Abstract
The Notch pathway is one of the major signaling pathways required for proper development in metazoans. Notch activity is regulated at numerous levels, and increasing evidence reveals the importance of “protein glycosylation” (modification of Notch receptors with sugars) for its regulation. In this review we summarize the significance of the Notch pathway in development and the players responsible for its glycosylation, and then discuss the molecular mechanisms by which protein glycosylation may regulate Notch function.
Keywords: Protein glycosylation, Notch, Development, Pofut1/Ofut1, Fringe, Rumi
1. Introduction
Notch signaling is essential for proper development in metazoans, and defects in this pathway result in a number of human diseases [1,2]. Notch is regulated at numerous overlapping levels, including endocytosis, ubiquitination, intracellular trafficking, degradation, and glycosylation [2–6]. Many genes impinge on this pathway, and the number of these genes continues to increase with the improved techniques for genome-wide analysis [7]. This review focuses on regulation of the Notch pathway by glycosylation.
1.1. Role of Notch in development and disease
The Notch phenotype was originally described in Drosophila nearly 100 years ago as an X-linked, dominant mutation which showed irregular “notches” at the tips of the wings [8]. Subsequent work demonstrated that Notch plays key roles in development of many tissues in flies, including formation of neurons and glial cells, leg segments, eyes, heart, muscles, and blood lineages [2,9,10]. Drosophila has a single Notch receptor, while mammals have four [1]. Targeted disruption of the four mouse Notch genes demonstrated that these genes play important roles in development of many tissues. Loss of mouse Notch1 results in an embryonic lethal phenotype with severe defects in somitogenesis [11,12]. Subsequent studies showed that Notch1 is also involved in neurogenesis and vasculogenesis [13,14]. Deletion of mouse Notch2 also results in an embryonic lethal phenotype with apoptotic cell death in a wide variety of tissues, especially neural tissues, from embryonic day 9.5 [15]. Notch3−/− mice are viable and fertile, but have defects in arterial differentiation and maturation of vascular smooth muscle [16]. Although Notch4−/− mice are viable and fertile [14], loss of Notch4 exacerbates the vascular remodeling defects observed in Notch1−/− embryo [14], suggesting partially overlapping function of Notch1 and 4 during embryogenesis. Aberrant Notch signaling leads to multiple human disorders [1,17]. Mutations of Notch and the components of this pathway are implicated in human developmental disorders such as Alagille Syndrome and Spondylocostal Dysostosis, adult onset diseases such as CADASIL and Multiple Scleorosis, and cancers such as T cell acute lymphoblastic leukemia (T-ALL) and colon cancer.
1.2. Notch basics
Notch receptors are large type I transmembrane proteins [2]. Their basic molecular structure is evolutionarily conserved and consists of three domains: an extracellular domain (ECD) with 29–36 tandem epidermal growth factor-like (EGF) repeats and a unique negative regulatory region (NRR) which consists of three Lin-12/Notch repeats and a heterodimerization domain; a single transmembrane domain; and an intracellular domain with an RBP-Jκ (recombination signal sequence-binding protein-Jκ) association module domain, several nuclear localization sequences, seven ankyrin repeats, and a transactivation domain that harbors proline/glutamic acid/serine/threonine-rich motifs responsible for rapid degradation. The mature receptor is a heterodimer with the ECD tethered to the transmembrane/intracellular domain (T/ICD) through non-covalent, calcium dependent interactions. The heterodimer is formed by cleavage of the nascent polypeptide at site 1 by a furin-like protease in the Golgi [18,19].
Notch ligands are also type I transmembrane proteins with a similar overall architecture: an ECD containing an N-terminal DSL (Delta/Serrate/LAG-2) motif, specialized tandem EGF repeats termed the DOS (Delta and OSM-11-like proteins) domain, and several tandem EGF repeats; a single transmembrane domain; and a small intracellular domain [20]. Drosophila has two ligands, Delta and Serrate, while mammals have three Delta-like ligands (Dll1, 3, and 4) and two Serrate homologues (Jagged1 and 2).
Notch activation is initiated by ligand binding, and accomplished through a proteolytic mechanism [21]. The first cleavage occurs at site 2 (S2), just outside the membrane on the T/ICD, and is catalyzed by a metalloprotease of the ADAM family. In the absence of ligand, S2 appears to be covered by the NRR, sterically blocking access of the ADAM protease to the site. Ligand binding results in a conformational change in the NRR, exposing the site and allowing cleavage [22–24]. Subsequently, cleavage at site 3 (S3) in the Notch transmembrane domain by the γ-secretase complex results in the release of the Notch intracellular domain (NICD), and translocation of the NICD into the nucleus [25]. Interaction between NICD and DNA binding proteins such as RBP-Jκ, activate target gene transcription [26].
