Abstract
The success of arsenic trioxide in the treatment of acute promyelocytic leukemia has renewed interest in the cellular targets of As(III) species. The effects of arsenicals are usually attributed to their ability to bind vicinal thiols or thiol-selenols in pre-folded proteins thereby compromising cellular function. The present studies suggest an additional, more pleiotropic, contribution to the biological effects of arsenicals. As(III) species, by avid coordination to the cysteine residues of unfolded reduced proteins, can compromise protein folding pathways. Three representative As(III) compounds (arsenite; monomethylarsenous acid, MMA; and an aryl arsenical, PSAO) have been tested with three reduced secreted proteins (lysozyme, ribonuclease A and riboflavin binding protein, RfBP). Using absorbance, fluorescence and pre-steady state methods, we show that arsenicals bind tightly to low micromolar concentrations of these unfolded proteins with stoichiometries of 1 As(III) per 2 thiols for MMA and PSAO and 1 As(III) for every 3 thiols with arsenite. Arsenicals, at 10 μM, strongly disrupt the oxidative folding of RfBP even in the presence of 5 mM reduced glutathione, a competing ligand for As(III) species. MMA catalyzes the formation of amyloid-like monodisperse fibrils using reduced RNase. These in vitro data show that As(III) species can slow, or even derail, protein folding pathways. In vivo, the propensity of As(III) species to bind to unfolded cysteine-containing proteins may contribute to oxidative and protein folding stresses that are prominent features of the cellular response to arsenic exposure.
While inorganic arsenicals have been used for millennia to treat a range of diseases, and organo-arsenicals were successful in the treatment of syphilis and trypanosomiasis, the medical uses of arsenicals declined with the advent of antibiotics and the rise of modern medicine. Recently, confirmation of the remarkable effectiveness of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (1–3) has led to a renewed interest in the use of As(III) compounds in medicine and in the biochemical and biological consequences of arsenic exposure.
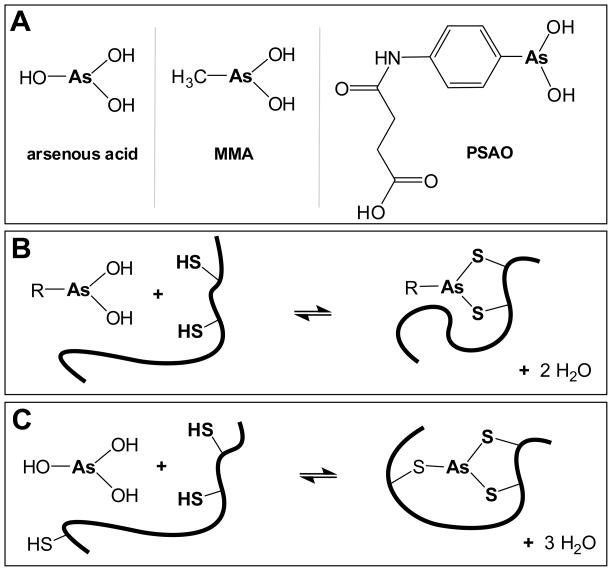
The coordination of arsenicals to thiol and selenol groups is well established (4–7). Monothiols bind arsenicals rather weakly (8), although the relatively high intracellular concentration of free GSH will strongly influence arsenic speciation in mammalian cells (see later). In contrast, protein dithiols bind monoalkyl or monoaryl As(III) species much more tightly via the chelate effect (Figure 1)(8, 9). Indeed, many of the suggested protein targets of arsenicals have pairs of thiols (or thiol-selenols) in close proximity (6, 10–12). Three representative arsenicals are used here (Figure 1A). Monomethylarsonous acid (MMA) is a toxic metabolite of As2O3 (12), and 4-(4-arsonophenyl-amino)-4-oxo-butanoate (PSAO) is a water-soluble analog for several biologically-active arsenicals (6, 13). Both can bind dithiols as in Figure 1B. Arsenite (predominantly arsenous acid at pH 7.5, pK1 = 9.2; Figure 1A (7)) is a hydrolysis product of As2O3 and can additionally form trivalent complexes of the type shown in Figure 1C (5).
FIGURE 1.
Three arsenicals and their reactions with di- and tri-thiol motifs. Panel A shows the fully protonated forms of the As(III) species used in this work. Panels B and C depict the reactions of alkyl/aryl As(III) species and of arsenous acid with di- and trithiols respectively.
The mammalian endoplasmic reticulum (ER) houses many proteins with vicinal redox-active thiols that are involved in the introduction and isomerization of disulfide bonds during oxidative protein folding. For example, the concentration of reduced CxxC motifs from the protein disulfide isomerases (PDI) alone is believed to approach mM (14). In addition, the oxidoreductases responsible for catalyzing the net generation of disulfide bridges in proteins, the flavin-dependent sulfhydryl oxidases Ero1p (15) and QSOX (16), also contain multiple vicinal dithiol motifs that might be potential sites of inhibition by arsenicals.
For this reason we investigated the effects of As(III) species on oxidative protein folding using a newly-developed in vitro model system (17). Surprisingly, while arsenicals strongly inhibited oxidative folding, their principal target was not the enzymatic machinery driving the acquisition of the native fold, but rather the unfolded reduced protein itself. We show here that arsenicals bind strongly, and extensively, to three representative reduced unfolded proteins. Our finding that MMA induces fibril formation in reduced RNase further suggests that arsenicals have the potential to disrupt the folding of susceptible proteins in vivo and that this effect may provide a source of cellular stress in addition to that caused by the inhibition of specific enzymes and receptors.
EXPERIMENTAL PROCEDURES
Reagents and Proteins
Potassium phosphate, sodium acetate, EDTA, sodium chloride, and tris were purchased from Fisher. Urea, guanidine hydrochloride, dithiothreitol (DTT), oxidized and reduced glutathione, and riboflavin were obtained from Sigma. TCEP hydrochloride was from Pierce and THP from Calbiochem. Sodium arsenite was from City Chemical. The arsenicals MMA and PSAO were synthesized as described earlier (13). All arsenicals should be handled with due caution. Bovine pancreatic ribonuclease A, chicken egg white lysozyme, and M. lysodeikticus cells were obtained from Sigma. A plasmid containing human PDI was generously provided by Dr. Jakob Winther and subcloned into a pTrcHisA plasmid to provide an N-terminal hexahistine tag for expression in E. coli BL21(DE3) cells (Invitrogen).
