Abstract
Background
The natural habitat of wild P. ginseng is currently found only in the Russian Primorye and the populations are extremely exhausted and require restoration. Analysis of the genetic diversity and population structure of an endangered species is a prerequisite for conservation. The present study aims to investigate the patterns and levels of genetic polymorphism and population structures of wild P. ginseng with the AFLP method to (1) estimate the level of genetic diversity in the P. ginseng populations in the Russian Primorsky Krai, (2) calculate the distribution of variability within a population and among populations and (3) examine the genetic relationship between the populations.
Methods
Genetic variability and population structure of ten P. ginseng populations were investigated with Amplified Fragment Length Polymorphism (AFLP) markers. The genetic relationships among P. ginseng plants and populations were delineated.
Results
The mean genetic variability within populations was high. The mean level of polymorphisms was 55.68% at the population level and 99.65% at the species level. The Shannon's index ranged between 0.1602 and 0.3222 with an average of 0.2626 at the population level and 0.3967 at the species level. The analysis of molecular variances (AMOVA) showed a significant population structure in P. ginseng. The partition of genetic diversity with AMOVA suggested that the majority of the genetic variation (64.5%) was within populations of P. ginseng. The inter-population variability was approximately 36% of the total variability. The genetic relationships among P. ginseng plants and populations were reconstructed by Minimum Spanning tree (MS-tree) on the basis of Euclidean distances with ARLEQUIN and NTSYS, respectively. The MS-trees suggest that the southern Uss, Part and Nad populations may have promoted P. ginseng distribution throughout the Russian Primorye.
Conclusion
The P. ginseng populations in the Russian Primorye are significant in genetic diversity. The high variability demonstrates that the current genetic resources of P. ginseng populations have not been exposed to depletion.
Background
Panax ginseng C.A. Meyer (Renshen, Asian ginseng) is a representative species of the Panax L. genus which is a relic of the Araliacea family [1]. Their natural stocks are over-exploited because they have the highest biological activities [2]. At the beginning of the twentieth century, wild P. ginseng spread over a vast territory including the Russian Primorsky Krai, Korea and China. Currently, wild P. ginseng can only be found in Russia; however, its populations are extremely exhausted and restoration is needed [1]. P. ginseng is listed in the Red Book of Primorsky Krai as an endangered species [3].
Analysis of the genetic diversity and population structure of an endangered species is a prerequisite for conservation [4]. Genetic variability is critical for a species to adapt to environmental changes and survive in the long term. A species with little genetic variability may suffer from reduced fitness in its current environment and may not have the evolutionary potential necessary for a changing environment [5]. Knowledge of genetic diversity within a population and among populations is important for conservation management, especially in identifying genetically unique structural units within a species and determining the populations that need protection.
A high level of polymorphism of a marker is a basic condition that must be assessed population genetics studies [6]. A study using allozyme analysis found a low level of polymorphism (7%) in wild ginseng [7]. Multi-locus DNA markers, e.g., Random Amplified Polymorphic DNA (RAPD), Inter Simple Sequence Repeat (ISSR) and Amplified Fragment Length Polymorphism (AFLP) would potentially produce higher values of polymorphism than allozyme analysis because non-coding DNA sequences, which mutate at a higher speed than coding sequences, would also be characterized [8]. RAPD polymorphisms in wild ginseng populations are low [7,9]. Results with RAPD markers corresponded with the lack of genetic variation demonstrated by isozyme gene loci in red pine [10]. In contrast, polymorphism in RAPD loci (about 46%) is high in cultivated P. ginseng [11]. Allozymes and RAPD markers are highly variable in populations of Panax quinquefolius (Xiyangshen, American ginseng) [12-16]. There are 62.5% polymorphic loci in populations of P. quinquefolius in the United States [16]. P. quinquefolius population from Ontario, Canada, has a polymorphism level of about 46% estimated with RAPD analysis [14].
As a reproducible and robust technique, AFLP [17] generates a large number of bands per assay and is best suited for analyzing genetic diversity. The fluorescence-based automated AFLP method demonstrated the highest resolving power as a multi-loci technique [18-20]. An automated DNA fingerprinting system utilizing fluorescently labeled primers and the laser detection technology associated with the automatic sequencer allowed the resolution of fragments that were undistinguishable by other methods. In a previous study, four fluorescently labeled AFLP primer pairs and 20 RAPD primers generated 645 and 170 polymorphic markers respectively [18]. In a study to characterize Miscanthus, three fluorescently labeled AFLP primer pairs generated 998 polymorphic markers, as opposed to only 26 polymorphic markers produced by two ISSR [20].
