Abstract
Current imaging methods have focused on evaluation of myocardial anatomy and function. However, since myocardial metabolism and function are interrelated, metabolic myocardial imaging techniques, such as positron emission tomography, single photon emission tomography, and magnetic resonance spectroscopy present novel opportunities for probing myocardial pathology and developing new therapeutic approaches. Potential clinical applications of metabolic imaging include hypertensive and ischemic heart disease, heart failure, cardiac transplantation, as well as cardiomyopathies. Furthermore, response to therapeutic intervention can be monitored using metabolic imaging. Analysis of metabolic data in the past has been limited, focusing primarily on isolated metabolites. Models of myocardial metabolism, however, such as the oxygen transport and cellular energetics model and constraint-based metabolic network modeling, offer opportunities for evaluation interactions between greater numbers of metabolites in the heart. In this review, the roles of metabolic myocardial imaging and analysis of metabolic data using modeling methods for expanding our understanding of cardiac pathology are discussed.
Keywords: Myocardial Metabolism, Modeling, Positron Emission Tomography, Single Photon Emission Tomography, Magnetic Resonance Spectroscopy
Introduction
Cardiac imaging plays an integral role in the diagnosis and management of heart disease. Coronary angiography, echocardiography, and single photon emission tomography (SPECT) are well established clinically, with defined indications and benefits. Newer imaging tools, including cardiac computed tomography (CT), magnetic resonance imaging (MRI), magnetic resonance spectroscopy (MRS), and positron emission tomography (PET) are increasing in importance and proving themselves to be valuable tools in disease management. Guidelines are now in place, for instance, for the application of cardiac CT in coronary artery disease [1]. MRI is currently being used in the evaluation of congenital heart disease, cardiac tumors, right ventricular dysplasia, and pericardial disease. Other uses for MRI, including measurement of myocardial perfusion and left ventricular function are being studied and show promise. Although of great clinical benefit from an anatomical perspective, coronary angiography, echocardiography, and cardiac CT do not deliver information regarding patient specific, clinically relevant metabolic processes. The application of hybrid technology, combining CT with metabolic PET and SPECT are currently under evaluation. Data obtained from quantitative metabolic imaging modalities such as PET add functional information to the anatomic imaging modalities and can aid with patient management.
An area of particular interest is the management of congestive heart failure (CHF), a disease process which leads to specific changes in cardiac function and metabolism. CHF is a common cause of morbidity and mortality and is increasing in prevalence. Despite advances in medical management, optimal treatment remains challenging. Applied metabolic imaging in heart failure patients may potentially contribute to improved treatment outcomes. Current research initiatives include evaluation of local myocardial conditions in ischemic cardiomyopathy associated with arrhythmogenesis and potential identification of responders to cardiac resynchronization therapy (CRT) [2]. Pre-procedure use of phase analysis has been able to identify responders by measuring LV dyssynchrony using conventional electrocardiogram-gated SPECT myocardial perfusion imaging, requiring no additional procedures [3]. This method shows promise as it appears to demonstrate both repeatability and reproducibility. Evaluation of metabolic parameters as markers of viability may also be linked to CRT response and deserves further attention [4]. Furthermore, quantitative metabolic imaging may play a role in tailoring medical management with metabolic modulators such as trimetazidine, which have recently generated interest in heart failure management [5, 6]. Application of such metabolic agents may allow for optimization of myocardial energy production. The effects on myocardial perfusion, free fatty acid, and oxidative metabolism can be directly assessed using PET imaging, thus potentially contributing to new pharmacologic therapy options in heart failure patients.
An eminent biochemical engineer Jay Bailey once enumerated several reasons for mathematical model construction as a worthwhile scientific endeavor, which included: organizing disparate information into a coherent whole to discover new strategies, to understand the essential/qualitative features, to think (and calculate) logically what components and interactions are important in a complex system, and to make important corrections to conventional wisdom [7]. These points become immediately clear when one considers the requirements to develop translational pipelines from the basic science bench to the hospital bedside. There has been a long and accomplished history of constructing models of various aspects of cardiac function from electrophysiology to metabolism during the past century. Here we briefly touch upon mathematical models on myocardial cellular energetics and metabolism and discuss potential applications of these models in the analysis of metabolic data obtained with PET, MRS, and SPECT myocardial imaging technologies as well as the Fick method and high-throughput metabolic profiling.
Overview of Myocardial Metabolism
Contractile shortening as well as maintenance of cellular energetics and the sarcoplasmic reticulum calcium pump of cardiomyocytes are fueled by oxidation of fatty acids and carbohydrate substrates. During contraction, regulation of substrate supply begins with an increase in the workload, which is governed by the Frank-Starling law under normal physiological conditions. A workload-dependent increase in the rate of the actomyosin ATPase results in the release of adenosine diphosphate (ADP) and phosphate and the signal is transmitted via the mitochondrial adenine nucleotide translocase, ultimately resulting in the activation of adenosine triphosphate (ATP) synthase and the utilization of the proton transmembrane electrochemical gradient [8]. This process leads to an increase in electron transfer to oxygen through the respiratory chain and NADH and FADH2 oxidation. Thus, increased workload decreases the NADH:NAD + ratio in the mitochondrial matrix, resulting in an increase in the rates of the dehydrogenase reactions in the citric acid cycle. NADH and FADH2 are produced in the citric acid cycle as well as in the fatty acid beta oxidation pathway, and to a lesser extent from glycolysis and the pyruvate dehydrogenase reaction [9].
The rates of oxygen consumption and free fatty acid oxidation increase linearly with elevation of the workload. If workload increases, ATP production and respiration are increased due to feedback signaling via the creatinine kinase system. Activation of the Krebs cycle results in a decrease in acetyl-CoA in the mitochondrial matrix. In the cytoplasm acetyl-CoA is converted by acetyl-CoA carboxylase (ACC) to malonyl-CoA, which is an inhibitor of carnitine palmitoyltransferase I (CPT I) and thus the transfer of acyl groups into mitochondria for beta oxidation. ACC is inhibited upon its phosphorylation by adenosine monophosphate activated protein kinase (AMPK), and this reaction sequence serves as the basis for the hypothesis advocated in recent works, according to which malonyl-CoA is a key regulator of fatty acid oxidation [8]. However, as an alternative, malonyl-CoA could be considered as an inhibitory signal, while a key regulator of respiration rate and fatty acid oxidation may be the actual energy demand and workload [8]. Thus, decrease in malonyl-CoA may not be the reason but rather the consequence of the increases in workload and fatty acid oxidation. Malonyl-CoA seems to be at the end of this sequence of events, and the AMPK signaling pathway apparently modifies only this last step of regulation under cellular stress conditions where AMP/ATP ratio increases significantly [8].
In the presence of both carbohydrate substrates and free fatty acids, approximately 60% to 90% of oxygen consumed is used for oxidation of free fatty acids. According to the Randle hypothesis, if glucose and free fatty acids are both present, free fatty acids inhibit transport of glucose across the plasma membrane and slow down oxidation of glucose and pyruvate. Furthermore, aerobic glycolysis is limited by the necessity to maintain a low NADH/NAD+ ratio in the cytoplasm necessary for a high steady-state flux through the glyceraldehyde 3-phosphate dehydrogenase that is accomplished by transfer of reducing equivalents into the mitochondrial matrix by the malate-aspartate shuttle [8]. The shuttle becomes the rate limiting step at medium workloads, while the fatty acid pathway does not have this type of limitation. Nevertheless, flexibility of myocardial substrate metabolism appears to be important for cardiac health. Loss of this metabolic plasticity with overdependence of metabolism on an individual substrate can be seen in pathologic conditions with predominance in fatty acid metabolism in diabetic heart disease and accelerated glucose use associated with pressure overload left ventricular hypertrophy [9], which we wish to evaluate with imaging methods.