Preparation of Reduced Unfolded Proteins
RfBP, a generous gift from Dr. Harold B. White was obtained as described previously (18). RfBP (7 mg) was incubated overnight with a 10-fold molar excess DTT per protein thiols in 0.5 mL of 50 mM potassium phosphate, pH 8.0, containing 1 mM EDTA and 6 M guanidine HCl in pH 8.0 at 37 °C. The reduced protein was gel-filtered at 4°C using a PD-10 column in the same buffer minus DTT. RNase was similarly reduced, but purified on a PD-10 column pre-equilibrated with 10 mM sodium acetate pH 4.0. Lysozyme was reduced in 100 mM Tris buffer, pH 8.0, with 100 mM NaCl, 1 mM EDTA, and 6 M guanidine HCl. Excess reductant was removed on a PD-10 column pre-equilibrated with 10 mM sodium acetate, pH 5.0, containing 1 mM EDTA and 3 M urea. Reduced proteins were quantitated using the following extinction coefficients: RNase, ε278 9,300 M−1 cm−1; RfBP, ε280 49,000 M−1cm−1 for reduced RfBP; and lysozyme ε280 34,000 M−1cm−1. Thiol titers were determined using DTNB (ε412 = 14,150 M−1cm−1) and reduced proteins were stored anaerobically at 4°C.
QSOX activity assays
QSOX was assayed in an oxygen electrode as reported earlier (19). Briefly, 60 nM avian QSOX was reacted with either 5 mM GSH or 5 mM TCEP in the presence of 10 μM concentrations of MMA, PSAO, and As(OH)3. All assays were conducted in 50 mM potassium phosphate, 1 mM EDTA, pH 7.5 at 25°C. Enzyme activities are reported as a percentage of a control reaction in the absence of arsenic (III) species.
Preparation of reduced PDI
Human PDI was reduced with a 40-fold molar excess of DTT for 1 h at 25°C in 50 mM phosphate buffer, pH 7.5, containing 1 mM EDTA. Excess reductant was removed by gel-filtration on a PD-10 gel filtration column (GE Healthcare) ensuring complete separation by assaying the thiol content of each. Reduced human PDI was quantitated with a molar extinction coefficient of 56,400 M−1 cm−1 at 280 nm (20).
Dissociation Constants for binding arsenicals to reduced PDI
The fluorescence data in Figures 3 and S2 were fit to the equation:
using GraphPad Prism 3.0, where Ao, Af and A are initial, saturating and current fluorescence readings, respectively. Pt and Lt are the total protein and added ligand concentration, respectively and Kd is the dissociation constant.
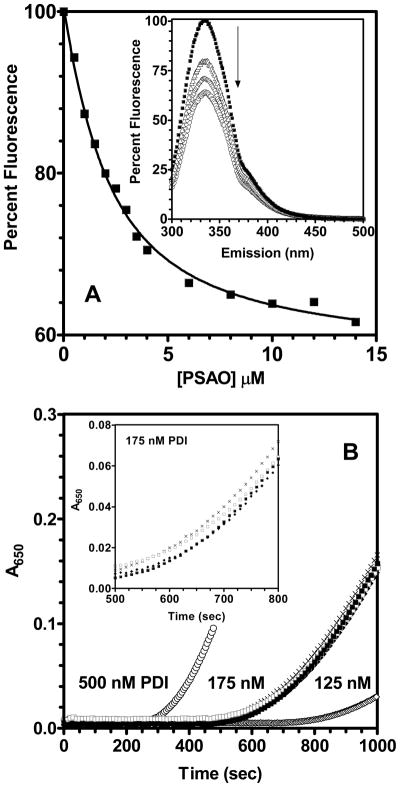
FIGURE 3.
Binding of PSAO to reduced PDI and the effect of As(III) species on the reductase activity of PDI. Panel A represents the changes in fluorescence emission intensity at 337 nm (exciting at 290 nm) on the addition of PSAO to 1 μM reduced human PDI (2 μM binding sites). The data were fit (solid line) to a Kd of 1.1 μM (see Experimental Procedures). The inset to panel A shows selected emission spectra (corresponding to 0, 2, 4, and 10 μM PSAO). Panel B: 10 μM arsenite (open square), MMA (cross) or PSAO (closed diamond) do not appreciably inhibit the activity of PDI in the insulin reduction assay (closed squares; in 50 mM potassium phosphate, pH 7.5, containing 1 mM EDTA, 5 mM GSH, and 100 μM insulin). For comparison, the traces labeled 125 and 500 nM PDI are controls in the absence of arsenicals (open diamond and circles respectively).
PDI Activity Assay
The reductase activity of PDI (21, 22) followed the increase in turbidity at 650 nm on reduction of insulin. Oxidized PDI (175 nM) (23) was mixed with 100 μM insulin in 50 mM potassium phosphate buffer, pH 7.5 containing 1 mM EDTA and 5 mM GSH.
Oxidative Refolding of RfBP
Reduced RfBP (1 μM) was refolded at 25°C in the presence of 30 nM QSOX, 30 μM reduced PDI, and 0.8 μM riboflavin in 50 mM phosphate, 1 mM EDTA, pH 7.5. Reactions were followed by quenching of riboflavin fluorescence (excitation: 450 nm, emission: 530 nm).
Arsenic Binding to Reduced Proteins
Protein titrations with arsenicals were monitored by UV-vis and fluorescence spectroscopy in the presence or absence of 5 mM GSH. To ensure equilibration each spectrum was recorded 7 min after addition of titrant. Titrations in the absence of GSH included 0.5 mM of the non-coordinating phosphine THP to ensure that the protein remains reduced throughout the experiments. Titrations with RNase and RfBP were run in 50 mM phosphate, 1 mM EDTA, pH 7.5; those with lysozyme used 100 mM Tris, pH 7.5, containing 100 mM NaCl, 1 mM EDTA, and 3 M urea. The starting fluorescence signal for all titrations was normalized and set to 100 percent fluorescence.
