Abstract
Tensegrity or tensional integrity is a property of a structure that relies on a balance between components that are either in pure compression or in pure tension for its stability [1,2]. Tensegrity structures exhibit extremely high strength-to-weight ratios and great resilience, and are therefore widely used in engineering, robotics and architecture [3,4]. Here we report nanoscale, prestressed, three-dimensional tensegrity structures in which rigid bundles of DNA double helices resist compressive forces exerted by segments of single-stranded DNA that act as tension-bearing cables. Our DNA tensegrity structures can self-assemble against forces up to 14 pN, which is twice the stall force of powerful molecular motors such as kinesin or myosin [5,6]. The forces generated by this molecular prestressing mechanism can be employed to bend the DNA bundles or to actuate the entire structure through enzymatic cleavage at specific sites. In addition to being building blocks for nanostructures, tensile structural elements made of single-stranded DNA could be used to study molecular forces, cellular mechanotransduction, and other fundamental biological processes.
Classic examples of tensegrity structures are the sculptures of Kenneth Snelson that suspend isolated rigid columns in mid-air by interconnecting them with a continuous tensile cable network that prestresses the whole system (Fig. 1a) [1], and the geodesic domes of Buckminster Fuller that utilize triangulation and minimal tensional paths between all pairs of neighboring vertices to maintain their stability [2]. Prestressed tensegrity structures are found at all size scales in living systems [7] and play a central role in cellular mechanotransduction [8]. A number of wireframe structures have been built from DNA [9, 10, 11, 12, 13]; however, these are relatively static shapes that do not display many of the novel mechanical features of prestressed tensegrity structures, such as the ability to globally reorient internal members and thereby strengthen in response to a local stress. Here we set out to use the DNA-origami method [14, 15] to build prestressed tensegrity structures on the nanoscale that exhibit integrated mechanical responses similar to those displayed by living cellular systems.
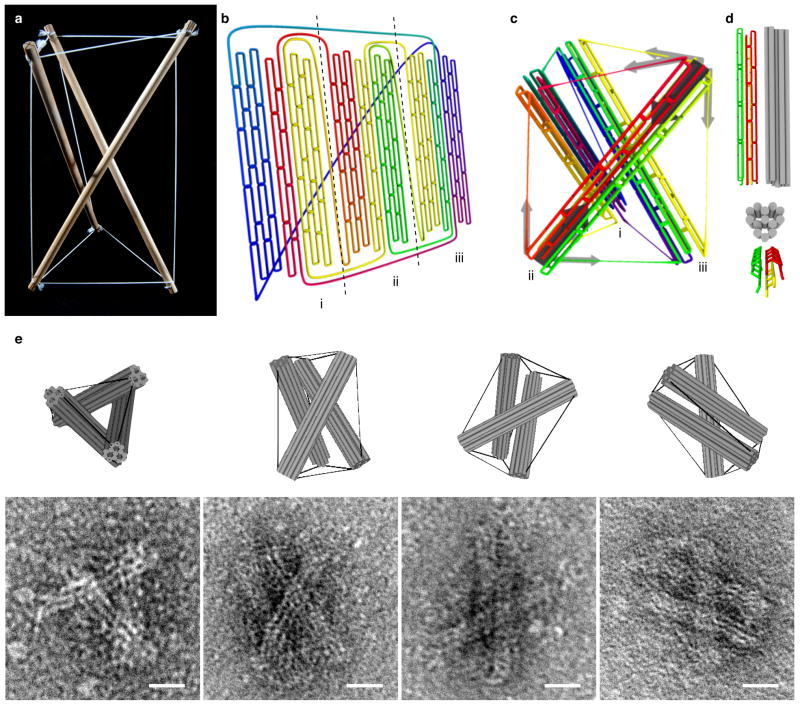
Figure 1. 3D prestressed DNA tensegrity.
a, Tensegrity prism constructed from wood and cord. b, Quasi-2D representation of the scaffold pathway through the prestressed tensegrity prism. The color code indicates the nucleotide (nt) index along the circular path. Red represents the first nt on the 8634-nt-long scaffold, violet the last nt. The three struts are labeled as i, ii, iii. c, 3D representation of the scaffold pathway for the assembled prism. Staple strands are omitted for clarity. Light grey arrows denote the contractile forces exerted by the ssDNA springs, while dark grey arrows indicate the sum of compressive forces along the axis of the 13-helix bundle. d, Cylinder and scaffold models of an individual 13-helix bundle. Every cylinder represents one double helix. e, Electron micrographs and cylinder models of DNA tensegrity prisms. Scale bars: 20 nm.
