Abstract
Impaired glucose tolerance (IGT) which precedes overt type 2 diabetes (T2DM) for decades is associated with multiple metabolic alterations in insulin sensitive tissues. In an UPLC-qTOF-mass spectrometry-driven non-targeted metabonomics approach we investigated plasma as well as spot urine of 51 non-diabetic, overnight fasted individuals aiming to separate subjects with IGT from controls thereby identify pathways affected by the pre-diabetic metabolic state. We could clearly demonstrate that normal glucose tolerant (NGT) and IGT subjects clustered in two distinct groups independent of the investigated metabonome. These findings reflect considerable differences in individual metabolite fingerprints, both in plasma and urine. Pre-diabetes associated alterations in fatty acid-, tryptophan-, uric acid-, bile acid-, and lysophosphatidylcholine-metabolism, as well as the TCA cycle were identified. Of note, individuals with IGT also showed decreased levels of gut flora-associated metabolites namely hippuric acid, methylxanthine, methyluric acid, and 3-hydroxyhippuric acid. The findings of our non-targeted UPLC-qTOF-MS metabonomics analysis in plasma and spot urine of individuals with IGT vs NGT offers novel insights into the metabolic alterations occurring in the long, asymptomatic period preceding the manifestation of T2DM thereby giving prospects for new intervention targets.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-010-0203-1) contains supplementary material, which is available to authorized users.
Keywords: Metabolomics, UPLC-qTOF-MS, Diabetes, Pre-diabetes, Impaired glucose tolerance
Introduction
Type 2 diabetes (T2DM), called the burden of the 21st century, is growing with an epidemic rate (Alberti et al. 2007). The risk to develop this metabolic disease is reflected by impaired glucose tolerance (IGT) (Unwin et al. 2002), which is diagnosed based on the results of an oral glucose tolerance test (oGTT) according to the threshold criteria defined by the World Health Organization/the International Diabetes Federation, or the American Diabetes Association (Nathan et al. 2007; WHO 2006). Global estimates of IGT are not available, but the number of individuals with IGT is likely to be even greater than the number with overt diabetes (Yach and Alberti 2003). Independent of an elevated risk to develop T2DM affected individuals experience an increased risk of cardiovascular events and mortality (Festa et al. 2004; Novoa et al. 2005; Petersen and McGuire 2005; Sorkin et al. 2005; Waugh et al. 2007). To gain novel insights in the metabolic traits reflecting a high risk to develop T2DM is an important prerequisite for the understanding of the pathophysiology of the long asymptomatic period of this heterogeneous metabolic disease and for the development of new intervention strategies to retard or even prevent the manifestation of overt T2DM.
It is well known that the metabolites in plasma, urine and cerebrospinal fluid reflect both normal variation and the pathophysiological impact of diseases. These differences in metabolite patterns can give insight into underlying molecular mechanisms. The generation of a metabolic fingerprint in body fluids may be a powerful tool to compare metabolic physiological and/or pathophysiological changes at a given time point. Metabonomics, recently introduced as the non-targeted analysis of metabolites (Holmes et al. 2008b; Lindon et al. 2003; Nicholson and Lindon 2008; van der Greef et al. 2004), has been proven in several recent human studies to fulfill these prerequisite aiming to elucidate biomarkers of dietary and cultural influences (Lenz et al. 2004), Parkinson’s disease (Bogdanov et al. 2008), coronary heart disease (Brindle et al. 2002), bowel disease (Marchesi et al. 2007), myocardial infarction (Lewis et al. 2008), kidney cancer (Kind et al. 2007), and prostate cancer (Sreekumar et al. 2009). Thus, metabonomics investigations of body fluids serve in two distinct but closely related modes: as a metabolic fingerprinting tool separating groups based on altered metabolic patterns and as a means to identify metabolites as potential biomarkers discriminating between normal and pathological states.
In the field of diabetes research recently a number of animal and a few human metabonomics studies had be published investigating the metabolic effects of an oral glucose challenge (Shaham et al. 2008; Wopereis et al. 2009; Zhao et al. 2009), insulin resistance (Chen et al. 2008; Plumb et al. 2006; Shaham et al. 2008; Shearer et al. 2008; Toye et al. 2007; Williams et al. 2006a, b), type 1 (Makinen et al. 2008; Zhang et al. 2008) or T2DM (Gipson et al. 2008; Huo et al. 2009; Salek et al. 2007; van Doorn et al. 2007; Zhang et al. 2009). However, currently only two studies investigated pre-diabetic metabolic pattern in humans (Shaham et al. 2008; Zhang et al. 2009). Shaham et al. investigated in detail the time-dependent response to a glucose challenge on distinct axes of insulin sensitivity by targeted metabolic profiling of 191 plasma metabolites (Shaham et al. 2008). In the study performed by Zhang et al. 1H-NMR was applied for non-targeted metabonomic analysis of serum samples from controls, pre-diabetic subjects with impaired glucose regulation and individuals with T2DM. A distinct clustering of controls and diabetic subjects was demonstrated, however the analysis of pre-diabetic individuals and controls resulted in no clear separation (Zhang et al. 2009).
Applying non-targeted metabonomics by UPLC-qTOF-MS we aimed to investigate metabolic fingerprints of pre-diabetic subjects vs normal glucose tolerant controls in plasma as well as spot urine. Based on the individual metabolic fingerprint, subjects with IGT were clearly separated from controls either in plasma or spot urine. We identified metabolites reflecting specific alterations of metabolic pathways. The detected pathway alterations offer new insights in the complex dysregulation of the metabolism in the pathogenesis of T2DM during the long asymptomatic period of IGT giving prospects for novel interventions targets like the gut flora or fatty acid metabolism.
Materials and methods
Human samples and oral glucose tolerance test
In total 51 subjects were included in the study group (age: 46.9 SE ± 11.9 years). All individuals underwent a 75 g oGTT according to the recommendations of the WHO/IDF to define normal (NGT) or IGT (WHO 2006). 39 were diagnosed to have NGT and 12 to have IGT. For the metabolomics investigations blood and spot urine samples were collected after an overnight fasting under standardized conditions and immediately stored in aliquots at −80°C. Analysis of routine laboratory parameters was performed as described recently (Schafer et al. 2007). The protocol of the study was approved by the Ethics Committee of the University Tuebingen conformed to the Declaration of Helsinki, and all subjects gave written informed consent. The investigation was conducted in accordance with the ethical principles of Good Clinical Practice.
Reversed-phase ultra performance liquid chromatography coupled to electrospray ionization quadrupole time of flight mass spectrometry (UPLC-qTOF-MS)
Plasma or urine sample aliquots were deproteinized with two volume parts of acetonitrile, centrifuged (13000 × rpm for 20 min), run to dryness in a vacuum centrifuge and stored at −20°C. For analysis, the plasma samples were reconstituted in 200 μl acetonitrile and water (4:1), and urine samples were reconstituted in 200 μl acetonitrile and water (1:4). The chromatographic separation was performed on a 100 × 2.1 mm ACQUITY 1.7 μm/C18 column at 35°C using an ACQUITY-UPLC system (Waters Corp, Milford, USA). We applied our recently reported UPLC-qTOF-MS approach, established and validated for the metabonomics analysis of serum and urine (Yin et al. 2008; Zhao et al. 2008). For plasma analysis the gradient program was 95% A (A = 0.1% formic acid in water) for 0.5 min, changed to 100% B (B = acetonitrile) linearly within 24 min and held for 4 min, finally back to 95% A (flow rate 0.35 ml/min). Urine samples were analyzed applying a gradient program starting at 98% A (A = 0.1% formic acid in water) for 0.5 min, changed to 70% B (B = acetonitrile) linearly within 25 min, then changed to 100% B and held for 3.5 min, finally back to 98% A (flow rate 0.35 ml/min). The UPLC system was coupled to a qTOF-MS (Micromass, Manchester, UK) equipped with an electrospray source operating in either positive or negative ion mode The metabolites were detected and identified following our recently published strategy for the identification of metabolite biomarkers (Chen et al. 2008).
