Abstract
Background/Aims
Knowledge regarding genetic factors that influence pancreatic cancer risk is currently limited. To identify novel pancreatic cancer susceptibility loci, we conducted a two-stage genome-wide association study.
Methods
The Affymetrix® Genome-Wide Human SNP Array 6.0 and DNA pooling were used in the screening stage. Twenty-six single-nucleotide polymorphisms (SNPs) were selected for follow-up. These 26 lead SNPs and additionally selected tagSNPs for the regions around the lead SNPs were evaluated by individual genotyping of the pooling population and an independent validation population.
Results
Of the lead SNPs, the strongest association was found with rs4820599 located in the γ-glutamyltransferase 1 (GGT1) gene. This SNP was significantly associated with pancreatic cancer risk in the validation population and the combined dataset (pallele-based = 0.019 and pallele-based = 0.003, respectively). Statistically significant associations were also observed with two GGT1 tagSNPs: rs2017869 and rs8135987. Lead SNP rs4820599 is in high linkage disequilibrium (LD; pairwise r2: 0.69) and tagSNP rs2017869 is in strong LD (pairwise r2: 0.96) with SNP rs5751901, which has been reported to be associated with increased GGT1 serum levels. GGT is expressed in the pancreas and plays a key role in glutathione metabolism.
Conclusion
Our results suggest that common variation in the GGT1 gene may affect the risk of pancreatic cancer.
Key Words: Pancreatic cancer, Genome-wide association, GGT1 gene, Single-nucleotide polymorphism
Introduction
Pancreatic cancer is a relatively rare disease but a major cause of cancer mortality. It is the fourth most common cause of cancer mortality in both men and women in the United States where an estimated 34,290 people will die of the disease in 2008 [1]. Similar to most other cancer types, early detection of pancreatic cancer provides the best chance of long-term survival. However, pancreatic adenocarcinomas are currently generally diagnosed after the cancer has already spread to regional lymph nodes or, worse, metastasized to more distant sites when treatment options are limited [1]. Median survival after diagnosis is only 6 months, and less than 5% of the patients are still alive after 5 years. To improve early detection, prevention and treatment practices, a better understanding of the factors involved in the development and progression of pancreatic cancer is needed.
Growing evidence suggests that genetic factors play an important role in the etiology of pancreatic cancer. Familial clustering of pancreatic cancer cases is well recognized [2,3], and epidemiological studies have shown that risk of pancreatic cancer increases with increasing numbers of affected first-degree relatives with pancreatic cancer [4,5]. Various known hereditary cancer syndromes, including hereditary pancreatitis and hereditary breast cancer, have also been found associated with increased risk of pancreatic cancer [6]. However, the majority of pancreatic cancer cases appear to be sporadic (i.e., are without a clear-cut inheritance pattern), and knowledge regarding common genetic variants that may affect pancreatic cancer risk in these individuals is yet limited. Further insight into the genetic factors involved in pancreatic carcinogenesis beyond that of the known hereditary cancer syndromes is important as it may contribute to the ability to effectively identify individuals at increased risk as well as to the identification of new targets for prevention and treatment.
Genome-wide association (GWA) studies are, unlike candidate gene studies, unbiased with regard to prior hypotheses about the biological pathways involved in disease development. They are an excellent but, unfortunately, expensive approach to identify genes and pathways involved in complex diseases. Pooling DNA in the first stage of a GWA study substantially reduces costs, and has been shown to be an efficient method to select candidate susceptibility loci for follow-up by individual genotyping [7,8,9,10,11,12,13]. To identify novel pancreatic cancer susceptibility loci, we conducted a two-stage GWA study using the Affymetrix® Genome-Wide Human SNP Array 6.0 (>900,000 single-nucleotide polymorphisms, SNPs) and DNA pooling in the screening stage. During the second stage, the most promising loci were evaluated by individual genotyping of the pooling population and an independent validation population. Our results indicate a role for the γ-glutamyltransferase 1 (GGT1) gene region in pancreatic carcinogenesis.
Methods
Study Participants
Pancreatic cancer cases in the pooling population were ascertained through the Pancreatic Adenocarcinoma Gene-Environment Risk (PAGER) study at the University of Pittsburgh and University of Pittsburgh Medical Center (Pittsburgh, Pa., USA) from February 2002 to October 2007 under Institutional Review Board-approved protocols. In total, 103 unrelated PAGER cases (41 females and 62 males) with histologically verified pancreatic cancer were included in the current study. Blood was available from all included cases, mean age at diagnosis was 63.6 (±10.5) years, 67% were ever smokers, and all were Caucasian.
