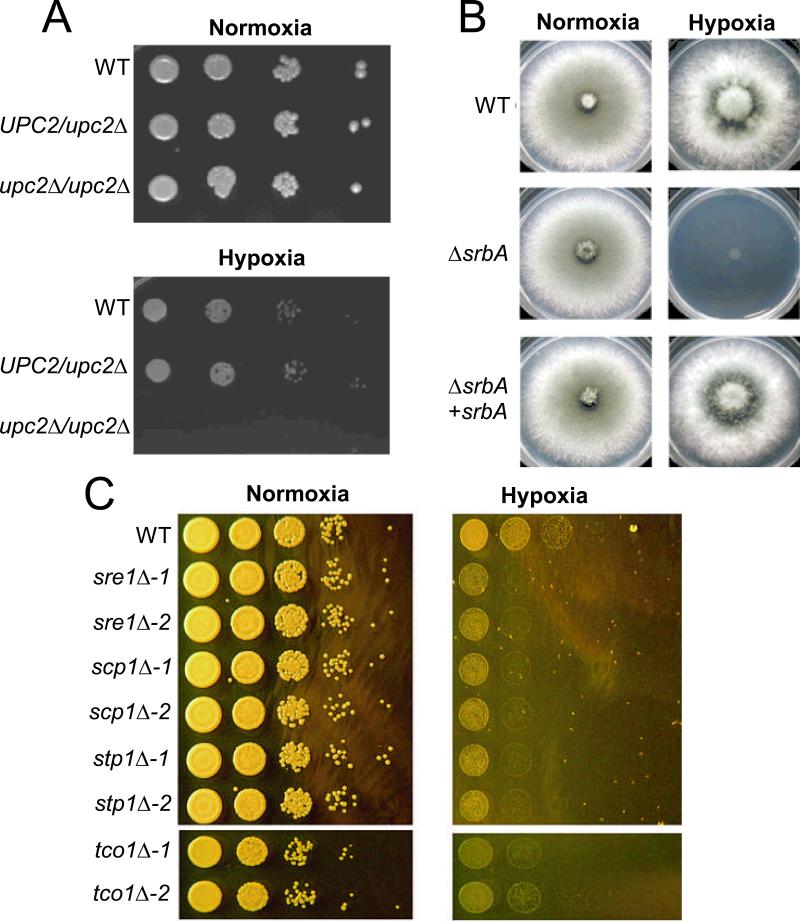
Fig. 2. Mutants in SREBP Pathway and Tco1 are sensitive to hypoxia.
Growth in normoxic and hypoxic conditions. A) Candida albicans: A heterozygous UPC2/upc2Δ and a homozygous upc2Δ/upc2Δ C. albicans strain were serially diluted and spotted on CSM plates and grown at 30°C. The top panel shows growth in aerobic conditions after 48h. The bottom panel shows growth in hypoxic conditions after 96h. Under hypoxic conditions the wild-type (WT) and the heterozygous strain showed comparable growth but the homozygous deletion strain did not demonstrate any detectable growth (Courtesy Chelsea Samaniego and Dr. Theodore C. White); B) Aspergillus fumigatus: 1×106 conidia were plated on GMM plates and incubated at 37°C under normoxic and hypoxic conditions for 48h. The wild-type and the reconstituted strain grew comparably under hypoxic conditions while no growth was detectable for the mutant strain (modified from Willger et al. (124)); C) Cryptococcus neoformans: C. neoformans cultures diluted to OD600nm = 0.6 were diluted serially in 10-fold increments prior to being spotted onto YPD plates. The plates were incubated in normoxic or hypoxic conditions in the dark at 37°C. Under hypoxic conditions all mutants in the SREBP pathway and the tco1Δ mutants showed reduced growth compared to the wild-type (modified from Chun et al. (100)).
Upc2p binds in vivo to the promoters of several ergosterol biosynthesis genes and other genes involved or predicted to be involved in lipid metabolism. Znaidi et al. (84) observed that up-regulation of ERG11 during hypoxia is strictly Upc2p dependent. Upc2p also binds the promoters of four genes encoding transcription factors (INO2, ACE2, SUT1, and UPC2 itself). One of them, Sut1p, controls sterol uptake in S. cerevisiae (57, 85) suggesting that in C. albicans Upc2p and Sut1p may interact in a sterol regulatory network (84).
Interestingly, Upc2p also binds to the promoter of CBP1, which was shown to encode a corticosteroid binding protein in C. albicans (86). C. albicans appears to take up steroids and possibly metabolic precursors from the host, and Upc2p seems to play a role in corticosteroid uptake from mammals and in adaptation of C. albicans to hypoxic conditions in the host (84). Thus, an emerging theme with studies in S. cerevisiae and C. albicans is the role of sterol homeostasis in adaptation to hypoxic microenvironments. This theme will also be expanded on in additional fungi discussed below with the discovery of SREBP orthologs. A summary of the known hypoxia regulation mechanisms in C. albicans is presented in Figure 1 and Table 1.
Fungi with SREBP orthologs
Schizosaccharomyces pombe
Recently, a novel mechanism of hypoxia adaptation mediated by a highly conserved family of transcription factors, the SREBPs, was characterized in Schizosaccharomyces pombe (87). S. pombe, also called “fission yeast”, is a non-pathogenic yeast that is used as a model organism in molecular and cell biology. SREBPs are a family of endoplasmic reticulum (ER) membrane bound transcription factors first identified in mammals as regulators of cholesterol and fatty acid synthesis (88-92). SREBPs contain two transmembrane segments and are inserted into ER membranes in a hairpin fashion such that the N- and C-terminal ends of the protein are in the cytosol. SREBP is synthesized as an inactive membrane-bound precursor that forms a complex with SCAP (SREBP Cleavage-Acting Protein), a multispan membrane protein that is a component of the sterol sensor (90). Under conditions with enough available sterols, the SREBP-SCAP complex is retained in the ER membrane through binding of SCAP to the resident ER protein Insig (93). In sterol-depleted cells, SCAP changes its confirmation, which releases the SREBP-SCAP complex from Insig (94). SCAP then escorts SREBP from the ER to the Golgi apparatus where SREBP is activated by two sequential proteolytic events catalyzed by site-1 and a site-2 proteases that release the N-terminal transcription factor domain from the membrane, allowing the transcription factor to enter the nucleus and direct the transcription of target genes (90, 95).