2. Regulation of Notch function with glycosylation
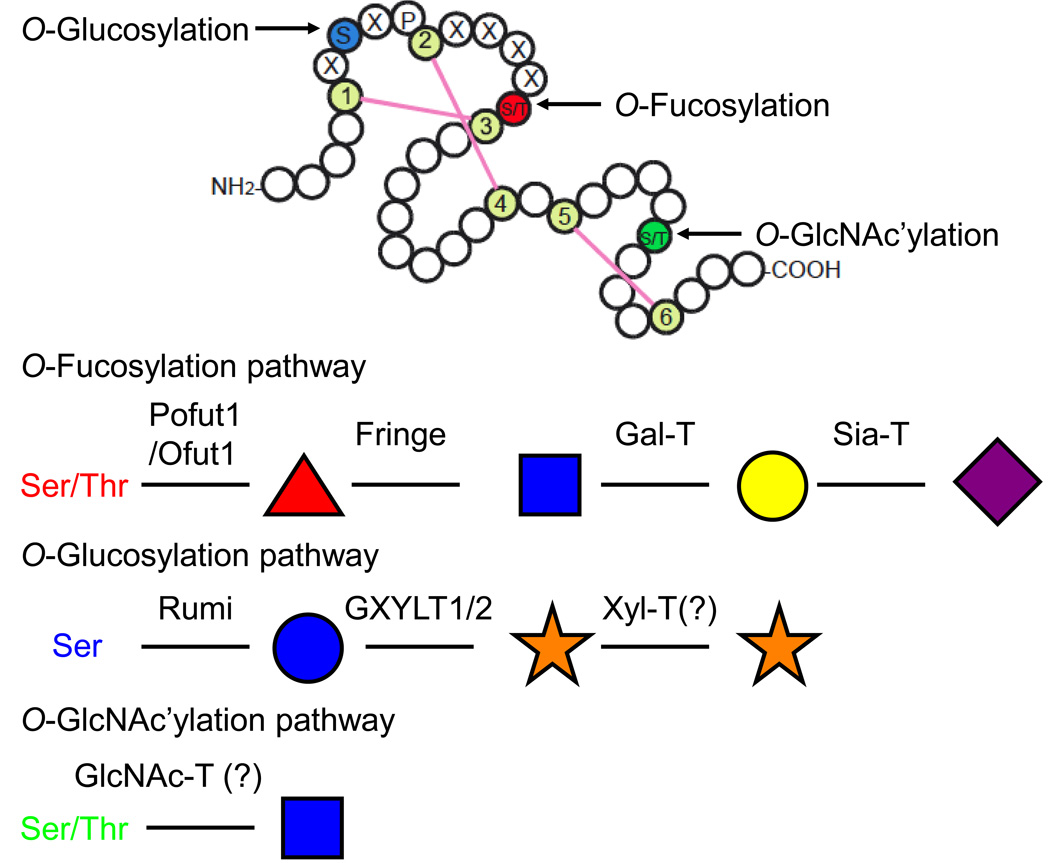
The discovery that Fringe, a known modulator of Notch activity, is a glycosyltransferase modifying O-fucose glycans on Notch EGF repeats [27,28], brought the study of Notch into the field of Glycobiology [29]. The EGF repeats of Notch are modified with three different types of O-linked glycosylation: O-fucosylation, O-glucosylation, and O-GlcNAc’ylation (Figure 1) [30–32]. Addition of O-fucose to Ser/Thr occurs within the consensus sequence C2-X-X-X-X-(S/T)-C3 (C, cysteine; X, any amino acid; S, serine; T, threonine) between the second and the third cysteines conserved in EGF repeats [33]. O-Fucose can be elongated by the addition of an N-acetylglucosamine (GlcNAc) [27,28,34]. Further elongation with a galactose and sialic acid occurs on mammalian Notch, but not in Drosophila (Figure 1) [30,35]. Notch ligands also have numerous EGF repeats in their ECDs which are modified with O-fucose glycans, but the functional significance of ligand O-fucosylation is unclear [36]. Similarly, addition of O-glucose occurs only at serine within the O-glucose consensus sequence C1-X-S-X-P-C2 (C, cysteine; X, any amino acid; S, serine; P, proline) between the first and the second cysteines conserved in EGF repeats [30,37]. O-glucose on the EGF repeats of mammalian Notch1 is elongated with two α1,3-linked xyloses [30,31], but our preliminary data suggest that O-glucose on Drosophila Notch may only be modified with a single xylose (Rana and Haltiwanger, unpublished observation). In contrast, O-GlcNAc seems to be a monosaccharide on the EGF repeats of Notch [32].
Figure 1. O-Glycosylation of EGF repeats.
Upper panel shows a single EGF repeat with the sites for addition of O-fucose, O-glucose, and O-GlcNAc. O-Fucose is attached to Ser/Thr in C2XXXX(S/T)C3 (red). O-Glucose is attached to Ser in C1XSXPC2 (blue). O-GlcNAc is attached to Ser/Thr between the fifth and sixth cysteines (green). Note that the consensus sequence of O-GlcNAc modification has not yet been proposed. Conserved cysteines are shown in light green. Disulfide bonds are shown by pink bars. Lower panel shows fully extended structures of O-fucose, O-glucose, and O-GlcNAc glycans and the glycosyltransferases responsible for their syntheses. Fucose (red triangle), GlcNAc (blue square), Galactose (yellow circle), Sialic acid (purple diamond), Glucose (blue circle), and Xylose (orange star). O-Fucose on Drosophila Notch has only been found as a disaccharide to date [35]. Xylosyltransferase(s) which adds a terminal xylose on O-glucose has not been cloned yet. GlcNAc-transferase(s) responsible for O-GlcNAc modfification of EGF repeats has not been cloned yet [32].
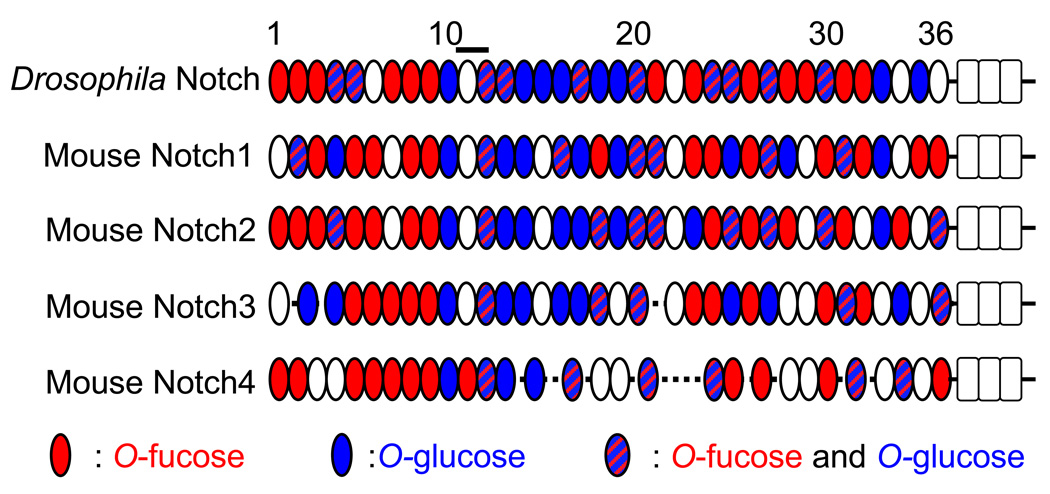
Most EGF repeats of Notch proteins contain consensus sequences for O-fucose and/or O-glucose (Figure 2). Mutations in Notch-related glycosyltransferase genes lead to aberrant Notch signaling, which clearly suggests that glycosylation is essential for Notch function. Although many other proteins bear, or are predicted to bear, these modifications [17,37], given the abundance of potential modification sites for these O-linked glycans on Notch and the biological importance of the Notch pathway, the functional significance of these modifications are predicted to be most evident in Notch. Although defects in the synthesis and transport of nucleotide sugars required for synthesis of the glycans found on Notch (e.g. GDP-fucose, UDP-GlcNAc, UDP-glucose, and UDP-xylose) also affect Notch signaling [27,29,38–43] (see reviews by Liu et al. and Freeze & Sharma in this issue), this review will focus on the Notch-related glycosyltransferases.
