Abstract
Atom transfer radical polymerization (ATRP) was employed to create isolated, metal-containing nanoparticles on the surface of non-porous polymeric beads with the goal of developing a new immobilized metal affnity chromatography (IMAC) stationary phase for separating prion peptides and proteins. Transmission electron microscopy was used to visualize nanoparticles on the substrate surface. Individual ferritin molecules were also visualized as ferritin–nanoparticle complexes. The column's resolving power was tested by synthesizing peptide analogs to the copper binding region of prion protein and injecting mixtures of these analogs onto the column. As expected, the column was capable of separating prion-related peptides differing in number of octapeptide repeat units (PHGGGWGQ), (PHGGGWGQ)2, and (PHGGGWGQ)4. Unexpectedly, the column could also resolve peptides containing the same number of repeats but differing only in the presence of a hydrophilic tail, Q → A substitution, or amide nitrogen methylation.
Keywords: Atom transfer radical polymerization (ATRP), Surface modification, Nanoscale, Nanoparticle, Prion, Immobilized metal affinity chromatography (IMAC), Transmissible spongiform encephalopathies
The cellular form of the prion protein (PrPC)1 is a glycosylphosphatidylinositol (GPI)-anchored glycoprotein expressed in abundance on the cell surface of neurons. The misfolded form of this protein, designated PrPSc,is responsible for a class of neurodegenerative disorders collectively known as the transmissible spongiform encephalopathies (TSEs) [1]. Examples of TSEs include mad cow disease, scrapie in sheep, chronic wasting disease in deer and elk, and Creutzfeldt–Jakob disease (CJD) in humans. PrPC contains a highly structured C-terminal “head” and a flexible, unstructured N-terminal “tail” [2]. The unstructured tail contains four or more tandem eight-residue repeats (PHGGGWGQ) called octarepeats [3]. Recent research demonstrates that this octarepeat domain binds Cu(II) in vivo and is highly selective for copper over other divalent metal ions. At full occupancy, each octapeptide segment takes up a single Cu(II) ion [4].
Immobilized metal affnity chromatography (IMAC) initially was developed for small molecule separations [5] and subsequently was adapted for protein separations [6]. Cu(II)–IMAC typically is used for His-tag protein purification [7]. It was found that native, nondenatured prion protein could be enriched from hamster brain by Cu(II)–IMAC [8]. On-column purification and refolding of recombinant prion protein has also been achieved [9]. Because the copper binding affnity of the prion protein tail is very high, it can be thought of as a “natural” His tag [9]. The copper binding affnity of PrPSc, however, was found to be significantly lower than that of PrPC. It has been reported that in Cu(II)–IMAC, PrPC is retained by Cu(II) and PrPSc elutes in the flow-through [10]. Similarly, fractionation of prion protein glycoforms by Cu(II)–IMAC has been achieved [11].
IMAC materials typically are made using low-effciency, low-pressure, 40- to 60-μm, high-capacity porous/gel-based substrates for protein or peptide capture/release applications. Fractions released from these cartridges are “enriched” with the protein/peptide of interest. Presumably, analyte resolution and peak effciency could be increased by using nonporous pressure stable particles, decreasing particle size, and optimizing surface modification chemistry [12]. Because IMAC is an affnity-based technique, resolution and effciency may also be increased by engineering a molecular recognition-type interaction where a single nanoscale protein interacts with a single nanoscale, copper-rich “interaction” site .
Traditional approaches to stationary-phase surface modification include bonding, encapsulation, and grafting, and they result in substrates where interaction sites are distributed on the substrate surface homogeneously and, therefore, cannot be used to produce surface-bound nanoscale architectures [12]. Synthetic approaches for controlling polymeric dimensions at the nanoscale include dendrimer synthesis [13], living polymerization [14], intra-molecular cross-linking of single polymers [15], cross-linking micelles [16], shell cross-linked knedle formation [17], and double-stranded polymer synthesis [18]. ATRP is a type of living polymerization that allows for control of the polymer molecular weight and molecular weight distribution [19]. Living polymerization has been used to graft substrates including silicon wafers [20], chromatographic beads [21], particles [22], and monoliths [23]. Grafting substrates by living polymerization typically is carried out by a “grafting from,” “grafting to,” or “grafting through” approach [24]. The “grafting from” approach involves polymerization from surface-bound initiator and results in living polymer grafts with high surface density [25]. The “grafting to” approach involves covalently bonding preformed polymers to the substrate surface through a reactive end group and typically results in dead polymer grafts with low surface density. The “grafting through” approach is a hybrid of “grafting from” and “grafting to” in that it results in living polymer grafts with low graft density.
The most common methods for analysis of small molecules involve the use of a reversed phase (RP), hydro-phobic interaction chromatography (HIC), and ion exchange chromatography (IEC) column. With choice of the proper eluent system, most proteins and peptides have the physical properties necessary to be retained and eluted from any one of these stationary phases. For complex biological samples, this situation is problematic. Even the highest effciency columns used with very long shallow gradients result in peaks with hundreds, if not thousands, of proteins and peptides. In IMAC, proteins and peptides are retained by coordination interaction. Because only a small number of peptides and proteins exhibit this type of interaction, most proteins flow through the column without being retained. This is especially important because if the analyte of interest has metal binding properties, that analyte can be enriched by IMAC. This enrichment results in a significant reduction of the complexity within the sample. Many commercially available IMAC substrates can achieve analyte enrichment by capture and release of analytes. Currently, there is a need for IMAC columns that are capable of capturing and separating analytes by strength of their coordination interaction.