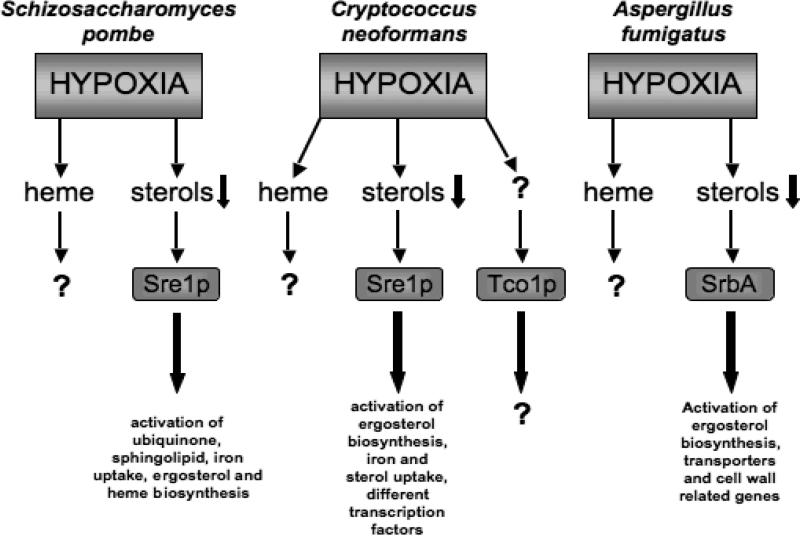
In S. pombe apparent orthologs of SREBP (SRE1), SCAP (SCP1) and Insig (INS1) have been identified and characterized. Sre1p is cleaved and activated in response to sterol depletion and hypoxia, and stimulates transcription of genes required for adaptation to hypoxia such as genes involved in heme, sphingolipid, ubiquinone, and ergosterol biosynthesis (Figure 3 and Table 1) (87, 96). Thus, in fission yeast, Sre1p and Scp1p appear to monitor-oxygen dependent sterol synthesis as an indirect measure of oxygen supply. Interestingly, there does not appear to be an impact of Ins1p on the SREBP pathway in fission yeast. In addition, Hughes and Espenshade (97) recently identified another component of this pathway, Ofd1p. Ofd1p is a prolyl 4-hydroxylase- like 2-oxoglutarate-Fe(II) dioxygenase that accelerates Sre1p degradation in the presence of oxygen. The N-terminal dioxygenase domain is an oxygen sensor that regulates the activity of the C-terminal degradation domain (97). Altogether, the SREBP pathway functions as an oxygen sensor and is required for adaptation to hypoxia in fission yeast. However, the critical function of Sre1 in allowing hypoxic adaptation and subsequent growth is not clearly defined. It seems likely that Sre1 is playing a pleiotropic role in regulating many different genes required for yeast cells to adapt and grow in hypoxia.
Fig. 3. Schematic of the oxygen sensing pathways in Schizosaccharomyces pombe, Cryptococcus neoformans and Aspergillus fumigatus.
The proteins are defined in the text.
Orthologs of the SREBP pathway were recently identified and characterized in the human fungal pathogens C. neoformans and A. fumigatus. Yet, the exact components and the mechanism behind SREBP regulation largely remain to be determined in these pathogenic fungi. Moreover, it appears that the SREBP pathway is similar in function to the Upc2p mediated pathway in S. cerevisiae and C. albicans, but the mechanisms behind the similarities and differences between these two pathways is currently not clear.
Cryptococcus neoformans
Unlike the Ascomycete yeast S. cerevisiae and C. albicans, the Basidiomycete yeas Cryptococcus neoformans is generally considered an obligate aerobe. Cryptococcus species cause the disease Cryptococcosis in both immunocompromised and apparently healthy hosts and are the most common cause of fungal meningitis (98). C. neoformans is primarily found in pigeon droppings and soil contaminated with avian guanos throughout the world (99). One would speculate that these environments are relatively oxygen poor suggesting that C. neoformans likely has evolved mechanisms to adapt to low oxygen microenvironments. In the laboratory, C. neoformans grows optimally under atmospheric oxygen conditions (21%), but oxygen concentrations in the human brain are drastically lower than in the atmosphere and vary significantly among anatomical sites (2). Thus, in order to establish an infection in the brain, it seems likely that C. neoformans must adapt to reduced oxygen levels during infection. Therefore, discovering the mechanisms utilized by C. neoformans to sense and adapt to low-oxygen conditions is an important area of research aimed towards understanding the pathobiology of this pathogenic yeast. Yet, until recently, the importance of hypoxia adaptation in C. neoformans biology and virulence have been largely un-studied.
Recent whole-genome microarray-based transcriptional profiling of C. neoformans in a hypoxic microenvironment has started to reveal genes and pathways regulated in response to hypoxia. Among them are genes involved in hexose uptake (sugar transporter and hexose transporter), ethanol production (pyruvate decarboxylase and alcohol dehydrogenases), and sterol metabolism (ergosterol biosynthesis genes) (100). The possibility of fermentation being important for hypoxic growth during infection is supported by another study, where ethanol was found in cerebral tissue of rats infected with C. neoformans (101). Yet the importance of fermentation pathways in this obligate aerobe's ability to cause disease and grow in hypoxic microenvironments is unknown. However, as with other yeasts we have discussed, and the filamentous mold A. fumigatus, sterol biosynthesis and homeostasis seems to be a common mechanism regulating adaptation to low oxygen environments in fungi.