Since the myocardium’s preference for a given fuel substrate can be manipulated, quantitative evaluation of myocardial metabolism can be particularly challenging. For example, after 5–24 h of fasting, when circulating free fatty acids became high and insulin concentrations became low, myocardial glucose utilization rates in normal volunteers averaged only 0.24 ± 0.17 umol/min gm while ingestion of carbohydrates with stimulation of insulin secretion raised glucose utilization to 0.69 ± 0.11 umol/min gm [10]. Therefore, investigators should attempt to standardize the metabolic environment, requiring a steady-state condition for quantitative analysis of myocardial metabolism. In this respect, either the fasting state or the euglycemic hyperinsulinemic clamp, which produce stable insulin and glucose concentrations in plasma, are suitable for maintaining a metabolic steady-state [11]. The oral glucose-loaded state, on the other hand, results in a gradual change in plasma-glucose and insulin levels, which alter the metabolic rate of glucose in the myocardium. The euglycemic hyperinsulinemic clamp technique requires a steady IV infusion of insulin to be administered in one arm. The serum glucose level is “clamped” at a normal fasting concentration by administering a variable IV glucose infusion in the other arm [12]. However, the clamp technique is a rather cumbersome procedure, requiring frequent measurements of plasma-glucose and insulin levels. The fasting state, on the other hand, seems to be a simple and reliable choice for keeping metabolism in a stable condition. Therefore, when myocardial models are applied to the metabolic data, investigators should acknowledge the metabolic environment in which the data was obtained and whether the study subject is glucose intolerant to ensure that comparisons between data sets are made under equivalent metabolic conditions.
In Vivo Quantification of Myocardial Metabolism
Positron Emission Tomography
PET has been utilized in clinical practice for assessment of myocardial perfusion and metabolism, using predominantly qualitative (Fig. 1) or semiquantitative approaches. However, PET can also be used to assess tissue metabolism quantitatively. PET measures spatiotemporal concentrations of radiopharmaceuticals, such as radiolabeled glucose or fatty acids, in organs in absolute units [13]. These measurements can be performed noninvasively in human subjects or in animals such as rats, monkeys, or knockout mice. The resolution of PET scanners can be as low as 3 mm for human imaging and 1.3 mm for imaging small animals using a microPET camera. Positron emitting radioisotopes are composed of unstable parent nuclei which contain a greater number of high-energy protons than the number of neutrons. An unstable proton decays to a neutron in the nucleus, and a positron and a neutrino are emitted. The positron typically travels a distance of several millimeters or more, losing some of its energy, until it undergoes annihilation with an electron, resulting in the production of two anti-parallel 511-keV photons. These photons can be detected with a PET camera, which contains high-density crystal detectors and coincidence circuitry, and the data is reconstructed to create tomographic images [14].
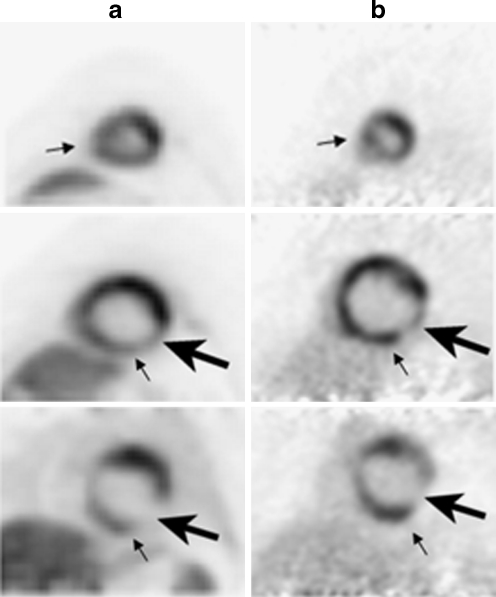
Fig. 1.
13N-ammonia axial PET myocardial perfusion images (column A) and 18F-FDG axial PET myocardial metabolism images (column B) in a 49-year-old male history of cardiomyopathy. Images demonstrate a mismatched pattern of decreased myocardial perfusion with preserved 18F-FDG metabolism (small arrows) in the inferoseptal and inferior walls, suggesting the presence of viable hibernating myocardium. Furthermore, there is a matched pattern of decreased myocardial perfusion and metabolism (large arrow) in the proximal to mid inferolateral wall, which is compatible with an infarct
One of the advantages of PET is that the positron emitting isotopes such as 15O, 13N, 11C, and 18F, which are generated in the cyclotron, can be directly substituted for oxygen, nitrogen, carbon, and hydrogen atoms in biological molecules. These PET radiopharmaceuticals, when injected into human subjects or animals at tracer concentrations, can be used to monitor biological processes without altering these processes. The sensitivity of PET for detection of radiotracers is very high, in the picomolar to femtomolar range. A radiotracer must be related to the process of concern and it is preferred that no labeled metabolites of the radiotracer accumulate in the tissue of interest. In general, the amount of the tracer should be at least several orders of magnitude smaller than that of the traced molecule. PET radiopharmaceuticals that have been useful in the evaluation of myocardial metabolism are listed in Table 1. Radiotracers can consist of radiolabeled natural substrates or chemical analogs. The advantage of using radiolabeled chemical analogs such as 18F-fluorodeoxyglucose (18F-FDG) as radiotracers is that they can be used to isolate selected portions of complex biochemical reaction sequences, such as the facilitated transport across cell membrane [14]. The advantage, on the other hand, of using a natural substrate such as glucose to create the radiotracer 11C-glucose is that there is no need for correction terms for different affinities of transporters and enzymes. However, 11C-glucose goes through many reaction steps in the glycolytic pathway before it is cleaved away as 11C-CO2 and soon found in other labeled compounds, making it a challenge to use this radiotracer to isolate a specific reaction sequence.
Table 1.
PET radiopharmaceuticals utilized in quantitative myocardial imaging, their physical half lives, and the biological parameters which the radiopharmaceuticals can be used to measure in heart tissue
| Radiopharmaceutical | Half life (min) | Parameter |
|---|---|---|
| 11C-acetate | 20 | Oxygen flux |
| 11C-glucose | 20 | Glucose flux |
| 18F-FDG | 110 | Glucose flux |
| 11C-palmitate | 20 | FFA flux |
| 18F-FTHA | 110 | FFA flux |
| 11C-lactate | 20 | Lactate flux |
| 15O-water | 2 | Blood flow |
| 13N-ammonia | 10 | Blood flow |
Quantitative biological reaction flux parameters, such as the metabolic rate of glucose, can be obtained with PET by performing (1) dynamic PET imaging, (2) arterial blood sampling, and (3) employing a kinetic model for a given radiopharmaceutical. First, however, a three-dimensional image of a phantom of known specific activity, containing radiotracer diluted in saline, is acquired to obtain a conversion factor, which is later used to convert activity data from tomographic images to absolute concentration units. Subsequently, the radiopharmaceutical of known specific activity is injected intravenously or inhaled as a bolus. Next, dynamic PET imaging is performed, which consists of an acquisition of many sequential data sets that are later used to generate tissue–time–activity curve (TAC) for a given volume of organ tissue outlined by a region of interest on the images. While dynamic images are being acquired, many sequential arterial blood samples are collected, which are later used to generate a plasma–time–activity curve. Finally, a kinetic compartment model or graphical analysis of data is used to obtain quantitative biological parameters such as flux levels of reactions. The plasma–time–activity curve serves as an input function for the kinetic compartment model, while the TAC serves as the output function [14]. Since arterial blood sampling is an invasive procedure, the input function may sometimes be approximated using data obtained through sampling of the mixed-venous blood or dynamic imaging of the radiotracer within the left ventricle. An ideal plasma–time–activity curve would only be composed of data from the authentic tracer; however, it is typically contaminated to various degrees with data from metabolized tracer. The degree of contamination is typically evaluated using HPLC column. Furthermore, it is important to identify the amount of radiotracer that persists in the plasma and the amount that is taken up by the red blood cells following intravenous or oral administration, in order to obtain an accurate input function.