Figure 2. Potential O-fucose and O-glucose modification sites in the ECDs of Notch receptors.
This is an updated version of the previously reported figure in [30] with current consensus sequences for O-fucose and O-glucose. Drosophila Notch (Swiss Prot #P07207), mouse Notch1 (Q01705), mouse Notch2 (O35516), mouse Notch3 (Q61982), and mouse Notch4 (P31695) are aligned based on homology between EGF repeats. Red ovals show EGF repeats with the O-fucose consensus sequence. Blue ovals show EGF repeats with the O-glucose consensus sequence. Blue and red shaded ovals show EGF repeats with both O-fucose and O-glucose consensus sequences. Open rectangles show Lin-12/Notch repeats. A black bar shows an essential domain for ligand binding.
2.1. O-Fucosylation
Even though O-fucosylation plays important roles in Notch signaling, the molecular mechanisms of Notch pathway regulation by O-linked fucose and its extended form are not fully understood. In this section we will summarize our current knowledge obtained from genetic and biochemical studies on the components of O-fucosylation machinery in mice and Drosophila and then discuss the mechanism of Notch pathway regulation by O-fucosylation.
2.1.1. Glycosyltransferases
Pofut1/Ofut1
Protein O-fucosyltransferase-1 (Pofut1 in mammals and Ofut1 in Drosophila) transfers O-fucose to Ser/Thr in the O-fucose consensus sequence of the EGF repeats (Figure 1). Pofut1 was first identified through conventional molecular cloning after biochemical purification of the enzymatic activity [44]. Ofut1/Pofut1 is a soluble protein retained in the ER by virtue of a C-terminal KDEL-like ER-retention sequence [39,45], and it only adds fucose to properly folded EGF repeats [46]. Together with its ER localization, the ability to distinguish folded from unfolded EGF repeats has led to the hypothesis that Pofut1 may play a role in quality control [45].
Pofut1 knockout mice showed embryonic lethality with defects in somitogenesis, vasculogenesis, cardiogenesis, and neurogenesis [47]. The phenotype of Pofut1−/− mice was similar to that in mice in which all Notch signaling pathways are blocked by lacking core components of Notch signaling (e.g. presenilins 1 and 2 [48,49] or RBP-Jκ knockouts [50–52]) and was more severe than that of mice lacking individual Notch receptors, suggesting that Pofut1 is required for proper function of all mammalian Notch receptors [47]. Similarly, knocking down of Ofut1 by RNAi in Drosophila revealed a cell-autonomous requirement of Ofut1 for Notch function in many cellular contexts [53]. A mutation in Drosophila Ofut1, neurotic, was independently identified as a critical component for Notch activity [54]. Importantly, these studies consistently showed that Pofut1/Ofut1 is required for Notch signaling in the signal-receiving cells. A spontaneous mouse mutation called “compact axial skeleton” (cax) in mice was recently demonstrated to be a hypomorphic Pofut1 allele that reduces its transcription and leads to decreased Notch signaling. cax mutant embryos show defective anterior-posterior somite patterning and axial skeleton development with virtually no defects in other Notch-regulated developmental processes, suggesting that the levels of Pofut1 required for proper Notch signaling depend on the cellular context, and that the somite patterning is highly sensitive to reduced Pofut1 levels [55].
Due to the essential nature of Pofut1 for function of all Notch receptors, several groups have taken advantage of knocking out Pofut1 in a tissue specific fashion to evaluate the significance of Notch signaling in specific contexts. Using conditional deletion of Pofut1 in mice, Pofut1 was shown to be dispensable for early cell fate specifications or for formation of the three germ layers [56], but indispensable for maintenance of enteric neural crest cells [57]. Conditional deletion of Pofut1 in the endoderm resulted in a lung phenotype similar to that seen in the absence of secretory Clara cells [58]. These mutants also showed airways populated by ciliated cells with an increase in neuroendocrine cells [58]. Intestine-specific deletion of Pofut1 resulted in a large increase in all intestinal secretory cell lineages, accompanied by alteration of the mucus-associated flora, resulting in enterocolitis [59]. Together, these studies suggest significance of Notch signaling in adult tissues as well as during development.
Fringe
Fringe was first described as a novel secreted protein required for wing formation in Drosophila [60]. Panin and Irvine identified an elegant mechanism for restriction of Notch activation to the dorsal/ventral boundary in the wing disc by demonstrating that Fringe makes Notch more sensitive to Delta and less sensitive to Serrate [61]. This Fringe effect is explained well by the fact that addition of GlcNAc to O-fucose by Fringe enhances Delta binding to Notch and decreases Serrate binding to Notch [35].
Mammals have three Fringes: Lunatic, Manic, and Radical [62,63]. Elimination of Lunatic Fringe in mice results in defects in somitogenesis, a Notch1-dependent event [64–66]. Consistent with this phenotype, a homozygous mutation in the Lunatic fringe gene was reported in a patient with Spondylocostal Dysostosis type 3, resulting in severe defects in vertebral segmentation [67]. Lunatic fringe plays an essential role in regulation of somitogenesis, and has been proposed to be a part of the “segmentation clock” [68]. Individual elimination of Radical or Manic fringe in mice did not show any obvious developmental phenotype [66,69].
A number of recent studies suggest more subtle roles for Fringes in regulating Notch in tissue-specific contexts. Lunatic fringe functions to enhance Notch signaling in myofibroblast precursor cells and is needed to coordinate differentiation and mobilization of myofibroblasts required for alveolar separation [70]. Similarly, Lunatic fringe inhibits angiogenic sprouting by modifying Notch activation in the retinal epithelium [71]. Distinct functions of all three fringes have been shown in bile duct growth and remodeling after birth [72]. The significance of Notch signaling has been well studied in the development of the immune system [73–75]. Lunatic and Manic fringe are involved in T cell development through regulating interactions between Notch1 on T cell progenitors and Dll4 on thymic epithelial cells [75]. Both fringes are also involved in B cell development in the spleen [75]. They modify Notch2 in marginal zone B cell progenitors, thereby enhancing the interaction with Dll1 on red pulp endothelial cells within the marginal zone [75]. The fact that Lunatic and Manic Fringe double mutations show a much more severe defect in the process of marginal zone B cell generation provides a specific function of Manic fringe [69,75]. Thus, Fringe plays roles in modulating Notch function in a wide variety of contexts.