The SREBP pathway in Cryptococcus neoformans
Orthologs of SREBP (SRE1), SCAP (SCP1) and a Site-2-protease (STP1) were identified and characterized in C. neoformans (100, 102). C. neoformans appears to lack an identifiable homologue of Insig, the ER retention-protein that controls ER-to-Golgi transport of SREBP-SCAP complex in mammalian cells. This finding is consistent with S. pombe data which suggests that Insig is not required for sterol-dependent regulation of Sre1p and Scp1p (87, 102). However, as in fission yeast, the SREBP pathway mediated by Sre1p and Scp1p in C. neoformans is crucial for adaptation to hypoxia and sterol biosynthesis (Figure 2). In addition, unlike in S. pombe, Sre1p controls low-oxygen expression of genes required for two different pathways of iron uptake (SIT1 and FRE7) (102), which might be crucial for survival under hypoxic conditions. Importantly, sre1Δ mutants fail to proliferate in host tissue, fail to cause fatal meningoencephalitis, and display hypersensitivity to the azole class of antifungal drugs (Table 1 and Figure 3) (100, 102). It is unclear if the virulence defect is due to deficiencies in iron homeostasis, a known virulence attribute of pathogenic fungi, or the inability of C. neoformans to grow in low oxygen microenvironments in the absence of Sre1p. The two phenotypes are likely not mutually exclusive given the importance of iron in ergosterol biosynthesis. However, it is clear that C. neoformans needs Sre1p activation to adapt to the host environment. Mechanisms linking ergosterol biosynthesis, iron homeostasis, and fungal virulence in this pathogenic yeast remain unknown. Identification and characterization of additional components of the SREBP pathway are likely to yield important insights into how this pathogenic yeast causes disease.
In order to identify novel Sre1p pathway components in C. neoformans, Lee et al. (103) observed that responses to cobalt chloride (CoCl2) in C. neoformans mimic certain aspects of hypoxia by targeting enzymes in the sterol biosynthesis pathway. CoCl2 has been widely used as a hypoxia-mimicking agent in mammalian systems (104-108), but the mechanisms by which it induces hypoxia-mimicking responses are not fully understood. However, Sre1p is required for adaptation to CoCl2 in C. neoformans. Upon CoCl2 treatment, Sre1p is likely activated in response to sterol defects caused by the inhibition of several enzymatic steps in the ergosterol biosynthetic pathway. CoCl2 treatment leads to increased levels of sterol intermediates, including the substrates of Erg25p, 4,4-dimethylfecosterol and 4-methylfecosterol, demonstrating that Sre1p regulates sterol homeostasis in response to CoCl2. CoCl2-induced sterol synthesis inhibition and Sre1p activation has also been observed in S. pombe, suggesting a conserved role for Sre1p in the adaptation to elevated levels of transition metals (103, 109).
Consequently, CoCl2 treatment has been used to screen for pathways involved in oxygen sensing in C. neoformans. In this context Ingavale et al. (109) observed that CoCl2 sensitivity and/or oxygen sensing and adaptation processes in C. neoformans have a complex nature. Importantly they identified several mutants with increased sensitivity to CoCl2 and they observed that most of the CoCl2 sensitive mutants are also sensitive to low oxygen concentrations. Mutants included genes involved in the sterol biosynthesis pathway such as SCP1, SRE1, ERG5, mutants in genes involved in mitochondrial function and energy metabolism such as H+ transporting ATP synthase, NADH:ubiquinone oxidoreductase or ATP:ADP anitporter, and various transporters and enzymes such as hexose transport related protein, seroheme synthase, amino acid transporter and myo-inositol oxygenase. The role of these genes and pathways in fungal virulence has yet to be explored, but the apparent role of the mitochondia in hypoxia adaptation in C. neoformans echoes the recent findings in S. cerevisiae discussed above.
The two-component like (Tco) system in Cryptococcus neoformans
Additional studies observed that the Sre1p pathway acts in parallel with a two-component signal transduction like pathway controlled by Tco1p in hypoxic adaptation of C. neoformans (100). Tco1p is a member of a highly conserved family of fungal-specific histidine kinases. Tco1p negatively regulates the expression of melanin formation and, redundantly with Tco2p, positively regulates the HOG MAPK pathway (which is dispensable for virulence) (110). Interestingly, it has been shown that Tco1p is required for growth under hypoxic conditions and for virulence of C. neoformans (Figure 2) (100, 110). However, it is unclear how this pathway is involved in hypoxic adaptation and fungal virulence (Figure 3). In contrast to mutants in SRE1, the tco1Δ mutant shows no detectable defects in the regulation of any of the known hypoxic genes ((100), unpublished data). As a result Chun et al. (100) hypothesize that the Tco1p pathway might act post-transcriptionally. However, this result may also indicate that novel pathways or altered function of existing known pathways regulated by Tco1p are involved in hypoxia adaptation. Clearly, however, these data suggest that oxygen sensing in C. neoformans is highly complex, and likely important for virulence of this organism.
Aspergillus fumigatus
A. fumigatus is a saprophytic, obligate aerobic filamentous fungus commonly found in soil and compost piles. Its primary ecological function is to recycle carbon and nitrogen through the environment (111-113). As with most fungi, it seems self-evident that these microenvironments would place significant oxygen related stress on the mold. While A. fumigatus is responsible for a number of clinically relevant diseases, invasive pulmonary aspergillosis (IPA) is the most lethal with mortality rates ranging from 60-90% (114-116). Interestingly, while IPA can be caused by several Aspergillus species, the majority of IPA cases are caused by A. fumigatus. This may suggest that A. fumigatus contains unique attributes that allow it to cause disease (117).