Pancreatic cancer cases in the validation population were recruited at Evanston Northwestern Healthcare (Evanston, Ill., USA) from June 2002 to June 2007 under protocols approved by their Institutional Review Board. In total, 75 unrelated ‘Evanston’ cases (40 females and 35 males) with histologically verified pancreatic cancer were included in the current study. Blood was available from all included cases, mean age at diagnosis was 69.8 (±11.5) years, 51% were ever smokers, and all were Caucasian. The PAGER cases were significantly younger at diagnosis (p = 0.0002) and more often ever smokers (p = 0.03) than the ‘Evanston’ cases.
All controls (Ntotal = 182) were selected from among Caucasian controls enrolled in the North American Pancreatitis Study 2 (NAPS2) for whom blood was available. Details of the NAPS2 study have been described elsewhere [14]. For the current study, controls were frequency matched to the PAGER cases and ‘Evanston’ cases by sex, race, age (5-year intervals) and smoking status (never, ever). Controls had never been diagnosed with pancreatic cancer and/or pancreatitis. Written informed consent was obtained from all study participants. For the current study, only coded (i.e. de-identified) private information and specimens were used.
DNA Extraction and Quantification, and Pool Construction
Genomic DNA was extracted from buffy coat samples using the Qiagen FlexiGene DNA Kit (Qiagen Inc., Valencia, Calif., USA). DNA concentrations were measured using spectroscopy (260 nm) and samples were diluted to 50 ng/μl. Each sample that was going to be pooled was subsequently quantified in triplicate by fluorimeter (Quant-IT PicoGreen™ method; Invitrogen Molecular Probes, Carlsbad, Calif., USA). If needed, the DNA concentration was adjusted. Fluorimeter measures were used to quantify the adjusted samples as well. DNA pools were constructed by combining equimolar amounts of DNA. Ten independent pools of DNA were created: 5 pools with DNA from PAGER cases (Ntotal = 103), 2 female only and 3 male only; 5 pools with DNA from frequency-matched controls (Ntotal = 103), 2 female only and 3 male only. Pools contained DNA from 20–21 subjects.
Genome-Wide Scan
All 10 DNA pools were genotyped in duplicate using the Affymetrix Genome-Wide Human SNP Array 6.0 according to the manufacturer's protocol. Genotyping was performed at the University of Pittsburgh Genomics and Proteomics Core Laboratories (GPCL). Raw probe intensity data (CEL files) were used to calculate relative allele signal (RAS) scores and estimate allele frequencies [15] (the program used to calculate RAS scores is available at: http://sgdp.iop.kcl.ac.uk/oleo/affy/).
SNP Selection and Individual Genotyping
Twenty-six SNPs, all high-ranked (see ‘Statistical Analysis’) and located in or near a different, known gene, were selected for follow-up. SNPs that were high-ranked but located in a genomic region containing no known genes were not followed up in this study. To evaluate the 26 candidate regions in more detail, we additionally selected tagSNPs for the regions around the 26 lead SNPs using data from the International HapMap project (www.hapmap.org; CEU population) and Haploview [16] (version 4.1) software. TagSNPs capturing common variants [minor allele frequency (MAF) >0.05] with pairwise correlation r2 >0.80 were chosen by Haploview's Tagger.
Individual genotyping was performed at the University of Pittsburgh GPCL using MassARRAY® iPLEX Gold (Sequenom, Inc., San Diego, Calif., USA). All SNP-specific and MassEXTEND oligonucleotides and all assays were designed using Sequenom RealSNP (www.realsnp.com) and MassARRAY Assay Design version 3.1 (Sequenom, Inc.). To monitor genotyping quality, a control DNA sample and a DNA-free (‘negative’) control were included, in duplicate, on every plate. No discrepant genotypes were detected. Cases and controls from the pooling population (103 PAGER cases, 103 frequency-matched controls) and from an independent validation population (75 ‘Evanston’ cases, 79 frequency-matched controls) were individually genotyped for a total of 26 lead SNPs and 196 tagSNPs. Twenty-six SNPs, including one of the lead SNPs (rs9397033 in MTHFD1L), failed the missingness test (i.e., more than 10% of the samples failed to be genotyped) in one or both of the study populations and were not included in the analyses. SNPs with MAF <0.01 were also not included (n = 7), leaving a total of 25 lead SNPs and 164 tagSNPs. Analyses were restricted to individuals with genotyping call rates of ≥85%. Six cases and 8 controls from the pooling population, and 12 cases from the validation population had call rates <85% and were excluded. Additionally, for 2 controls from the pooling population there was not enough DNA available for individual genotyping, leaving a total of 97 cases and 93 controls in the pooling population and 63 cases and 79 controls in the validation population available for analyses.