Quantitative biologic parameters such as metabolic reaction rates can be obtained from PET data and arterial sampling data using (A) model driven methods or (B) data-driven methods. Model driven methods use compartments to describe the behavior of the tracer and to obtain micro system parameters, such as rate constants. Examples of biologic compartments include the plasma compartment, interstitial space and the intracellular compartment (Fig. 2). A compartment model can be reasonably analyzed with standard mathematical methods if a first-order process is assumed, where rate constants are independent of concentrations and time [14]. Processes involving PET imaging are typically pseudo first-order, since the concentration of one reactant, the radiotracer, is very small compared to the others, and equations simplify to the same form as for a first-order process. This modeling approach also assumes that the dynamic biological process is evaluated in a steady state, meaning that the rate of a process is not changing with time and the amount of traced molecule is constant during evaluation period. The radiotracer is considered to be distributed uniformly within a compartment and separate compartments can be combined to form a single compartment if the transport rates between them are not limiting. A compartment model is described in terms of linear first-order ordinary differential equations (Fig. 2). Compartment parameter estimates such as k 1, k 2, and k 3 rate constants are obtained by fitting the compartment models to measured data [14].
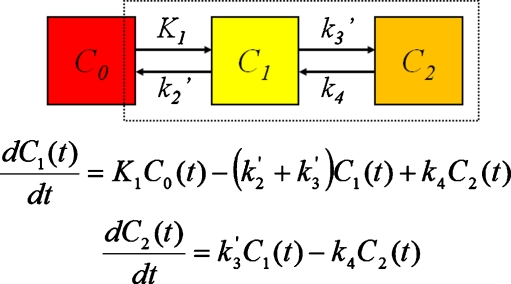
Fig. 2.
Two tissue compartment model described in terms of differential equations. C 0 represents radiotracer concentration in the blood pool compartment, C 1 in interstitial space, and C 2 in the intracellular compartment. K 1, k 2, k 3, and k 4 represent intercompartmental rate constants
Data-driven methods, on the other hand, such as graphical analysis or spectral analysis derive a macro system parameter, such as the influx rate constant K i, which is a composite of the k 1, k 2, and k 3 rate constants, from a less constrained description of the kinetic processes. The actual influx rate of a substrate can be obtained by multiplying the influx rate constant by the arterial concentration of the substrate. Data-driven methods require no a priori decision about the most appropriate model structure. The graphical methods, such as the Patlak plot for irreversible uptake (Fig. 3), employ a transformation of the data such that the linear regression of the transformed data yields to macro system parameter interest [14]. These methods are attractive and elegant due to their simplicity. However, they require the determination of when the plot becomes linear, which may be biased by statistical noise, and they do not return information about compartmental structure.
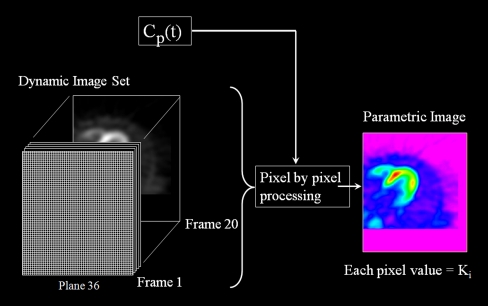
Fig. 3.
Dynamic image set of the myocardium obtained with 18F-FDG PET and the plasma–time–activity curve C p(t) are used to generate a parametric horizontal long axis image of the heart, where the value of each pixel represents the magnitude of the influx contestant K i. Pixel by pixel processing was performed using the Patlak graphical analysis
One of the disadvantages of PET is that all radioisotopes decay with photons having the same energy of 511 keV. Therefore, it is not possible to image several radiotracers simultaneously at different energy windows in order to evaluate myocardial metabolism of several different substrates. Instead, it is necessary to evaluate the metabolism of each substrate separately by performing sequential studies on a given subject. Once a subject has been injected with the first radiopharmaceutical, it is necessary to wait for that radiopharmaceutical to decay before another radiotracer can be injected. Therefore, it is advantageous for the radioisotopes to have short half lives for sequential metabolic myocardial PET imaging. An imaging protocol has been developed which takes advantage of the short half lives of the 15O and 11C radioisotopes to sequentially obtain quantitative measurements of myocardial blood flow (MBF) with 15O-water (imaging time of 5 min); myocardial oxygen consumption (MVO(2)) with 11C-acetate (30 min); myocardial glucose uptake and utilization (MGU) with 11C-glucose (60 min); and myocardial fatty acid uptake, utilization (MFAU), and oxidation (MFAO) with 11C-palmitate (30 min) in human subjects [15]. This protocol has been applied by Dávila-Román et al. [15], for instance, to quantitatively evaluate myocardial metabolism in patients with idiopathic dilated cardiomyopathy (IDCM). In this study, seven patients with IDCM and normal controls underwent positron emission tomography for measurements of MBF, MVO(2), MGU, MFAU, and MFAO. Dávila-Román et al. [15] demonstrated that patients with IDCM exhibit alterations in myocardial metabolism characterized by decreased fatty acid metabolism and increased myocardial glucose metabolism, a pattern similar to that shown in animal models of HF. Soto et al. [16] also utilized this serial quantitative myocardial imaging protocol to study the effect of endurance exercise training (EET) on myocardial substrate metabolism in older men and women. The authors found that EET in older individuals improves the catecholamine response of myocardial glucose metabolism.
Although this serial imaging protocol enables quantification of myocardial metabolism of several important substrates, it does not evaluate the metabolism of lactate, another important substrate for the heart. Noninvasive quantitative measurements of myocardial lactate metabolism have been difficult to perform in humans. Herrero et al. performed a pilot study on dogs, showing that under conditions of net lactate extraction, L-3-(11)C-lactate faithfully traces myocardial metabolism of exogenous lactate and that measurements of lactate metabolism are feasible with PET using myocardial clearance analysis (oxidation) or compartmental modeling [17]. A large body of evidence has demonstrated that an uncoupling of glycolysis from glucose oxidation resulting in myocardial acidosis can contribute to a number of cardiac pathologies. Another limitation of PET has been the inability to directly separate out rates of glycolysis from glucose oxidation with currently available tracers, although recently it has been demonstrated that PET with 11C-glucose can be utilized to estimate rates of glycogen synthesis, glycolysis, and glucose oxidation [18].