Stopped-Flow Kinetics
A SF-61 DX2 stopped-flow spectrometer (Hi-Tech Scientific) was used for rapid mixing kinetics experiments in both absorbance and fluorescence modes. For kinetic protection studies, reduced RNase, in the absence or presence of 1.1 disulfide-equivalents of As(OH)3, was mixed with DTNB to give final concentration of 5 μM protein and 2 mM DTNB). Analogous experiments were performed with RfBP in the presence or absence of MMA or arsenite. Binding of MMA to reduced RfBP was followed by fluorescence exciting at 295 nm. The traces were multiphasic and fit to a three-exponential decay dominated by a rapid phase accounting for about 60% of the total change. While these data (see Supplementary Materials) illustrate the rapidity of arsenic coordination to reduced unfolded RfBP, the complexity of these fluorescence changes make a detailed analysis of these kinetics unwarranted (see Results).
Oxidative Refolding of Lysozyme
Reduced lysozyme (2 μM) was incubated at 25°C with 1 mM GSH and 0.2 mM GSSG in 100 mM Tris, pH 8.0, containing 100 mM NaCl, 1 mM EDTA, and 3 M urea (24). Aliquots (20 μL) were mixed with 80 μL of a suspension of 0.15 mg/mL M. lysodeikticus cells in 50 mM phosphate, pH 7.5 and the initial rates calculated from the first 10 sec of absorbance decrease at 450 nm.
TEM Imaging of RNase A
Bright field images were obtained at an accelerating voltage of 120 kV on a FEI Technai-12 Transmission Electron Microscope with a Gatan CCD camera. Reduced RNase (50 μM) was mixed with 10 μM MMA in phosphate buffer, pH 7.5, containing 1 mM EDTA and 0.5 mM THP. (Precipitation was also found using 5 μM RNase and 1 μM MMA). Aliquots (4 μL) of suspensions were placed on carbon-coated Cu grids (400 mesh) and excess liquid wicked with filter paper. An equal volume of 1 w/v of aqueous uranyl acetate was added to the grid, excess removed with filter paper, and the samples dried in air for at least 1 h prior to imaging.
RESULTS
Arsenicals inhibit oxidative folding of a 9-disulfide-containing protein
Figure 2A presents the model oxidative folding system used here (17). Riboflavin binding protein (RfBP) contains 9 disulfides (25) corresponding to >34 million possible disulfide isomers for fully oxidized protein. The reduced protein (1 μM, 18 μM free thiols) is incubated with a slight sub-stoichiometric level of riboflavin in the presence of both 30 nM Quiescin-sulfhydryl oxidase (QSOX), to generate disulfide bonds (19), and 30 μM reduced PDI, to isomerize incorrect pairings (Figure 2A). The formation of active binding protein is monitored by the progressive quenching of the strong fluorescence of free riboflavin ((26), Figure 2B, control). At a concentration of 10 μM, arsenite, MMA and PSAO (Figure 1A) all inhibit riboflavin binding very strongly (Figure 2B). This concentration was chosen because it approximates peak plasma levels of arsenite attainable in the treatment of acute promyelocytic leukemia (27, 28) and because it corresponds to a slight molar excess over the 9 μM dithiol pairs used to form 9 disulfides in 1 μM RfBP.
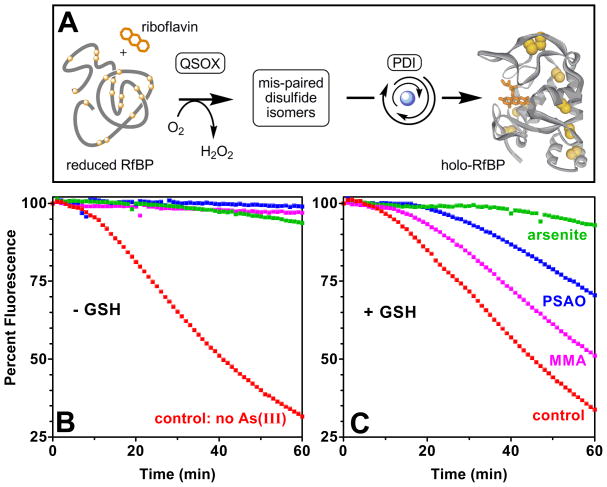
FIGURE 2.
As(III) species inhibit oxidative protein folding of riboflavin binding protein. Panel A shows the oxidative folding system for reduced RfBP. QSOX (30 nM) introduces disulfide bonds into the reduced unfolded protein, and reduced PDI (30 μM) isomerizes incorrectly paired disulfide bridges. The fluorescence of riboflavin (0.8 μM) is quenched on binding to the native apo-RfBP allowing oxidative folding to be followed continuously (control). Panel B and C depict the inhibition of oxidative folding by 10 μM arsenite (green), MMA (pink) and PSAO (blue) in the absence or presence of 5 mM GSH, respectively. Red curves are controls in the absence of arsenicals. The starting fluorescence is set arbitrarily to 100 percent in these experiments.
A key consideration for in vitro models of arsenic toxicity is that they should account for the abundance of reduced glutathione (GSH) in mammalian cells. GSH is typically present at about 5 mM intracellularly (29) and can sequester arsenic species by mono-, di- and, for arsenite, trivalent coordination (8). Figure 2C shows that inclusion of 5 mM glutathione had comparatively little effect on the rate of oxidative folding of RfBP in the absence of arsenic but significantly attenuates inhibition by MMA and PSAO. However, arsenite remains a potent inhibitor in Figure 2C consistent with prior speciation studies (8). Figure S1 in Supporting Information shows that 5 mM GSH almost completely sequesters 10 μM MMA (> 99.8%) but only about 60% of the arsenite.
We next examined the effects of arsenicals on the individual components of the protein folding system shown in Figure 2A. QSOX was found to be relatively insensitive to these inhibitors at 10 μM when assayed using TCEP, a model substrate (30) which does not coordinate arsenicals (Table S1 in Supporting Information). Here, arsenite, MMA and PSAO resulted in 0%, 3 and 10% inhibition respectively. Similarly, less than 10% inhibition was observed using 5 mM GSH as a substrate of the oxidase (Table S1). The abundance of reduced PDI in the mammalian ER (14) make it an obvious potential target of As(III) binding. PDI has two WCGHC motifs with rather similar properties (31–34). The binding of arsenicals, followed by quenching of tryptophan fluorescence (Figure 3A), could be fit to a single binding isotherm as shown for PSAO in the inset (yielding a Kd of 1.1 μM in the absence of GSH; see Experimental Procedures). Comparable titrations with MMA and arsenite showed Kd values of 25, and 18 μM, respectively (Figure S2 in Supporting Information). Such relatively modest Kd values, and the sequestration of arsenicals by 5 mM GSH, lead to insignificant inhibition of PDI in the insulin reductase assay shown in Figure 3B. These experiments suggest that typical intracellular concentrations of GSH would minimize the direct inhibition of PDI by the arsenicals used here. However, outside the cell the concentration of competing glutathione is typically much lower (in the 5–50 μM range (35)) rationalizing the effectiveness of phenylarsine oxide as an inhibitor of surface-bound PDI (36, 37).