To accomplish this goal, we moved beyond current DNA-origami methods used to create nanostructures that rely only on paired bases to provide structural integrity, and instead, we incorporated stretched ssDNA segments as nanoconstruction elements that act in solution as entropic springs whose behavior can be described over a wide range of loads using a modified freely-jointed-chain model (mFJC) that accounts for stretchable Kuhn segments [16] (Supp. Note S9). We designed and fabricated prestressed DNA tensegrity structures consisting of a 8634-nucleotide (nt) M13mp18-based ‘scaffold strand’ and hundreds of oligodeoxyribonucleotide ‘staple strands’ that self-assemble into tensed structures despite kinetic barriers imposed by the prestress. The assembly process for prestressed origami objects is, as for DNA-origami objects in general, a one-pot reaction where the scaffold strand, the staple strands, and buffer containing Mg2+ ions are heated to 80°C and then cooled down over the course of 72 hours to room temperature to allow each staple strand to find its unique position on the scaffold sequence and hence achieve the correct assembly of the structure. The staple sequences were designed, using the software caDNAno [17], to promote self-assembly of rigid columns or struts composed of bundles of multiple DNA double helices. Importantly, this design differed from those used for previously reported DNA-origami structures because we also incorporated loops of hundreds of unpaired bases on the scaffold strand that connect the ends of multiple individual struts and act as the single-stranded DNA (ssDNA) springs. Two stretches of the scaffold DNA whose sequences are prone to hairpin formation were incorporated into the rigid struts. A simplifying assumption of our model is that secondary-structure formation in the ssDNA springs can be ignored. Future experiments could employ structure-free ssDNA (e.g. scaffold segments programmed to consist primarily of the bases A, C, and T) as the springs to make this assumption more valid. The energy necessary for tightening of these ssDNA springs is provided during the assembly process by the base pairing of the double helices that form the compression-resistant struts, and thereby prestress the entire integrated DNA structure.
As a proof-of-principle for this strategy, we designed a 3D “tensegrity prism”, which is composed of three compression-resistant, 57-nm–long, 13-helix bundles held in place by nine tensed ssDNA springs (each 226 bases long) (Fig. 1b and c). Each strut is constructed from three segments of the scaffold that are far apart in the primary sequence, providing five, three, and five of the 13 helices, respectively (Fig. 1d). Transmission electron microscopic (TEM) analysis of gel-purified structures self-assembled in this manner, and comparison of these 2D images with predicted 3D computer models, confirmed that this self-assembly process resulted in 3-strut tensegrity prisms (Figs 1e, Supp. Fig. S1), and similar results were obtained by constructing another tensegrity prism using 6-helix bundles as struts (Supp. Fig. S2).
Towards investigating the influence of ssDNA spring length, and thus tension, on the structure and assembly success of DNA tensegrity structures, we created a two-dimensional, prestressed ‘kite’ structure (Fig. 2a). The ends of two 92-nm–long, 12-helix bundles were connected through four unpaired regions of a 8634-nt scaffold strand; no DNA strands cross over between the two 12-helix bundles at the point of intersection. Hence, the structural integrity is solely provided by the tension of the four DNA springs balanced by the compression imposed on the struts to which they connect. In a perfect square arrangement, the distances between pairs of neighboring ends of the struts are 65 nm each. Stretched over this distance in solution at room temperature, an idealized ssDNA spring of 220 nt exerts a contractile force between 6 and 7 pN. We calculated the force with two models that estimate extension-force relations for polymers in solution: the mFJC model (6.4 pN) and the wormlike chain (WLC) model [18], which implies continuous bending elasticity (6.9 pN) (Fig. S3, Supp. Notes S9–11). To investigate the structural rigidity and the maximum force achievable with this approach, the kite structure was assembled in nine versions with spring lengths of 520, 420, 320, 220, 190, 170, 160, 150, and 140 nt. The different numbers of bases in the single-stranded regions were achieved by exchanging a small subset of staple strands, which led to spooling of the desired number of unused bases. The enlarged schematics in Fig. 2a show the scaffold and staple paths at the end of a 12-helix bundle before and after spooling.