Although radioisotopes with short half lives allow for sequential quantitative metabolic myocardial PET imaging of several different substrates, the short half lives prevent these tracers from being used in clinical practice at sites where a cyclotron is not available. For this reason, radiotracers with longer half lives have been implemented, such as 18F-fluorodeoxyglucose to analyze myocardial glucose metabolism and 18F-fluoro-6-thia-heptadecanoic acid (18F-FTHA) to assess myocardial fatty acid metabolism. Taylor et al. [19], for instance, conducted an evaluation of myocardial fatty acid and glucose uptake using PET with 18F-FTHA and 18F-FDG in patients with congestive heart failure. From the dynamic image data, the fractional uptake rates (K i) were determined for 18F-FTHA and 18F-FDG. Subsequently, serum free fatty acid and glucose concentrations were used to calculate the myocardial free fatty acid and glucose uptake rates. In contrast to results obtained by Dávila-Román et al. for patients with IDCM, Taylor et al. found that myocardial fatty acid uptake rates in heart failure are higher than expected for the normal heart, whereas myocardial glucose uptake rates are lower. This shift in myocardial substrate use may be an indication of impaired energy efficiency in the failing heart, providing a target for therapies directed at improving myocardial energy efficiency. Furthermore, the authors demonstrated that myocardial free fatty acid and glucose use in heart failure can be quantitatively assessed using PET with 18F-FTHA and 18F-FDG. However, since 18F-FDG is a chemical analog it may not be as accurate at evaluating the metabolism of the traced molecule glucose as the radiolabeled natural substrate 11C-glucose. Indeed, Herrero et al. [20] have demonstrated that over a wide range of conditions, PET-derived measurements of glucose uptake rate are obtained more accurately with 11C-glucose than with 18F-FDG.
Magnetic Resonance Spectroscopy
While MRI provides information on the spatial location and local chemical environment of protons, MRS provides a biochemical analysis of the myocardial tissue, making it akin to a virtual biopsy of the heart. The basis for nuclear magnetic resonance (NMR) is the quantum interaction between a nuclear spin of nuclei such as 1H, 31P, 23Na, and 13C, and an external magnetic field. These nuclei release a frequency signal in the range of normal radio waves when perturbed by a radiofrequency pulse [21]. The strength of the signal depends on the strength of the external magnetic field. The greater the field strength, the stronger is the observed signal and the better the signal to noise ratio of the observed spectrum. (31)P-MRS allows for the detection of adenosine triphosphate, phosphocreatine (PCr), inorganic phosphate (Pi), and intracellular pH. In addition to the measurement of 31P metabolites, MRS 1H, and 13 C MRS have also been utilized in animal studies [22, 23]. Szczepaniak et al. [22] demonstrated for the noninvasive in vivo 1H-MRS measurement of intracellular TG is feasible within myocytes at 1.5-T field strengths and is comparable in accuracy to biochemical measurement. The additional application of 1H-MRS has allowed for the detection of total creatine, enabling an in depth examination of the creatine kinase system [24]. All these compounds are involved in the regulation of the available energy from ATP hydrolysis via the creatine kinase (CK) reaction [25]. PCr forms the primary ATP buffer in the cell via the CK reaction and is involved in transporting the chemical energy from the ATP-producing mitochondria to the ATP-consuming contractile proteins [24].
The MR signal of 31P tends to be relatively weak, similar to that of 1H-MRS for compounds other than water and lipids containing 1H. The low signal of 31P can be partially compensated by using techniques such as nuclear overhauser enhancement, which increase the signal of 31P by pre-excitation of the adjacent protons. The “scarcity” of the MR signal of 31P limits spatial resolution. Several different techniques can be used to obtain MRS spectral data, including point-resolved spectroscopy and stimulated echo acquisition mode, which are more typically applied with 1H-MRS, and depth-resolved spectroscopy. Image-selected in vivo spectroscopy demonstrates high spatial resolution and reduced contamination but greater sensitivity to motion artifacts and cardiac gating. Chemical-shift imaging is capable of obtaining multiple spectra relative to the voxels of a single slice (two dimensional) or several slices (three dimensional) [25].
MRS can provide metabolic insights into the role of cardiac metabolism, in particular, cardiac energetics, in various conditions, including hypertensive, valvular, and ischemic heart disease; heart failure; cardiac transplantation; and cardiomyopathies [26]. Furthermore, response to therapeutic intervention can be monitored using this method. To study the metabolic changes that occur in heart failure, (31)P- and (1)H-MRS have been applied in both patients and experimental animal studies. MRS examination of the heart failure has revealed that the PCr/CK system is impaired. PCr levels as well as total creatine levels are reduced in patients and experimental models and in severe heart failure ATP is also reduced. These findings have led to the concept that the heart is energy starved [24]. PCr/ATP ratios correlate with the clinical severity of heart failure and are a prognostic indicator of mortality. Experimental studies suggest that these changes can result in increased free ADP levels when the failing heart is stressed. Increased free ADP levels, in turn, result in a reduction in the available free energy of ATP hydrolysis, which may directly contribute to contractile dysfunction. MRS can also be used to identify and assess viable myocardium, although measurement of absolute concentrations of high-energy phosphates is required to assess the extent of viable tissue loss, as necrosis and scar formation result in reduced levels of phosphocreatine and ATP. Accordingly, in patients with fixed 201-Thallium defects, the absolute content of myocardial ATP as measured by 31P-MRS is reduced, whereas it remains constant in those with viable myocardium [27].
MRS is generally more suitable for the evaluation of myocardium with a spatially homogeneous pathology, such as dilated cardiomyopathy. However, application of a technique called spectral localization with optimum point spread function (SLOOP) allows for the examination of voxel of any shape, in contrast to conventional rectangular voxels. Matching voxel shape to the curvature of the heart helps to increase the signal to noise ratios and avoid contamination from myocardial tissue outside the region of interest and from skeletal muscle and blood. The latter is particularly relevant for 31P MRS, since blood gives rise to a signal for 2,3-diphosphoglycerate, which resonates at a frequency close to Pi, making Pi quantification as well as determination of the intracellular pH difficult. The application of SLOOP allows for the measurements of absolute concentrations of PCr and ATP. Using (31)P saturation transfer MRS techniques it is possible to measure flux through the CK reaction in the intact heart [24]. The application of this technique has proven that flux is reduced in failing myocardium in both experimental models and in patients. The study of transgenic animal models by MRS has also been performed and has led to further insights into the role of energy metabolism in heart failure, suggesting that an intact creatine/CK system is critical for situations of cardiac stress [25]. Nevertheless, MRS is currently used primarily as a research tool, since low spatial and temporal resolution and low reproducibility preclude its diagnostic use in clinical practice. Future technical developments and use of higher magnetic field strengths, such as the 7 Tesla systems, will enable improvements in resolution and reproducibility that may take cardiac MRS into the clinical realm [28]. It is hoped that enrichment studies using C-13 labeled precursors, which have been performed in animals, may also be performed in humans to measure parameters such as the metabolic rate of glucose in vivo.