These data suggest that a direct inhibition of QSOX or PDI is insufficient to explain the marked slowing of oxidative folding seen in Figure 2. We next describe three approaches to show that arsenicals can target cysteine residues in reduced proteins. Reduction of structural disulfides in secreted proteins typically leads to a highly mobile unfolded state that could present a multiplicity of potential As(III) binding sites analogous to the range of disulfide pairings in oxidatively misfolded proteins. Indeed, the –S-As-S- linkage could be regarded as a lengthened and conformationally more accommodating pseudo-disulfide crosslink (with a minimal distance between participating S atoms of 3.6 Å compared to 2.0 A for a disulfide (Figure S3 in Supporting Information)). Arsenite provides the additional possibility of restraining up to three cysteine residues (Figure 1C).
The increased absorbance generated when sulfur replaces oxygen as a ligand for As(III) species (8, 13) provides one measure of the coordination of arsenicals to reduced proteins. This approach is only useful in the absence of glutathione (it cannot distinguish between coordination to GSH or reduced protein). Figure 4A shows that PSAO and MMA bind almost quantitatively to reduced unfolded RfBP (with a sharp inflexion between 8 and 9 As(III)/RfBP: a value that approximates the disulfide count for this secreted protein). With arsenite, the absorbance changes, while non-linear, are essentially complete after the addition of ~6 arsenite ions (Figure 3A), suggesting some degree of trivalent coordination. Overall these absorbance results suggest tight binding of all three arsenicals to the reduced protein.
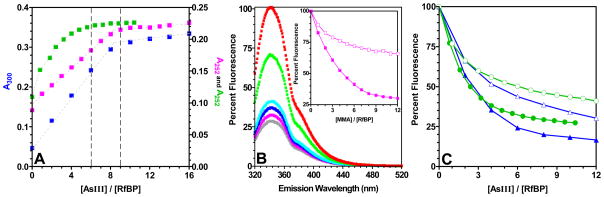
FIGURE 4.
Absorbance and fluorescence measurements of the binding of As(III) species to reduced RfBP. Panel A: a solution of reduced RfBP (5 μM in 50 mM phosphate buffer, pH 7.5, 25°C; see Methods) was titrated with the arsenicals and followed by absorbance increase at 300 nm (PSAO: blue) or 252 nm (arsenite and MMA: green and pink respectively). Dashed lines are drawn at 6 and 9 equivalents per RfBP. Panel B: MMA quenches the fluorescence emission of 5 μM reduced RfBP (exciting at 295 nm; emission 344 nm) in the absence of 5 mM GSH. The inset compares fluorescence quenching with or without GSH (open and closed symbols respectively). The absence of an inner-filter effect was confirmed by repeating the titration using 1 μM RfBP (not shown). Panel C: corresponding fluorescence titrations for arsenite (green) and PSAO (blue).
Arsenicals strongly quench the protein fluorescence of reduced RfBP. This is illustrated for MMA in Figure 4B. The inset shows that the fluorescence changes are substantially complete by the addition of 9 equivalents of arsenical and that 5 mM glutathione attenuates, but does not eliminate, the fluorescence quenching resulting from the binding of MMA. Here, GSH is expected to lower the concentration of free MMA by >500-fold (Table S1 in Supporting Information) and so binding of MMA to the reduced protein is evidently avid. The interaction between As(III) compounds and reduced proteins reflect binding events of considerable complexity that are not amenable to a detailed quantitative description. At early stages of the titration, multiple dithiol pairs can compete for limiting As(III) species. Later, preexisting arsenic-dithiol chelates may yield to new pairings as additional constraints on the structures accumulate. A further difficulty in extracting dissociation constants from these experiments is that the extent of quenching of the 6 tryptophan residues in RfBP is likely to be a non-linear function of the extent of As(III) loading. For example, almost one-half of the total change in fluorescence is accomplished after the addition of 3 out of 9 equivalents of MMA (Figure 4B).
Rapid reaction studies confirm the expected complexity of the interaction between MMA and reduced RfBP. With excess MMA, fluorescence quenching was multiphasic in the stopped-flow instrument, with an initial fast phase accounting for approximately 60% of the total decrease (Figure S4 in Supporting Information). Under pseudo first-order conditions this phase yields a second-order association rate constant of 2350 M−1s−1. While far from diffusion controlled, this bimolecular rate constant is within an order of magnitude of those observed when PSAO binds to a range of small di-cysteinyl peptides (4, 13).
Figure 4C shows that PSAO and arsenite also bind to reduced RfBP with a strong fluorescence quenching that is substantially complete before the addition of 9-equivalents of arsenical. In these cases, the quenching appears less sensitive to glutathione compared to titrations with MMA (Figure 4B). In summary, the binding of arsenicals to RfBP is weakened, but still significant, in the presence of 5 mM glutathione.
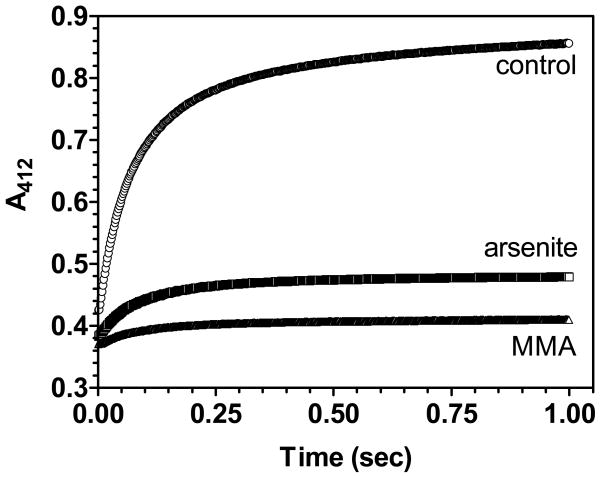
As an independent measure of the interaction between reduced protein and As(III) species, we compared the kinetic reactivity of the cysteine residues in RfBP in the presence and absence of arsenicals. A precedent for this approach comes from a method to measure the concentration of monothiols in the presence of dithiothreitol (DTT) that exploits the sluggish reactivity of As-DTT complexes towards 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (38, 39). As a control, Figure 5 shows that most of the 18 free cysteines of reduced RfBP react with the thiol-specific reagent DTNB in less than 1 sec consistent with a protein that is extensively unfolded. When the reduced protein is mixed with a 1.1-fold molar excess of MMA or arsenite (over dithiols), profound slowing of the reactivity towards (DTNB) is observed. These kinetic data corroborate the spectroscopic titrations: arsenicals bind tightly and dissociate slowly from reduced RfBP.