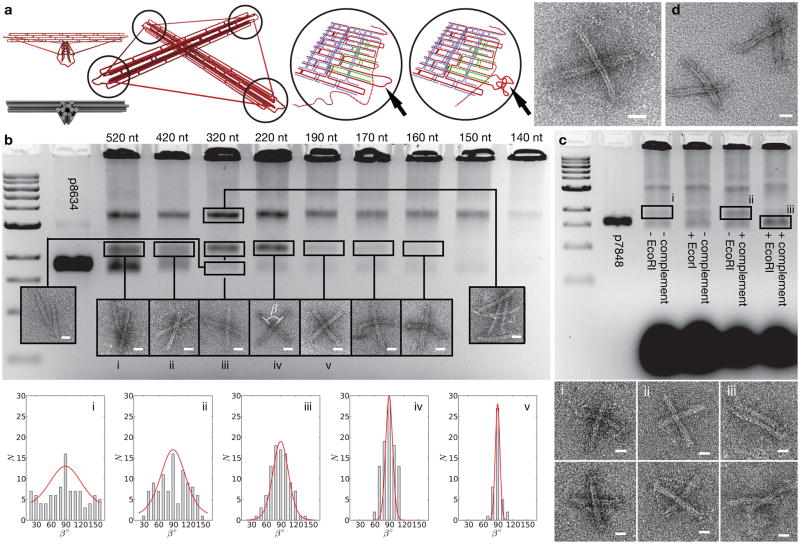
Figure 2. DNA tensegrity-structure kite.
a, 3D scaffold path representation for the prestressed tensegrity kite and transmission electron micrograph of assembled object with 220 nt-long springs. Changes of the spring lengths of a DNA tensegrity structure can be achieved by spooling of the scaffold DNA. Staple strands that have to be exchanged to achieve a shortening of a spring are depicted in green. b, Gel and TEM analysis of kites equipped with varying spring lengths between the ends of the struts. Left to right: 2-Log DNA ladder, p8634 scaffold, 520 nt, 420 nt, 320 nt, 220 nt, 190 nt, 170 nt, 160 nt, 150 nt, 140 nt. The bands containing the various objects were extracted and investigated with TEM. Below: Histograms of observed angles β between the struts. c, A 12-helix–bundle kite with four springs of equal length (300 nt) was designed such that the restriction site for EcoRI is located in one of the 300-nt–spring regions. Gel and TEM analysis shows that the enzymes only cut the springs when the complementary sequence to the restriction site is present during folding. Species cut out of gel and analyzed by TEM are indicated as i, ii, iii. d, TEM image of kites with one short spring (140 nt) and three longer springs (320 nt). All scale bars: 20 nm.
Sample analysis by gel electrophoresis after annealing resulted in a series of bands (Fig. 2b). The second fastest band contained structures with the desired kite configuration, where shorter spring lengths lead to tighter distributions of crossing angles β (Fig. 2b, lower panel). The red lines overlaying the histograms represent the equilibrium angle distributions at room temperature calculated from the mFJC model. (for a WLC model fit, see Fig. S3) The next slower migrating band was composed of dimers, i.e. two sets of staple strands sharing two scaffold strands. With spring lengths that generate forces higher than 14 pN (160 nt), no desired kite structures formed, indicating that a twelve-helix bundle can fold into its full length against forces up to 14 pN acting on each end. However, the yield of correctly folded objects for this particular geometry was highest (~15% efficiency) at forces between 4 and 7 pN (320 nt and 220 nt, respectively).
In all lanes, we detected a faint fast moving lower most band that contained objects with a parallel arrangement of the two struts when visualized using TEM. We attribute these defective particles to the occasional rupturing of the scaffold strand in one of the four springs. We also were able to mechanically actuate this structural transition between a square-kite configuration to the parallel conformation in a controlled manner, and with high efficiency, by clipping specific sites in the springs with a restriction endonuclease. After clipping, the band of the kite in square-arrangement disappears and the band of the struts arranged in parallel gains intensity (Fig. 2c).