Currently, we have a limited understanding of the in vivo growth mechanisms of A. fumigatus during IPA (118). Given that the lung is the primary site of infection for this mold, it may be counter-intuitive to think that low oxygen levels would be a critical component of the pathophysiology of IPA. However, during infection, A. fumigatus causes significant damage to host tissue through invasive growth by hyphae and subsequent recruitment of immune effector cells (depending on the immune system status of the host). Thus, infection generates significant inflammation and necrosis in lung tissue that can be visualized by histopathology. These pathologic lesions also likely represent areas of poor oxygen availability to the pathogen and host. Thus, it is likely that to cause disease A. fumigatus must adapt to hypoxic conditions.
In general, mechanisms of hypoxia adaptation in molds have gone largely unstudied. Tarrand et al. (119, 120) hypothesized that the low rate of Aspergillus recovery from clinical material is due to adaptation by the fungus to the physiologic temperature and hypoxic milieu found in vivo. However, recent studies with A. fumigatus suggest that it cannot grow in anaerobic environments (121). Interestingly, studies with the relatively non-pathogenic model mold A. nidulans observed that while the mold could not proliferate without oxygen, ethanol fermentation was required for its long-term survival in anaerobic conditions (122, 123). Indeed, analyses of Aspergillus genome sequences have revealed numerous potential fermentation pathways in these molds. Yet, as obligate aerobes, it remains unclear what function these pathways may serve in hypoxia adaptation and fungal virulence. Recent studies in our laboratory have identified ethanol fermentation during A. fumigatus infection in a murine model of IPA indicating that fermentation may be a component of the virulence arsenal of this mold (Grahl et al. unpublished data). However, our knowledge about the mechanisms by which A. fumigatus, an obligate aerobe, adapts to hypoxic environments remains extremely limited.
The SREBP pathway in Aspergillus fumigatus
Recently, our laboratory identified and characterized an SREBP (Sre1p) ortholog, SrbA, in A. fumigatus (124). As in C. neoformans, SrbA is crucial for adaptation to hypoxia, mediates resistance to the azole class of antifungal drugs and is involved in sterol biosynthesis in A. fumigatus (Figure 2 and Figure 3). In addition, unlike C. neoformans, but similar to S. pombe, transcriptional profiling of the SrbA null mutant suggested that SrbA does not appear to be involved in iron uptake or homeostasis in A. fumigatus. However, these studies may have been limited by the type of media utilized, and direct studies regarding the role of SrbA in iron homeostasis in this pathogenic mold are ongoing.
Importantly, srbA null mutants are almost avirulent in two distinct murine models of IPA. While our results strongly suggest that the virulence defect is due to the inability of this mutant to grow in hypoxia due to the loss of hypoxia adaptation mechanisms regulated by SrbA, as with C. neoformans, other potential hypotheses may explain the attenuation of virulence (124). For example, SrbA plays an important role in maintenance of cell polarity in A. fumigatus (124). We hypothesize that the accumulation of sterol intermediates leads to dysfunction in the formation or localization of sterol microdomains known to be required for maintaining cell polarity. Thus, the alteration of cell polarity may inhibit the ability of the SrbA mutant to cause disease. Yet, the mutant displays a normal growth rate in vitro suggesting that the altered cell polarity does not alter growth in these conditions. Altogether, these data promote the hypothesis that hypoxia plays a key role in the pathophysiology of IPA. At the least, it is apparent that SREBPs are critical components of fungal virulence in both pathogenic yeast and molds.
So far the molecular mechanism behind SrbA regulation and activation in molds is unclear. In the yeast S. pombe and C. neoformans, it seems evident that Sre1p is regulated post-translationally in response to sterol biosynthesis perturbation that occurs in low oxygen environments. Indeed, Hughes et al. (125) have identified 4-methyl sterols as the primary activating agent of Sre1p in S. pombe and C. neoformans. Thus, our finding that the SrbA null mutant in A. fumigatus accumulates 4-methyl sterols may also suggest that these sterols are the trigger for SrbA activation in A. fumigatus (124).
While many of the phenotypes observed in the A. fumigatus srbA mutant may suggest that SrbA is regulated in a similar manner as Sre1p in yeast, our results may also suggest an alternative model in molds. For example, despite intense bioinformatic analyses, we have been unable to identify clear homologs of SCAP or the proteases required for Sre1p activation. We have, however, identified a potential Insig1 homolog (insA) and we are currently characterizing a possible role for InsA in SREBP signaling in filamentous fungi. Yet, given the conservation of SCAP across many organisms, it is surprising that an ortholog does not appear to be present in the Aspergilli. It may be that another protein with a divergent sequence performs a similar function as SCAP, or it may suggest that a novel mechanism of SREBP regulation and activation exists in molds. Studies to examine these potential mechanisms are ongoing in our laboratory.
Conclusion
In this review we have attempted to survey the known mechanisms utilized by fungi to regulate adaptation to hypoxic microenvironments. It is clear that we are just beginning to understand the mechanisms human fungal pathogens use to survive in vivo during infection. With the possible exception of SREBPs, the molecular mechanisms utilized by pathogenic fungi to adapt to hypoxic microenvironments found at sites of infection remain to be elucidated. A master regulator of hypoxia adaptation, such as HIF1 found in mammals, has not been identified in fungi. It remains to be seen whether one exists, or, if as suggested by current data, fungi rely on multiple mechanisms to sense oxygen levels and adapt to low oxygen environments.