The present study aims to investigate the patterns and levels of genetic polymorphism and population structures of wild P. ginseng with the AFLP method to (1) estimate the level of genetic diversity in the P. ginseng populations in the Russian Primorsky Krai, (2) calculate the distribution of variability within a population and among populations and (3) examine the genetic relationship between the populations.
Methods
Sampled populations
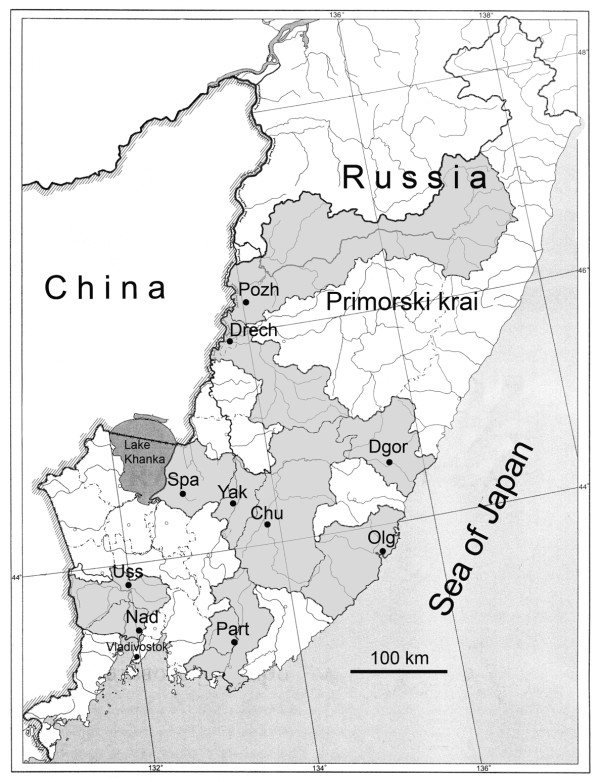
One hundred and sixty-seven (167) P. ginseng individuals were collected from the ten administrative areas of Primorsky Krai (Figure 1) and transferred to a collection nursery. The study populations were coded with the names of the areas. Twenty (20) P. ginseng individuals were collected from the Chuguevsk area (Chu), 19 from the Spassk area (Spa), 16 from the Ussuriisk area (Uss), 13 from the Dalnerechensk area (Drech), 16 from the Dalnegorsk area (Dgor), 15 from the Olginsk area (Olg), 15 from the Pozharsk area (Pozh), 24 from the Nadezhdinsk area (Nad), 19 from the Partizansk area (Part) and 10 from the Yakovlevsk area (Yak).
Figure 1.
The administrative areas in the territory of the Russain Primorskiy Krai where Panax ginseng plants were collected.
DNA extraction
Total genomic DNA was extracted from fresh leaf tissue according to Echt et al. [21]. The extracted DNA was purified according to the Murray and Thompson method [22].
AFLP procedure
AFLP genotyping was performed according to Vos et al. [17] using EcoRI and MseI restriction enzymes. Pre-amplification reactions utilized AFLP primers with two selective nucleotides. EcoRI and MseI selective amplification primers contained three and four selective nucleotides, respectively (Table 1). AFLP adapters and primers were purchased from Syntol (Russia). All the EcoRI-NNN selective primers were labeled with fluorescent 6-carboxy fluorescein (6-FAM) at the 5' end. The AFLP fragments were analyzed on an ABI Prism 3100 automated capillarity system with GeneScan Analysis Software (Applied Biosystems, USA). All unambiguous peaks including monomorphic peaks between 50-500 base pairs (bp) were analyzed and the scoring results were exported as a presence/absence matrix.
Table 1.