Statistical Analysis
Analysis of the initial pooled data for the purpose of selecting candidate SNPs for follow-up was done by t test (comparing allele frequency estimates in case and control pools) and also by a modified ANOVA method (S.Y.C. and E.F., unpublished) that corrects the t test to account for pooling variance. The results of the two methods were similar in terms of ranking of priority of SNPs for follow-up.
Differences in characteristics between cases and controls, and between PAGER cases and ‘Evanston’ cases were assessed using t tests for continuous and χ2 tests for categorical variables. For individual genotyping results, deviation from Hardy-Weinberg equilibrium was examined in the control population for each SNP using the χ2 goodness-of-fit test. Associations between alleles and genotypes and pancreatic cancer risk were evaluated using Pearson χ2 test and Cochran-Armitage trend test, respectively. Per-allele odds ratios (ORs) and corresponding 95% confidence intervals (95% CIs) were also calculated for each SNP. p values of <0.05 were considered statistically significant. Analyses were performed using PLINK [17] (version 1.03; http://pngu.mgh.harvard.edu/purcell/plink/), R [18], and SAS (version 9.2; SAS Institute Inc., Cary, N.C., USA) software. Pairwise linkage disequilibrium (LD) was estimated by r2 in Haploview [16] (version 4.1).
Results
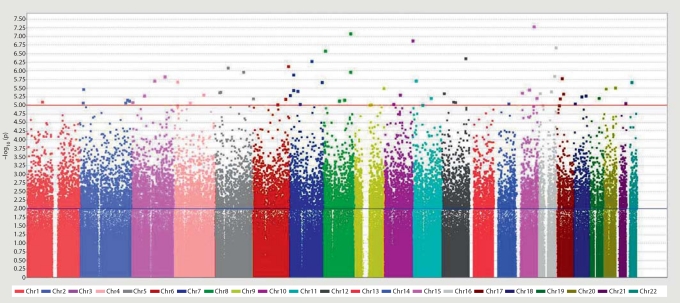
In the screening stage, 10 independent DNA pools (5 pools of case subjects; 5 pools of control subjects; 20–21 subjects/pool) were genotyped in duplicate using the Affymetrix Genome-Wide Human SNP Array 6.0 (>900,000 SNPs). Allele frequencies were estimated and differences between case and control frequencies were ranked using several different statistical procedures (see ‘Methods’). Figure 1 shows the pooling-based GWA results. The p values, shown as –log10 (p) in figure 1, are for ranking purposes only. They are based on a t test analysis that overstates the statistical significance. From among the SNPs that were consistently the most different between cases and controls (p value threshold: 1 × 10−5), 26 SNPs, all located in or near a different, known gene, were selected for follow-up by individual genotyping. The 26 selected lead SNPs are listed in table 1.
Fig. 1.
Pooling-based GWA results. Each chromosome (Chr) is depicted as a different color. The p values shown are for ranking purposes only; they are based on a t test analysis (comparing allele frequency estimates in case and control pools) that overstates the statistical significance. The upper horizontal line (red in the online version) indicates the p value threshold (1 × 10−5) used for selecting SNPs for follow-up.
Table 1.
Individual genotyping results for the 26 lead SNPs
| dbSNP ID | CHR | BP | Gene | Minor allele | Major allele | Validation population (63 cases, 79 controls) |
Pooling population (97 cases, 93 controls) |
Combined (160 cases, 172 controls) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF (cases/pa controls) | Ptrendb | MAF (cases/pa controls) | Ptrendb | MAF (cases/pa controls) | Ptrendb | |||||||||
| rs10497950 | 2 | 212399950 | ERBB4 | A | C | 0.17/0.19 | 0.729 | 0.726 | 0.17/0.21 | 0.349 | 0.368 | 0.17/0.20 | 0.359 | 0.368 |