Galactosyltransferase and Sialyltransferase
Fully extended O-fucose glycans on Notch differ between Drosophila and mammals [30,35]. O-Fucose on Notch produced in Drosophila S2 cells is elongated with a GlcNAc, while the O-fucose disaccharide is further elongated with a galactose and a sialic acid in mammals, forming a tetrasaccharide. A novel O-fucose trisaccharide (GlcA-β1,4(GlcNAc-β1,3)-Fuc) has been reported in total extracts of Drosophila embryos, suggesting that a different type of elongation may occur on O-fucose in flies, but this trisaccharide has not yet been detected on Notch [76].
Studies in Chinese hamster ovary (CHO) cells defective in addition of galactose revealed that the minimum structure of O-fucose glycans that exhibits a Fringe effect on Jagged1-induced Notch signaling is the trisaccharide, Galβ1-4GlcNAcβ1-3Fuc, and that out of six known β4-Galactosyltransferases in CHO cells, β4-Galactosyltransferase-1 is required for addition of Gal to the disaccharide, GlcNAcβ1-3Fuc [77]. Subtle defects in Notch signaling were observed during somitogenesis in embryos lacking the gene encoding β4-Galactosyltransferase-1, consistent with a role for the trisaccharide in Fringe-mediated modulation of Notch signaling [78].
The sialic acid can be linked either α2,3 or α2,6 to the galactose, and can be added by the corresponding sialyltransferases [30,77,79]. Studies using Lec2 cells with defects in addition of sialic acids (mutation in the transporter for CMP-sialic acids) demonstrated that the sialic acid is not essential for Fringe to inhibit Jagged1-dependent Notch activation, suggesting that the sialic acid is not necessary for Fringe to modulate Notch [77].
2.1.2. Molecular mechanisms for effects of O-fucosylation on Notch signaling
Models for O-fucosylation
Extensive research conducted by several groups has resulted in several models describing how O-fucosylation regulates Notch activation. Three major effects of O-fucose have been proposed: 1. Ofut1 is a chaperone required for proper Notch folding and O-fucose is required as a substrate for Fringe, 2. O-fucose is essential for ligand binding/Notch function, and 3. Ofut1 is required for proper Notch localization. Not all of these functions are seen in all contexts, suggesting that some may be cell or species specific. Data supporting each model is summarized below.
Data supporting an ER chaperone activity for Ofut1/Pofut1 comes mainly from studies in Drosophila [39,41,80]. An early observation in flies lacking Ofut1 was the reduction in cell surface expression of Notch [39,53]. Okajima and Irvine then demonstrated that cell surface expression could be rescued by overexpression of an enzymatically inactive form of Ofut1 (Ofut1R245A) suggesting that O-fucosylation is not necessary for proper folding and cell surface expression. This mutant form of Ofut1 also rescued the Ofut1 null Notch neurogenic phenotype [39]. The phenotype of embryos lacking endogenous Ofut1 but overexpressing Ofut1R245A from a genomic transgene was similar with that of Fringe mutants, suggesting that the major function of the O-fucose is to serve as a substrate for Fringe. This conclusion was supported by studies with gmd mutants, which lack GDP-fucose (and thus all fucosylation) [39,41]. Gmd mutants do not show a neurogenic Notch-phenotype but do show a Fringe phenotype. Functional Notch was detected on the cell surface in these mutants. Decreased cell-surface Notch has also been observed in somites in Pofut1−/− mice, providing further support for this view [81]. These results suggest that O-fucose may only be required for Notch signaling events that are regulated by Fringe and that Ofut/Pofut1 has a separate chaperone-like activity.
Data supporting a direct role for O-fucose in ligand binding comes mainly from in vitro studies using mammalian cells. Although cell surface expression of Notch is unaffected in CHO cells lacking GDP-fucose (Lec13), both ligand binding and Notch activation is reduced, suggesting the importance of O-fucose for Notch function [27,77,82]. Similarly, embryonic stem cells lacking Pofut1 show cell surface expression of Notch proteins at similar levels with wild type [82]. However, ligand binding and activation of Notch was severely compromised in these cells [82]. Interestingly, Notch activity was partially restored by overexpression of an enzymatically inactive Pofut1 (equivalent to the R245A mutant of Ofut1) as well as an unrelated ER protein, α-glucosidase I [82]. These results suggest that O-fucose is required for optimal ligand binding and Notch activation, and that the chaperone activity is not specific for Pofut1, but that overexpression of other ER proteins has similar effects.
Finally, several reports support a role for Ofut1 in transport and localization of Notch in Drosophila. As mentioned above, loss of Ofut1 in flies results in decreased cell surface expression of Notch [39,53]. Matsuno and coworkers showed that Ofut1 interacts with the Notch ECD and is required for the constitutive endocytic trafficking of Notch from the plasma membrane to the early endosome independent of its O-fucosyltransferase activity [80]. They also found Ofut1 promoted turnover of Notch, thereby downregulating Notch signaling [80]. They performed further dissection of roles of Ofut1 for trafficking/localization of Notch in Drosophila wing discs epithelial cells [83]. In their analyses, Notch was delivered to the apical plasma membrane and adherence junctions independently of Ofut1. However, transcytosis (re-localization step of Notch from the apical region of the plasma membrane to subapical complex and adherence junctions) depended on O-fucosylation of Notch by Ofut1. These results suggest a role for Ofut1 in subcellular trafficking of Notch.
Additional work needs to be done to resolve what appear to be inconsistencies between these models. For instance, much of the data supporting the chaperone effect of Ofut1/Pofut1 relies on overexpression data. Further work needs to be done to resolve whether the chaperone effect is specific for Ofut1/Potut1 or just a general chaperone effect of overexpressing proteins in the ER. Similarly, cell-type or species dependent differences could be due to differences in expression patterns of chaperones in the ER of individual cells. Finally, little or no biochemical evidence for how these manipulations (overexpression or deletion of Ofut1/Pofut1) affect the carbohydrate modifications on Notch exists.
Models for how elongation of O-fucose by Fringe affects Notch activity
In vitro reconstitution studies using purified components of Drosophila Notch signaling showed that the addition of GlcNAc on O-fucose is sufficient to enhance Notch binding to the Delta ligand and to inhibit Notch binding to the Serrate ligand [35]. Further addition of a galactose did not affect Notch-ligand binding detectably in vitro. Thus, the effect of Fringe on Notch-ligand binding is solely explained by addition of GlcNAc in the fly system [35]. The data obtained in mammalian system suggests a more complex situation partly due to the increased number of Notch receptors, ligands, and Fringes [5]. Many groups have shown that Fringe modifications alter binding between mammalian Notches and ligands [28,82], but exceptions exist. For example, the Weinmaster group showed that Lunatic fringe does not appear to inhibit binding of Jagged1 to Notch1, even though it inhibits Jagged1-mediated Notch1 signaling in cell-based assays [84,85]. Therefore, some details are still unclear for how elongation of O-fucose by Fringe affects the interaction between Notch and its ligands in mammals.