In any case, we feel that this area of pathogenic fungal physiology has been ignored for too long. Certainly, some mechanisms of hypoxia adaptation, perhaps such as heme biosynthesis, are likely to be conserved between S. cerevisiae and the human pathogenic fungi. However, the different life-styles and selection pressures on nonpathogenic and pathogenic fungi likely have resulted in unique mechanisms of hypoxia adaptation Thus, solely relying on S. cerevisiae as a model for how pathogenic regulate adaptation to hypoxic microenvironments is likely not appropriate. Regardless, it seems clear that mechanisms of hypoxia adaptation have important implications for fungal virulence and how we manage and treat invasive fungal infections. Therefore, future studies on discovering the conserved and unique pathways utilized by the major fungal pathogens of humans to adapt to hypoxia are likely to yield important insights into sterol metabolism, fungal growth, mechanisms of drug resistance, and fungal virulence.
Acknowledgements
RAC is currently supported by funding from the National Institutes of Health, COBRE grant RR020185, and the Montana State University Agricultural Experiment Station.
Footnotes
Disclosure of Conflict of Interest
The authors have no conflict of interest to report.
References
- 1.McNeil MM, Nash SL, Hajjeh RA, et al. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin Infect Dis. 2001;33(5):641–7. doi: 10.1086/322606. [DOI] [PubMed] [Google Scholar]
- 2.Erecinska M, Silver IA. Tissue oxygen tension and brain sensitivity to hypoxia. Respir Physiol. 2001;128(3):263–76. doi: 10.1016/s0034-5687(01)00306-1. [DOI] [PubMed] [Google Scholar]
- 3.Carlsson PO, Palm F, Andersson A, Liss P. Markedly decreased oxygen tension in transplanted rat pancreatic islets irrespective of the implantation site. Diabetes. 2001;50(3):489–95. doi: 10.2337/diabetes.50.3.489. [DOI] [PubMed] [Google Scholar]
- 4.Studer L, Csete M, Lee SH, et al. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J Neurosci. 2000;20(19):7377–83. doi: 10.1523/JNEUROSCI.20-19-07377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West JB. Respiratory Physiology - The Essentials. 3 edn Williams & Wilkins; Baltimore, MD, USA: 1985. [Google Scholar]
- 6.Warn PA, Sharp A, Guinea J, Denning DW. Effect of hypoxic conditions on in vitro susceptibility testing of amphotericin B, itraconazole and micafungin against Aspergillus and Candida. J Antimicrob Chemother. 2004;53(5):743–9. doi: 10.1093/jac/dkh153. [DOI] [PubMed] [Google Scholar]
- 7.Matherne GP, Headrick JP, Coleman SD, Berne RM. Interstitial transudate purines in normoxic and hypoxic immature and mature rabbit hearts. Pediatr Res. 1990;28(4):348–53. doi: 10.1203/00006450-199010000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Van Belle H, Goossens F, Wynants J. Formation and release of purine catabolites during hypoperfusion, anoxia, and ischemia. Am J Physiol. 1987;252(5 Pt 2):H886–93. doi: 10.1152/ajpheart.1987.252.5.H886. [DOI] [PubMed] [Google Scholar]
- 9.Dewhirst MW. Concepts of oxygen transport at the microcirculatory level. Semin Radiat Oncol. 1998;8(3):143–50. doi: 10.1016/s1053-4296(98)80040-4. [DOI] [PubMed] [Google Scholar]
- 10.Arnold F, West D, Kumar S. Wound healing: the effect of macrophage and tumour derived angiogenesis factors on skin graft vascularization. Br J Exp Pathol. 1987;68(4):569–74. [PMC free article] [PubMed] [Google Scholar]
- 11.Simmen HP, Battaglia H, Giovanoli P, Blaser J. Analysis of pH, pO2 and pCO2 in drainage fluid allows for rapid detection of infectious complications during the follow-up period after abdominal surgery. Infection. 1994;22(6):386–9. doi: 10.1007/BF01715494. [DOI] [PubMed] [Google Scholar]
- 12.Schofield CJ, Ratcliffe PJ. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun. 2005;338(1):617–26. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 13.Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17(1):71–7. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. Faseb J. 2002;16(10):1151–62. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell PH, Dachs GU, Gleadle JM, et al. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1997;94(15):8104–9. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiel M, Chouker A, Ohta A, et al. Oxygenation inhibits the physiological tissue-protecting mechanism and thereby exacerbates acute inflammatory lung injury. PLoS Biol. 2005;3(6):e174. doi: 10.1371/journal.pbio.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brock M, Jouvion G, Droin-Bergere S, et al. Bioluminescent Aspergillus fumigatus, a new tool for drug efficiency testing and in vivo monitoring of invasive aspergillosis. Appl Environ Microbiol. 2008;74(22):7023–35. doi: 10.1128/AEM.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkhofer S, Jost D, Dierich MP, Lass-Florl C. Susceptibility testing of anidulafungin and voriconazole alone and in combination against conidia and hyphae of Aspergillus spp. under hypoxic conditions. Antimicrob Agents Chemother. 2008;52(5):1873–5. doi: 10.1128/AAC.01572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry CV, Zitomer RS. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc Natl Acad Sci U S A. 1984;81(19):6129–33. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56(1):1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi JY, Stukey J, Hwang SY, Martin CE. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J Biol Chem. 1996;271(7):3581–9. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]
- 22.Evangelista CC, Jr., Rodriguez Torres AM, Limbach MP, Zitomer RS. Rox3 and Rts1 function in the global stress response pathway in baker's yeast. Genetics. 1996;142(4):1083–93. doi: 10.1093/genetics/142.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sertil O, Cohen BD, Davies KJ, Lowry CV. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene. 1997;192(2):199–205. doi: 10.1016/s0378-1119(97)00028-0. [DOI] [PubMed] [Google Scholar]