Data pretreatment
For an efficient evaluation of the metabolic variability between NGT and IGT subjects, mass spectra were digitally analyzed using the Micromass MarkerLynx Applications Manager version 4.0 (Waters Ltd, Manchester, UK). The data were combined into a single matrix by aligning peaks with the same mass and retention time together from each data file in the data set. The intensity for each peak was normalized to the sum of the peak intensities for each data set. Metabolites which did not exist in 80% of NGT or IGT subjects were filtered. After that, the data were exported into Soft Independent Modelling of Class Analogy (SIMCA)-P (version 11.0, Umetrics AB, Umea, Sweden) for analysis and visualization by multivariate statistical methods.
Multivariate statistical analysis
The pre-processed UPLC-qTOF-MS data were exported into Soft Independent Modelling of Class Analogy (SIMCA)-P (version 11.0, Umetrics AB, Umea, Sweden) for analysis and visualization by multivariate statistical methods. After Pareto scaling and OSC-filtering according to Wold et al. (Wold et al. 1998), partial least squares-discriminant analysis (PLS-DA) was applied. The predictive ability of the model was assessed by internal validation using 7-fold cross-validation and response permutation testing. For the detection of metabolite ion masses with major influence on the group membership the S-plot was used according to (Wiklund et al. 2008). The metabolite heat map was generated using MultiExperiment View V4.1 (www.tm4.org). The Wilcoxon Rank Sum test for the comparison of clinical chemical and oGTT data of the IGT vs NGT individuals were computed using the statistical software packet JMP (SAS Institute, Inc., Cary, NC). Statistical significance was set at P < 0.05.
Results
Analysis of the individual metabolite fingerprint in fasting plasma and spot urine by UPLC-qTOF-MS in normal and impaired glucose tolerant subjects
Each subject (n = 51) was characterized by clinic check-up and the analysis of clinical chemical routine parameters in blood and urine to exclude subjects with disease of the cardiovascular system, the kidney or the liver, as well as severe inflammation (Table 1). Furthermore, an oGTT was performed to detect potential diabetic individuals and moreover to identify individuals with IGT. The comparison between 39 control and 12 IGT subjects showed, besides parameters of glucose metabolism, also a significant difference in the inflammatory marker C-reactive protein (CRP). This is not an unexpected finding since low-grade inflammation, i.e. within the reference interval for CRP, is well-known in the pathogenesis of pre-diabetes and diabetes (Cefalu 2009). No difference was detected in the parameters reflecting kidney function, namely creatinine and uric acid in plasma, total protein and albumin in urine (see Table 1), which is an important prerequisite for the non-targeted metabonomics analysis of urine and plasma since renal dysfunction may greatly influence the metabolite composition in these body fluids. The body mass index was also not significantly different between the NGT and the IGT group (30.76 ± 1.15 vs 34.60 ± 2.0; P = 0.1121).
Table 1.
Routine laboratory parameters of the study group
| Normal glucose tolerance (n = 39) mean ± SEM | Impaired glucose tolerance (n = 12) mean ± SEM | Reference valuesa | P-values | |
|---|---|---|---|---|
| Blood parameter | ||||
| 2 h plasma glucose [mmol/l] | 6.19 ± 0.17 | 9.35 ± 0.38 | <7.8b | <0.0001 |
| Glucose (fasting) [mmol/l] | 5.10 ± 0.07 | 5.68 ± 0.16 | <5.6 | 0.0004 |
| Insulin (fasting) [pmol/l] | 62.08 ± 7.8 | 94.89 ± 15.70 | <185 | 0.052 |
| HbA1c [%] | 5.85 ± 0.05 | 6.13 ± 0.15 | <6.1 | 0.026 |
| Triglycerides [mg/dl] | 107.9 ± 9.6 | 123.3 ± 10.5 | <150 | 0.405 |
| Cholesterol [mg/dl] | 190.3 ± 5.8 | 186.6 ± 8.5 | <190 | 0.746 |
| LDL-cholesterol [mg/dl] | 121.9 ± 5.1 | 120.8 ± 8.5 | <160 | 0.916 |
| HDL-cholesterol [mg/dl] | 52.1 ± 2.0 | 46.2 ± 2.6 | >35 | 0.136 |
| C-reactive protein [mg/dl] | 0.20 ± 0.03 | 0.41 ± 0.09 | <0.5 | 0.007 |
| Leukocytes [1/μl] | 6348 ± 249 | 5955 ± 273 | 4000–9500 | 0.412 |
| Total protein [mg/dl] | 7.0 ± 0.06 | 6.9 ± 0.11 | 6.5–8.5 | 0.800 |
| Creatinine [mg/dl] | 0.99 ± 0.02 | 0.99 ± 0.03 | <1.1 | 0.913 |
| Uric acid [mg/dl] | 5.5 ± 0.26 | 6.2 ± 0.31 | 3.4–7.0 | 0.178 |
| Homocysteine [μmol/l] | 10.0 ± 0.56 | 9.3 ± 0.55 | <15 | 0.500 |
| Urine parameter | ||||
| Total protein [mg/g creatinine] | 143.7 ± 16.1 | 124.8 ± 19.9 | <150 | 0.543 |
| Albumin [mg/g creatinine] | 10.0 ± 1.2 | 17.8 ± 6.6 | <20 | 0.068 |
| Creatinine [g/l] | 0.85 ± 0.09 | 0.98 ± 0.17 | 0.8–2.0 | 0.446 |
| pH-value | 6.0 ± 0.15 | 6.1 ± 0.19 | 4.8–7.4 | 0.661 |
Based on the 2 h blood glucose value individuals were divided in groups of normal and impaired glucose tolerance according to the recommendations of the WHO/IDF (WHO 2006). Data shown as (average) mean ± standard deviation (Standard Error of the Mean = SEM)
aFrom the University Hospital of Tuebingen, Germany
bAccording to the definition of the WHO
For analysis of the individual metabolite fingerprint, we applied our recently reported UPLC-qTOF-MS approach (Yin et al. 2008; Zhao et al. 2008). The relative standard deviation (RSD) of retention times of the main peaks was less than 1%, and the RSD of peak area was less than 10%. These data represented a good repeatability of retention times and a satisfactory precision of quantitation. Therefore the method was reliable for the subsequent analysis of metabolite fingerprints in human body fluids.
For each sample, reversed-phase UPLC-qTOF-MS analysis was applied in the positive and negative ESI mode to increase the number of detected metabolite ions. Figure 1a shows a typical base peak intensity chromatogram (BPC) of plasma recorded in the positive ionization mode, and Fig. 1b shows the BPC of plasma recorded in the negative ionization mode. The BPCs of urine are given in Fig. 1c and d, respectively. After peak alignment, the datasets of plasma contained 2122 metabolite ions (1161 in positive and 961 in negative ionization mode) and of spot urine 2574 metabolite ions (1060 in positive and 1514 in negative ionization mode). The pre-processed UPLC-qTOF-MS data were further investigated using multivariate statistical analyses.
Fig. 1.