| rs13079652 | 3 | 2977447 | CNTN4 | A | G | 0.07/0.12 | 0.184 | 0.177 | 0.06/0.11 | 0.108 | 0.127 | 0.07/0.11 | 0.034 | 0.038 |
| rs1878014 | 3 | 58170414 | DNASE1L3 | G | A | 0.31/0.26 | 0.420 | 0.373 | 0.27/0.23 | 0.335 | 0.332 | 0.29/0.24 | 0.234 | 0.213 |
| rs9397033c | 6 | 151456785 | MTHFD1L | A | T | |||||||||
| rs1l49517 | 7 | 16767337 | TSPAN13 | A | G | 0.49/0.50 | 0.892 | 0.256 | 0.50/0.50 | 1.000 | NA | 0.50/0.50 | 0.935 | 0.297 |
| rs3778884 | 7 | 38445745 | AMPH | T | C | 0.31/0.28 | 0.593 | 0.565 | 0.28/0.42 | 0.007 | 0.010 | 0.30/0.36 | 0.106 | 0.106 |
| rs12705284 | 7 | 104284914 | LHFPL3 | C | T | 0.43/0.49 | 0.312 | 0.294 | 0.49/0.40 | 0.066 | 0.077 | 0.47/0.44 | 0.486 | 0.489 |
| rs971462 | 8 | 75357268 | JPH1 | G | A | 0.36/0.45 | 0.143 | 0.150 | 0.46/0.38 | 0.126 | 0.152 | 0.42/0.41 | 0.819 | 0.827 |
| rs13277433 | 8 | 124794509 | ANXA13 | T | G | 0.19/0.28 | 0.105 | 0.106 | 0.23/0.39 | 0.001 | 0.001 | 0.22/0.34 | 0.001 | 0.001 |
| rs10869335 | 9 | 70618966 | PIP5K1B | T | C | 0.48/0.46 | 0.737 | 0.733 | 0.35/0.52 | 0.001 | 0.001 | 0.40/0.49 | 0.023 | 0.021 |
| rs7903091 | 10 | 73811045 | CBARA1 | T | C | 0.43/0.34 | 0.141 | 0.145 | 0.34/0.47 | 0.008 | 0.003 | 0.37/0.41 | 0.306 | 0.281 |
| rs7921512 | 10 | 84045997 | NRG3 | A | G | 0.36/0.39 | 0.690 | 0.686 | 0.37/0.20 | 0.0004 | 0.0003 | 0.37/0.29 | 0.033 | 0.031 |
| rs443193 | 11 | 2893422 | SLC22A18 | T | C | 0.36/0.47 | 0.086 | 0.079 | 0.39/0.49 | 0.048 | 0.054 | 0.38/0.48 | 0.010 | 0.010 |
| rs7928379 | 11 | 83055714 | DLG2 | T | C | 0.38/0.37 | 0.923 | 0.922 | 0.29/0.45 | 0.002 | 0.003 | 0.33/0.41 | 0.022 | 0.024 |
| rs2682724 | 12 | 63321126 | RASSF3 | C | T | 0.42/0.36 | 0.357 | 0.362 | 0.37/0.44 | 0.137 | 0.165 | 0.39/0.41 | 0.623 | 0.638 |
| rs2240189 | 12 | 111887880 | OAS3 | T | C | 0.25/0.30 | 0.318 | 0.300 | 0.20/0.29 | 0.041 | 0.038 | 0.22/0.30 | 0.024 | 0.021 |
| rs622959 | 15 | 25002683 | GABRG3 | C | T | 0.21/0.20 | 0.780 | 0.776 | 0.14/0.22 | 0.036 | 0.040 | 0.17/0.21 | 0.158 | 0.160 |
| rs10851648 | 15 | 57397045 | MYOIE | T | G | 0.16/0.12 | 0.265 | 0.241 | 0.17/0.24 | 0.113 | 0.115 | 0.17/0.18 | 0.645 | 0.641 |
| rs12439920 | 15 | 78591796 | ARNT2 | A | G | 0.11/0.14 | 0.491 | 0.455 | 0.08/0.14 | 0.094 | 0.092 | 0.10/0.14 | 0.081 | 0.069 |
| rs17760312 | 16 | 7651989 | A2BP1 | C | G | 0.12/0.14 | 0.652 | 0.637 | 0.08/0.12 | 0.177 | 0.166 | 0.10/0.13 | 0.172 | 0.157 |
| rs17688192 | 16 | 20587471 | ACSM1 | A | G | 0.17/0.23 | 0.189 | 0.230 | 0.19/0.32 | 0.005 | 0.004 | 0.18/0.28 | 0.004 | 0.005 |
| rs8062058 | 16 | 83039135 | ATP2C2 | T | C | 0.16/0.11 | 0.306 | 0.266 | 0.15/0.13 | 0.667 | 0.664 | 0.15/0.12 | 0.313 | 0.294 |
| rs16581 | 17 | 28401656 | ACCN1 | T | C | 0.13/0.20 | 0.103 | 0.102 | 0.18/0.23 | 0.215 | 0.217 | 0.16/0.22 | 0.057 | 0.058 |
| rs1719953 | 18 | 5515037 | EPB41L3 | T | C | 0.20/0.16 | 0.344 | 0.339 | 0.13/0.21 | 0.024 | 0.020 | 0.16/0.19 | 0.277 | 0.268 |
| rs12152040 | 21 | 40990938 | DSCAM | C | T | 0.17/0.17 | 1.000 | 1.000 | 0.18/0.06 | 0.0004 | 0.0003 | 0.18/0.11 | 0.016 | 0.010 |
| rs4820599 | 22 | 23320213 | GGTl | G | A | 0.35/0.23 | 0.019 | 0.022 | 0.36/0.27 | 0.071 | 0.077 | 0.36/0.25 | 0.003 | 0.004 |
CHR = Chromosome; BP = base pair.