- 24.Kwast KE, Burke PV, Staahl BT, Poyton RO. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc Natl Acad Sci U S A. 1999;96(10):5446–51. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ter Linde JJ, Liang H, Davis RW, et al. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J Bacteriol. 1999;181(24):7409–13. doi: 10.1128/jb.181.24.7409-7413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zitomer RS, Carrico P, Deckert J. Regulation of hypoxic gene expression in yeast. Kidney Int. 1997;51(2):507–13. doi: 10.1038/ki.1997.71. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Hach A. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell Mol Life Sci. 1999;56(5-6):415–26. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hon T, Dodd A, Dirmeier R, et al. A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem. 2003;278(50):50771–80. doi: 10.1074/jbc.M303677200. [DOI] [PubMed] [Google Scholar]
- 29.Davies BS, Rine J. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics. 2006;174(1):191–201. doi: 10.1534/genetics.106.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbe-Bois R, Labbe P. Tetrapyrrole and heme biosynthesis in the yeast Saccharomyces cerevisiae. In: HA Dailey., editor. Biosynthesis of Heme and Chlorophylls. McGraw-Hill Publishing Co.; New York: 1990. pp. 235–285. [Google Scholar]
- 31.Creusot F, Verdiere J, Gaisne M, Slonimski PP. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988;204(2):263–76. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer K, Kim KS, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. Embo J. 1995;14(2):313–20. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeifer K, Arcangioli B, Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- 35.Cerdan ME, Zitomer RS. Oxygen-dependent upstream activation sites of Saccharomyces cerevisiae cytochrome c genes are related forms of the same sequence. Mol Cell Biol. 1988;8(6):2275–9. doi: 10.1128/mcb.8.6.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Guarente L. The yeast activator HAP1--a GAL4 family member--binds DNA in a directly repeated orientation. Genes Dev. 1994;8(17):2110–9. doi: 10.1101/gad.8.17.2110. [DOI] [PubMed] [Google Scholar]
- 37.Lowry CV, Zitomer RS. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8(11):4651–8. doi: 10.1128/mcb.8.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balasubramanian B, Lowry CV, Zitomer RS. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol Cell Biol. 1993;13(10):6071–8. doi: 10.1128/mcb.13.10.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deckert J, Perini R, Balasubramanian B, Zitomer RS. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics. 1995;139(3):1149–58. doi: 10.1093/genetics/139.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang HA, Schwelberger HG, Hershey JW. The two genes encoding protein synthesis initiation factor eIF-5A in Saccharomyces cerevisiae are members of a duplicated gene cluster. Mol Gen Genet. 1992;233(3):487–90. doi: 10.1007/BF00265449. [DOI] [PubMed] [Google Scholar]
- 41.Zagorec M, Labbe-Bois R. Negative control of yeast coproporphyrinogen oxidase synthesis by heme and oxygen. J Biol Chem. 1986;261(6):2506–9. [PubMed] [Google Scholar]
- 42.Klinkenberg LG, Mennella TA, Luetkenhaus K, Zitomer RS. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryot Cell. 2005;4(4):649–60. doi: 10.1128/EC.4.4.649-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Hach A, Wang C. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol Cell Biol. 1998;18(7):3819–28. doi: 10.1128/mcb.18.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abramova N, Sertil O, Mehta S, Lowry CV. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J Bacteriol. 2001;183(9):2881–7. doi: 10.1128/JB.183.9.2881-2887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sertil O, Kapoor R, Cohen BD, Abramova N, Lowry CV. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31(20):5831–7. doi: 10.1093/nar/gkg792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abramova NE, Cohen BD, Sertil O, et al. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics. 2001;157(3):1169–77. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sertil O, Vemula A, Salmon SL, Morse RH, Lowry CV. Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol Cell Biol. 2007;27(6):2037–47. doi: 10.1128/MCB.02297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vik A, Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(19):6395–405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Germann M, Gallo C, Donahue T, et al. Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J Biol Chem. 2005;280(43):35904–13. doi: 10.1074/jbc.M504978200. [DOI] [PubMed] [Google Scholar]
- 50.Lewis TL, Keesler GA, Fenner GP, Parks LW. Pleiotropic mutations in Saccharomyces cerevisiae affecting sterol uptake and metabolism. Yeast. 1988;4(2):93–106. doi: 10.1002/yea.320040203. [DOI] [PubMed] [Google Scholar]
- 51.Crowley JH, Leak FW, Jr., Shianna KV, Tove S, Parks LW. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1998;180(16):4177–83. doi: 10.1128/jb.180.16.4177-4183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilcox LJ, Balderes DA, Wharton B, et al. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. 2002;277(36):32466–72. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- 53.Marie C, Leyde S, White TC. Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet Biol. 2008;45(10):1430–8. doi: 10.1016/j.fgb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies BS, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol Cell Biol. 2005;25(16):7375–85. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwast KE, Lai LC, Menda N, et al. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol. 2002;184(1):250–65. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bourdineaud JP, De Sampaio G, Lauquin GJ. A Rox1-independent hypoxic pathway in yeast. Antagonistic action of the repressor Ord1 and activator Yap1 for hypoxic expression of the SRP1/TIR1 gene. Mol Microbiol. 2000;38(4):879–90. doi: 10.1046/j.1365-2958.2000.02188.x. [DOI] [PubMed] [Google Scholar]
- 57.Ness F, Bourot S, Regnacq M, et al. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur J Biochem. 2001;268(6):1585–95. [PubMed] [Google Scholar]
- 58.Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. Embo J. 1997;16(9):2179–87. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rachidi N, Martinez MJ, Barre P, Blondin B. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol Microbiol. 2000;35(6):1421–30. doi: 10.1046/j.1365-2958.2000.01807.x. [DOI] [PubMed] [Google Scholar]