Here we describe the preparation and demonstration of an IMAC column that is capable of capturing metal binding peptides and separating them in the same chromatographic run. To achieve this column, we engineered nanoscale features into the surface of a chromatographic stationary phase using “grafting through” ATRP. Polymer chain collapse was used, resulting in covalently bound metal/polymer nanoparticles with low surface density. This stationary phase was then used to separate very similar synthetic peptides derived from the tail region of prion protein.
Materials and methods
General methods
Reagents were obtained from Aldrich and used without further purification. Stationary-phase substrate with hydrophilic layer substrate was prepared in-house. Chromatography experiments were performed using a Dionex instrument equipped with a Dionex AD20 detector, a Dionex GP50 pump, and Chromeleon software. Transmission electron microscopy (TEM) measurements were performed on a Zeiss EM109 instrument. Cross-sectioned samples were embedded in Spurr low-viscosity epoxy resin (Ted Pella) and cured at 60 °C for 8 h. Cross-sectioning was carried out on an LKB Nova ultramicrotome with a Diatome diamond knife.
Preparation of glycidyl methacrylate grafted resin
Glycidyl methacrylate monomer (60.0 g, 422.1 mmol), anhydrous 2-butanone (60.0 g), and hydrophilic layer-modified substrate (30.0 g) were combined in a 250-ml round-bottom flask and bubbled with nitrogen for 2 h. In a separate round bottom, the copper complex was assembled under nitrogen by combining copper(I) bromide (0.75 g, 5.28 mmol), 2,2′-bipyridine (1.65 g, 10.56 mmol), and anhydrous 2-butanone (2 ml). The catalyst solution was added to the monomer/resin mixture and bubbled with nitrogen for 30 min. Ethyl 2-bromoisobutyrate (1.03 g, 5.28 mmol) was added. The reaction mixture was sealed, transferred to an oil bath at 50 °C, and allowed to react for 4 h. The resulting material was diluted with acetone (250 ml), vacuum filtered, and washed with acetone (250 ml). The resulting glycidyl methacrylate grafted resin was dried at 60 °C in an oven overnight.
Functionalization with iminodiacetate
Iminodiacetic acid (IDA, 30 g, 0.23 mol) and sodium carbonate (30 g) were dissolved in deionized water (90 g). The mixture was titrated to pH 10.5 with 50% sodium hydroxide. Glycidyl methacrylate grafted resin (30 g) was added and allowed to stir at 50 °C for 24 h. The resulting mixture was diluted with deionized water, vacuum filtered, and washed with 1 M NaCl.
Peptide synthesis and purification
Peptides were synthesized on Rink amide MBHA resin by the N-(9-fluorenyl)methoxycarbonyl (Fmoc) protection strategy using 1-hydroxybenzotriazole (HOBt)/O-benzotriazolyl-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU)/diisopropylethylamine (DIEA) for coupling (20 min) and 4% 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) and 8% piperidine (20 min) for deprotection. Unless otherwise mentioned, all reactions were carried out in dimethyl formamide (DMF). Methylated glycine was introduced by the coupling of bromoacetic acid (1 eq) to the preceding glycine using diisopropylcarbodiimide (DIC)/DIEA in dichloromethane for 30 min, followed by reaction with methylamine in tetrahydrofuran (THF) for 30 min. The next amino acid was coupled directly. All peptides were acetylated prior to cleavage. They were purified by reversed-phase HPLC on a C-18 column using a water/acetonitrile gradient containing 0.06% trifluoroacetic acid (TFA).
Chromatographic conditions
Chromatographic analysis was carried out on a Dionex HPLC instrument with UV detection (280 nm) and a Pro-Pac IMAC column. The flow rate was 0.5 ml/min. Eluent components and gradients were optimized for each analysis (see figure captions).