To demonstrate the versatility of prestressed DNA tensegrity structures, a geometrical variant of the kite was designed and nanofabricated through self-assembly: one of the four springs was fixed to a length of 140 nt while the remaining three springs were set to a length of 320 nt (Fig. 2d). Interestingly, when we engineered similar structures using thinner, 128-nm long, six-helix bundles [19] as compression-resistant elements, we observed buckling of these struts (Fig. 3, Supp. Figs S5). Using shorter 42-nt–long six-helix bundle segments introduced into each spring, the ssDNA length between the ends of a six-helix–bundle kite structure was fixed to 486 bases. After thermal annealing, gel purification, and TEM imaging, most of these objects displayed two bent six-helix bundles, which can be explained as follows: the critical force Fc at which a strut of length L buckles can be expressed as Fc = π2·P·kBT/L2. If we assume a persistence length of P = 1.6 ± 0.6 μm for our gel-purified six-helix-bundles (c.f. next paragraph and Supp. Fig. S5), Fc = 3.9±1.5 pN. Since the expected force exerted by the ssDNA on the straight struts of 4.7±1 pN is above this critical force, the buckling of the 6-helix bundles is not surprising. The mean distance d between ends of neighboring struts on the TEM grid was 72±9 nm, which corresponds to an average force of 2.5±0.5 pN along each spring. These forces sum up to a resulting force of 3.5±1 pN along the axis of each strut, which in turn coincides within the error margins with the predicted critical buckling force of 3.9±1.5 pN. The measured distances and the resulting forces are estimated under the simplifying assumption, that our DNA objects adhere to the grids without further equilibrating movements during drying and staining and that the restoring force of a buckling strut is constant. Thereby, we are ignoring effects of a potentially damaged or distorted cross section of the strut.
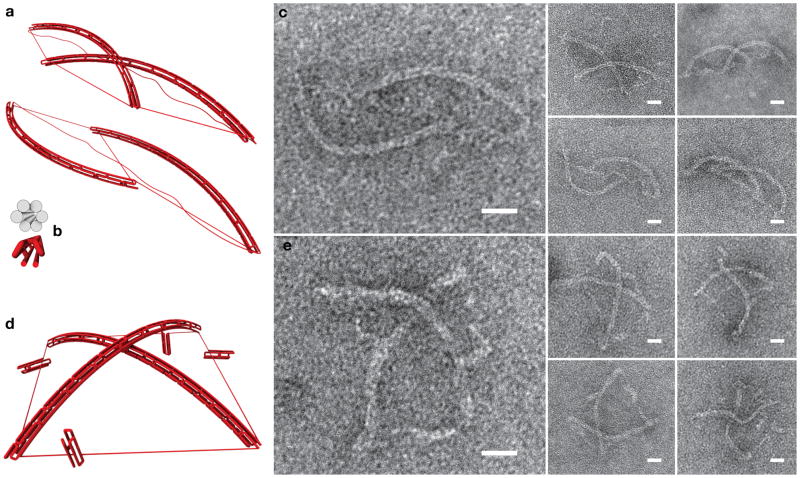
Figure 3. Force generation with DNA tensegrity objects.
Six-helix bundles under compression buckle if the compressive force exceeds the critical Euler force Fc. a, Models of distorted tensegrity kites with 128 nm long six-helix-bundle struts, three short ssDNA springs that are 273-nt long, and a long fourth spring that is 2207-nt long. b, Profile views of the design of the struts, cylinder model and scaffold path representation. c, TEM micrographs of gel purified objects of the type shown in (a). d, Model of a buckling kite with four springs of equal length (486 nt each, length adjusted with four six-helix-bundle clamps). e, TEM micrographs of gel purified buckling kites. By measuring the end-to-end distance, c, of multiple six-helix bundles like in (c) and (e), the resultant force acting on the six-helix bundle ends, Fs, could be estimated to be 4 pN. Equating the obtained value with Fc leads to an estimated persistence length P of the six-helix bundle of 1.4 μm. This is in good agreement with a P-value obtained for gel-purified, ethidium-bromide-soaked six-helix bundles by independent measurements (cf. Fig. S7). All scale bars: 20 nm.