Representative UPLC-ESI-qTOF-MS base peak intensity chromatograms (BPC) (m/z 100–700; 0–25 min) from a human plasma sample analyzed in positive (a) and in negative ionization mode (b), and from a spot urine in positive (c) and in negative ionization mode (d). Methodological details are given in the “Method” section
Comparison of the individual metabolite fingerprints by multivariate data analysis
To determine whether the metabolite fingerprints in fasting plasma and/or spot urine differs between NGT and IGT in our metabonomics approach, we constructed two component PLS-DA models. NGT and IGT individuals are clearly divided in two distinct clusters in the PLSA-DA scores plot independent if plasma (Fig. 2a) or urine (Fig. 2b) was used. To ensure that the calculated models are reliable and the observed clustering is not due to chance, we performed an internal validation using 7-fold cross-validation (Eriksson et al. 2001). The calculated goodness of fit (R²Y) was 0.99 for plasma, and 0.95 for urine and the goodness of prediction (Q²Y) of 0.96 for plasma and 0.76 for urine, respectively which underlines the robustness of the model. In addition a response permutation test (Eriksson et al. 2001) was used to confirm the significance of the predictive ability. The separation is more pronounced in plasma in comparison to the non-invasively collected urine samples, which may be a hint to more evident alterations in the metabolite pattern of plasma but may be also a consequence of the higher sample variability in urine (Lenz et al. 2003, 2004).
Fig. 2.
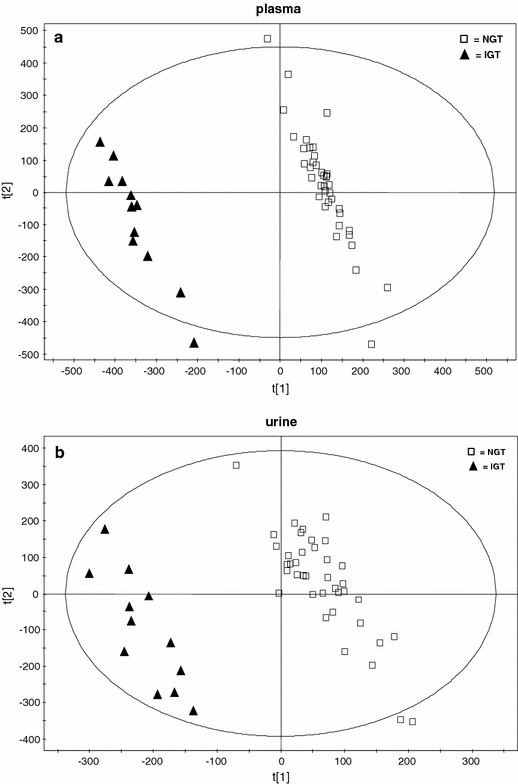
Two dimensional scores plot of a partial least squares-discriminant analysis (PLS-DA) with orthogonal signal correction (OSC) data filter of the first and second PLS component of (a) plasma and (b) spot urine. Normal glucose tolerant (NGT) individuals are labelled by open squares and impaired glucose tolerant (IGT) subjects are labelled with triangles
Since the population sizes of 39 NGT and 12 IGT subjects are different, which may affect the multivariate data analysis and consequently the clustering of the groups, we repeated the data evaluation by randomly separating the control (NGT) group into three subgroups, each consisting out of 13 individuals. In the score plots of these two-component PLS-DA models (Fig. S1A, S1B, S1C for plasma, and Fig. S2A, S2B, S2C for urine, shown in the supporting information of the supplementary material) it became obvious that each NGT subgroup can be clearly separated from the IGT group. For plasma, the calculated goodness of fit (R²Y) was 0.995, 0.986 and 0.995, and the goodness of prediction (Q²Y) was 0.948, 0.915 and 0.939, respectively. For urine, the calculated goodness of fit (R²Y) was 0.998, 0.996 and 0.993, and the goodness of prediction (Q²Y) was 0.729, 0.715 and 0.590, respectively. These values are comparable to the data achieved in the two-component PLS-DA model constructed from 39 NGT and 12 IGT individuals (Fig. 2). Thus, the results show that unbalance of the two groups has no effect on the classification. Therefore we continued using 39 NGT subjects as control group for further data analysis.
Identification of plasma and urinary metabolites related to pre-diabetic alterations of metabolic pathways
For mining the complex data to propose which metabolites are potentially biochemical interesting compounds we used the S-plot of SIMCA-P according to literature (Wiklund et al. 2008). It is a scatter plot which combines the covariance and correlation loading profiles resulting from the predictive component of the OSC-filtered PLS-DA scores plot (Wiklund et al. 2008). The S-plots given in Fig. 3 show the m/z values of the most altered metabolite ions in plasma and spot urine of IGT subjects. The metabolite ions with the greatest influence on the clustering in the PLS-DA score plots (Fig. 2) are located furthest away from the center of the S-plot shown in Fig. 3a for plasma and Fig. 3b for urine.
Fig. 3.
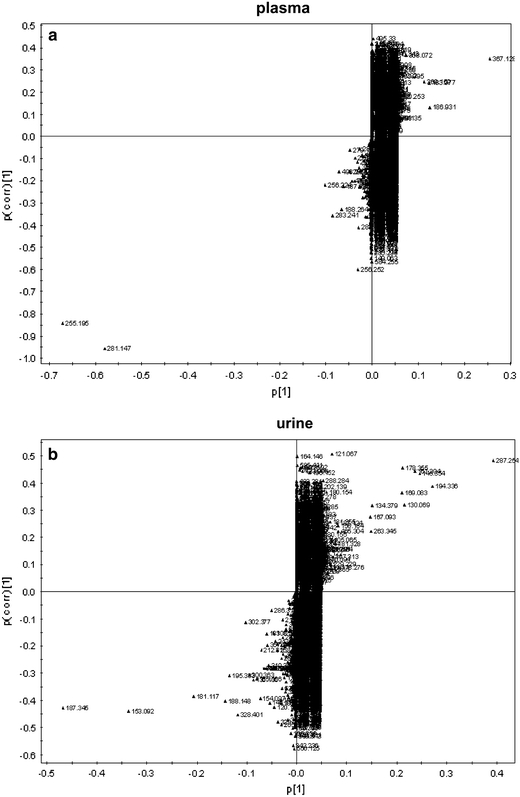
S-plot of (a) plasma and (b) spot urine. The variables are labeled with m/z values. Potential metabolic biomarkers including the corresponding m/z values are presented in Tables 2 and 3
The metabolites were identified following our recently published strategy (Chen et al. 2008). Accurate mass and mass spectrometric fragmentation pattern was utilized to search databases including KEGG, PubChem compound, METLIN, and HMDB database. Standard samples were used for the confirmation of the identity of ambiguous compounds. A summary of identified metabolites is given in Table 2 (for plasma) and Table 3 (for urine).
Table 2.