AFLP selective primers used in the study of the population genetics of Panax ginseng
| Eco RI primer | Mse I primer | Number of loci |
|---|---|---|
| E-ACA | M-CCGG | 149 |
| E-ACA | M-CCTG | 133 |
| Total | 282 |
Data analysis
Parameters of genetic variability and genetic mutual relations of populations were calculated with the POPGEN32 (POPGENE v. 1.31, Centre for International Forestry Research, University of Alberta and Tim Boyle, Canada) [23] and ARLEQUIN (Arlequin v.3.11, Excoffier L. Zoological Institute, University of Berne, Switzerland). As AFLPs were dominant markers, Shannon's information measure (IS) [24] was used to quantify the degree of the within-population diversity. Analysis of molecular variance (AMOVA) [25] was conducted to calculate the variance components and significance levels of variation within a population and among populations. AMOVA derived genetic differentiation values (FST) between pairs of populations (analogous to traditional F statistics) were calculated. Gene flow between pairs of populations (Nm = (1-FST)/4FST) was calculated from FST values [26]. We reconstructed the Minimum Spanning tree (MS-tree) between representatives of P. ginseng and populations from a matrix of squared Euclidean distances using ARLEQUIN (Arlequin v.3.11, Excoffier L. Zoological Institute, University of Berne, Switzerland) and NTSYS (NTSYS-pc v.1.70, Applied Biostatistics, Inc, USA) respectively.
Results
Nine (9) AFLP primer pairs were tested, namely Eco(ACG)/Mse(CCTC), Eco(ACG)/Mse(CCTT), Eco(ACA)/Mse(CCTG), Eco(ACA)/Mse(CCGG), Eco(ACA)/Mse(CCAC), Eco(ACT)/Mse(CCGA), Eco(ACT)/Mse(CCTA), Eco(ACC)/Mse(CCAG), and Eco(ACC)/Mse(CCGC). Using two of the primer pairs Eco (ACA)/Mse(CCTG) and Eco(ACA)/Mse(CCGG) (Table 1), we detected polymorphic bands among the various samples of P. ginseng in this study. Among the scored 282 fragments, 281 were polymorphic across all ten populations (Table 2). Genetic variability was high within populations (Table 2). The highest genetic diversity values (approximately 70%) were obtained in the Chu, Nad, Olg and Pozh populations, whereas the lowest values (approximately 40%) were found in the Uss and Dgor populations. The mean level of polymorphisms was 55.68% at the population level and 99.65% at the species level. The Shannon's index ranged between 0.1602 and 0.3222 with an average of 0.2626 at the population level and 0.3967 at the species level. The intra-population genetic polymorphisms ranged from 38.65% (Uss) to 69.15% (Chu) with an average of 55.68% (Table 2).
Table 2.
Sample size and genetic variability parameters of Panax ginseng populations calculated from AFLP data for 282 fragments
| Population number |
Population code |
Number of plants (order numbers of plants) |
Shannon's index (IS) |
Polymorphic loci | |
|---|---|---|---|---|---|
| Number | % (P) | ||||
| 1 | Spa | 19 (17-35) | 0.2972 | 163 | 57.80 |
| 2 | Yak | 10 (158-167) | 0.2487 | 145 | 51.42 |
| 3 | Drech | 13 (36-48) | 0.2614 | 138 | 48.94 |
| 4 | Pozh | 15 (100-114) | 0.3222 | 186 | 65.96 |
| 5 | Uss | 16 (1-16) | 0.1602 | 109 | 38.65 |
| 6 | Nad | 24 (115-138) | 0.2840 | 190 | 67.38 |
| 7 | Chu | 20 (49-68) | 0.3169 | 195 | 69.15 |
| 8 | Dgor | 16 (69-84) | 0.1821 | 114 | 40.43 |
| 9 | Olg | 15 (85-99) | 0.3195 | 188 | 66.67 |
| 10 | Part | 19 (139-157) | 0.2335 | 142 | 50.35 |
| Population average | 17 | 0.2626 | 157 | 55.68 | |
| Species-level value | 167 | 0.3967 | 281 | 99.65 | |
All pair wise FST between populations, obtained with AMOVA, were significant (P = 0.0000) and varied from 0.09180 (Pozh-Nad) to 0.60506 (Drech-Uss) (Table 3). The non-hierarchical AMOVA analyses revealed that 35.54% of the total variation was attributed to the variability among the populations, whereas 64.46% was accumulated within the populations (Table 4). The average number of migrants (Nm) between populations based on AMOVA (FST = 0.355) was 0.45.
Table 3.