- 60.Castello PR, David PS, McClure T, Crook Z, Poyton RO. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab. 2006;3(4):277–87. doi: 10.1016/j.cmet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Castello PR, Woo DK, Ball K, et al. Oxygen-regulated isoforms of cytochrome c oxidase have differential effects on its nitric oxide production and on hypoxic signaling. Proc Natl Acad Sci U S A. 2008;105(24):8203–8. doi: 10.1073/pnas.0709461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta KJ, Stoimenova M, Kaiser WM. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J Exp Bot. 2005;56(420):2601–9. doi: 10.1093/jxb/eri252. [DOI] [PubMed] [Google Scholar]
- 63.Nohl H, Staniek K, Sobhian B, et al. Mitochondria recycle nitrite back to the bioregulator nitric monoxide. Acta Biochim Pol. 2000;47(4):913–21. [PubMed] [Google Scholar]
- 64.Nohl H, Staniek K, Kozlov AV. The existence and significance of a mitochondrial nitrite reductase. Redox Rep. 2005;10(6):281–6. doi: 10.1179/135100005X83707. [DOI] [PubMed] [Google Scholar]
- 65.Planchet E, Jagadis Gupta K, Sonoda M, Kaiser WM. Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 2005;41(5):732–43. doi: 10.1111/j.1365-313X.2005.02335.x. [DOI] [PubMed] [Google Scholar]
- 66.Tischner R, Planchet E, Kaiser WM. Mitochondrial electron transport as a source for nitric oxide in the unicellular green alga Chlorella sorokiniana. FEBS Lett. 2004;576(1-2):151–5. doi: 10.1016/j.febslet.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 67.Dirmeier R, O'Brien KM, Engle M, et al. Exposure of yeast cells to anoxia induces transient oxidative stress. Implications for the induction of hypoxic genes. J Biol Chem. 2002;277(38):34773–84. doi: 10.1074/jbc.M203902200. [DOI] [PubMed] [Google Scholar]
- 68.Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J Mol Biol. 2006;361(3):399–411. doi: 10.1016/j.jmb.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 69.Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol. 2001;21(7):2496–505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becerra M, Lombardia-Ferreira LJ, Hauser NC, et al. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol Microbiol. 2002;43(3):545–55. doi: 10.1046/j.1365-2958.2002.02724.x. [DOI] [PubMed] [Google Scholar]
- 71.Ernst JF, Tielker D. Responses to hypoxia in fungal pathogens. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01259.x. [DOI] [PubMed] [Google Scholar]
- 72.Doedt T, Krishnamurthy S, Bockmuhl DP, et al. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell. 2004;15(7):3167–80. doi: 10.1091/10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo HJ, Kohler JR, DiDomenico B, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–49. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 74.Mulhern SM, Logue ME, Butler G. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot Cell. 2006;5(12):2001–13. doi: 10.1128/EC.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelly MT, MacCallum DM, Clancy SD, et al. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53(3):969–83. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 76.MacCallum DM, Findon H, Kenny CC, et al. Different consequences of ACE2 and SWI5 gene disruptions for virulence of pathogenic and nonpathogenic yeasts. Infect Immun. 2006;74(9):5244–8. doi: 10.1128/IAI.00817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lan CY, Rodarte G, Murillo LA, et al. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53(5):1451–69. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 78.Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53(4):1209–20. doi: 10.1111/j.1365-2958.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- 79.Kaplan J, McVey Ward D, Crisp RJ, Philpott CC. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta. 2006;1763(7):646–51. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 80.Sosinska GJ, de Groot PW, Teixeira de Mattos MJ, et al. Hypoxic conditions and iron restriction affect the cell-wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology. 2008;154(Pt 2):510–20. doi: 10.1099/mic.0.2007/012617-0. [DOI] [PubMed] [Google Scholar]
- 81.MacPherson S, Akache B, Weber S, et al. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother. 2005;49(5):1745–52. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell. 2004;3(6):1391–7. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoot SJ, Oliver BG, White TC. Candida albicans UPC2 is transcriptionally induced in response to antifungal drugs and anaerobicity through Upc2p-dependent and -independent mechanisms. Microbiology. 2008;154(Pt 9):2748–56. doi: 10.1099/mic.0.2008/017475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Znaidi S, Weber S, Al-Abdin OZ, et al. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot Cell. 2008;7(5):836–47. doi: 10.1128/EC.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bourot S, Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165(1):97–102. doi: 10.1016/0378-1119(95)00478-o. [DOI] [PubMed] [Google Scholar]
- 86.Malloy PJ, Zhao X, Madani ND, Feldman D. Cloning and expression of the gene from Candida albicans that encodes a high-affinity corticosteroid-binding protein. Proc Natl Acad Sci U S A. 1993;90(5):1902–6. doi: 10.1073/pnas.90.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120(6):831–42. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 88.Briggs MR, Yokoyama C, Wang X, Brown MS, Goldstein JL. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268(19):14490–6. [PubMed] [Google Scholar]
- 89.Wang X, Briggs MR, Hua X, et al. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem. 1993;268(19):14497–504. [PubMed] [Google Scholar]
- 90.Rawson RB. The SREBP pathway--insights from Insigs and insects. Nat Rev Mol Cell Biol. 2003;4(8):631–40. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 91.Espenshade PJ. SREBPs: sterol-regulated transcription factors. J Cell Sci. 2006;119(Pt 6):973–6. doi: 10.1242/jcs.02866. [DOI] [PubMed] [Google Scholar]