Pearson χ2 test (1 d.f.).
Cochran-Armitage trend test.
Not included in the analyses because genotyping failed in >10% of the samples.
We genotyped the 26 lead SNPs in the pooling population and in an independent validation population. Results are presented in tables 1 and 2. Lead SNP rs9397033 located in MTHFD1L (6q25.1) was not included in the analyses because genotyping failed in >10% of the samples. With the exception of rs1149517 (TSPAN13, 7p21.1), all remaining lead SNPs were in Hardy-Weinberg equilibrium (data not shown). The strongest association was observed for SNP rs4820599 located in the GGT1 gene on chromosome 22q11.23. This SNP was significantly associated with pancreatic cancer risk in the validation population (pallele-based = 0.019, ptrend = 0.022, per-allele OR: 1.86, 95% CI: 1.11–3.15). Allele frequencies for rs4820599 were comparable between the two study populations, and the association with pancreatic cancer risk was significant in the combined dataset (pallele-based = 0.003, ptrend = 0.004, per-allele OR: 1.66, 95% CI: 1.18–2.32); borderline significant in the pooling population (pallele-based = 0.071, ptrend = 0.077, per-allele OR: 1.50, 95% CI: 0.96–2.34).
Table 2.
Per-allele odds ratios (OR) and corresponding 95% CIs for the 26 lead SNPs
| dbSNP ID | CHR | Gene | Validation population (63 cases, 79 controls) |
Pooling population (97 cases, 93 controls) |
Combined (160 cases, 172 controls) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | L95 | U95 | OR | L95 | U95 | OR | L95 | U95 | |||
| rs10497950 | 2 | ERBB4 | 0.90 | 0.48 | 1.67 | 0.78 | 0.46 | 1.32 | 0.83 | 0.56 | 1.24 |
| rs13079652 | 3 | CNTN4 | 0.57 | 0.25 | 1.31 | 0.55 | 0.26 | 1.15 | 0.55 | 0.32 | 0.96 |
| rs1878014 | 3 | DNASE1L3 | 1.24 | 0.74 | 2.09 | 1.26 | 0.79 | 2.03 | 1.24 | 0.87 | 1.75 |
| rs9397033a | 6 | MTHFD1L | |||||||||
| rs1l49517 | 7 | TSPAN13 | 0.97 | 0.60 | 1.55 | 1.00 | 0.66 | 1.51 | 0.99 | 0.72 | 1.35 |
| rs3778884 | 7 | AMPH | 1.15 | 0.69 | 1.93 | 0.55 | 0.35 | 0.85 | 0.76 | 0.55 | 1.06 |
| rs12705284 | 7 | LHFPL3 | 0.78 | 0.49 | 1.26 | 1.48 | 0.97 | 2.24 | 1.12 | 0.82 | 1.53 |
| rs971462 | 8 | JPH1 | 0.70 | 0.43 | 1.13 | 1.38 | 0.91 | 2.09 | 1.04 | 0.76 | 1.42 |
| rs13277433 | 8 | ANXA13 | 0.62 | 0.35 | 1.11 | 0.47 | 0.29 | 0.74 | 0.54 | 0.38 | 0.77 |
| rs10869335 | 9 | PIP5K1B | 1.09 | 0.67 | 1.74 | 0.50 | 0.33 | 0.76 | 0.70 | 0.51 | 0.95 |
| rs7903091 | 10 | CBARA1 | 1.44 | 0.89 | 2.33 | 0.57 | 0.37 | 0.86 | 0.85 | 0.62 | 1.16 |
| rs7921512 | 10 | NRG3 | 0.91 | 0.56 | 1.47 | 2.29 | 1.43 | 3.64 | 1.43 | 1.03 | 1.98 |
| rs443193 | 11 | SLC22A18 | 0.65 | 0.40 | 1.06 | 0.66 | 0.43 | 1.00 | 0.66 | 0.48 | 0.91 |
| rs7928379 | 11 | DLG2 | 1.02 | 0.63 | 1.66 | 0.51 | 0.33 | 0.78 | 0.69 | 0.50 | 0.95 |
| rs2682724 | 12 | RASSF3 | 1.26 | 0.77 | 2.05 | 0.73 | 0.48 | 1.11 | 0.92 | 0.67 | 1.27 |
| rs2240189 | 12 | OAS3 | 0.76 | 0.45 | 1.30 | 0.61 | 0.38 | 0.98 | 0.67 | 0.47 | 0.95 |
| rs622959 | 15 | GABRG3 | 1.09 | 0.61 | 1.95 | 0.56 | 0.33 | 0.97 | 0.75 | 0.51 | 1.12 |
| rs10851648 | 15 | MYOIE | 1.47 | 0.74 | 2.93 | 0.66 | 0.40 | 1.11 | 0.91 | 0.60 | 1.37 |
| rs12439920 | 15 | ARNT2 | 0.78 | 0.38 | 1.59 | 0.57 | 0.29 | 1.11 | 0.65 | 0.40 | 1.06 |
| rs17760312 | 16 | A2BP1 | 0.85 | 0.42 | 1.72 | 0.62 | 0.31 | 1.24 | 0.71 | 0.43 | 1.16 |
| rs17688192 | 16 | ACSM1 | 0.67 | 0.36 | 1.22 | 0.50 | 0.31 | 0.81 | 0.57 | 0.39 | 0.83 |