Matrix of pairwise differences (FST) among Panax ginseng populations calculated with AMOVA
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.00000 | |||||||||
| 2 | 0.41235 | 0.00000 | ||||||||
| 3 | 0.27212 | 0.53153 | 0.00000 | |||||||
| 4 | 0.30808 | 0.26936 | 0.47046 | 0.00000 | ||||||
| 5 | 0.35629 | 0.52259 | 0.60506 | 0.36954 | 0.00000 | |||||
| 6 | 0.30464 | 0.25556 | 0.49335 | 0.09180 | 0.36057 | 0.00000 | ||||
| 7 | 0.18200 | 0.42200 | 0.21348 | 0.35356 | 0.40031 | 0.35103 | 0.00000 | |||
| 8 | 0.21894 | 0.48054 | 0.54275 | 0.32029 | 0.27451 | 0.31409 | 0.33000 | 0.00000 | ||
| 9 | 0.38764 | 0.25381 | 0.42708 | 0.24434 | 0.49424 | 0.30650 | 0.35041 | 0.46318 | 0.00000 | |
| 10 | 0.34993 | 0.16600 | 0.52375 | 0.27691 | 0.42249 | 0.15721 | 0.38540 | 0.39335 | 0.36194 | 0.00000 |
P value = 0.00000
Table 4.
AMOVA analysis of genetic variances within and among populations of Panax ginseng (Level of significance is based on 1000 iterations)
| Source of variation | Degree of freedom | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among populations | 9 | 2413.140 | 14.55557 | 35.54 |
| Within populations | 157 | 4145.651 | 26.40542 | 64.46 |
| Total | 166 | 6558.790 | 40.96099 |
Fixation index FST = 0.35535
P value = 0.0000
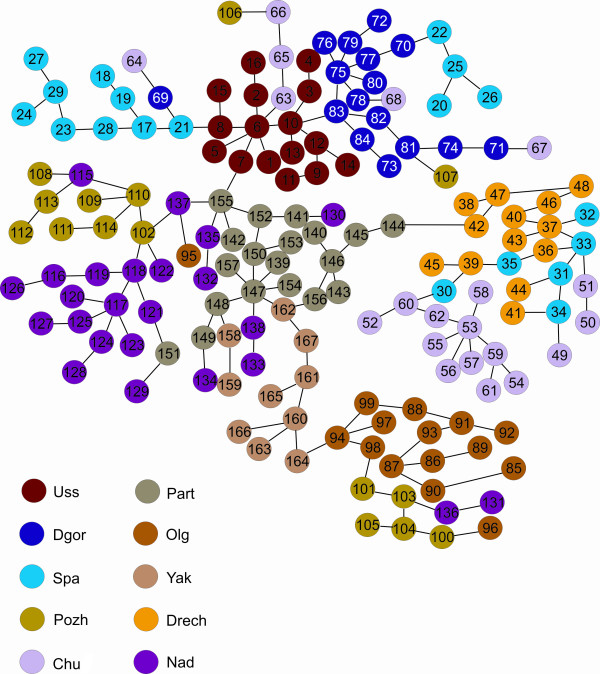
The MS-tree showed the genetic relationships among P. ginseng plants (Figure 2). Calculated in AMOVA on the basis of Euclidean distances, the length of the lines connecting the representatives inside the populations and between the populations reflects the intra- and inter-population genetic distances respectively (Table 5).
Figure 2.
MS-tree representing phylogenetic relationships among representative Panax ginseng populations. Length of lines is proportional to the Euclidean distances among plants. Length of scale line is equal to 50 units of Euclidean distances
Table 5.