Molecular details for how Fringe-mediated elongation of O-fucose alters Notch-ligand interactions are still sparse. Fringe modifies O-fucose on many EGF repeats of Notch [35,86], but it is not clear whether all of these sites participate in effects on Notch-ligand interactions. Using deletion mutants of Drosophila Notch, EGF repeats 11–12 were shown to be necessary and sufficient for ligand binding [87]. Consistent with this notion, the Hanford group showed that a fragment of human Notch1 containing just EGF repeats 11–13 expressed in bacteria is capable of physical interaction with DSL domains of ligands in vitro [88,89]. Interestingly, they also showed that addition of EGF repeat 10 modulates ligand binding [90]. The Irvine group showed that ligand binding is stronger when larger portions of the Notch ECD are used [91]. Thus, EGF repeats 11–12 may be the essential core for ligand binding, but many other EGF repeats may regulate ligand binding. The O-fucose at the EGF repeat 12 is elongated by Fringe on both mouse Notch1 and Drosophila Notch [35,86]. Mutation of the O-fucose site of EGF12 decreases mouse Notch1 activity in both cell-based assays [92] and in mice [93]. Elimination of the O-fucose site in EGF repeat 12 of Drosophila Notch led to a hyperactive response to Serrate even in the presence of Fringe in overexpression studies, but did not affect the response to Delta [94]. Recent modeling studies based on three dimensional structures of EGF repeats 11–13 of human Notch1 and the DSL-region of human Jagged1 showed that the O-fucose modification on EGF repeat 12 faces away from Jagged1, suggesting that the effects of Fringe elongation at this site may be indirect [89]. Mutations of O-fucose sites outside of the ligand binding region (e.g. on EGF repeats 26 and 27, also Fringe targets) also affect Notch1 activation in cell-based assays, suggesting that Fringe-modification of additional regions of the Notch ECD also participate in the effects of Fringe [92]. Some regions of the Notch ECD are more flexible than others [88,91]. Thus, Fringe modification may exert their effects on Notch-ligand interactions by altering the overall structure of the Notch ECD [91,92].
2.2. O-Glucosylation
Our understanding of the biological importance of O-glucosylation has lagged behind that of the O-fucosylation. O-Glucose glycans were initially reported on several blood coagulation factors [95–97] and more recently on Notch [30]. The recent identification of Rumi as a protein O-glucosyltransferase led to the demonstration that O-glucose modifications are also essential for Notch function [98].
2.2.1. Glycosyltransferases
Rumi (Protein O-glucosyltransferase: Poglut)
The gene encoding the enzyme responsible for addition of O-glucose to EGF repeats (protein O-glucosyltransferase, Poglut, Figure 1) was identified in a mutant screen to identify novel genes that affect adult bristle development (a Notch dependent process) in Drosophila [98]. One of the complementation groups, called rumi, showed severe defects in formation of bristle in clones that had been raised at 25°C, but not at 18°C. The gene responsible for this temperature-sensitivity encoded a protein with a predicted signal peptide, a CAP10 domain, and a C-terminal KDEL ER-recycling signal. CAP proteins are involved in the formation of a capsule consisting of sugar polymers in Cryptococcus neoformans, suggesting that rumi may encode a glycosyltransferase [99,100]. Indeed, Rumi showed the Poglut activity in vitro. These results demonstrated that Rumi is a Poglut, and that mutations in rumi result in temperature sensitive, Notch-like phenotypes in flies.
Xylosyltransferases
Xylosyltransferases elongate O-glucose to the mature trisaccharide (Figure 1). Two distinct mammalian genes (GXYLT1 and GXYLT2) encoding the first xylosyltransferase were recently identified based on homology with UDP-glucose: glycoprotein α3-glucosyltransferase [101]. Both catalyze addition of an α1,3-linked xylose to O-glucose, but not to xylose. The second xylosyltransferase has not yet been identified. The functional importance of elongation with xyloses on O-glucose of Notch remains to be elucidated.
2.2.2. Molecular mechanisms for effects of O-glucosylation on Notch signaling
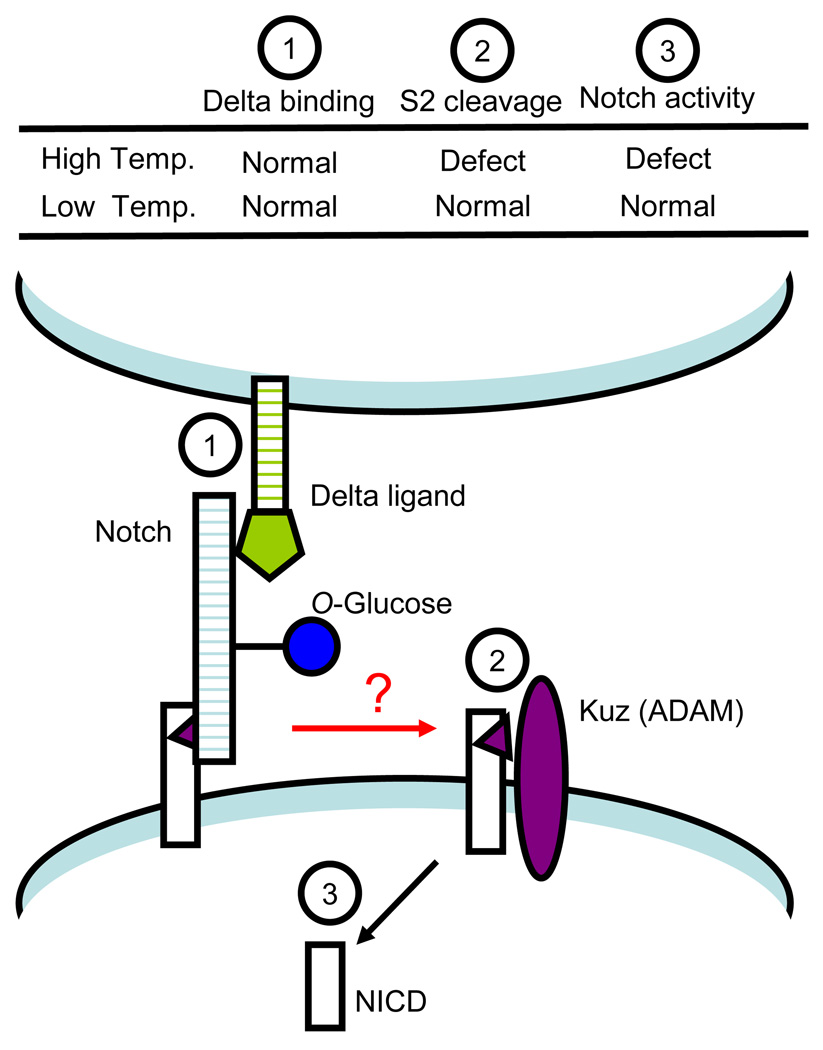