Single Photon Emission Tomography
Single photon emission tomography has been extensively used in clinical practice for evaluation of relative myocardial perfusion and function. Therefore, SPECT cameras are widely available in clinical and research settings. Unlike PET radioisotopes, many SPECT radioisotopes decay at different energies and, therefore, several different biological processes can theoretically be imaged simultaneously with SPECT at different energy windows. Furthermore, unlike PET, which utilizes coincidence circuitry to detect two anti-parallel photons, SPECT cameras are designed to detect single photons. Only photons traveling parallel to the detector crystal are detected and all other photons are discarded by a collimator. Since SPECT cameras typically do not contain a transmission source, SPECT is typically able to identify only relative changes in concentration of the radiopharmaceutical rather than absolute concentrations. In this respect, the role of SPECT in the quantitation of myocardial metabolism is limited. Table 2 compares the advantages and disadvantages of PET, MRS, and SPECT for quantification of myocardial metabolism. Despite these limitations, Okizaki et al. were able to perform a compartment model analysis of myocardial fatty acid metabolism in patients with hypertrophic cardiomyopathy using SPECT with 123I-beta-methyl-iodophenylpentadecanoic acid (123I-BMIPP), a fatty acid analog and 99mTc-tetrofosmin perfusion radiotracer [29]. Using the myocardial and blood pool time-activity curves, 123I-BMIPP pharmacokinetics were analyzed through a two-compartment model. k 1 and k 2 were defined as influx and outflux rate constants between blood and myocardial reversible component, and k 3 as the specific uptake rate constant between myocardial reversible and irreversible compartments. The authors concluded that k 3 might be a sensitive predictor for early detection of hypertrophic cardiomyopathy, and k 1/k 2 could be a useful index to evaluate its progression. Yazaki et al. performed a study to determine the clinical and prognostic value of identifying metabolic abnormalities of myocardial fatty acid metabolism in idiopathic dilated cardiomyopathy using 123I-BMIPP in 32 patients [30]. They found that the extent of the abnormality of myocardial fatty acid metabolism in idiopathic dilated cardiomyopathy reflected the severity of hemodynamic deterioration and histopathological changes. However, it should be noted that the actual kinetics of 123I-BMIPP are complex and have not been validated in heart failure. Another disadvantage of SPECT is that there are currently no available SPECT radiotracers to measure myocardial glucose metabolism, although a SPECT camera may theoretically be fitted with an appropriate collimator to image18F-FDG.
Table 2.
Advantages and disadvantages of PET, MRS, and SPECT in evaluation of myocardial metabolism
| Advantages | Disadvantages | |
|---|---|---|
| PET | Quantitative metabolic and blood flow measurements; high sensitivity; radiotracers as natural substrates and chemical analogs; many metabolic radiotracers available | Cyclotron needed on site due to short half lives; only one process can be evaluated at a time; difficulty with separating reaction steps in a pathway |
| MRS | Simultaneous measurements of multiple metabolic pathways and function; ease in performing serial measurements, no ionizing radiation; higher field strength in small animal imaging | Poor sensitivity (millimolar); low signal to noise ratio; intravoxel signal contamination; long acquisition times; measurement limited to anterior wall in humans |
| SPECT | High sensitivity; widely available; long half lives facilitate radiopharmaceutical delivery; simultaneous imaging of two radiopharmaceuticals with different energies; ECG gating for myocardial function | Inability to quantify processes with lack of attenuation correction; poor temporal and spatial resolution; few radiopharmaceuticals available for evaluation of metabolism |
Fick Method and Metabolic Profiling
The Fick method was first devised as a technique for measuring cardiac output; however, its underlying principles may be also applied to measure uptake and secretion rates of substrates by the myocardium. Myocardial substrate uptake rates are calculated by taking the product of myocardial perfusion and the difference of substrate concentrations in arterial blood and venous blood from the coronary sinus. Myocardial perfusion can be obtained with the 133-Xenon washout technique, which utilizes a scintigraphic detector, or approximated via the thermodilution technique [31]. Unlike PET, SPECT, and MRS, the Fick method is an invasive procedure, but can still be carried out in human subjects in vivo. While imaging technologies are able to quantify only several metabolic parameters in the myocardium at a time, the Fick method can provide a more complete evaluation of metabolism. Wisneski et al. used the Fick method to demonstrate that the circulating level of free fatty acids plays a major role in determining the amount of glucose extracted and oxidized by the normal human myocardium [32]. In this study coronary sinus and arterial catheters were placed in 23 healthy male volunteers and 6-14C glucose and U-13C lactate were infused. Simultaneous blood samples were obtained for chemical analyses of glucose, lactate, and free fatty acids and for the isotopic analyses of glucose and lactate.
The Fick has been used to quantify a large number metabolic processes in the myocardium at a time, including fluxes of oxygen, glucose, fatty acids, lactate, pyruvate, beta-hydroxybutyrate, and amino acids such as glutamate, alanine, and aspartate. Since these data sets are more extensive than those obtained with imaging methods, they may be more amendable to mathematical models such as constraint-based metabolic network modeling. Vanky et al. [31] applied the Fick method to study myocardial uptake of oxygen and energy substrates before and after valve replacement for aortic stenosis. In this study, preoperative and postoperative metabolic adaptation with substantial uptake of glutamate, previously claimed to be due to chronic or repetitive ischemia, was demonstrated.
Recent advances in the application of the Fick Method include high-throughput approaches such as mass spectrometry to perform metabolic profiling of the myocardial substrate metabolism in vivo. A study by Turer et al. [33], for instance, applied a mass spectrometry-based platform to profile 63 intermediary metabolites in serial paired peripheral arterial and coronary sinus blood effluents obtained from 37 patients undergoing cardiac surgery, stratified by presence of coronary artery disease or left ventricular dysfunction. The authors found that the dysfunctional ventricle is characterized by global suppression of metabolic fuel uptake and limited myocardial metabolic reserve and flexibility after global ischemia/reperfusion stress in the setting of cardiac surgery. Altered metabolic profiles after ischemia/reperfusion were associated with postoperative hemodynamic course, suggesting a role for perioperative metabolic monitoring and targeted optimization in cardiac surgical patients. Nevertheless, one of the disadvantages of the Fick method is that it only provides quantitative metabolic flux measurements of the entire myocardium, while the tomographic imaging methods can provide information on regional myocardial metabolism and evaluate its heterogeneity.
Modeling of Myocardial Metabolism
Constraint-Based Metabolic Network Model
Constraint-based modeling developed out of the availability of fully sequenced genomes in the mid-1990s, when it became possible to compile complete parts lists initially for bacterial organisms [34] and eventually humans [35, 36]. The generation of these large volumes of data created the need to organize this vast biological information. Hence, a systems approach was required that could be used to construct genome or organelle-scale models using large, yet still incomplete data sets. This information, coupled to the rich biochemistry literature, notable for fundamental work elucidating biochemical mechanisms of action and catalysis, enabled the definition of metabolic stoichiometric networks and the subsequent ability to formulate the networks in terms of a linear programming problem [37]. The underlying principle of this modeling approach is to balance the inputs and outputs of a system at a non-equilibrium steady state (Fig. 4). This approach has many benefits, including the flexibility for incorporating and integrating various data types and incomplete data sets. A challenge resulting from these benefits is the fact that there exist many points that satisfy the solution space. This problem can be alleviated in part through the choice of an appropriate metabolic objective function, such as biomass production in bacteria grown in the laboratory or ATP synthesis in the cardiomyocyte. Evaluation of existing objective functions and investigation of new and alternative functions is an active area of development in this field [38]. Additionally, constraint-based modeling problems can be carried out without specifying an objective function through randomly sampling the feasible solution space, thus calculating flux probability distributions for every reaction within the network [39]. There are numerous other methods that can be applied to the network and newer ones continue to be developed as part of the constraint-based modeling approach [40].