We also determined that our 6-helix bundles have a persistence length of 2.4±0.6 μm, which agrees well with the theoretically expected value of 2.7 μm (Supp. Figs. S6, S7). For 6-helix bundles purified from an agarose gel, the measured persistence length drops to 1.6±0.6 μm, most likely due to incorporation of ethidium bromide (Supp. Fig. S8) and mechanical damage during gel electrophoresis and extraction (Supp. Figs. S8). Consistent with the observation that gel purification lowers the persistence length, we found that unpurified 6-helix-bundle kites exhibit straight struts, and therefore display an average distance of 89±3 nm between the neighboring strut ends (Supp. Fig. S4). Hence, unpurified, 128-nm, 6-helix bundles that experience a compression force of 4.7±1 pN do not buckle. This observation is consistent with theory, since > 6 pN force is required to buckle unpurified 6-helix bundles of this length, with a persistence length of 2.4 μm. Thus our results are roughly consistent with the mFJC model for ssDNA and a compressive Young’s modulus for dsDNA of 3 · 108 Pa [20, 21]. In the future, more complex models (e.g. one that considers the ionic strength of the solution) might yield a more complete picture and will have to be used in the low-force regime (< 2 pN) to obtain accurate predictions [22]. However, the active bending of DNA origami objects provides a visual demonstration of our ability to generate controllable forces during the process of DNA self-assembly. By applying known nanoscale forces via this structural approach, it might be possible to quantitatively probe and analyze biophysical mechanisms at the molecular level in the future.
Application of ssDNA as spring elements in DNA structures could give rise to the fabrication of flexible and mechanically responsive DNA structures spanning great areas or volumes while using relatively small amounts of material. Current efforts to use dsDNA as a scaffold source [23], and the potential of hierarchical assembly [12, 15], may facilitate the creation of such large biocompatible constructs. We have shown that the inclusion of specific cleavage sequences in the DNA sequence enables ligand-inducible mechanical actuation. In combination with the coupling of aptamer-based enzymatic activity into the DNA tensegrity structures, this could provide a mechanism to generate mechanochemical conversion mechanisms similar to those observed in living cells [8] and that might be useful in origin of life studies to model hierarchical, self assembling, tensegrity structures that have been proposed to contribute to the emergence of the first living cells [24]. These programmable biomimetic nanostructures could be employed as nanoscale solution-based force sensors [25] and as platforms for biophysical experiments where DNA of defined length under controlled tension is desirable. Furthermore, they could serve as artificial extracellular matrices with controllable mechanics or as cytoskeletal-network mimics that could aid in the study of fundamental cellular processes such as cellular mechanotransduction that involves stress-dependent control of molecular biochemistry and gene expression.
Methods
Folding and purification of Prestressed DNA Tensegrity structures
For the self-assembling process, a solution containing 10 nM scaffold strand, 50 nM of each staple strand (reverse-phase cartridge purified, Bioneer Inc.), 5 mM Tris + 1 mM EDTA (pH 7.9 at 20 °C), 16 mM MgCl2 was heated to 80 °C for 4 min, cooled down to 60°C over the course of 80 min, and cooled further down to 24 °C over the course of 72 h. The folded objects were electrophoresed on a 1.5% agarose gel containing 45 mM Tris borate + 1 mM EDTA and 11 mM MgCl2 at 70 V for 3 h. To prevent the origami structures from denaturation during electrophoresis, the gel-box was cooled in an ice-water bath. The gel-band containing the structures was physically extracted from the gel and run through spin columns (Freeze’n’Squeeze Spin Columns, Biorad) at 5000 rcf. The DNA tensegrity objects were then imaged with TEM after negative staining with uranyl formate on a FEI Tecnai T12 BioTWIN at 80 kV.