List of plasma metabolites most influential in discriminating between NGT and IGT individuals in the S-plot shown in Fig. 3a
| Retention time (min) | m/z | Ionization modea | Mass accuracy (ppm) | Metabolite | Changeb |
|---|---|---|---|---|---|
| 4.56 | 186.9711 | ESI− | ui | ↑ | |
| 12.05 | 448.3051 | ESI− | 2.6 | Glycochenodeoxycholic acid | ↑ |
| 19.66 | 253.2159 | ESI− | 3.7 | FFA C16:1 [M − 1]− | ↑ |
| 19.88 | 303.2315 | ESI− | 3.1 | FFA C20:4 [M − 1]− | ↑ |
| 20.18 | 329.2466 | ESI− | 4.6 | FFA C22:5 [M − 1]− | ↑ |
| 20.19 | 279.2314 | ESI− | 3.5 | FFA C18:2 [M − 1]− | ↑ |
| 20.81 | 305.2472 | ESI− | 2.9 | FFA C20:3 [M − 1]− | ↑ |
| 21.37 | 331.2624 | ESI− | 3.9 | FFA C22:4 [M − 1]− | ↑ |
| 21.50 | 255.2313 | ESI− | 4.2 | FFA C16:0 [M − 1]− | ↑ |
| 21.51 | 256.2395 | ESI− | 3.2 | Isotope of FFA C16:0 [M − 1]− | ↑ |
| 21.73 | 282.2553 | ESI− | 2.4 | Isotope of FFA C18:1 [M − 1]− | ↑ |
| 21.73 | 281.2475 | ESI− | 2.2 | FFA C18:1 [M − 1]− | ↑ |
| 22.04 | 307.2627 | ESI− | 3.1 | FFA C20:2 [M − 1]− | ↑ |
| 23.37 | 283.2627 | ESI− | 3.4 | FFA C18:0 [M − 1]− | ↑ |
| 9.05 | 367.1643 | ESI− | ui | ↓ | |
| 14.49 | 504.3154 | ESI− | 2.7 | LPC C18:2 [M − CH3]− | ↓ |
| 14.49 | 564.3284 | ESI− | 3.1 | LPC C18:2 [M + HCOO]− | ↓ |
| 14.54 | 520.3392 | ESI+ | 2.1 | LPC C18:2 [M + 1]+ | ↓ |
| 14.54 | 184.0733 | ESI+ | 3.5 | Fragment of LPC C18:2 | ↓ |
| 14.77 | 476.3046 | ESI− | ui | ↓ | |
| 14.84 | 504.3148 | ESI− | 3.9 | LPC C18:2 [M − CH3]− | ↓ |
| 15.30 | 480.3069 | ESI− | 4.5 | LPC C16:0 [M − CH3]− | ↓ |
| 15.31 | 540.3284 | ESI− | 3.2 | LPC C16:0 [M + HCOO]− | ↓ |
| 15.39 | 184.0733 | ESI+ | 3.1 | Fragment of LPC C16:0 | ↓ |
| 15.75 | 480.3073 | ESI− | 3.7 | LPC C16:0 [M − CH3]− | ↓ |
| 15.75 | 540.3286 | ESI− | 2.8 | LPC C16:0 [M + HCOO]− | ↓ |
| 15.85 | 506.3237 | ESI− | 1.9 | LPC C16:1 [M − CH3]− | ↓ |
| 15.85 | 566.3444 | ESI− | 2.4 | LPC C18:1 [M + HCOO]− | ↓ |
| 15.93 | 496.3393 | ESI+ | 2.1 | LPC C16:0 [M + H]+ | ↓ |
| 15.93 | 184.0733 | ESI+ | 3.2 | Fragment of LPC C16:0 | ↓ |
| 16.21 | 506.3233 | ESI− | 2.8 | LPC C18:1 [M − CH3]− | ↓ |
| 16.86 | 494.3236 | ESI− | 1.9 | ui | ↓ |
| 17.57 | 568.3602 | ESI− | 2.1 | LPC C18:0 [M + HCOO]− | ↓ |
| 17.64 | 184.0735 | ESI+ | 2.4 | Fragment of LPC C18:0 | ↓ |
| 18.03 | 568.3601 | ESI− | 2.2 | LPC C18:0 [M + HCOO]− | ↓ |
The metabolites were divided in groups of increasing and decreasing signal intensities and sorted by retention time
aESI+ means positive electrospray ionization mode and ESI− means negative electrospray ionization mode, respectively
bRelative metabolite level in plasma of IGT subjects in comparison to NGT individuals
Table 3.
List of metabolites in the S-plot most influential in discriminating between NGT and IGT individuals in urine shown in Fig. 3b
| Retention time (min) | m/z | Ionization modea | Mass accuracy (ppm) | Metabolite | Change(b) |
|---|---|---|---|---|---|
| 1.91 | 153.0399 | ESI+ | 8.9 | Xanthine [M + 1]+ | ↑ |
| 6.65 | 195.0311 | ESI− | ui | ↑ | |
| 6.82 | 188.0702 | ESI+ | 4.2 | Fragment of tryptophan | ↑ |
| 19.24 | 300.1803 | ESI+ | 2.5 | C8:2-OH carnitine [M + 1]+ | ↑ |
| 23.09 | 328.2117 | ESI+ | 2.1 | C10:2-OH carnitine [M + 1]+ | ↑ |
| 1.00 | 169.0351 | ESI+ | 6.3 | Uric acid [M + 1]+ | ↓ |
| 1.03 | 167.0198 | ESI− | 4.3 | Uric acid [M − 1]− | ↓ |
| 2.37 | 167.0482 | ESI+ | 5.1 | 7-methylxanthine [M + 1]+ | ↓ |
| 2.45 | 183.0511 | ESI+ | 3.9 | Methyluric acid M + 1]+ | ↓ |
| 2.50 | 181.0431 | ESI− | 4.7 | Methyluric acid [M − 1]− | ↓ |
| 2.77 | 167.0483 | ESI+ | 4.9 | 3-methylxanthine [M + 1]+ | ↓ |
| 3.10 | 167.0481 | ESI+ | 5.8 | 1-methylxanthine [M + 1]+ | ↓ |
| 4.97 | 181.0671 | ESI+ | 4.9 | ui | ↓ |
| 6.14 | 194.0446 | ESI− | 3.7 | 3-hydroxyhippuric acid [M − 1]− | ↓ |
| 8.62 | 105.0334 | ESI+ | 5.3 | Fragment of hippuric acid | ↓ |
| 8.67 | 134.0602 | ESI− | 6.1 | Fragment of hippuric acid | ↓ |
| 8.67 | 178.0500 | ESI− | 2.4 | Hippuric acid [M − 1]− | ↓ |
| 9.77 | 287.0998 | ESI+ | 3.6 | Phenylacetyl-glutamine [M + Na]+ | ↓ |
| 11.7 | 187.0031 | ESI− | ui | ↓ |
The metabolites were divided in groups of increasing and decreasing signal intensities and sorted by retention time
aESI+ means positive electrospray ionization mode and ESI− means negative electrospray ionization mode, respectively
bRelative metabolite level in urine of IGT subjects in comparison to NGT individuals
The most discriminative plasma metabolites separating IGT and NGT subjects were the saturated free fatty acid (FFA) palmitate, the unsaturated FFA oleate, and the saturated FFA stearate. Additionally, the FFAs C18:2, C16:1, C20:4 and C22:4 were also detected to be increased in plasma of IGT subjects (Table 2). Furthermore, the bile acid glycochenodeoxycholic acid, as well as lyso-phosphatidylcholines (LPCs) are discriminative plasma metabolite biomarkers classifying normal and impaired glucose tolerant individuals. The pre-diabetic metabolic state resulted in increased relative plasma levels of free fatty acids (C16:0, C18:0, C18:1, C18:2, etc.) and bile acid (glycochenodeoxycholic acid), as well as decreased plasma levels of LPC (C16:0 and C18:2 etc.; Table 2). Since reversed-phase chromatography was used highly polar potential biomarkers, like glucose, could not be detected in our approach.
In the S-plot of urine, we detected decreased levels of hippuric acid, uric acid, 3-hydroxyhippuric acid, phenylacetyl-glutamine (PAGN), methyluric acid and methylxanthine, whereas the urinary excretion of C8:2-OH carnitine, C10:2-OH carnitine, xanthine and tryptophan was increased (Table 3).