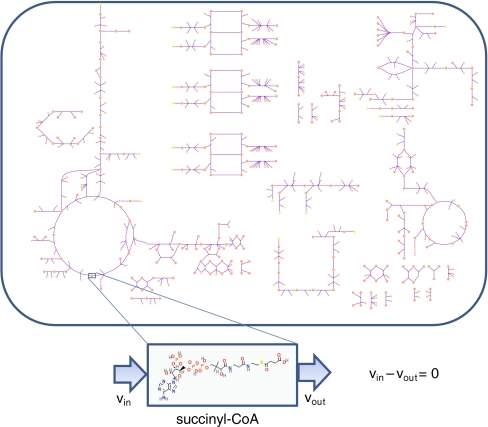
Fig. 4.
Human cardiomyocyte mitochondria; a bird’s eye view of the cardiomyocyte mitochondria metabolic network. Each node represents a chemical species and each line represents an enzymatic (or spontaneous) reaction converting substrates into products. Elementally charge and mass balanced equations are specified for every reaction in the network. Application of steady-state conditions amounts to balancing each element constituent of the chemical species
The flexibility of incorporating disparate data sets that attempt to be comprehensive, but are known to be incomplete, is a strong advantage of constraint-based network modeling [41]. Proteomic data has, for example, been used in conjunction with biochemical data in the literature (the bibliome) to generate a reconstruction of the human cardiomyocyte. Initial analyses using data from the literature enabled the postulation of alternate mechanisms for regulation of key enzyme fluxes, such as pyruvate dehydrogenase in diabetic conditions [42]. The use of gene–protein–reaction relationships [43] in genome or organelle-scale models enables mapping of various high-throughput data on networks for analysis. This can be used to analyze systemic consequences of genetic mutations and genotype–phenotype relationships. For example, the analysis of multiple causal single nucleotide polymorphisms within the network can be assessed to evaluate the influence of genetic mutations on different reactions within the cardiomyocyte mitochondria, and to subsequently carry out correlations among the different reactions to find a smaller, unique set of predicted phenotypes [44]. MRS data, which is becoming increasing available, also has applications for analysis of genome scale models. Studies have recently been carried out using 1H NMR spectroscopy data investigating systemic effects of hypoxia in Drosophila [45]. The use of 13C-isotopomer-labeled metabolic substrates, such as glucose and long-chain fatty acids, can be used to trace fluxes of molecules within network and to analyze them under mass conservation constraints [46]. Analysis of isotopomer uptake experiments from perfused mouse hearts via an in silico cardiomyocyte model enabled the identification of fluxes through various pathways. A study of the mouse cardiomyocyte using 13C-labeled glucose and fatty acids suggested a small role for anapleurotic fluxes, a predominant uptake of glucose over the 18 carbon fatty acids, but approximately 1/2 of the contribution to ATP production originating from glucose when compared to oleic acid [47]. While the development of constraint-based genome or cell scale models is still in relatively early stages, developments made to date, particularly in bacterial organisms, show great promise and potential in aiding the study of complex multi-cellular systems. Imaging technologies such as MRS and PET, coupled with blood flow measurements, may aid in the determination of multiple small metabolite input/output fluxes for metabolic network models. These measured parameters can serve as constraints on the networks and enable a more comprehensive systems analysis, with clinical applications spanning from classification and diagnosis to potentially serving as prognostic measures, provided that correlations to clinical phenotypes are performed.
Oxygen Transport and Cellular Energetics Model
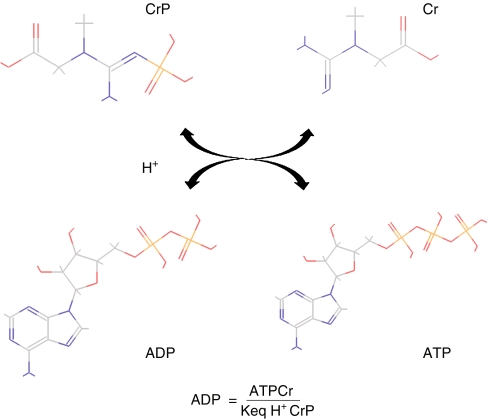
From a metabolic perspective, cardiac function is to pump blood to the pulmonary and systemic circulation and to perfuse all tissues, thus providing oxygen and substrates and removing waste products. High-energy phosphate bonds serve as the cells’ energy currency, which is stored in the adenosine phosphate and creatine phosphate pools in heart tissue. The chemical reaction depicted in Fig. 5 summarizes core energy transfer in cardiac tissue, coupled to the ATP hydrolysis reaction. The significance and potential practical applications of 31P MRS have long been recognized as a means for carrying out measurements on intact tissue [48]. Under the assumption of constant total creatine during the experimental course, the ADP concentration can be calculated, and other phosphate-bound metabolites can be identified and quantified from the MRS spectra. This enables assessment of in vivo energy states of the cardiomyocyte and comparisons can be carried out under different experimental conditions investigating the effects of hypoxia and/or ischemia. Some initial observations, such as the constancy of ATP, inorganic phosphate, and creatine phosphate levels as well as a two- to threefold increase in oxygen demand [49] accompanied by changes in ratios of the metabolites (e.g., ATP/ADP ratio) with ischemia all highlight that complex control mechanisms are involved. These observations stimulated the development of more complex models [50–52] for exploring mechanisms of regulation of metabolism in normal as well as pathophysiological conditions. Recent studies suggest that inorganic phosphate may be a critical factor in feedback and prediction of cardiac capacity under normal and ischemic conditions [53]. More recently the size of the adenine nucleotide pool has been shown to be an important parameter for characterizing left ventricular hypertrophic heart failure in canines [54]. This finding is consistent with earlier statements that key metabolite moiety pool sizes are important in determining cellular function [55]. Interestingly, the pool size of nucleotide precursors has been identified as an important indicator of function in other tissues as well [56]. Developments such as these may provide investigators with potential biomarkers for qualitative or even quantitative metabolic assessment of cardiac status in the clinical setting.
Fig. 5.