FIGURE 5.
Reactivity of the cysteine residues of reduced RfBP towards DTNB. The reduced protein (5 μM, 90 μM thiols with or without 50 μM arsenite or MMA) was mixed with an equal volume of 4 mM DTNB in phosphate buffer, pH 7.5, 25°C. The increase in absorbance at 412 nm was followed in the stopped-flow. The initial absorbance readings of about 0.38 reflect background DTNB absorbance.
Arsenic binding to reduced lysozyme
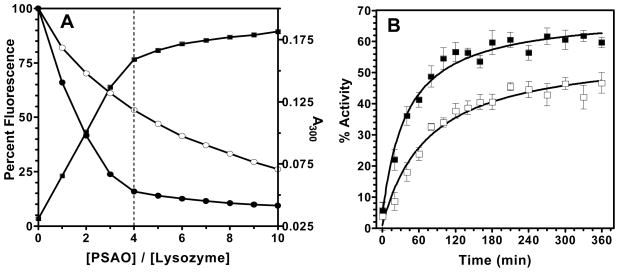
RfBP shows a more complex disulfide connectivity than proteins typically employed for studies of oxidative protein folding. We next describe comparable experiments with two simpler 4-disulfide proteins: hen egg lysozyme and bovine pancreatic RNase A. Following earlier practice (40), experiments with lysozyme were conducted in urea to avoid precipitation of unfolded or mis-folded proteins. Figure 6A shows tight binding of PSAO to reduced lysozyme followed by absorbance increase at 300 nm. The increase in absorbance after the inflexion point of 4 PSAO/lysozyme largely reflects the contribution of free PSAO as it accumulates in solution (13). Similarly fluorescence measurements in the absence of glutathione suggest that binding is substantially complete after the addition of 4-equivalents of PSAO. Binding is weakened by 5 mM glutathione, but remains significant (Figure 6A). Corresponding titrations for MMA and arsenite are presented in Supporting Information (Figure S5): both show the expected interaction between lysozyme thiols and arsenicals.
FIGURE 6.
Reduced avian lysozyme binds As(III) species. Panel A: PSAO titrations of 5 μM protein (in 100 mM Tris buffer, pH 7.5, containing 100 mM NaCl, 1 mM EDTA and 3 M urea) followed by absorbance (closed squares) and fluorescence (excitation 295 nm; emission 348 nm) in the absence and presence of 5 mM GSH (closed and open squares, respectively). Panel B: regain of enzyme activity during oxidative refolding of 2 μM reduced lysozyme (closed squares; in the same buffer adjusted to pH 8.0 plus 1 mM GSH and 0.2 mM GSSH) is impaired by 10 μM arsenite (open squares).
The ability of reduced lysozyme to be oxidatively refolded in a glutathione redox buffer allows the impact of arsenical to be evaluated in the absence of PDI or QSOX. (This approach cannot be applied to RfBP because glutathione redox buffers are ineffective in the absence of PDI (17)). With lysozyme, we used a mixture of 1 mM GSH and 0.2 mM GSSG frequently used in the refolding protocols of small proteins (14, 40). In the absence of arsenite both the refolding rate and the yield of active lysozyme (Figure 6B) are comparable to those obtained earlier (40). The addition of 10 μM arsenite slows the initial regain of activity by about 2-fold and lowers the recovery of folded enzyme. These effects are comparatively modest and may reflect the relative simplicity of disulfide connectivity in lysozyme compared to RfBP (see Discussion).
Arsenic binding to reduced RNase
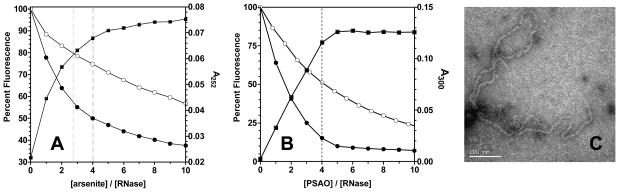
Figure 7A demonstrates the binding of arsenite to reduced bovine pancreatic RNase using absorbance and fluorescence quenching. Dotted lines, corresponding to the theoretical equivalence point for di- or trivalent coordination of arsenicals (4 and 2.7 As(III)/RNase, respectively), show that substantial binding is observed at sub-stoichiometric levels of arsenite even in the presence of 5 mM glutathione. As observed for RfBP, arsenite also provides strong kinetic protection against the reactivity of reduced RNase with DTNB (Figure S6 in Supporting Information). All 8 thiols react with DTNB in less than 250 msec (as expected for its reaction with a small unfolded protein). However, in the presence of a slight molar excess of arsenite (in terms of divalent coordination) only about 30% of these thiols react rapidly with DTNB. The remaining thiols are sequestered very tightly by arsenicals. PSAO binds to reduced RNase more tightly than arsenite and yields clear endpoints at the expected 4 equivalents of arsenical (Figure 7B). Significant fluorescence quenching is again retained in the presence of glutathione.
FIGURE 7.
Arsenicals and reduced RNase. Panels A and B: arsenite and PSAO binding to 5 μM reduced RNase measured by absorbance (closed squares) and fluorescence emission (excitation 276 nm; emission 303 nm) in the absence and presence of 5 mM GSH (closed and open circles, respectively). Panel C: sub-stoichiometric concentrations of MMA (here 10 μM) cause the aggregation of 50 μM reduced RNase. The precipitate was collected and found to be formed of monodisperse fibrils by transmission electron microscopy (see Experimental Procedures).