Since FFAs and LPCs were the dominating biomarkers detected a metabolite heat map was generated to study the alterations of the signal intensity of FFAs and LPCs in plasma of the 51 subjects (Fig. 4a). Plasma levels were represented by colors in the heat map, and each FFA and LPC was represented by a single row of colored boxes, whereas columns represented different subjects. The yellow labels show increase, and the blue labels means decrease intensities relative to the median metabolite levels. The heat map revealed considerable differences between NGT and IGT subjects. In IGT subjects with a 2-h glucose level above 7.8 mmol/l (marked by a red line) increased FFAs and decreased LPC levels are visible. In contrast to that in the heat map region of subjects with 2-h glucose levels less than 6.4 mmol/l FFAs showed decreasing and LPCs increasing levels. In subjects with a 2-h glucose below 5 mmol/l the pattern is inverse to the IGT group. In region of 2-h plasma glucose between 6.4 and 7.8 mmol/l a transition pattern was detected. Based on these findings student’s t test was performed to evaluate statistically significances in FFA and LPC levels (Fig. 4b and c). In the NGT group a tendency for lower plasma levels of the saturated fatty acids palmitate (P = 0.08) and stearate (P = 0.07) were detected, but not for the mono- and poly-unsaturated FFAs oleate and arachidonic acid. LPCs (C16:0; C18:0, C18:1; C18:2) all tended to be increased in the NGT group. Based on the signatures in the heat map we performed also a comparison of the IGT and subjects with 2-h glucose below 6.4 mmol/l which resulted in significant differences (P < 0.05) in all LPCs and palmitate as well as stearate.
Fig. 4.
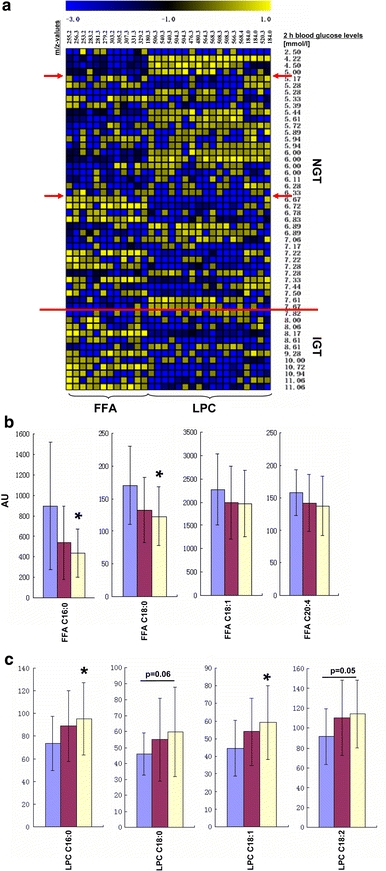
(a) Heat map of plasma free fatty acids (FFA) and lysophsphatidylcholines (LPCs) of 39 healthy subjects with normal glucose tolerance (NGT) and 12 individuals with impaired glucose tolerance (IGT). The row above the heat map shows the m/z values of these metabolites corresponding to Table 2. Individual 2-h oGTT plasma glucose level in increasing order are given in the column to the right. Cells are colored based on the signal intensity measured in plasma. Yellow represents high plasma levels and blue showed low signal intensity (see color scale above the heat map). A red line is drawn at a 2-h plasma glucose concentration of 7.8 mmol/l representing the WHO definition for the separation of NGT and IGT. Furthermore, arrows are drawn at the 2-h plasma glucose >5 mmol/l and >6.4 mmol/l where the heat map pattern shows distinct changes. (b) and (c) differences in FFA and LPCs levels between controls and pre-diabetic subjects were examined using Student’s t-test. (b) comparison of the plasma signal intensities of free fatty acids FFA (mean peak area ± standard error; * P < 0.05, IGT vs. the subgroup of NGT with a 2-h plasma glucose level below 6.4 mmol/l) and (c) lysophosphatidylcholines LPC (mean peak area ± standard error; * P < 0.05, IGT vs. subgroup of NGT with a 2-h plasma glucose level below 6.4 mmol/l). blue filled square IGT group (n = 12), red filled square NGT group (n = 39), yellow filled square subgroup of NGT with a 2-h plasma glucose level below 6.4 mmol/l (n = 22). (Color figure online)
Discussion
Impaired glucose tolerance is a polygenetic and environmentally determined metabolic disease. To study these pre-diabetic metabolic traits we applied an UPLC-qTOF-MS-based non-targeted metabonomics approach for the investigation of plasma and spot urine of subjects with IGT vs controls. Independent from the body fluid used the individual metabolite fingerprints led to a clear discrimination of pre-diabetic individuals from the control group with normal glucose tolerance. Identification of discriminating metabolites revealed alterations of various metabolic pathways and a metabolite heat map of plasma free fatty acids and lysophosphytidylcholines revealed a close relationship of changes of the plasma heat map pattern of FFA and LPCs to the 2-h oGTT glucose levels.
The most discriminative metabolites in plasma showing an increased level in pre-diabetic individuals were the free fatty acids, including e.g. C16:0, C18:0, C18:1 and C20:4 (Table 2). Since insulin effects are impaired in IGT subjects, reduced insulin-mediated inhibition of lipolysis and attenuated stimulation of lipid storage in adipose tissue as well as increased hepatic output leads to increased circulating free fatty acids and triglycerides (Montecucco et al. 2008; Stumvoll et al. 2000). These increased blood levels may result in enhanced β-oxidation in liver and in skeletal muscle. This assumption is supported by our finding of increased urinary excretion of acylcarnitines (Table 3), which reflect the substrate flux through β-oxidation and are therefore “readouts” of it (Koves et al. 2008) and commonly used to screen for inborn metabolic disorders (Van Hove et al. 1993). Hence, the findings of our LC–MS-driven non-targeted approach are well in line with a current model discussed by the group of Muoio where lipid oversupply induced insulin resistance in muscle is explained by “overload” of mitochondrial lipid oxidation, accumulation of acylcarnitines, and depletion of TCA intermediates which may lead to mitochondrial stress activating currently unknown signaling pathways that interfere with insulin action (Bain et al. 2009; Koves et al. 2005; Koves et al. 2008). A scheme, given in Fig. 5, illustrates the concept of increased lipolysis in adipose tissue and hepatic and muscular lipid oversupply in the pre-diabetic state. As a consequence the triglyceride storage in the liver and the hepatic output of very-low-density lipoproteins is increased. In skeletal muscle the augmented mitochondrial lipid oxidation may lead to accumulation of β-oxidation intermediates and depletion of TCA intermediates which then may be reflected by the detected increased urinary excretion of acyl-carnitines and decreased excretion of phenylacetyl-glutamine in our study (Fig. 5). Though we find most of detected free fatty acids increased in IGT individuals in the heat map (Fig. 4a), saturated free fatty acids (SFA; C16:0 and C18:0) increased more pronounced than mono-unsaturated free fatty acids (C16:1, C18:1) and poly-unsaturated fatty acids (C18:2, C20:4, C22:4 etc.; Fig. 4a). Compared with the NGT group (2 h oGTT glucose less than 7.8 mmol/l), mean peak area of SFA C16:0 and C18:0 in the IGT group increased 66% and 26%, respectively (Fig. 4b), but this tendency was not significant. A significant difference was detected when IGT subjects and individuals with 2-h oGTT glucose levels <6.4 mmol/l were compared. SFA have been shown to exert lipotoxic effects like the induction of apoptosis, inflammation, and endoplasmatic reticulum stress that are associated with metabolic diseases (Borradaile et al. 2006; Eizirik et al. 2008; Peter et al. 2008; Staiger et al. 2004; Stefan et al. 2008; Weigert et al. 2004).