The creatine kinase reaction allows exchange between creatine phosphate and ATP for energy storage via the high-energy phosphate bond
Integration of Data and Organization of Efforts
Integration is the central theme of Systems Biology. However, given the complexity of biological systems, “integration” may refer to several different concepts, such as the integration of data across spatial hierarchies, temporal hierarchies, or various biological processes. Developments in the integration of metabolism with cardiac function include incorporation of excitation–contraction coupling, ATP generation [57], and molecular contraction mechanisms [58]. As more and more complex data sets became available and a wide range of models describing biological functions (from excitation–contraction coupling to metabolism to mechanical contraction) were developed, it was recognized that there would be a need to organize these efforts. Anticipating these developments, a number of organizations and initiatives have been underway to meet these new developments and to maximize the utility of these efforts [59–61]. The Human Physiome Project, launched by the International Union of Physiological Sciences in 1997, was one of the earlier efforts recognizing the importance of incorporating data obtained with imaging modalities, such as PET and MRS, in future applications of myocardial metabolism modeling. Aside from the technical challenges of experimental tools and methods to develop these projects, there is a concurrent need to maintain a set of standards. This has been recognized as a challenge in mathematical modeling fields [62] with regards to criteria for developing and assessing model quality, as well as availability of formats that are accessible to the scientific community. Fortunately, these issues have been recognized and are addressed in the Human Physiome Project [63]. While the challenges are notable, the prospects of even small advances warrant further efforts in the integration of in vivo imaging data and mathematical models of myocardial metabolism.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.AHA Scientific Statement Assessment of coronary artery disease by cardiac computed tomography A scientific statement from the American Heart Association Committee on Cardiovascular Imaging and Intervention, Council on Cardiovascular Radiology and Intervention, and Committee on Cardiac Imaging, Council on Clinical Cardiology. Circulation. 2006;114:1761–1791. doi: 10.1161/CIRCULATIONAHA.106.178458. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell J, Link J, Levy W, Poole J, Stratton J. Evidence for pre- to post synaptic mismatch of the cardiac sympathetic nervous system in ischemic congestive heart failure. Journal of Nuclear Medicine. 2008;49(2):234–241. doi: 10.2967/jnumed.107.044339. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Bax J, Henneman M, Boogers M, Ernest VG. Is nuclear imaging a viable alternative technique to assess dyssynchrony? Europace. 2008;10(Supplement3):iii101–iii105. doi: 10.1093/europace/eun221. [DOI] [PubMed] [Google Scholar]
- 4.Ukkonen H, Sundell J, Knuuti J. Effects of CRT on myocardial innervations, perfusion and metabolism. Europace. 2008;10(Supplement 3):iii114–iii117. doi: 10.1093/europace/eun228. [DOI] [PubMed] [Google Scholar]
- 5.Fragasso G, Palloshi A, Puccetti P, Silipigni C, Rossodivita A, Pala M, Calori G, Alferi O, Margonato A. A randomized clinical trial of trimetazidine, a partial free fatty acid oxidation inhibitor, in patients with heart failure. JACC. 2006;48:5. doi: 10.1016/j.jacc.2006.03.060. [DOI] [PubMed] [Google Scholar]
- 6.Tuunanen H, Engblom E, Naum A, Nagren K, Scheinin M, Hesse B, Airaksinen J, Nuutila P, Ioo P, Ukkonen H, Opie L, Knuuti J. Trimetazidine, a metabolic modulator, has cardiac and extracardiac benefits in idiopathic dilated cardiomyopathy. Circles. 2008;118:000–000. doi: 10.1161/CIRCULATIONAHA.108.778019. [DOI] [PubMed] [Google Scholar]
- 7.Bailey JE. Mathematical modeling and analysis in biochemical engineering: Past accomplishments and future opportunities. Biotechnology Progress. 1998;14(1):8–20. doi: 10.1021/bp9701269. [DOI] [PubMed] [Google Scholar]
- 8.Valdur S. Molecular system bioenergetics: Energy for life. Weinheim: WILEY-VCH; 2007. pp. 367–405. [Google Scholar]
- 9.Stanely WC, et al. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 10.Schelbert HR, Henze E, Sochor H, Grossman RG, Huang SC, Barrio JR, Schwaiger M, Phelps ME. Effects of substrate availability on myocardial C-11 palmitate kinetics by positron emission tomography in normal subjects and patients with ventricular dysfunction. American Heart Journal. 1986;111(6):1055–1064. doi: 10.1016/0002-8703(86)90006-2. [DOI] [PubMed] [Google Scholar]
- 11.Tamaki N, Yonekura Y, Kawamoto M, Magata Y, Sasayama S, Takahashi N, Nohara R, Kambara H, Kawai C, Konishi J. Simple quantification of regional myocardial uptake of fluorine-18-deoxyglucose in the fasting condition. Journal of Nuclear Medicine. 1991;32(11):2152–2157. [PubMed] [Google Scholar]
- 12.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: A method for quantifying insulin secretion and resistance. American Journal of Physiology. 1979;237(3):E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 13.Bengel FM, Higuchi T, Javadi MS, Lautamäki R. Cardiac positron emission tomography. Journal of the American College of Cardiology. 2009;54(1):1–15. doi: 10.1016/j.jacc.2009.02.065. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt KC, Turkheimer FE. Kinetic modeling in positron emission tomography. Quarterly Journal of Nuclear Medicine. 2002;46(1):70–85. [PubMed] [Google Scholar]
- 15.Dávila-Román VG, Vedala G, Herrero P, de las Fuentes L, Rogers JG, Kelly DP, Gropler RJ. Altered myocardial fatty acid and glucose metabolism in idiopathic dilated cardiomyopathy. Journal of the American College of Cardiology. 2002;40(2):271–277. doi: 10.1016/S0735-1097(02)01967-8. [DOI] [PubMed] [Google Scholar]
- 16.Soto PF, Herrero P, Schechtman KB, Waggoner AD, Baumstark JM, Ehsani AA, Gropler RJ. Exercise training impacts the myocardial metabolism of older individuals in a gender-specific manner. American Journal of Physiology Heart and Circulatory Physiology. 2008;295(2):H842–H850. doi: 10.1152/ajpheart.91426.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero P, Dence CS, Coggan AR, Kisrieva-Ware Z, Eisenbeis P, Gropler RJ. L-3-11C-lactate as a PET tracer of myocardial lactate metabolism: A feasibility study. Journal of Nuclear Medicine. 2007;48(12):2046–2055. doi: 10.2967/jnumed.107.044503. [DOI] [PubMed] [Google Scholar]
- 18.Herrero P, et al. PET measurements of myocardial glucose metabolism with 1-11C-glucose and kinetic modeling. Journal of Nuclear Medicine. 2007;48(6):955–964. doi: 10.2967/jnumed.106.037598. [DOI] [PubMed] [Google Scholar]
- 19.Taylor M, Wallhaus TR, Degrado TR, Russell DC, Stanko P, Nickles RJ, Stone CK. 18F-FTHA An evaluation of myocardial fatty acid and glucose uptake using PET with [18F]fluoro-6-thia-heptadecanoic acid and [18F]FDG in patients with congestive heart failure. Journal of Nuclear Medicine. 2001;42(1):55–62. [PubMed] [Google Scholar]
- 20.Herrero P, Sharp TL, Dence C, Haraden BM, Gropler RJ. Comparison of 1-(11)C-glucose and (18)F-FDG for quantifying myocardial glucose use with PET. Journal of Nuclear Medicine. 2002;43(11):1530–1541. [PubMed] [Google Scholar]
- 21.Sardanelli F, Quarenghi M. MR spectroscopy of the heart. Radiologia Medica. 2006;111(8):1025–1034. doi: 10.1007/s11547-006-0102-8. [DOI] [PubMed] [Google Scholar]
- 22.Szczepaniak LS, et al. Measurement of intracellular triglyceride stores by H spectroscopy: Validation in vivo. American Journal of Physiology. 1999;276(5 Pt 1):E977–E989. doi: 10.1152/ajpendo.1999.276.5.E977. [DOI] [PubMed] [Google Scholar]
- 23.Lewandowski ED. Cardiac carbon 13 magnetic resonance spectroscopy: On the horizon or over the rainbow? Journal of Nuclear Cardiology. 2002;9(4):419–428. doi: 10.1067/mnc.2002.125811. [DOI] [PubMed] [Google Scholar]