Notably, titrations of reduced RNase with MMA were repeatedly thwarted by the formation of protein precipitates. This effect was not observed in any of the other combinations of 3 arsenicals and 3 test proteins. Turbidity was evident before 1 dithiol equivalent of MMA had been added to 5 μM reduced RNase using conditions identical to those described throughout this work. The resulting RNase precipitates were insoluble after washing in buffer in the presence or absence of a large excess of DTT (a reagent that is effective at both reducing disulfides and sequestering arsenicals). Substoichiometric levels of MMA (10 μM arsenical in the presence of 50 μM reduced RNase) leads to a marked increase in the fluorescence of Thioflavin T (Figure S7A in Supporting Information) consistent with the formation of amyloid (41, 42). The precipitates were also shown to be Congo Red positive (Figure S7B) and were found to exhibit a distinct fibrillar appearance. A transmission electron microscope image of uranyl acetate-stained material is shown in Figure 7C. These monodisperse fibrils have widths of 110 ± 22 Å (Figure S7C in Supporting Information). Control experiments showed that reduced RNase did not precipitate, or yield fibrillar images in the absence of MMA, and these aggregates were absent if 5 mM glutathione was added before MMA. While a domain-swapped version of RNase has been shown to form amyloid-like fibrils (43), the present results appear to be the first induction of fibril formation using a native RNase sequence. A detailed characterization of the role of arsenic in fibrillization is beyond the scope of this contribution, but the present data show that metalloid binding can seriously subvert folding pathways.
DISCUSSION
Several factors are likely to influence the extent to which arsenicals interact with unfolded proteins in vivo. One aspect is the competition from monothiols such as glutathione. While the extracellular concentration of GSH is believed to be very low (35), the much higher intracellular concentration of GSH would be expected to selectively moderate the effectiveness of arsenicals. For example, arsenite is a much better inhibitor than MMA during the refolding of RfBP in the presence of 5 mM GSH, whereas this trend is reversed in the absence of GSH (Figure 2). A second factor modulating binding of arsenicals is likely to be the density and placement of cysteine residues in unfolded proteins.
Of the three secreted proteins studied, the refolding of RfBP is by far the most strongly inhibited by As(III) species. Oxidative folding pathways for proteins with millions of potential disulfide pairings may be more prone to derailment by arsenicals than those for proteins with much simpler disulfide connectivities. In addition, while all three proteins contain between 6.2 and 8.5 % cysteine, 8 out of 18 cysteine residues in RfBP are separated by an average of just 3.9 amino acids. In contrast, the 8 cysteine residues in lysozyme and RNase are more uniformly distributed along the primary sequence, with a corresponding separation averaging 11.0 and 16.3 residues respectively. The proximity of pairs of cysteines in the sequence would, a priori, be expected to increase the strength of divalent or trivalent coordination to the unfolded state. There is a further interesting difference in the placement of disulfide bridges in these three proteins. While lysozyme and RNase have disulfides bonds rather evenly distributed throughout their 3D structures, pairs of disulfides are found almost within van der Waals contact at two locations in RfBP (Supporting Information Figure S8). Reduction of each disulfide pair might provide multiple opportunities for effective coordination of arsenic in native-like conformations.
The absorbance titrations in Figure 4, 6, 7 and S5 show that MMA and PSAO can form complexes whose stoichiometry corresponds to the number of disulfides originally present in RfBP, lysozyme and RNase. This extensive coordination, in hindsight, is perhaps not surprising. First, arsenicals bind avidly to di-cysteine motifs and can overwhelm the secondary structural preferences of helical and beta-hairpin peptides (4, 13). Second, unfolded proteins are conformationally mobile and structurally accommodating (44). Third, exhaustive pairing of cysteine residues can occur in many proteins even when two or more of these disulfides are incorrectly paired. While such “scrambled” protein forms are prepared under denaturing conditions, the titrations with RfBP and RNase were conducted in the absence of chaotrophes: hence at least some of the cysteines coordinated to arsenic might reflect native pairings.
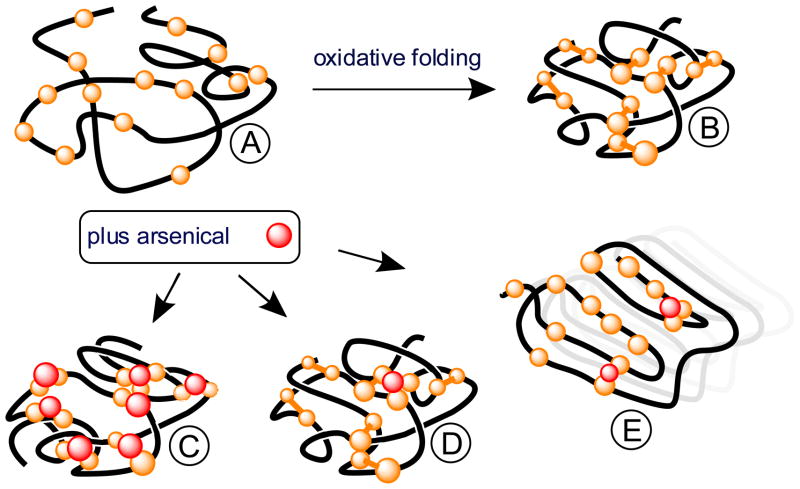
The extension of this argument: that arsenic bridging might stabilize regions of a native fold in lieu of disulfides is not new: it has been demonstrated by partial reduction of proteins and using arsenicals to capture the liberated dithiol pairs (45, 46). Further, hydrid proteins containing both disulfide and arsenic-dicysteinyl bridges appear plausible outcomes of an oxidative folding system exposed to arsenicals in vivo. For example, an avid As-binding site in a nascent chain might be subsequently encapsulated and trapped via the introduction of additional native disulfide pairings as oxidative maturation progresses (Figure 8 structure D). The small size of MMA and arsenite might favor generation of such hybrid proteins that, while they might be compromised in terms of biological function, may appear folded to the quality control system of the ER. Finally the formation of insoluble fibers of reduced RNase in the presence of the low concentrations of MMA (less than 1 arsenical/RNase molecule) suggests that arsenicals might in some instances subvert protein folding pathways entirely (Figure 8 structure E) leading to the formation of fibrillar structures.
FIGURE 8.
Schematic depiction of the oxidative folding of a 14-cysteine containing protein and outcomes of concurrent exposure to a monoalkyl/aryl arsenic (III) species. Forms A and B are unfolded reduced and native 7-disulfide folded protein respectively. Capture of A before disulfide bond formation generates species C with a full complement of bound arsenical. D represents a hybrid protein containing both arsenical and disulfide crosslinks. E depicts arsenic-induced formation of ordered aggregates following the precedent set in Figure 7C.