The length of lines on MS-tree characterizing the Euclidean genetic distances among plants in populations and among populations of Panax ginseng
| Among plants in population | Among populations | |||
|---|---|---|---|---|
| Population | Range of length | Average length | Population pair | Length |
| Uss | 8-41 | 17.33 | Uss - Spa | 24 |
| Spa | 9-30 | 22.57 | Uss - Dgor | 22 |
| Dgor | 15-38 | 23.0 | Uss - Part | 33 |
| Pozh | 14-43 | 28.0 | Part - Drech | 50 |
| Nad | 21-36 | 29.07 | Drech - Chu | 25 |
| Part | 12-30 | 22.06 | Part - Yak | 19 |
| Yak | 22-57 | 35.13 | Yak - Olg | 35 |
| Chu | 15-44 | 25.0 | Pozh -Nad | 24 |
| Drech | 12-51 | 24.88 | Pozh - Part | 27 |
| Olg | 14-52 | 36.5 | ||
| Average | 26.35 | 28.78 | ||
According to values of genetic distances, all of the studied ginseng plants on the MS-tree formed two groups (Figure 2, Table 5), the first group consisting of the Drech and Chu populations and the second group the Part, Yak, Olg, Nad, Pozh, Uss, Dgor and Spa populations. These two groups were divided by a genetic distance of 50 units of Euclidean distance (Figure 2, Table 5). The Spa, Uss, Dgor and Part, Yak, Nad, Pozh populations formed two subgroups divided by a genetic distance of 33 Euclidean distance units. The plants of the Olg population were distanced from the Part, Yak, Nad, Pozh subgroup by 35 Euclidean distance units (Figure 2, Table 5).
The location of a P. ginseng on the MS-tree was dependent on the population it belonged to; however, such clustering was not strict and some populations partially overlapped (Figure 2). For example, some plants of the Pozh population were grouped with those of the Olg population while some plants of the Spa population were with the Dgor and Drech populations. The plants of the Nad population were partially mixed with those of the Part and Pozh populations. Moreover, the plants of the Chu population were mixed with those of the Uss, Drech and Dgor populations.
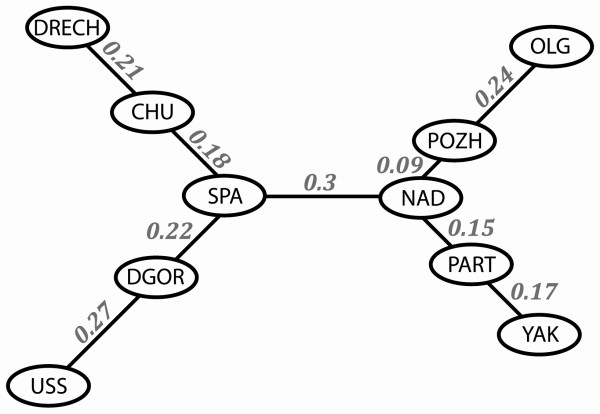
The arrangement of the populations on the MS-tree did not always correspond to their geographical areas. For example, the Pozh population was geographically distant from the Nad and Part populations but was genetically close to them (Figure 2 and 3, Table 5). By contrast, populations that are geographically close, such as Uss and Nad, were genetically distant and therefore belonged to different subgroups (Figure 2) or groups (Figure 3).
Figure 3.
MS-tree representing phylogenetic relationships among Panax ginseng populations. The numbers on lines show the genetic FST distances among populations.
The Uss population was characterized by the smallest average value of Euclidean genetic distances between plants (17.33 units), whereas the Olg population was characterized by the highest value (36.5 units). The average value of Euclidean genetic distances between the plants of different populations (28.78 units) was higher than that of intra-population genetic distances (26.35 units) (Table 5).
Discussion
P. ginseng populations located in Primorsky Krai have a low level of genetic polymorphisms (approximately 7%) by allozyme and RAPD [7,9,27-29] which means effective conservation strategies would be difficult to implement.
High genetic variability in P. ginseng was revealed by the AFLP method. While genetic diversity is theoretically higher in large populations, the Uss population was small in size but appeared to have suffered from the loss of a genetic diversity more than other populations. Several populations (Spa, Pozh, Nad, Chu and Olg) were distinguished by having higher levels of variability. For these populations, the average value of polymorphisms was 65.39%. At the species level, the percentage of polymorphisms was 99.65%. The high level of variability may be due to cross-pollination; however, P. ginseng's capability for cross-pollination is yet to be established [30]. A large number of the insects visiting P. ginseng inflorescences are potential pollinators [1]. In Panax notoginseng, four pairs of fluorescently labeled AFLP primers produced 312 fragments, of which 240 (76.9%) were polymorphic [31]. In Panax stipuleanatus, the same primers revealed 346 loci, of which 334 (96.5%) were polymorphic [31].