Little is known about the molecular mechanisms by which Rumi affects Notch activity. Although rumi mutants show accumulation of Notch inside cells, Notch also accumulates on the cell surface [98], suggesting an effect of Rumi on trafficking and/or stability of Notch. However, the presence of Notch on the surface of the rumi mutant cells suggests that there is no defect in the cell surface expression of Notch at the restrictive temperature [98]. The rumi79 allele results from a single point mutation (G189E), resulting in a temperature-sensitive loss of Notch activity similar to that seen in rumi alleles where Rumi expression is totally lost. Rumi-G189E is expressed at normal levels in rumi79 flies, suggesting that G189E mutation does not impair Rumi expression or stability [98], but Rumi-G189E has no Poglut activity [98]. The data indicate that addition of O-glucose is essential for Notch activity at high temperatures, and that preventing the addition of O-glucose to Notch results in temperature sensitive Notch phenotypes. These data do not support a chaperone-like role for Rumi, unlike that reported for Ofut1. Since temperature sensitivity is generally associated with changes in the protein structure, the O-glucose glycans may hold the ECD of Notch in a stable conformation essential for proper function at higher temperature (Figure 3). Preliminary data suggests that reduction in O-glucose on Notch does not affect binding to Delta in cell-based assays but does affect S2-mediated cleavage of Notch in Drosophila wing discs [98]. Thus, the O-glucose may function in maintaining a proper conformation of Notch to allow ligand binding to lead to proteolysis (Figure 3).
Figure 3. S2 cleavage defect in Drosophila rumi mutants.
rumi shows a temperature-sensitive defect in Notch signaling. Preliminary data suggests that O-glucose does not affect cell-surface presentation of Notch or ligand binding, but does affect the S2 cleavage of Notch at high temperatures [98]. Thus, O-glucose may function to hold the Notch ECD in a conformation required to link ligand binding to the conformational changes necessary for S2 cleavage.
Much work remains to be done on the function of O-glucosylation. A mammalian Rumi homologue has been identified as an active Poglut (Takeuchi, Fernandez-Valdivia, Jafar-Nejad, and Haltiwanger, manuscript in preparation). The O-glucose consensus sites are well conserved in all four Notch receptors in mammals (Figure 2) [4]. Future studies will focus on whether and how mammalian Rumi regulates Notch signaling through modifying all four Notch receptors.
2.3. Other types of glycosylation of Notch
Notch proteins contain multiple consensus sequences for N-linked glycans, but studies in CHO cell glycosylation mutants suggest that alterations in N-glycans have no effect on Notch activity [27,77]. Drosophila Notch was recently reported to be modified with O-GlcNAc on several EGF repeats [32]. O-GlcNAc has previously only been found on nuclear and cytoplasmic proteins [102]. Comparison of the sites of O-GlcNAc modification revealed that the Ser/Thr modified with O-GlcNAc is located between the fifth and sixth cysteines in the EGF repeats (Figure 1). It will be interesting to see how the O-GlcNAc modification affects Notch function.
3. Conclusions
Evidence for the importance of carbohydrate modifications on Notch for signaling is largely based on genetic studies. While unidentified sugars may yet exist on Notch, most of the genes encoding the enzymes responsible for the synthesis of the known structures have been identified. The potential sites for O-fucosylation and O-glucosylation on the ECD of Notch are well conserved among species (Figure 2), suggesting a distinct pattern of each modification on the entire ECD of Notch. Such conservation suggests that this pattern of modifications will play an important role in Notch function.
A full understanding of how these carbohydrates affect Notch function requires structural analysis. Methods need to be developed to examine how these carbohydrate structures change temporally and spatially in various tissues and throughout development. This is one of the ultimate goals of Glycobiology. Although initial models of Notch-ligand interactions exist [89], these involve small regions of the Notch ECD. Future studies need to examine the structure of the entire ECD and determine how carbohydrate modifications alter this structure. Finally, a great deal is still unknown regarding the specific details of how these sugar modifications affect specificity between the four mammalian Notch isoforms and five ligands. For a better understanding of how the sugars affect Notch function such as ligand binding and the subsequent activation, a great deal of rigorous biochemical and structural analysis is required.
Acknowledgement
We would like to thank Dr. Kelly Ten Hagen for giving us an opportunity to write this manuscript, and Drs. Bernadette C. Holdener, Hamed Jafar-Nejad and Haltiwanger lab members for helpful comments. Primary work was supported by NIH grant GM061126 (to R.S.H.) and the research grant from Mizutani Foundation for Glycoscience (to H.T.).