Fig. 5.
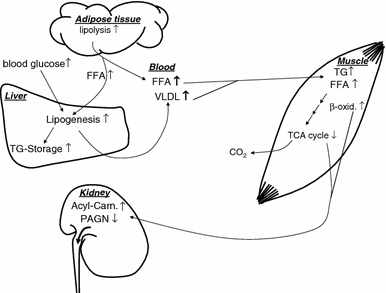
Suggestion for a mechanistic model of metabolic alterations in the pre-diabetic state with a special focus on lipid metabolism. Insulin mediated inhibition of lipolysis is attenuated in adipose tissue in the pre-diabetic metabolic state. Consequently circulating plasma free fatty acids (FFA) are increased leading to a hepatic and muscular lipid oversupply. Hyperlipidemia, hyperglycemia and hepatic insulin resistance augments the triglyceride (TG) storage in the liver and the output of very-low-density lipoproteins (VLDL). In skeletal muscle the overload of mitochondrial lipid oxidation results in accumulation of β-oxidation intermediates and depletion of TCA intermediates which may lead to increased urinary excretion of acyl-carnitines and decreased excretion of phenylacetyl-glutamine
An increased level of the bile acid glycochenodeoxycholic acid (GCDCA, m/z 448.29 in ESI−) was also detected in the fasting plasma of the pre-diabetic individuals in our study. Bile acids have recently evolved from detergents that emulsify nutrient lipids and remove cholesterol from the body to systemic signaling molecules (Thomas et al. 2008) and novel functions of bile acids as metabolic integrators of whole body energy homeostasis that influence glucose and lipid metabolism have been discovered (Thomas et al. 2008). Whether GCDCA reflects a general alteration of the bile acid metabolism under the pathophysiological conditions or if it is solely discriminating between NGT and IGT under fasting conditions needs to be proven. However, using a multi-analytical platform approach Wilson’ group detected taurocholic acid in obese Zucker (fa/fa) rats—an important disease model to study obesity and onset of insulin resistance, as one of the discriminating biomarkers, being increased in plasma (Plumb et al. 2006; Williams et al. 2006a) and decreased in urine (Williams et al. 2006b). Furthermore, one of the most striking changes during a 120 min oGTT in healthy humans was the dramatic increases of bile acids, very recently detected in a targeted metabolic profiling approach studying the profile of 191 plasma metabolites (Shaham et al. 2008) and by our group in a non-targeted metabolomics study (Zhao et al. 2009). Of note, applying a model relating fasting insulin to 120 min metabolite change in individuals with IGT Shamam et al. could show that pre-diabetic individuals had a greater change in the excursion of GCDCA compared with controls (Shaham et al. 2008). The excursion of lactate, valine, methionine, β-hydroxybutyrate and leucine/isoleucin were also different in the pre-diabetics (Shaham et al. 2008).
Another group of metabolites identified in our study to reflect pathophysiological alterations in IGT were the lysophosphatidylcholines (LPCs), showing decreased plasma levels in the heat map (Fig. 4a) and in the comparison of signal intensities of selected LPCs (Fig. 4c). These findings are similar to our previous investigation in individuals with manifest T2DM (Wang et al. 2005).
Of note in urine, we detected that decreased urinary levels of gut microbiota-associated metabolite biomarkers i.e. hippuric acid, methylxanthine, methyluric acid and 3-hydroxyhippuric acid are related to IGT. In humans the role of gut flora in health is well recognized (Nicholson et al. 2005) and accumulating evidence indicates that the gut microbiota is instrumental in energy metabolism and immune function of the host (Jia et al. 2008). Recent reports from the group of J.K. Nicholson demonstrated the association of diet and gut microbial activities with the human metabolic phenotype (Li et al. 2008) and the blood pressure of individuals (Holmes et al. 2008a). Furthermore the group of Gordon demonstrated in animal and human studies by sequencing faecal bacterial 16S rRNA genes that variation in gut microbial composition is an important factor in the aetiology of obesity and insulin resistance (Ley et al. 2006; Turnbaugh et al. 2006; Turnbaugh et al. 2009). Thus, our metabonomics results may be an additional hint supporting the relevant role of the gut microbiota in the pathogenesis of the pre-diabetic state.
Furthermore we detected in urine increased levels of xanthine, and tryptophan, while the level of phenylacetylglutamine (PAGN) was decreased. The decreased excretion of PAGN in IGT subjects may reflect differences in gluconeogenesis and/or TCA cycle intermediates. PAGN is formed in the liver of humans and primates from glutamine which is in equilibrium with glutamate and α-ketoglutarate and phenylacetate (Yang and Brunengraber 2000). Metabonomics investigations of pre-diabetic animal models as well as human studies of manifest T2DM are well in line with our findings of alterations in TCA cycle intermediates (Salek et al. 2007; Shearer et al. 2008; Williams et al. 2006b; Zhang et al. 2009). In human serum of individuals with manifest T2DM a recent non-targeted 1H-NMR metabolomics study revealed decreased citrate levels (Zhang et al. 2009). Salek et al. compared metabolite profiles in urine from unmedicated humans with T2DM, db/db mice and fa/fa rats (Salek et al. 2007). They found decreased urinary α-ketoglutarate excretion in the diabetic state. Another major finding of this study was that changes in metabolites involved in nucleotide, methylamine metabolism and TCA cycle intermediates were common between the animal models and humans (Salek et al. 2007). We conclude that not only in the diabetic stage, but also in the latent pre-diabetic phase, i.e. long before the manifestation of diabetes, a dysregulation of the TCA cycle occurs.
Concluding remarks
Impaired glucose tolerance is widely present throughout the general population (Abdul-Ghani et al. 2006) and studies suggest that this pre-diabetic state is associated with insulin resistance in muscle, liver and fat as well as a defective insulin secretion resulting in multiple metabolic traits (Abdul-Ghani et al. 2006). Although IGT is not a clinical entity, it is a risk factor for future diabetes and/or adverse outcomes (e.g. cardiovascular disease) (WHO 2006). A metabonomics approach based on UPLC qTOF MS has been used to analyze individual metabolite fingerprints in plasma as well as in spot urine from overnight fasted individuals. The investigation of a single body fluid sample, either plasma or spot urine led to a clear separation of the clusters of pre-diabetic subjects and controls. The identification of discriminating metabolites revealed pathways affected by the pre-diabetic metabolic state. Alterations in fatty acid-, tryptophan-, uric acid-, bile acid-, lysophosphatidylcholine-metabolism, and the TCA cycle were identified. The alterations in the levels of saturated free fatty acids are more pronounced than mono-unsaturated and poly-unsaturated free fatty acids. Of note, also decreased levels of the gut flora-associated metabolites hippuric acid, methylxanthine, methyluric acid, and 3-hydroxyhippuric acid were detected in pre-diabetic individuals. The present study shows in fasting plasma and urine metabolic pathway alterations reflecting metabolic traits in individuals at high risk to develop T2DM.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank S. Herbert, I. Riedlinger, A. Bury, B. Horrer and H. Luz for excellent technical assistance. This study was supported in part by grants from the “Kompetenznetz Diabetes mellitus” (Competence Network for Diabetes mellitus) funded by the Federal Ministry of Education and Research (FKZ 01GI0803-04), the foundation from National Natural Science Foundation of China (No. 20675082), the Sino-German Center for Research Promotion (DFG and NSFC, GZ 364), the National Basic Research Program of China (No. 2006CB503902) and National Key Project of Scientific and Technical Supporting Programs (No. 2006BAK02A12) from State Ministry of Science & Technology of China.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Guowang Xu, Phone: +86-411-84379559, FAX: +86-411-84379559, Email: xugw@dicp.ac.cn.