Analysis of molecular variance (AMOVA) of the AFLP data showed a significant population pattern of the wild Russian P. ginseng. FST, estimates of inter-population variability, varied from 0.09180 to 0.60506 (Table 3), indicating that all populations may be different from each other. The partition of genetic diversity with AMOVA suggested that the majority of the genetic variation (64.5%) was within populations of P. ginseng. The inter-population variability was approximately 36% of the total variability (Table 4). The value of gene flow (Nm) was 0.45; therefore, wild P. ginseng has a relatively high genetic differentiation value among populations and a relatively low level of gene flow. In cultivated P. ginseng, inter-population RAPD variability ranged from 1.77% to 42.01% [11] and was 31% in another study [32]. The fluorescence-based automated AFLP method demonstrated that over 40% of the genetic variation of wild P. stipuleanatus was among the populations [31]. P. ginseng' FST values are consistent with estimates of inter-population variability, which were obtained with AMOVA and AFLP markers for plant species with mixed type of propagation (FST = 0.35) [33]. According to Nybom [33], P. ginseng is a species with mixed type of propagation. The ability of P. ginseng to produce seeds via autogamy, out-crossing or agamospermy without pollination was demonstrated earlier [30]. The high level of genetic variation and high proportion of variation within populations in P. ginseng suggest that human activities (e.g. overexploitation, habitat destruction, urbanization, pollution) are the major contributor that threatens the survival of the wild P. ginseng populations.
Six populations (Uss, Part, Olg, Yak, Dgor and Drech) clustered together and four populations (Spa, Chu, Pozh and Nad) were partially mixed with other populations (Figure 2). We believe that the spread of wild P. ginseng seeds by humans, animals and birds is the main factor contributing to the population re-mixing.
The MS-tree arrangement of populations did not always correspond to their geographical areas, which may be due to converging common selection forces in geographically disparate populations [34]. Future research with greater numbers of AFLP loci coupled with other high variable markers (SSR) is warranted to confirm the factors that shaped the genetic structures of P. ginseng in Russia.
The finding that the average value of inter-population genetic distances is higher that of intra-population genetic distances (Table 5) is consistent with the AMOVA conclusion that reveals the population genetic structures of wild P. ginseng.
The Uss population was characterized by the least average value of genetic distances between plants (Table 5), which was consistent with the low parameters of variability calculated in POPGENE for this population (Table 2),. On the other hand, the Olg population demonstrated the highest genetic distances (Table 5). The Olg population is, therefore, the most genetically diverse population according to the MS-tree, suggesting that it should be conserved first.
The central node position on the MS-tree is occupied by a plant (No. 6) that belongs to the Uss population and the genetic communications spread to the Spa and Dgor populations, and to a cluster of the rest of the P. ginseng populations (Part, Nad, Yak, Olg, Chu, Drech and Pozh), suggesting the ancestral status of the Uss population. The Part population, also at the central position on the MS-tree, may have the same ancestral status as the Uss population (Figure 2); Nad and Spa populations may be ancestors as well (Figure 3). The absence of a strong Spa population cluster on the MS-tree (Figure 2) may be evidence for its ancestral origin.
The MS-trees suggest that the southern Uss, Part and Nad populations may have promoted P. ginseng distribution throughout the Russian Primorye. This result supports the assumption that Sikhote-Alin was re-colonized by P. ginseng when thermophilic plants spread from the south to the north during the early Holocene warm period [27].
Future studies may focus on (1) using AMOVA to investigate whether genetically differentiated regions exists for P. ginseng and whether P. ginseng is adapted for heterogeneous conditions; (2) whether a positive correlation between genetic and geographical distances among P. ginseng populations may be established; and (3) using the multi-locus mating system program (MLTR) to estimate the level of inbreeding and cross-pollination in wild P. ginseng populations.
Conclusion
The P. ginseng populations in the Russian Primorye contain a significant level of genetic diversity and are essentially differentiated. The gene flow of the populations was less than one (Nm = 0.45) which indicates continued divergence among populations [26]. The current high level of variability demonstrates that the genetic resources of P. ginseng populations have not been exposed to depletion.