Abbreviations
- Cax
compact axial skeleton
- CHO
Chinese hamster ovary
- CMP
cytidine monophosphate
- Dll
Delta-like ligand
- DSL
Delta/Serrate/LAG-2
- DOS
Delta and OSM-11-like proteins
- ECD
extracellular domain
- EGF
epidermal growth factor-like
- ER
endoplasmic reticulum
- Fuc
fucose
- Gal
galactose
- GDP
guanosine diphosphate
- Glc
glucose
- GlcA
glucuronic acid
- GlcNAc
N-acetylglucosamine
- GMD
GDP-mannose 4,6-dehydratase
- NICD
Notch intracellular domain
- NRR
negative regulatory region
- Pofut1
Protein O-fucosyltransferase-1
- Poglut
Protein O-glucosyltransferase
- RBP-Jκ
recombination signal sequence-binding protein-Jκ
- S2
site 2
- S3
site 3
- T-ALL
T cell acute lymphoblastic leukemia
- T/ICD
transmembrane/intracellular domain
- UDP
uridine diphosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: emerging roles for beta-arrestins in developmental signaling pathways. Dev Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haines N, Irvine KD. Glycosylation Regulates Notch Signaling. Nat. Rev. Mol. Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 5.Luther KB, Haltiwanger RS. Role of unusual O-glycans in intercellular signaling. Int J Biochem Cell Biol. 2009;41:1011–1024. doi: 10.1016/j.biocel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanley P. Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr OL. Character changes caused by mutation of an entire region of a chromosome in Drosophila. Genetics. 1919;4:275. doi: 10.1093/genetics/4.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 11.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 12.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 13.de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 14.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- 16.Krebs LT, Xue Y, Norton CR, Sundberg JP, Beatus P, Lendahl U, et al. Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis. 2003;37:139–143. doi: 10.1002/gene.10241. [DOI] [PubMed] [Google Scholar]
- 17.Rampal R, Luther KB, Haltiwanger RS. Notch signaling in normal and disease States: possible therapies related to glycosylation. Curr Mol Med. 2007;7:427–445. doi: 10.2174/156652407780831593. [DOI] [PubMed] [Google Scholar]
- 18.Lake RJ, Grimm LM, Veraksa A, Banos A, Artavanis-Tsakonas S. In vivo analysis of the Notch receptor S1 cleavage. PLoS One. 2009;4:e6728. doi: 10.1371/journal.pone.0006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon WR, Vardar-Ulu D, L'Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 22.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 23.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling--a structural and biochemical perspective. J Cell Sci. 2008;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, et al. A presenilin-1-dependent γ-secretase-like protease medaites release of Notch intracellular fragment. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 26.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, et al. Fringe is a Glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 28.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 29.Haltiwanger RS, Lowe JB. Role of Glycosylation in Development. Annu.Rev.Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 30.Moloney DJ, Shair L, Lu FM, Xia J, Locke R, Matta KL, et al. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on Epidermal Growth Factor-like modules. J Biol Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 31.Whitworth GE, Zandberg WF, Clark T, Vocadlo DJ. Mammalian Notch is modified by D-Xyl-{alpha}1-3-D-Xyl-{alpha}1-3-D-Glc-{beta}1-O-Ser: Implementation of a method to study O-glucosylation. Glycobiology. 2009 doi: 10.1093/glycob/cwp173. [DOI] [PubMed] [Google Scholar]
- 32.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, et al. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 33.Shao L, Haltiwanger RS. O-fucose modifications of epidermal growth factor-like repeats and thrombospondin type 1 repeats: unusual modifications in unusual places. Cell Mol Life Sci. 2003;60:241–250. doi: 10.1007/s000180300019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, et al. Lunatic Fringe, Manic Fringe, and Radical Fringe Recognize Similar Specificity Determinants in O-Fucosylated Epidermal Growth Factor-like Repeats. J Biol Chem. 2005;280:42454–42463. doi: 10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- 35.Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS, et al. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. J Biol Chem. 2007;282:35153–35162. doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- 36.Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. Notch ligands are substrates for EGF protein O-fucosyltransferase and Fringe. J Biol Chem. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- 37.Harris RJ, Spellman MW. O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology. 1993;3:219–224. doi: 10.1093/glycob/3.3.219. [DOI] [PubMed] [Google Scholar]
- 38.Smith PL, Myers JT, Rogers CE, Zhou L, Petryniak B, Becker DJ, et al. Conditional control of selectin ligand expression and global fucosylation events in mice with a targeted mutation at the FX locus. J Cell Biol. 2002;158:801–815. doi: 10.1083/jcb.200203125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307:1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Li LW, Yan Q, Petryniak B, Man Y, Su C, et al. Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood. 2008;112:308–319. doi: 10.1182/blood-2007-11-115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okajima T, Reddy B, Matsuda T, Irvine KD. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 2008;6:1. doi: 10.1186/1741-7007-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haltiwanger RS. Fucose is on the TRAIL of colon cancer. Gastroenterology. 2009;137:36–39. doi: 10.1053/j.gastro.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse C, Johnson S, Phillipson M, Zbytnuik L, Petri B, Kelly M, et al. Secretory Cell Hyperplasia and Defects in Notch Activity in a Mouse Model of Leukocyte Adhesion Deficiency Type II. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.10.049. (in press) [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, et al. Modification of Epidermal Growth Factor-like Repeats with O-Fucose: Molecular Cloning of a Novel GDP-Fucose: Protein O-Fucosyltransferase. J.Biol.Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 45.Luo Y, Haltiwanger RS. O-fucosylation of Notch occurs in the endoplasmic reticulum. J Biol Chem. 2005;280:11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Spellman MW. Purification and Characterization of a GDP-fucose: Polypeptide Fucosyltransferase from Chinese Hamster Ovary Cells. J.Biol.Chem. 1998;273:8112–8118. doi: 10.1074/jbc.273.14.8112. [DOI] [PubMed] [Google Scholar]
- 47.Shi S, Stanley P. Protein O-fucosyltransferase I is an essential component of Notch signaling pathways. Proc.Natl.Acad.Sci.USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donoviel DB, Hadjantonakis AK, Ikeda M, Zheng H, Hyslop PS, Bernstein A. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, et al. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc Natl Acad Sci U S A. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 51.De la Pompa J, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 52.Barrantes IB, Elia AJ, Wunsch K, Hrabe de Angelis MH, Mak TW, Rossant J, et al. Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- 53.Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893-–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 54.Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, et al. neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- 55.Schuster-Gossler K, Harris B, Johnson KR, Serth J, Gossler A. Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC Dev Biol. 2009;9:6. doi: 10.1186/1471-213X-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi S, Stahl M, Lu L, Stanley P. Canonical Notch signaling is dispensable for early cell fate specifications in mammals. Mol Cell Biol. 2005;25:9503–9508. doi: 10.1128/MCB.25.21.9503-9508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okamura Y, Saga Y. Notch signaling is required for the maintenance of enteric neural crest progenitors. Development. 2008;135:3555–3565. doi: 10.1242/dev.022319. [DOI] [PubMed] [Google Scholar]