The folding pathways of proteins from non-ER locations may also be susceptible to intervention by the tight binding of arsenicals to reduced proteins that we have demonstrated here. While they typically contain lower cysteine contents than proteins that reside or transit the ER, there are numerous examples from the human proteome of non-ER proteins with cysteine contents above 10% (reaching to greater than 30% for members of the keratin associated protein family (47)). Cytosolic proteins with more typical cysteine contents may also exhibit clusters of thiols that render them particularly sensitive to arsenicals during folding.
Finally, we suggest that the facility with which As(III) species bind unfolded proteins in vitro may rationalize aspects of the induction of heat shock genes (48), and the upregulation of polyubiquitinated proteins and proteasome subunits that are prominent features of cells treated with arsenic (49–51). Ron and coworkers write: “… exposure to arsenic promotes a significant burden of potentially toxic misfolded proteins or proteotoxins” (52). While this work was being prepared for publication Sharma et al. reported that Cd2+, Hg2+, and Pb2+, are potent inhibitors of the folding of several non-ER proteins with modest cysteine contents: luciferase and lactate, malate, and glucose-6-phosphate dehydrogenases (53). Proteins undergoing folding may represent under-appreciated targets for interception by toxic metals/metalloids as well as additional opportunities for pharmaceutical intervention.
Supplementary Material
Acknowledgments
We thank Stephanie A. Schaefer for help with the RNase experiments and Dr. David Ron for stimulating discussions. Dr. Hugo Monaco is acknowledged for the coordinates of RfBP and Dr. Hal White for a gift of riboflavin binding protein. We thank an anonymous reviewer for helpful suggestions. This work was supported in part by NIH grants GM26643 (C.T.) and 1-T32-GM08550 (P.C.R.).
Abbreviations
- DTNB
5,5′-dithiobis(2-nitrobenzoate)
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- PDI
protein disulfide isomerase
- QSOX
Quiescin-sulfhydryl oxidase
- RfBP
riboflavin binding protein
- RNase
ribonuclease A
- TCEP
tris(2-carboxyethyl)phosphine
- THP
tris(hydroxypropyl)phosphine
Footnotes
This work was supported in part by National Institutes of Health Grant (GM26643) and USPHS Training Grant 1-T32-GM08550 (P.C.R.). The content of this work is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
SUPPORTING INFORMATION AVAILABLE
Table S1 and Figures S1-S8 provide supplementary data and analysis for the effects of arsenite, MMA, and PSAO on oxidative protein folding. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ, Corso D, DeBlasio A, Gabrilove J, Scheinberg DA, Pandolfi PP, Warrell RP., Jr Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. New Engl J Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 2.Miller WH, Jr, Schipper HM, Lee JS, Singer J, Waxman S. Mechanisms of action of arsenic trioxide. Cancer Res. 2002;62:3893–3903. [PubMed] [Google Scholar]
- 3.Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 4.Ramadan D, Cline DJ, Bai S, Thorpe C, Schneider JP. Effects of As(III) binding on beta-hairpin structure. J Amer Chem Soc. 2007;129:2981–2988. doi: 10.1021/ja067068k. [DOI] [PubMed] [Google Scholar]
- 5.Touw DS, Nordman CE, Stuckey JA, Pecoraro VL. Identifying important structural characteristics of arsenic resistance proteins by using designed three-stranded coiled coils. Proc Natl Acad Sci U S A. 2007;104:11969–11974. doi: 10.1073/pnas.0701979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nidhubhghaill OM, Sadler PJ. The Structure and Reactivity of Arsenic Compounds - Biological-Activity and Drug Design. Structure and Bonding. 1991;78:129–190. [Google Scholar]
- 7.NRC. Arsenic in Drinking Water. National Academy Press; Washington: 1999. [Google Scholar]
- 8.Spuches AM, Kruszyna HG, Rich AM, Wilcox DE. Thermodynamics of the As(III)-thiol interaction: arsenite and monomethylarsenite complexes with glutathione, dihydrolipoic acid, and other thiol ligands. Inorg Chem. 2005;44:2964–2972. doi: 10.1021/ic048694q. [DOI] [PubMed] [Google Scholar]
- 9.Donoghue N, Yam PT, Jiang XM, Hogg PJ. Presence of closely spaced protein thiols on the surface of mammalian cells. Protein Sci. 2000;9:2436–2445. doi: 10.1110/ps.9.12.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Chew EH, Holmgren A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc Natl Acad Sci U S A. 2007;104:12288–12293. doi: 10.1073/pnas.0701549104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowles FC. Reactions of Lipoamide Dehydrogenase and Glutathione-Reductase with Arsonic Acids and Arsonous Acids. Arch Biochem Biophys. 1985;242:1–10. doi: 10.1016/0003-9861(85)90472-2. [DOI] [PubMed] [Google Scholar]
- 12.Aposhian HV, Aposhian MM. Arsenic toxicology: five questions. Chem Res Toxicol. 2006;19:1–15. doi: 10.1021/tx050106d. [DOI] [PubMed] [Google Scholar]
- 13.Cline DJ, Thorpe C, Schneider JP. Effects of As(III) Binding on alpha-Helical Structure. J Amer Chem Soc. 2003;125:2923–2929. doi: 10.1021/ja0282644. [DOI] [PubMed] [Google Scholar]
- 14.Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- 15.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 16.Heckler EJ, Rancy PC, Kodali VK, Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rancy PC, Thorpe C. Oxidative Protein Folding in vitro: a Study of the Cooperation between Quiescin-sulfhydryl Oxidase and Protein Disulfide Isomerase. Biochemistry. 2008 doi: 10.1021/bi801604x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller MS, White HB., 3rd Isolation of avian riboflavin-binding protein. Methods Enzymol. 1986;122:227–234. doi: 10.1016/0076-6879(86)22174-6. [DOI] [PubMed] [Google Scholar]
- 19.Hoober KL, Sheasley SS, Gilbert HF, Thorpe C. Sulfhydryl oxidase from egg white: a facile catalyst for disulfide bond formation in proteins and peptides. J Biol Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 20.Darby NJ, Creighton TE. Functional properties of the individual thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:11725–11735. doi: 10.1021/bi00037a009. [DOI] [PubMed] [Google Scholar]