Abbreviations
AFLP: Amplified Fragment Length Polymorphism; ISSR: Inter Simple Sequence Repeat; AFLP: Amplified Fragment Length Polymorphism; Chu: Chuguevsk area; Spa: Spassk area; Uss: Ussuriisk area; Drech: Dalnerechensk area; Dgor: Dalnegorsk area; Olg: Olginsk area; Pozh: Pozharsk area; Nad: Nadezhdinsk area; Part: Partizansk area; Yak: Yakovlevsk area; bp: base pairs; AMOVA: Analysis of molecular variance; MS-tree: Minimum Spanning tree; 6-FAM: 6-carboxy fluorescein
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YNZ and GDR designed the research. GDR and ILK performed the research and analyzed the data. TIM collected the plants. GDR wrote the manuscript. AAB contributed to the data acquisition. YNZ helped in writing the manuscript and coordinating the study. All authors read and approved the final version of the manuscript.
Contributor Information
Yuri N Zhuravlev, Email: zhuravlev@ibss.dvo.ru.
Galina D Reunova, Email: reunova@ibss.dvo.ru.
Irina L Kats, Email: kats@ibss.dvo.ru.
Tamara I Muzarok, Email: muzarok@ibss.dvo.ru.
Alexander A Bondar, Email: alex.bondar@mail.ru.
Acknowledgements
We thank Drs VL Semerikov and EV Brenner for their kind assistance in the AFLP analysis. We are grateful to Dr GN Chelomina for the discussion of the results. This work was supported by grants from the Russian Academy of Sciences (No. 09-I-P23-06; No. 09-I-OBN-02), by the Russian Fund for Fundamental Investigations (No. 08-04-99132-r_ofi; 09-04-90309-Viet-a) and by the Grant Program "Molecular and Cell Biology" of the Russian Academy of Sciences and the "Leading Schools of Thought" grant from the President of the Russian Federation (No. NSH 1635-2008.4).
References
- Zhuravlev YN, Kolyada AS. Araliaceae: Ginseng and Others. Vladivostok: Dalnauka; 1996. [In Russian] [Google Scholar]
- Leem K, Kim SC, Yang CH, Seo J. Genetic identification of Panax ginseng and Panax quinquefolius by pyrosequencing methods. Biosci Biotechnol Biochem. 2005;69:1771–1773. doi: 10.1271/bbb.69.1771. [DOI] [PubMed] [Google Scholar]
- Red Book of Primorsky Kray: Plants. Vladivostok: Apelsin; 2008. [In Russian] [Google Scholar]
- Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv Biol. 2003;17:230–237. doi: 10.1046/j.1523-1739.2003.01236.x. [DOI] [Google Scholar]
- Crawford DL. Molecular markers for the study of genetic variation within and between populations of rare plants. Opera Botanica. 1997;132:149–157. [Google Scholar]
- Zhuravlev YN, Koren OG, Kozyrenko MM, Reunova GD, Artyukova EV, Krylach TY, Muzarok TI. In: Biodiversity and Allelopathy: From Organism to Ecosystems in the Pacific. Chou CH, Waller GR, Reinhardt C, editor. Taipei: Academia Sinica; 1999. Use of molecular markers to design the reintroduction strategy for Panax ginseng; pp. 183–192. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Zhuravlev YN, Reunova GD, Kozyrenko MM, Artyukova EV, Muzarok TI. Genetic variation of wild ginseng populations (RAPD analysis) Mol Biol (Moscow) 1998;32:910–914. [Google Scholar]
- Mosseler A, Egger KN, Hughes GA. Low levels of genetic diversity in red pine confirmed by random amplified polymorphic DNA markers. Can J For Res. 1992;22:1332–1337. doi: 10.1139/x92-177. [DOI] [Google Scholar]
- Ma XJ, Wang XQ, Xu ZX, Xiao PG, Hong DY. RAPD variation within and among populations of ginseng cultivars. Acta Bot Sinica. 2000;42:587–590. [Google Scholar]
- Schluter C, Punja ZK. Genetic diversity among natural and cultivated field populations and seed lots of american ginseng (Panax quinquefolius L.) in Canada. Int J Plant Sci. 2002;163:427–439. doi: 10.1086/339512. [DOI] [Google Scholar]
- Grubbs HJ, Case MA. Allozyme variation in American ginseng (Panax quinquefolius L.): Variation, breeding system, and implications for current conservation practice. Conserv Genet. 2004;5:13–23. doi: 10.1023/B:COGE.0000014064.44592.bc. [DOI] [Google Scholar]