Rainer Lehmann, Phone: +49-7071-2983193, FAX: +49-7071-295348, Email: rainer.lehmann@med.uni-tuebingen.de.
References
- Abdul-Ghani MA, Jenkinson CP, Richardson DK, Tripathy D, DeFronzo RA. Insulin secretion and action in subjects with impaired fasting glucose and impaired glucose tolerance: Results from the Veterans Administration Genetic Epidemiology Study. Diabetes. 2006;55:1430–1435. doi: 10.2337/db05-1200. [DOI] [PubMed] [Google Scholar]
- Alberti KG, Zimmet P, Shaw J. International Diabetes Federation: A consensus on Type 2 diabetes prevention. Diabetic Medicine. 2007;24:451–463. doi: 10.1111/j.1464-5491.2007.02157.x. [DOI] [PubMed] [Google Scholar]
- Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, et al. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain. 2008;131:389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- Borradaile NM, Han X, Harp JD, Gale SE, Ory DS, Schaffer JE. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. Journal of Lipid Research. 2006;47:2726–2737. doi: 10.1194/jlr.M600299-JLR200. [DOI] [PubMed] [Google Scholar]
- Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HW, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nature Medicine. 2002;8:1439–1444. doi: 10.1038/nm802. [DOI] [PubMed] [Google Scholar]
- Cefalu WT. Inflammation, insulin resistance, and type 2 diabetes: Back to the future? Diabetes. 2009;58:307–308. doi: 10.2337/db08-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhao X, Fritsche J, Yin P, Schmitt-Kopplin P, Wang W, et al. Practical approach for the identification and isomer elucidation of biomarkers detected in a metabonomic study for the discovery of individuals at risk for diabetes by integrating the chromatographic and mass spectrometric information. Analytical Chemistry. 2008;80:1280–1289. doi: 10.1021/ac702089h. [DOI] [PubMed] [Google Scholar]
- Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocrine Reviews. 2008;29:42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- Eriksson, L., Johansson, E., Kettaneh-Wold, N., & Wold, S. (2001). Multi- and megavariate data analysis principals and applications Umetrics, Umea, Sweden.
- Festa A, D’Agostino R, Jr, Hanley AJ, Karter AJ, Saad MF, Haffner SM. Differences in insulin resistance in nondiabetic subjects with isolated impaired glucose tolerance or isolated impaired fasting glucose. Diabetes. 2004;53:1549–1555. doi: 10.2337/diabetes.53.6.1549. [DOI] [PubMed] [Google Scholar]
- Gipson GT, Tatsuoka KS, Ball RJ, Sokhansanj BA, Hansen MK, Ryan TE, et al. Multi-platform investigation of the metabolome in a leptin receptor defective murine model of type 2 diabetes. Molecular Biosystems. 2008;4:1015–1023. doi: 10.1039/b807332e. [DOI] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Huo T, Cai S, Lu X, Sha Y, Yu M, Li F. Metabonomic study of biochemical changes in the serum of type 2 diabetes mellitus patients after the treatment of metformin hydrochloride. Journal of Pharmaceutical and Biomedical Analysis. 2009;49:976–982. doi: 10.1016/j.jpba.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: A potential new territory for drug targeting. Nature Reviews Drug Discovery. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancer. Analytical Biochemistry. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, et al. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. The Journal of Biological Chemistry. 2005;280:33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metabolism. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Lenz EM, Bright J, Wilson ID, Hughes A, Morrisson J, Lindberg H, et al. Metabonomics, dietary influences and cultural differences: A 1H NMR-based study of urine samples obtained from healthy British and Swedish subjects. Journal of Pharmaceutical and Biomedical Analysis. 2004;36:841–849. doi: 10.1016/j.jpba.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lenz EM, Bright J, Wilson ID, Morgan SR, Nash AF. A 1H NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. Journal of Pharmaceutical and Biomedical Analysis. 2003;33:1103–1115. doi: 10.1016/S0731-7085(03)00410-2. [DOI] [PubMed] [Google Scholar]
- Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M, et al. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. Journal of Clinical Investigation. 2008;118:3503–3512. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon JC, Holmes E, Nicholson JK. So what’s the deal with metabonomics? Analytical Chemistry. 2003;75:384A–391A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- Makinen VP, Soininen P, Forsblom C, Parkkonen M, Ingman P, Kaski K, et al. 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Molecular Systems Biology. 2008;4:167. doi: 10.1038/msb4100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. Journal of Proteome Research. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- Montecucco F, Steffens S, Mach F. Insulin resistance: a proinflammatory state mediated by lipid-induced signaling dysfunction and involved in atherosclerotic plaque instability. Mediators of Inflammation. 2008;2008:767623. doi: 10.1155/2008/767623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DM, Davidson MB, DeFronzo RA, Heine RJ, Henry RR, Pratley R, et al. Impaired fasting glucose and impaired glucose tolerance: Implications for care. Diabetes Care. 2007;30:753–759. doi: 10.2337/dc07-9920. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nature Reviews. Microbiology. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- Novoa FJ, Boronat M, Saavedra P, Diaz-Cremades JM, Varillas VF, La Roche F, et al. Differences in cardiovascular risk factors, insulin resistance, and insulin secretion in individuals with normal glucose tolerance and in subjects with impaired glucose regulation: The Telde Study. Diabetes Care. 2005;28:2388–2393. doi: 10.2337/diacare.28.10.2388. [DOI] [PubMed] [Google Scholar]
- Peter A, Weigert C, Staiger H, Rittig K, Cegan A, Lutz P, et al. Induction of stearoyl-CoA desaturase protects human arterial endothelial cells against lipotoxicity. American Journal of Physiology, Endocrinology and Metabolism. 2008;295:E339–E349. doi: 10.1152/ajpendo.00022.2008. [DOI] [PubMed] [Google Scholar]
- Petersen JL, McGuire DK. Impaired glucose tolerance and impaired fasting glucose—a review of diagnosis, clinical implications and management. Diabetes and Vascular Disease Research. 2005;2:9–15. doi: 10.3132/dvdr.2005.007. [DOI] [PubMed] [Google Scholar]
- Plumb RS, Johnson KA, Rainville P, Shockcor JP, Williams R, Granger JH, et al. The detection of phenotypic differences in the metabolic plasma profile of three strains of Zucker rats at 20 weeks of age using ultra-performance liquid chromatography/orthogonal acceleration time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2006;20:2800–2806. doi: 10.1002/rcm.2655. [DOI] [PubMed] [Google Scholar]
- Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, Cheeseman M, et al. A metabolic comparison of urinary changes in type 2 diabetes in mouse, rat and man. Physiological Genomics. 2007;29:99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- Schafer S, Kantartzis K, Machann J, Venter C, Niess A, Schick F, et al. Lifestyle intervention in individuals with normal versus impaired glucose tolerance. European Journal of Clinical Investigation. 2007;37:535–543. doi: 10.1111/j.1365-2362.2007.01820.x. [DOI] [PubMed] [Google Scholar]
- Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Molecular Systems Biology. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer J, Duggan G, Weljie A, Hittel DS, Wasserman DH, Vogel HJ. Metabolomic profiling of dietary-induced insulin resistance in the high fat-fed C57BL/6J mouse. Diabetes, Obesity and Metabolism. 2008;10:950–958. doi: 10.1111/j.1463-1326.2007.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin JD, Muller DC, Fleg JL, Andres R. The relation of fasting and 2-h postchallenge plasma glucose concentrations to mortality: Data from the Baltimore Longitudinal Study of Aging with a critical review of the literature. Diabetes Care. 2005;28:2626–2632. doi: 10.2337/diacare.28.11.2626. [DOI] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Staiger H, Staiger K, Stefan N, Wahl HG, Machicao F, Kellerer M, et al. Palmitate-induced interleukin-6 expression in human coronary artery endothelial cells. Diabetes. 2004;53:3209–3216. doi: 10.2337/diabetes.53.12.3209. [DOI] [PubMed] [Google Scholar]
- Stefan N, Peter A, Cegan A, Staiger H, Machann J, Schick F, et al. Low hepatic stearoyl-CoA desaturase 1 activity is associated with fatty liver and insulin resistance in obese humans. Diabetologia. 2008;51:648–656. doi: 10.1007/s00125-008-0938-7. [DOI] [PubMed] [Google Scholar]
- Stumvoll M, Jacob S, Wahl HG, Hauer B, Loblein K, Grauer P, et al. Suppression of systemic, intramuscular, and subcutaneous adipose tissue lipolysis by insulin in humans. Journal of Clinical Endocrinology and Metabolism. 2000;85:3740–3745. doi: 10.1210/jc.85.10.3740. [DOI] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature Reviews Drug Discovery. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Toye AA, Dumas ME, Blancher C, Rothwell AR, Fearnside JF, Wilder SP, et al. Subtle metabolic and liver gene transcriptional changes underlie diet-induced fatty liver susceptibility in insulin-resistant mice. Diabetologia. 2007;50:1867–1879. doi: 10.1007/s00125-007-0738-5. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Unwin N, Shaw J, Zimmet P, Alberti KG. Impaired glucose tolerance and impaired fasting glycaemia: the current status on definition and intervention. Diabetic Medicine. 2002;19:708–723. doi: 10.1046/j.1464-5491.2002.00835.x. [DOI] [PubMed] [Google Scholar]
- van der Greef J, Stroobant P, van der HR. The role of analytical sciences in medical systems biology. Current Opinion in Chemical Biology. 2004;8:559–565. doi: 10.1016/j.cbpa.2004.08.013. [DOI] [PubMed] [Google Scholar]
- van Doorn M, Vogels J, Tas A, van Hoogdalem EJ, Burggraaf J, Cohen A, et al. Evaluation of metabolite profiles as biomarkers for the pharmacological effects of thiazolidinediones in Type 2 diabetes mellitus patients and healthy volunteers. British Journal of Clinical Pharmacology. 2007;63:562–574. doi: 10.1111/j.1365-2125.2006.02816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove JL, Zhang W, Kahler SG, Roe CR, Chen YT, Terada N, et al. Medium-chain acyl-CoA dehydrogenase (MCAD) deficiency: diagnosis by acylcarnitine analysis in blood. The American Journal of Human Genetics. 1993;52:958–966. [PMC free article] [PubMed] [Google Scholar]
- Wang C, Kong H, Guan Y, Yang J, Gu J, Yang S, et al. Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Analytical Chemistry. 2005;77:4108–4116. doi: 10.1021/ac0481001. [DOI] [PubMed] [Google Scholar]
- Waugh N, Scotland G, McNamee P, Gillett M, Brennan A, Goyder E, et al. Screening for type 2 diabetes: Literature review and economic modelling. Health Technology Assessment. 2007;11:1–125. doi: 10.3310/hta11170. [DOI] [PubMed] [Google Scholar]
- Weigert C, Brodbeck K, Staiger H, Kausch C, Machicao F, Haring HU, et al. Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappaB. The Journal of Biological Chemistry. 2004;279:23942–23952. doi: 10.1074/jbc.M312692200. [DOI] [PubMed] [Google Scholar]
- WHO. (2006). Definition and diagnosis of diabetes mellitus and intermediate hyperglycemia: Report of WHO/IDF consultation. WHO Press, Geneva.
- Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical Chemistry. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- Williams RE, Lenz EM, Rantalainen M, Wilson ID. The comparative metabonomics of age-related changes in the urinary composition of male Wistar-derived and Zucker (fa/fa) obese rats. Molecualr Biosystems. 2006;2:193–202. doi: 10.1039/b517195d. [DOI] [PubMed] [Google Scholar]
- Williams R, Lenz EM, Wilson AJ, Granger J, Wilson ID, Major H, et al. A multi-analytical platform approach to the metabonomic analysis of plasma from normal and Zucker (fa/fa) obese rats. Molecular Biosystems. 2006;2:174–183. doi: 10.1039/b516356k. [DOI] [PubMed] [Google Scholar]
- Wold S, Antti H, Lindgren F, Ohman J. Orthogonal signal correction of near-infrared spectra. Chemometrics and Intelligent Laboratory Systems. 1998;44:175–185. doi: 10.1016/S0169-7439(98)00109-9. [DOI] [Google Scholar]
- Wopereis S, Rubingh CM, van Erk MJ, Verheij ER, van Vliet T, Cnubben NH, et al. Metabolic profiling of the response to an oral glucose tolerance test detects subtle metabolic changes. PLoS ONE. 2009;4:e4525. doi: 10.1371/journal.pone.0004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yach, D., & Alberti, G. (2003). Screening for type 2 diabetes. Marketing and Dissemination, World Health Organization, Geneva.
- Yang D, Brunengraber H. Glutamate, a window on liver intermediary metabolism. Journal of Nutrition. 2000;130:991S–994S. doi: 10.1093/jn/130.4.991S. [DOI] [PubMed] [Google Scholar]
- Yin P, Mohemaiti P, Chen J, Zhao X, Lu X, Yimiti A, et al. Serum metabolic profiling of abnormal savda by liquid chromatography/mass spectrometry. Journal of Chromatography B. 2008;871:322–327. doi: 10.1016/j.jchromb.2008.05.043. [DOI] [PubMed] [Google Scholar]
- Zhang S, Nagana Gowda GA, Asiago V, Shanaiah N, Barbas C, Raftery D. Correlative and quantitative (1)H NMR-based metabolomics reveals specific metabolic pathway disturbances in diabetic rats. Analytical Biochemistry. 2008;383:76–84. doi: 10.1016/j.ab.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y, Hao F, Zhou X, Han X, Tang H, et al. Human serum metabonomic analysis reveals progression axes for glucose intolerance and insulin resistance statuses. Journal of Proteome Research. 2009;8:5188–5195. doi: 10.1021/pr900524z. [DOI] [PubMed] [Google Scholar]
- Zhao X, Peter A, Fritsche J, Elcnerova M, Fritsche A, Haring HU, et al. Changes of the plasma metabolome during an oral glucose tolerance test: is there more than glucose to look at? American Journal of Physiology, Endocrinology and Metabolism. 2009;296:E384–E393. doi: 10.1152/ajpendo.90748.2008. [DOI] [PubMed] [Google Scholar]
- Zhao X, Zhang Y, Meng X, Yin P, Deng C, Chen J, et al. Effect of a traditional Chinese medicine preparation Xindi soft capsule on rat model of acute blood stasis: A urinary metabonomics study based on liquid chromatography-mass spectrometry. Journal of Chromatography B. 2008;873:151–158. doi: 10.1016/j.jchromb.2008.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