- 21.Lundstrom J, Holmgren A. Protein disulfide-isomerase is a substrate for thioredoxin reductase and has thioredoxin-like activity. J Biol Chem. 1990;265:9114–9120. [PubMed] [Google Scholar]
- 22.Smith AM, Chan J, Oksenberg D, Urfer R, Wexler DS, Ow A, Gao L, McAlorum A, Huang SG. A high-throughput turbidometric assay for screening inhibitors of protein disulfide isomerase activity. J Biomol Screen. 2004;9:614–620. doi: 10.1177/1087057104265292. [DOI] [PubMed] [Google Scholar]
- 23.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 24.van den Berg B, Chung EW, Robinson CV, Mateo PL, Dobson CM. The oxidative refolding of hen lysozyme and its catalysis by protein disulfide isomerase. EMBO J. 1999;18:4794–4803. doi: 10.1093/emboj/18.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monaco HL. Crystal structure of chicken riboflavin-binding protein. EMBO J. 1997;16:1475–1483. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becvar J, Palmer G. The binding of flavin derivatives to the riboflavin-binding protein of egg white. A kinetic and thermodynamic study. J Biol Chem. 1982;257:5607–5617. [PubMed] [Google Scholar]
- 27.Shen ZX, Chen GQ, Ni JH, Li XS, Xiong SM, Qiu QY, Zhu J, Tang W, Sun GL, Yang KQ, Chen Y, Zhou L, Fang ZW, Wang YT, Ma J, Zhang P, Zhang TD, Chen SJ, Chen Z, Wang ZY. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood. 1997;89:3354–3360. [PubMed] [Google Scholar]
- 28.Shen Y, Shen ZX, Yan H, Chen J, Zeng XY, Li JM, Li XS, Wu W, Xiong SM, Zhao WL, Tang W, Wu F, Liu YF, Niu C, Wang ZY, Chen SJ, Chen Z. Studies on the clinical efficacy and pharmacokinetics of low-dose arsenic trioxide in the treatment of relapsed acute promyelocytic leukemia: a comparison with conventional dosage. Leukemia. 2001;15:735–741. doi: 10.1038/sj.leu.2402106. [DOI] [PubMed] [Google Scholar]
- 29.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brohawn SG, Rudik I, Thorpe C. Avian sulfhydryl oxidase is not a metalloenzyme: adventitious binding of divalent metal ions to the enzyme. Biochemistry. 2003;42:11074–11082. doi: 10.1021/bi0301385. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta-Prot Proteom. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 32.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 33.Appenzeller-Herzog C, Ellgaard L. In vivo reduction-oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signal. 2008;10:55–64. doi: 10.1089/ars.2007.1837. [DOI] [PubMed] [Google Scholar]
- 34.Freedman RB, Klappa P, Ruddock LW. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic Biol Med. 1999;27:922–935. doi: 10.1016/s0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 36.Raturi A, Vacratsis PO, Seslija D, Lee L, Mutus B. A direct, continuous, sensitive assay for protein disulphide-isomerase based on fluorescence self-quenching. Biochem J. 2005;391:351–357. doi: 10.1042/BJ20050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett TA, Edwards BS, Sklar LA, Rogelj S. Sulfhydryl regulation of L-selectin shedding: phenylarsine oxide promotes activation-independent L-selectin shedding from leukocytes. J Immunol. 2000;164:4120–4129. doi: 10.4049/jimmunol.164.8.4120. [DOI] [PubMed] [Google Scholar]
- 38.Zahler WL, Cleland WW. A specific and sensitive assay for disulfides. J Biol Chem. 1968;243:716–719. [PubMed] [Google Scholar]
- 39.Le M, Means GE. A procedure for the determination of monothiols in the presence of dithiothreitol--an improved assay for the reduction of disulfides. Anal Biochem. 1995;229:264–271. doi: 10.1006/abio.1995.1411. [DOI] [PubMed] [Google Scholar]
- 40.Wain R, Smith LJ, Dobson CM. Oxidative refolding of amyloidogenic variants of human lysozyme. J Mol Biol. 2005;351:662–671. doi: 10.1016/j.jmb.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 41.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 44.Ptitsyn OB. Molten globule and protein folding. Adv Prot Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 45.Happersberger HP, Glocker MO. A mass spectrometric approach to the characterization of protein folding reactions. Eur Mass Spectrom. 1998;4:209–214. [Google Scholar]
- 46.Happersberger HP, Przybylski M, Glocker MO. Selective bridging of bis-cysteinyl residues by arsonous acid derivatives as an approach to the characterization of protein tertiary structures and folding pathways by mass spectrometry. Anal Biochem. 1998;264:237–250. doi: 10.1006/abio.1998.2836. [DOI] [PubMed] [Google Scholar]
- 47.Rogers MA, Langbein L, Winter H, Beckmann I, Praetzel S, Schweizer J. Hair keratin associated proteins: characterization of a second high sulfur KAP gene domain on human chromosome 21. J Invest Dermatol. 2004;122:147–158. doi: 10.1046/j.0022-202X.2003.22128.x. [DOI] [PubMed] [Google Scholar]
- 48.Johnston D, Oppermann H, Jackson J, Levinson W. Induction of four proteins in chick embryo cells by sodium arsenite. J Biol Chem. 1980;255:6975–6980. [PubMed] [Google Scholar]
- 49.Del Razo LM, Quintanilla-Vega B, Brambila-Colombres E, Calderon-Aranda ES, Manno M, Albores A. Stress proteins induced by arsenic. Toxicol Appl Pharmacol. 2001;177:132–148. doi: 10.1006/taap.2001.9291. [DOI] [PubMed] [Google Scholar]
- 50.Zheng PZ, Wang KK, Zhang QY, Huang QH, Du YZ, Zhang QH, Xiao DK, Shen SH, Imbeaud S, Eveno E, Zhao CJ, Chen YL, Fan HY, Waxman S, Auffray C, Jin G, Chen SJ, Chen Z, Zhang J. Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc Natl Acad Sci U S A. 2005;102:7653–7658. doi: 10.1073/pnas.0502825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bond U, Agell N, Haas AL, Redman K, Schlesinger MJ. Ubiquitin in stressed chicken embryo fibroblasts. J Biol Chem. 1988;263:2384–2388. [PubMed] [Google Scholar]
- 52.Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart CM, Kong XP, Ron D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol Cell. 2006;23:875–885. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 53.Sharma SK, Goloubinoff P, Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem Biophys Res Commun. 2008;372:341–345. doi: 10.1016/j.bbrc.2008.05.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.