| rs8062058 | 16 | ATP2C2 | 1.44 | 0.72 | 2.87 | 1.14 | 0.62 | 2.09 | 1.26 | 0.80 | 1.99 |
| rs16581 | 17 | ACCN1 | 0.58 | 0.30 | 1.12 | 0.73 | 0.44 | 1.21 | 0.68 | 0.46 | 1.01 |
| rs1719953 | 18 | EPB41L3 | 1.34 | 0.73 | 2.48 | 0.53 | 0.30 | 0.92 | 0.80 | 0.53 | 1.20 |
| rs12152040 | 21 | DSCAM | 1.00 | 0.53 | 1.89 | 3.44 | 1.68 | 7.03 | 1.73 | 1.11 | 2.72 |
| rs4820599 | 22 | GGT1 | 1.86 | 1.11 | 3.15 | 1.50 | 0.96 | 2.34 | 1.66 | 1.18 | 2.32 |
Not included in the analyses because genotyping failed in >10% of the samples.
Our individual genotyping results suggest that several other lead SNPs (i.e., those in CNTN4, ANXA13, and SLC22A18) may potentially be of interest as well. No statistically significant associations with pancreatic cancer risk were observed for these 3 lead SNPs in the validation population. However, their allele frequencies were comparable between the two different study populations, and statistically significant associations were observed in the pooling population and/or in the combined dataset (tables 1 and 2).
The cases and controls from the pooling population and the validation population were also genotyped for 196 additionally selected tagSNPs covering the regions around the 26 lead SNPs. Online supplementary tables 1 and 2 (www.karger.com/doi/10.1159/000236023) show results for all the 164 tagSNPs that met the quality criteria (see ‘Methods’). Here, we focus on results for tagSNPs in the GGT1 region because rs4820599 was the only lead SNP for which statistically significant associations with pancreatic cancer risk were observed in the independent validation population. The results for rs2017869 and rs8135987, located respectively 7.1 and 22.6 kb upstream of lead SNP rs4820599 in GGT1, support a role for this region in pancreatic carcinogenesis. Both SNPs were significantly associated with pancreatic cancer risk in the validation population (rs2017869: pallele-based = 0.001, ptrend = 0.001, per-allele OR: 2.38, 95% CI: 1.44–3.93; rs8135987: pallele-based = 0.001, ptrend = 0.001, per-allele OR: 2.33, 95% CI: 1.40–3.89) and in the combined dataset (rs2017869: pallele-based = 0.001, ptrend = 0.001, per-allele OR: 1.71, 95% CI: 1.24–2.35; rs8135987: pallele-based = 0.002, ptrend = 0.002, per-allele OR: 1.69, 95% CI: 1.21–2.35). In the pooling population, results were in line with those observed in the validation population but nonsignificant (rs2017869: pallele-based = 0.184, ptrend = 0.188, per-allele OR: 1.33, 95% CI: 0.87–2.02; rs8135987: pallele-based = 0.220, ptrend = 0.225, per-allele OR: 1.31, 95% CI: 0.85–2.03). Both GGT1 tagSNPs were in Hardy-Weinberg equilibrium (data not shown). For the three evaluated SNPs in GGT1, no significant differences in genotype frequencies were observed between males and females, and between ever smokers and never smokers (data not shown).
The correlation (pairwise r2) between lead SNP rs4820599 and tagSNPs rs2017869 and rs8135987 was, respectively, 0.61 and 0.37 in the combined dataset; correlation between rs2017869 and rs8135987 was 0.67. SNP rs2017869 is in strong LD (pairwise r2: 0.96; HapMap CEU population data) with rs5751901, which was recently reported to be associated with increased GGT1 serum levels by Melzer et al. [19]. Lead SNP rs4820599 is in high LD (pairwise r2: 0.69; HapMap CEU population data) with this SNP.