Ligand-induced mechanical actuation of the 12-helix-bundle kite
A 12-helix–bundle kite with four springs of equal length (300 nt) was designed such that the restriction site for EcoRI is located in one of the 300-nt–spring regions. The assembled kites with a double-stranded region at the EcorRI restriction site (GAATTC + 10 bases or more overhang on both sides of the sequence) were incubated for one hour with the enzymes (20 μl of 10 nM scaffold, 80 nM each staple, 1xNEB-buffer 2, 20 units of enzyme, New England Biolabs, Inc.). Uncut and cut tensegrity kites were electrophoresed on a 1.5% agarose gel. The gel-bands were physically extracted and imaged with TEM after negative staining with uranyl formate.
Supplementary Material
Acknowledgments
We thank Oskar Hallatschek, Richard Neher, Hendrik Dietz, and Shawn Douglas for helpful discussions and advice. This work was funded by the Wyss Institute for Biologically Inspired Engineering, Deutscher Akademischer Austauschdienst (DAAD) to T.L, Swedish Science Council (Vetenskapsrådet) Fellowship to B.H., and Claudia Adams Barr Program Investigator and NIH New Innovator (1DP2OD004641-01) grants to W.M.S.
Footnotes
Author contributions T.L., D.E.I, and W.M.S. conceived and designed the research, T.L. designed the DNA shapes, T.L., B.H. and J.T. performed the experiments, T.L. and B.H. analyzed the data, T.L., B.H., D.E.I., and W.M.S. co-wrote the paper.
Additional Information Supplementary information accompanies this paper at www.nature.com/naturenanotechnology. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Snelson K. Snelson on the tensegrity invention. Int J Space Struct. 1996;11:43–48. [Google Scholar]
- 2.Fuller B. Synergetics-Explorations in the Geometry of Thinking. I & II. New York: Macmillan Publishing Co; 1975, 1979. [Google Scholar]
- 3.Skelton RE, Adhikari R, Pinaud JP, Chan W. An Introduction to the Mechanics of Tensegrity Structures. IEEE Decis Contr P. 2001;5:4254–4259. [Google Scholar]
- 4.Sultan C, Corless M, Skelton RE. The Prestressabilty Problem of Tensegrity Structures: Some Analytical Solutions. Int J Solids Struct. 2001;38:5223–5252. [Google Scholar]
- 5.Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 6.Clemen AE, Vilfan M, Jaud J, Zhang J, Bärmann M, Rief M. Force-dependent stepping kinetics of myosin-V. Biophys J. 2005;88:4402–4410. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingber DE. The Architecture of Life. Sci Am. 1998;278:48–57. doi: 10.1038/scientificamerican0198-48. [DOI] [PubMed] [Google Scholar]
- 8.Ingber DE. Cellular Mechanotransduction: Putting all the Pieces together again. FASEB J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 9.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 10.Liu D, Wang MS, Deng ZX, Walulu R, Mao CD. Tensegrity: Construction of Rigid DNA Triangles with Flexible Four-Arm DNA Junctions. J Am Chem Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Su M, He Y, Zhao X, Fang PA, Ribbe AE, Jiang W, Mao CD. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc Natl Acad Sci. 2008;31:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao CD. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 13.Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, Ginell SL, Mao C, Seeman NC. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:287–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 15.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: The Elastic Response of Individual Double-Stranded and Single-Stranded DNA Molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 17.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Rapid prototyping of three-dimensional DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759. [Google Scholar]
- 19.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc Natl Acad Sci USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland: Sinauer Associates; 2001. [Google Scholar]
- 21.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 22.Saleh OA, McIntosh DB, Pincus P, Ribeck N. Nonlinear Low-Force Elasticity of Single-Stradned DNA Molecules. Phys Rev Let. 2009;102:068301-1-4. doi: 10.1103/PhysRevLett.102.068301. [DOI] [PubMed] [Google Scholar]
- 23.Högberg B, Liedl T, Shih WM. Folding DNA origami from a double-stranded source of scaffold. J Am Chem Soc. 2009;131:91544–94155. doi: 10.1021/ja902569x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingber DE. The origin of cellular life. Bioessays. 2000;22:1160–1167. doi: 10.1002/1521-1878(200012)22:12<1160::AID-BIES14>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, Liphardt J. Biocompatible Force Sensor with Optical Readout and Dimensions of 6 nm3. Nano Lett. 2005;5:1509–1514. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.