- 92.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–27. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 93.Yang T, Espenshade PJ, Wright ME, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110(4):489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 94.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10(2):237–45. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 95.DeBose-Boyd RA, Brown MS, Li WP, et al. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99(7):703–12. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 96.Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol. 2006;26(7):2817–31. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. Embo J. 2008;27(10):1491–501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen S, Sorrell T, Nimmo G, et al. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin Infect Dis. 2000;31(2):499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 99.Kwon-Chung KJ, Bennett JE. Medical Mycology. Lea & Febiger; Philadelphia, PA: 1992. [Google Scholar]
- 100.Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3(2):e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Himmelreich U, Dzendrowskyj TE, Allen C, et al. Cryptococcomas distinguished from gliomas with MR spectroscopy: an experimental rat and cell culture study. Radiology. 2001;220(1):122–8. doi: 10.1148/radiology.220.1.r01jl25122. [DOI] [PubMed] [Google Scholar]
- 102.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64(3):614–29. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 103.Lee H, Bien CM, Hughes AL, et al. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol Microbiol. 2007;65(4):1018–33. doi: 10.1111/j.1365-2958.2007.05844.x. [DOI] [PubMed] [Google Scholar]
- 104.Goldberg MA, Glass GA, Cunningham JM, Bunn HF. The regulated expression of erythropoietin by two human hepatoma cell lines. Proc Natl Acad Sci U S A. 1987;84(22):7972–6. doi: 10.1073/pnas.84.22.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A. 1993;90(9):4304–8. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang G, Hazra TK, Mitra S, Lee HM, Englander EW. Mitochondrial DNA damage and a hypoxic response are induced by CoCl(2) in rat neuronal PC12 cells. Nucleic Acids Res. 2000;28(10):2135–40. doi: 10.1093/nar/28.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang Y, Du KM, Xue ZH, et al. Cobalt chloride and low oxygen tension trigger differentiation of acute myeloid leukemic cells: possible mediation of hypoxia-inducible factor-1alpha. Leukemia. 2003;17(11):2065–73. doi: 10.1038/sj.leu.2403141. [DOI] [PubMed] [Google Scholar]
- 108.Grasselli F, Basini G, Bussolati S, Bianco F. Cobalt chloride, a hypoxia-mimicking agent, modulates redox status and functional parameters of cultured swine granulosa cells. Reprod Fertil Dev. 2005;17(7):715–20. doi: 10.1071/rd05059. [DOI] [PubMed] [Google Scholar]
- 109.Ingavale SS, Chang YC, Lee H, et al. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog. 2008;4(9):e1000155. doi: 10.1371/journal.ppat.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bahn YS, Kojima K, Cox GM, Heitman J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17(7):3122–35. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Millner PD, Marsh PB, Snowden RB, Parr JF. Occurrence of Aspergillus fumigatus during composting of sewage sludge. Appl Environ Microbiol. 1977;34(6):765–72. doi: 10.1128/aem.34.6.765-772.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tekaia F, Latge JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol. 2005;8(4):385–92. doi: 10.1016/j.mib.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 113.Wilson DM, Mubatanhema W, Jurjevic Z. Biology and ecology of mycotoxigenic Aspergillus species as related to economic and health concerns. Adv Exp Med Biol. 2002;504:3–17. doi: 10.1007/978-1-4615-0629-4_2. [DOI] [PubMed] [Google Scholar]
- 114.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12(2):310–50. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perfect JR, Cox GM, Lee JY, et al. The impact of culture isolation of Aspergillus species: a hospital-based survey of aspergillosis. Clin Infect Dis. 2001;33(11):1824–33. doi: 10.1086/323900. [DOI] [PubMed] [Google Scholar]
- 116.Latge JP. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 2001;9(8):382–9. doi: 10.1016/s0966-842x(01)02104-7. [DOI] [PubMed] [Google Scholar]
- 117.Rhodes JC. Aspergillus fumigatus: growth and virulence. Med Mycol. 2006;44(Suppl 1):S77–81. doi: 10.1080/13693780600779419. [DOI] [PubMed] [Google Scholar]
- 118.Hohl TM, Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell. 2007;6(11):1953–63. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tarrand JJ, Lichterfeld M, Warraich I, et al. Diagnosis of invasive septate mold infections. A correlation of microbiological culture and histologic or cytologic examination. Am J Clin Pathol. 2003;119(6):854–8. doi: 10.1309/EXBV-YAUP-ENBM-285Y. [DOI] [PubMed] [Google Scholar]
- 120.Tarrand JJ, Han XY, Kontoyiannis DP, May GS. Aspergillus hyphae in infected tissue: evidence of physiologic adaptation and effect on culture recovery. J Clin Microbiol. 2005;43(1):382–6. doi: 10.1128/JCM.43.1.382-386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Taubitz A, Bauer B, Heesemann J, Ebel F. Role of respiration in the germination process of the pathogenic mold Aspergillus fumigatus. Curr Microbiol. 2007;54(5):354–60. doi: 10.1007/s00284-006-0413-y. [DOI] [PubMed] [Google Scholar]
- 122.Lockington RA, Borlace GN, Kelly JM. Pyruvate decarboxylase and anaerobic survival in Aspergillus nidulans. Gene. 1997;191(1):61–7. doi: 10.1016/s0378-1119(97)00032-2. [DOI] [PubMed] [Google Scholar]
- 123.Kelly JM, Drysdale MR, Sealy-Lewis HM, Jones IG, Lockington RA. Alcohol dehydrogenase III in Aspergillus nidulans is anaerobically induced and post-transcriptionally regulated. Mol Gen Genet. 1990;222(2-3):323–8. doi: 10.1007/BF00633836. [DOI] [PubMed] [Google Scholar]
- 124.Willger SD, Puttikamonkul S, Kim KH, et al. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4(11):e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hughes AL, Lee CY, Bien CM, Espenshade PJ. 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J Biol Chem. 2007;282(33):24388–96. doi: 10.1074/jbc.M701326200. [DOI] [PMC free article] [PubMed] [Google Scholar]