Discussion
In this GWA study of pancreatic cancer, we used pooled DNA in the screening stage and evaluated 26 candidate susceptibility loci in the second stage by individual genotyping of the pooling population and an independent validation population. Of the lead SNPs, the strongest association with pancreatic cancer risk was observed for rs4820599 located in GGT1 on chromosome 22q11.23. Additional fine-mapping results supported a role for the GGT1 region in pancreatic carcinogenesis. Interestingly, rs4820599 is in high LD and one of the evaluated GGT1 tagSNPS, rs2017869, is in strong LD with SNP rs5751901 which has been found associated with increased GGT1 serum levels in a recent GWA study [19].
When using a pooled DNA approach, separate DNA pools of case and control samples are constructed, and allele frequencies for the SNPs are estimated by genotyping these pools instead of individual samples. Pooling DNA substantially reduces the number of genotyping assays and, thus, also the costs of the study. However, compared to individual genotyping, DNA pooling does add extra experimental error (e.g. due to errors in pool construction) to the allele frequency measurements which may affect the ability to detect a true association. Therefore, validation of the results is necessary. Most pooling-based GWA studies use, like us in the current study, a two-stage design in which the most promising SNPs identified in the screening stage are followed up by individual genotyping. The effectiveness of this method has been successfully demonstrated by the identification of previously published as well as novel susceptibility loci [8,9,10,11,12].
GGT, the protein encoded by the GGT1 gene, is a membrane-bound extracellular enzyme that is highly expressed in tissues with secretory or absorptive functions such as liver, kidney and pancreas [20]. Experimental data suggest that regulation of GGT expression is mediated through the Ras signal transduction pathway [21,22], one of the core signaling pathways in human pancreatic cancer [23]. GGT is also present in bodily fluids, and serum GGT levels are clinically commonly used as an indicator of liver disease or marker of excessive alcohol intake. However, elevated GGT levels have also been found associated with various other diseases, including cardiovascular disease, and serum GGT has been proposed as a marker of oxidative stress [24]. Importantly, Strasak et al. [25,26] recently reported that elevated GGT serum levels were significantly associated with increased cancer risk in men and women in a large population-based cohort. Associations between serum GGT levels and risk of pancreatic cancer specifically have, to our knowledge, not yet been investigated.
The relationship between serum GGT and membrane-bound GGT is unknown, but increased serum levels plausibly reflect increased expression. Membrane-bound GGT catalyzes the transfer of the γ-glutamyl moiety from glutathione and other γ-glutamyl compounds to acceptors. As such, it plays a key role in maintaining intracellular glutathione levels which is critical for phase II reactions and the detoxification of reactive metabolites and xenobiotics [20]. Paradoxically, in the presence of iron or other transition metals, the extracellular cleavage of glutathione by GGT can also result in the generation of reactive oxygen species, free radicals, and oxidative damage [27,28]. Moreover, (over)expression of GGT may promote tumor progression and drug resistance by protecting cells from oxidative stress-associated apoptosis [29]. Thus, functionally relevant polymorphisms in GGT1 may well affect the risk of developing cancer in tissues that express GGT.
In summary, our data suggest that common variation in the GGT1 gene may affect risk of pancreatic cancer. It should be noted that our study populations were small and, hence, power to identify and validate pancreatic cancer risk loci was relatively low. Additionally, it is possible that susceptibility loci were not identified due to the limited number of SNPs selected for follow-up by individual genotyping and to selecting only SNPs located in or near a known gene. Likely, there are additional pancreatic cancer susceptibility loci to be detected. With that being said, the associations with GGT1 found in this pooling-based GWA study of pancreatic cancer were statistically significant, reproducible and, as discussed above, are biologically plausible. Additional studies are needed to identify causal GGT1 variants, evaluate clinical relevance, and to investigate the processes underlying the effects of these variants on pancreatic cancer risk.
Supplementary Material
Supplementary information, Diergaarde et al. Pooling-based genome-wide association study implicates gamma-glutamyltransferase 1 (GGT1) gene in pancreatic carcinogenesis.