- 24.Ten Hove M, Neubauer S. The application of NMR spectroscopy for the study of heart failure. Current Pharmaceutical Design. 2008;14(18):1787–1797. doi: 10.2174/138161208784746743. [DOI] [PubMed] [Google Scholar]
- 25.Ten Hove M, Neubauer S. MR spectroscopy in heart failure—Clinical and experimental findings. Heart Failure Reviews. 2007;12(1):48–57. doi: 10.1007/s10741-007-9003-8. [DOI] [PubMed] [Google Scholar]
- 26.Hudsmith LE, Neubauer S. Magnetic resonance spectroscopy in myocardial disease. JACC Cardiovasc Imaging. 2009;2(1):87–96. doi: 10.1016/j.jcmg.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Yabe T, Mitsunami K, Inubushi T, Kinoshita M. Quantitative measurements of cardiac phosphorus metabolites in coronary artery disease by 31P magnetic resonance spectroscopy. Circulation. 1995;92(1):15–23. doi: 10.1161/01.cir.92.1.15. [DOI] [PubMed] [Google Scholar]
- 28.Hudsmith LE, Neubauer S. Detection of myocardial disorders by magnetic resonance spectroscopy. Nat Clin Pract Cardiovasc Med. 2008;5(Suppl 2):S49–S56. doi: 10.1038/ncpcardio1158. [DOI] [PubMed] [Google Scholar]
- 29.Okizaki A, Shuke N, Sato J, Sasaki T, Hasebe N, Kikuchi K, Aburano T. A compartment model analysis for investigation of myocardial fatty acid metabolism in patients with hypertrophic cardiomyopathy. Nuclear Medicine Communications. 2007;28(9):726–735. doi: 10.1097/MNM.0b013e32828da1c7. [DOI] [PubMed] [Google Scholar]
- 30.Yazaki Y, et al. Assessment of myocardial fatty acid metabolic abnormalities in patients with idiopathic dilated cardiomyopathy using 123I BMIPP SPECT: Correlation with clinicopathological findings and clinical course. Heart. 1999;81(2):153–159. doi: 10.1136/hrt.81.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanky FB, Hakanson E, Szabó Z, Jorfeldt L, Svedjeholm R. Myocardial metabolism before and after valve replacement for aortic stenosis. Journal of Cardiovascular Surgery (Torino) 2006;47(3):305–313. [PubMed] [Google Scholar]
- 32.Wisneski JA, et al. Metabolic fate of extracted glucose in normal human myocardium. Journal of Clinical Investigation. 1985;76(5):1819–1827. doi: 10.1172/JCI112174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turer AT, Stevens RD, Bain JR, Muehlbauer MJ, van der Westhuizen J, Mathew JP, Schwinn DA, Glower DD, Newgard CB, Podgoreanu MV. Circulation. 2009;119(13):1736–1746. doi: 10.1161/CIRCULATIONAHA.108.816116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 35.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Venter JC, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 37.Schilling CH, Edwards JS, Palsson BO. Toward metabolic phenomics: Analysis of genomic data using flux balances. Biotechnology Progress. 1999;15(3):288–295. doi: 10.1021/bp9900357. [DOI] [PubMed] [Google Scholar]
- 38.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3:119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schellenberger J, Palsson BO. Use of randomized sampling for analysis of metabolic networks. Journal of Biological Chemistry. 2009;284(9):5457–5461. doi: 10.1074/jbc.R800048200. [DOI] [PubMed] [Google Scholar]
- 40.Palsson BO. Systems biology: Determining the capabilities of reconstructed networks. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 41.Vo TD, Greenberg HJ, Palsson BO. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. Journal of Biological Chemistry. 2004;279(38):39532–39540. doi: 10.1074/jbc.M403782200. [DOI] [PubMed] [Google Scholar]
- 42.Thiele I, Price ND, Vo TD, Palsson BØ. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. Journal of Biological Chemistry. 2005;280(12):11683–11695. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 43.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nature Reviews Cancer. 2006;7(2):130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 44.Jamshidi N, Palsson BØ. Systems biology of SNPs. Mol Syst Biol. 2006;2:38. doi: 10.1038/msb4100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feala JD, Coquin L, McCulloch AD, Paternostro G. Flexibility in energy metabolism supports hypoxia tolerance in Drosophila flight muscle: Metabolomic and computational systems analysis. 2007;3:99. doi: 10.1038/msb4100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiechert W, de Graaf AA. In vivo stationary flux analysis by 13C labeling experiments. Advances in Biochemical Engineering/Biotechnology. 1996;54:109–154. doi: 10.1007/BFb0102334. [DOI] [PubMed] [Google Scholar]
- 47.Vo TD, Palsson BO. Isotopomer analysis of myocardial substrate metabolism: A systems biology approach. Biotechnology and Bioengineering. 2006;95(5):972–983. doi: 10.1002/bit.21063. [DOI] [PubMed] [Google Scholar]
- 48.Burt CT, Cohen SM, Barany M. Analysis with intact tissue with 31P NMR. Annu Rev Biophys Bioeng. 1979;8:1–25. doi: 10.1146/annurev.bb.08.060179.000245. [DOI] [PubMed] [Google Scholar]
- 49.Balaban RS, et al. Relation between work and phosphate metabolite in the in vivo paced mammalian heart. Science. 1986;232(4754):1121–1123. doi: 10.1126/science.3704638. [DOI] [PubMed] [Google Scholar]
- 50.Cortassa S, et al. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophysical Journal. 2003;84(4):2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korzeniewski B, Noma A, Matsuoka S. Regulation of oxidative phosphorylation in intact mammalian heart in vivo. Biophysical Chemistry. 2005;116(2):145–157. doi: 10.1016/j.bpc.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Wu F, et al. Computer modeling of mitochondrial tricarboxylic acid cycle, oxidative phosphorylation, metabolite transport, and electrophysiology. Journal of Biological Chemistry. 2007;282(34):24525–24537. doi: 10.1074/jbc.M701024200. [DOI] [PubMed] [Google Scholar]
- 53.Wu F, et al. Phosphate metabolite concentrations and ATP hydrolysis potential in normal and ischaemic hearts. Journal of Physiology. 2008;586(Pt 17):4193–4208. doi: 10.1113/jphysiol.2008.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu F, Zhang J, Beard DA. Experimentally observed phenomena on cardiac energetics in heart failure emerge from simulations of cardiac metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7143–7148. doi: 10.1073/pnas.0812768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reich J, Selkov E. Energy metabolism of the cell: A theoretical treatise. New York: Academic Press; 1981. [Google Scholar]
- 56.Jamshidi N, Palsson BO. Using in silico models to simulate dual perturbation experiments: Procedure development and interpretation of outcomes. BMC Syst Biol. 2009;3:44. doi: 10.1186/1752-0509-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuoka S, et al. Simulation of ATP metabolism in cardiac excitation-contraction coupling. Progress in Biophysics and Molecular Biology. 2004;85(2–3):279–299. doi: 10.1016/j.pbiomolbio.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Campbell SG, et al. Mechanisms of transmurally varying myocyte electromechanics in an integrated computational model. Philos Transact A Math Phys Eng Sci. 2008;366(1879):3361–3380. doi: 10.1098/rsta.2008.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bassingthwaighte J, Hunter P, Noble D. The cardiac physiome: Perspectives for the future. Experimental Physiology. 2009;94(5):597–605. doi: 10.1113/expphysiol.2008.044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fenner JW, et al. The EuroPhysiome, STEP and a roadmap for the virtual physiological human. Philos Transact A Math Phys Eng Sci. 2008;366(1878):2979–2999. doi: 10.1098/rsta.2008.0089. [DOI] [PubMed] [Google Scholar]
- 61.Kohl P, Noble D. Systems biology and the virtual physiological human. Mol Syst Biol. 2009;5:292. doi: 10.1038/msb.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Novere N, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nature Biotechnology. 2005;23(12):1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 63.Smith NP, et al. Computational biology of cardiac myocytes: Proposed standards for the physiome. Journal of Experimental Biology. 2007;210(Pt 9):1576–1583. doi: 10.1242/jeb.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]