- 58.Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guilmeau S, Flandez M, Bancroft L, Sellers RS, Tear B, Stanley P, et al. Intestinal Deletion of Pofut1 in the Mouse Inactivates Notch Signaling and Causes Enterocolitis. Gastroenterology. 2008;135:8499–8460. doi: 10.1053/j.gastro.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irvine KD, Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595-–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 61.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates notch ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 62.Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, et al. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nature Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- 63.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 64.Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- 65.Zhang N, Gridley T. Defects in somite formation in lunatic fringe-deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- 66.Zhang N, Norton CR, Gridley T. Segmentation defects of Notch pathway mutants and absence of a synergistic phenotype in lunatic fringe/radical fringe double mutant mice. Genesis. 2002;33:21–28. doi: 10.1002/gene.10081. [DOI] [PubMed] [Google Scholar]
- 67.Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet. 2006;78:28–37. doi: 10.1086/498879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aulehla A, Pourquie O. Oscillating signaling pathways during embryonic development. Curr Opin Cell Biol. 2008;20:632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Tan JB, Xu K, Cretegny K, Visan I, Yuan JS, Egan SE, et al. Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches. Immunity. 2009;30:254–263. doi: 10.1016/j.immuni.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 70.Xu K, Nieuwenhuis E, Cohen B, Wang W, Canty AJ, Danska J, et al. Lunatic Fringe-mediated Notch signaling is required for lung alveogenesis. Am J Physiol Lung Cell Mol Physiol. 2010;298:L45–L56. doi: 10.1152/ajplung.90550.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 72.Ryan MJ, Bales C, Nelson A, Gonzalez DM, Underkoffler L, Segalov M, et al. Bile duct proliferation in Jag1/fringe heterozygous mice identifies candidate modifiers of the Alagille syndrome hepatic phenotype. Hepatology. 2008;48:1989–1997. doi: 10.1002/hep.22538. [DOI] [PubMed] [Google Scholar]
- 73.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 74.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 75.Stanley P, Guidos CJ. Regulation of Notch signaling during T- and B-cell development by O-fucose glycans. Immunol Rev. 2009;230:201–215. doi: 10.1111/j.1600-065X.2009.00791.x. [DOI] [PubMed] [Google Scholar]
- 76.Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, et al. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem. 2008;283:30385–30400. doi: 10.1074/jbc.M804925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J, Moloney DJ, Stanley P. Fringe modulation of Jagged1-induced Notch signaling requires the action of β4Galactosyltransferase-1. Proc.Natl.Acad.Sci. USA. 2001;98:13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen J, Lu L, Shi S, Stanley P. Expression of Notch signaling pathway genes in mouse embryos lacking beta4galactosyltransferase-1. Gene Expr Patterns. 2006;6:376–382. doi: 10.1016/j.modgep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 79.Nishimura H, Takao T, Hase S, Shimonishi Y, Iwanaga S. Human factor IX has a tetrasaccharide O-glycosidically linked to serine 61 through the fucose residue. J.Biol.Chem. 1992;267:17520–17525. [PubMed] [Google Scholar]
- 80.Sasamura T, Ishikawa HO, Sasaki N, Higashi S, Kanai M, Nakao S, et al. The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of Notch in Drosophila. Development. 2007;134:1347–1356. doi: 10.1242/dev.02811. [DOI] [PubMed] [Google Scholar]
- 81.Okamura Y, Saga Y. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mech Dev. 2008;125:663–673. doi: 10.1016/j.mod.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 82.Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem. 2008;283:13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sasaki N, Sasamura T, Ishikawa HO, Kanai M, Ueda R, Saigo K, et al. Polarized exocytosis and transcytosis of Notch during its apical localization in Drosophila epithelial cells. Genes Cells. 2007;12:89–103. doi: 10.1111/j.1365-2443.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- 84.Hicks C, Johnston SH, DiSibio G, Collazo A, Vogt TF, Weinmaster G. Jagged1 and Delta1 mediated Notch signaling is differentially regulated by Lunatic Fringe. Nature Cell Biology. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 85.Yang LT, Nichols JT, Yao C, Manilay JO, Robey EA, Weinmaster G. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol Biol Cell. 2005;16:927–942. doi: 10.1091/mbc.E04-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shao L, Moloney DJ, Haltiwanger RS. Fringe Modifies O-Fucose on Mouse Notch1 at Epidermal Growth Factor-like Repeats within the Ligand-binding Site and the Abruptex Region. J Biol Chem. 2003;278:7775–7782. doi: 10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- 87.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF Repeats of Notch Mediate Interactions with Delta and Serrate: Implications for Notch as a Multifunctional Receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 88.Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, et al. Structural and functional properties of the human notch-1 ligand binding region. Structure (Camb) 2004;12:2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 89.Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cordle J, Redfield C, Stacey M, van der Merwe PA, Willis AC, Champion BR, et al. Localisation of the Delta-like-1 binding site in human notch-1 and its modulation by calcium affinity. J Biol Chem. 2008 doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- 91.Xu A, Lei L, Irvine KD. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J Biol Chem. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. [DOI] [PubMed] [Google Scholar]
- 92.Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. J Biol Chem. 2005;280:32133–32140. doi: 10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ge C, Stanley P. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci U S A. 2008;105:1539–1544. doi: 10.1073/pnas.0702846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lei L, Xu A, Panin VM, Irvine KD. An O-fucose site in the ligand binding domain inhibits Notch activation. Development. 2003;130:6411–6421. doi: 10.1242/dev.00883. [DOI] [PubMed] [Google Scholar]
- 95.Nishimura H, Kawabata S, Kisiel W, Hase S, Ikenaka T, Takao T, et al. Identification of a disaccharide (Xyl-Glc) and a trisaccharide (Xyl2-Glc) O-glycosidically linked to a serine residue in the first epidermal growth factor-like domain of human factors VII and IX and protein Z and bovine protein Z. J.Biol.Chem. 1989;264:20320–20325. [PubMed] [Google Scholar]
- 96.Hase S, Kawabata S, Nishimura H, Takeya H, Sueyoshi T, Miyata T, et al. A New Trisaccharide Sugar Chain Linked to a Serine Residue in Bovine Blood Coagulation Factors VII and IX. J.Biochem.(Tokyo) 1988;104:867–868. doi: 10.1093/oxfordjournals.jbchem.a122571. [DOI] [PubMed] [Google Scholar]
- 97.Hase S, Nishimura H, Kawabata S, Iwanaga S, Ikenaka T. The structure of (xylose)2glucose-O-serine 53 found in the first epidermal growth factor-like domain of bovine blood clotting factor IX. J.Biol.Chem. 1990;265:1858–1861. [PubMed] [Google Scholar]
- 98.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang YC, Kwon-Chung KJ. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J Bacteriol. 1999;181:5636–5643. doi: 10.1128/jb.181.18.5636-5643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okabayashi K, Hasegawa A, Watanabe T. Microreview: capsule-associated genes of Cryptococcus neoformans. Mycopathologia. 2007;163:1–8. doi: 10.1007/s11046-006-0083-0. [DOI] [PubMed] [Google Scholar]
- 101.Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch EGF repeats. J Biol Chem. 2010;285:1582–1586. doi: 10.1074/jbc.C109.065409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]