Acknowledgements
We thank Erin Fink and Drs. Herbert Zeh, Arthur James Moser, Malcolm Bilimoria and Karen Kaul for assistance in patient recruitment, and all study participants for their cooperation. This research was supported by the Wayne Fusaro Pancreatic Cancer Research Fund, Gott Family Foundation, Frieda G. and Saul F. Shapira BRCA Cancer Research Program, James F. Walsh Foundation and the National Pancreas Foundation. Evanston Northwestern Healthcare provided institutional support for the pancreatic cancer biospecimen repository. The NAPS2 study is supported by NIH grant R01 DK061451. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ries LAG, et al., editors. SEER Cancer Statistics Review. Bethesda, MD: National Cancer Institute; 1975–2005. seer.cancer.gov/csr/1975_2005/, based on November 2007 SEER data submission, posted to the SEER web site, 2008. [Google Scholar]
- 2.Lynch HT, et al. Familial pancreatic cancer: clinicopathologic study of 18 nuclear families. Am J Gastroenterol. 1990;85:54–60. [PubMed] [Google Scholar]
- 3.Hruban RH, et al. Familial pancreatic cancer. Ann Oncol. 1999;10(suppl 4):69–73. [PubMed] [Google Scholar]
- 4.Tersmette AC, et al. Increased risk of incident pancreatic cancer among first-degree relatives of patients with familial pancreatic cancer. Clin Cancer Res. 2001;7:738–744. [PubMed] [Google Scholar]
- 5.Klein AP, et al. Prospective risk of pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 6.Greer JB, Whitcomb DC, Brand RE. Genetic predisposition to pancreatic cancer: a brief review. Am J Gastroenterol. 2007;102:2564–2569. doi: 10.1111/j.1572-0241.2007.01475.x. [DOI] [PubMed] [Google Scholar]
- 7.Sham P, Bader JS, Craig I, O'Donovan M, Owen M. DNA pooling: a tool for large scale association studies. Nat Rev Gen. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- 8.Craig DW, et al. Identification of disease causing loci using an array-based genotyping approach on pooled DNA. BMC Genomics. 2005;6:138. doi: 10.1186/1471-2164-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steer S, et al. Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes and Immunity. 2007;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- 10.Pearson JV, et al. Identification of the genetic basis for complex disorders by use of pooling-based genome-wide single-nucleotide-polymorphism association studies. Am J Hum Genet. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum AE, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shifman S, et al. Genome-wide association identifies a common variant in the Reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Wang S. Optimal DNA pooling-based two-stage designs in case-control association studies. Hum Hered. 2009;67:46–56. doi: 10.1159/000164398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitcomb DC, et al. Multicenter approach to recurrent acute and chronic pancreatitis in the United States: the North American Pancreatitis Study 2 (NAPS2) Pancreatology. 2008;8:520–531. doi: 10.1159/000152001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meaburn E, Butcher LM, Schalkwyk LC, Plomin R. Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucl Acids Res. 2006;34:e28. doi: 10.1093/nar/gnj027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 19.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitfield JB. Gamma glutamyl transferase. Crit Rev Clin Lab Sci. 2001;38:263–355. doi: 10.1080/20014091084227. [DOI] [PubMed] [Google Scholar]
- 21.Pankiv S, Møller S, Bjørkøv G, Moens U, Huseby NE. Radiation-induced upregulation of gamma-glutamyltransferase in colon carcinoma cells is mediated through the Ras signal transduction pathway. Biochim Biophys Acta. 2006;1760:151–157. doi: 10.1016/j.bbagen.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Pandur S, Pankiv S, Johannessen M, Moens U, Huseby NE. Gamma-glutamyl transferase is upregulated after oxidative stress through the Ras signal transduction pathway in rat colon carcinoma cells. Free Radic Res. 2007;41:1376–1384. doi: 10.1080/10715760701739488. [DOI] [PubMed] [Google Scholar]
- 23.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee DH, Blomhoff R, Jacobs DR., Jr Is serum gamma glutamyltransferase a marker of oxidative stress? Free Radic Res. 2004;38:535–539. doi: 10.1080/10715760410001694026. [DOI] [PubMed] [Google Scholar]
- 25.Strasak AM, et al. Association of gamma-glutamyltransferase and risk of cancer incidence in men: a prospective study. Cancer Res. 2008;68:3970–3977. doi: 10.1158/0008-5472.CAN-07-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strasak AM, et al. Prospective study of the association of gamma-glutamyltransferase with cancer incidence in women. Int J Cancer. 2008;123:1902–1906. doi: 10.1002/ijc.23714. [DOI] [PubMed] [Google Scholar]
- 27.Stark AA, Zeiger E, Pagano DA. Glutathione metabolism by gamma-glutamyltranspeptidase leads to lipid peroxidation: characterization of the system and relevance to hepatocarcinogenesis. Carcinogenesis. 1993;14:183–189. doi: 10.1093/carcin/14.2.183. [DOI] [PubMed] [Google Scholar]
- 28.Drozdz R, et al. Gamma-glutamyltransferase dependent generation of reactive oxygen species from a glutathione/transferring system. Free Radic Biol Med. 1998;22:786–792. doi: 10.1016/s0891-5849(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 29.Hanigan MH, Gallagher BC, Townsend DM, Gabarra V. Gamma-glutamyl transpeptidase accelerates tumor growth and increases the resistance of tumors to cisplatin in vivo. Carcinogenesis. 1999;20:553–559. doi: 10.1093/carcin/20.4.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Diergaarde et al. Pooling-based genome-wide association study implicates gamma-glutamyltransferase 1 (GGT1) gene in pancreatic carcinogenesis.