- Bai D, Brandke J, Reeleder R. Genetic diversity in North American ginseng (Panax quinquefolius L.) grown in Ontario detected by RAPD analysis. Genome. 1997;40:111–115. doi: 10.1139/g97-015. [DOI] [PubMed] [Google Scholar]
- Boehm CL, Harrison HC, Jung G, Nienhuis J. Organization of American and Asian ginseng germplasm using randomly amplified polymorphic DNA (RAPD) markers. J Am Soc Hortic Sci. 1999;124:252–256. [Google Scholar]
- Cruse-Sanders JM, Hamrick JL. Genetic diversity in harvested and protected populations of wild American ginseng, Panax quinquefolius L. (Araliaceae) Am J Bot. 2004;91:540–548. doi: 10.3732/ajb.91.4.540. [DOI] [PubMed] [Google Scholar]
- Vos PR, Hogers M, Bleeker M, Reijans M, van der Lee T, Hornes M, Frijters A, Pot J, Paleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JH, Mattes M, Arnold GM, Edwards KJ, Ahman I, Larsson S, Karp A. Characterization of genetic diversity in potential biomass willows (Salix spp) by RAPD and AFLP analyses. Genome. 1999;42:173–183. doi: 10.1139/gen-42-2-173. [DOI] [PubMed] [Google Scholar]
- McGregor CE, Lambert CA, Greyling MM, Louw JH, Warnick L. A comparative assessment of DNA fingerprinting techniques (RAPD, ISSR, AFLP and SSR) in tetraploid potato (Solanum tuberosum L.) germplasm. Euphytica. 2000;113:135–144. doi: 10.1023/A:1003925620546. [DOI] [Google Scholar]
- Hodkinson TR, Chase MW, Renvoize SA. Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropogoneae, Poaceae) using AFLP and ISSR PCR. Ann of Bot. 2002;89:627–636. doi: 10.1093/aob/mcf091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echt CS, Erdahl LA, McCoy TJ. Genetic segregation of random amplified polymorphic DNA in diploid cultivated alfalfa. Genome. 1992;35:84–87. doi: 10.1139/g92-014. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot. 1997;129:157. [Google Scholar]
- Lewontin RC. The apportionment of human diversity. Evol Biol. 1972;6:381–398. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetic structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Koren OG, Potenko VV, Zhuravlev YN. Inheritance and variation of allozymes in Panax ginseng C.A. Meyer (Araliaceae) Int J Plant Sci. 2003;164:189–195. doi: 10.1086/344758. [DOI] [Google Scholar]
- Zhuravlev YN, Koren OG, Reunova GD, Artyukova EV, Kozyrenko MM, Muzarok TI, Kats IL. Ginseng conservation program in Russian Primorye: genetic structure of wild and cultivated populations. J Ginseng Res. 2004;28:60–66. doi: 10.5142/JGR.2004.28.1.060. [DOI] [Google Scholar]
- Zhuravlev YN, Kozyrenko MM, Artyukova EV, Reunova GD, Muzarok TI, Elyakov GB. Genetic typing of Panax ginseng by use RAPD-PCR. Dokl RAS. 1996;349:111–114. [PubMed] [Google Scholar]
- Zhuravlev YN, Koren OG, Reunova GD, Muzarok TI, Gorpenchenko TY, Kats IL, Khrolenko YA. Panax ginseng natural populations: their past, current state and perspectives. Acta Pharmacol Sin. 2008;29:1127–1136. doi: 10.1111/j.1745-7254.2008.00866.x. [DOI] [PubMed] [Google Scholar]
- Zhou SL, Xiong GM, Li ZY, Wen J. Loss of genetic diversity of domesticated Panax notoginseng F H Chen as evidenced by ITS sequence and AFLP polymorphism: a comparative study with P. stipuleanatus H T et K M Feng. J Integr Plant Biol. 2005;47:107–115. [Google Scholar]
- Kim C, Choi H-K. Genetic diversity and relationship in Korean Ginseng (Panax schinseng) based on RAPD analysis. Korean J Genet. 2003;25:181–188. [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Bonin A, Taberlet P, Miaud C, Pompanon F. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frog (Rana temporaria) Mol Biol Evol. 2006;23:773–783. doi: 10.1093/molbev/msj087. [DOI] [PubMed] [Google Scholar]