Results and discussion
Phase design
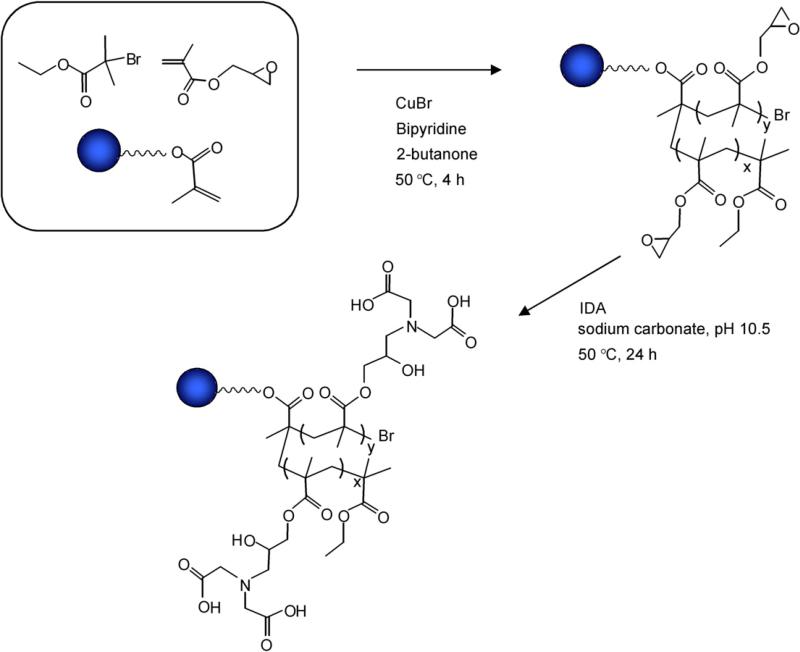
Scheme 1 shows the synthetic steps for preparation of the ProPac IMAC phase. We designed this stationary phase so that a single protein could interact with a single nanoparticle on the surface of the bead. In this way, the captured protein is isolated from all other components in the matrix (e.g., proteins, peptides, DNA) and isolated from components bound to neighboring nanoparticles.
Scheme 1.
Reaction scheme for preparation of IMAC phase.
Synthetic approach
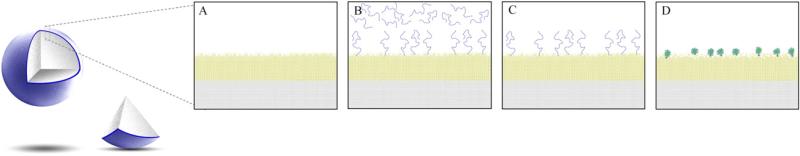
The synthetic approach for preparation of this stationary phase is shown in Fig. 1 and involved encapsulation of highly effcient, nonporous, 10-μm diameter, polystyrene divinylbenzene (PS–DVB) beads in a highly cross-linked, hydrophilic polymeric network containing residual double bonds; grafting the hydrophilic layer with reactive polymers using a “grafting through” ATRP approach; removing the ungrafted polymer from the beads by vacuum filtration; reacting the polymer grafts with chelating groups (IDA) so that each monomer in the polymer contained a chelating group; and inducing chain collapse by the introduction of copper ions to form metal–polymer composite nanoparticles. This approach allowed for control of nano-particle size by adjusting the monomer/initiator ratio and, therefore, optimization of the nanoscale surface architecture for optimal protein chromatographic performance.
Fig. 1.
Diagram of IMAC stationary phase preparation and polymer chain collapse.
Microscopy
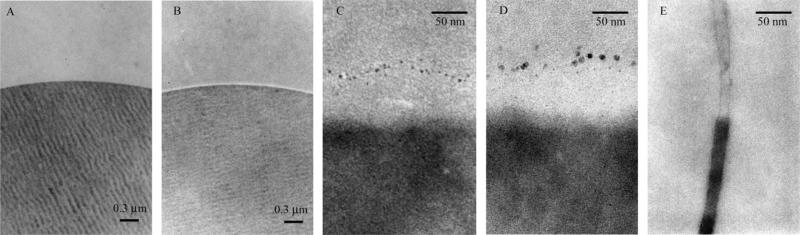
Figs. 2A and B show the TEM images of the raw substrate and hydrophilic layer-modified substrate, respectively. Fig. 2C shows a high-magnification TEM image of the cross section of an IMAC bead loaded with copper. The dark lower part of the picture is the PS–DVB substrate. The light layer on top of that is the hydrophilic layer. Copper–polymer nanoparticles can be seen as 2- to 5-nm diameter dark spots on the surface of the hydrophilic layer. Gel permeation chromatography of the polymer isolated from the ATRP reaction of glycidyl methacrylate had a degree of polymerization of 73 and a polydispersity index of 1.31. In the above TEM image, nanoparticle size corresponds well with the theoretical size of the polymer graft.
Fig. 2.
TEM images of bead cross sections of raw substrate (A), substrate with hydrophilic layer (B), copper-loaded IMAC resin (C), ferritin-stained, copper-loaded IMAC resin (D), and copper-loaded IMAC resin (E).
ATRP conditions were tuned to result in nanoparticles with size dimensions similar to those of proteins. Because the diameters of many proteins fall between 2 and 10 nm, we expect that a single protein may interact with a single nanoparticle on the surface of the bead. Fig. 2D is a high-magnification TEM image of the cross section of an IMAC bead loaded with copper and stained with ferritin. We chose ferritin for its well-defined protein shell, which contains surface-exposed histidines, and iron core for TEM visualization [26]. The individual ferritin proteins can be seen interacting with the bead surface. The ferritin does not intercalate into the hydrophilic layer. This suggests that the hydrophilic layer is highly cross-linked.
The TEM of the cross section in Fig. 2C shows a number of surface-bound nanoparticles. Without a measurement of the thickness of the cross section, nanoparticle density could not be calculated. To measure the thickness of the cross section, we embedded it in resin and cut a new cross section perpendicular to the plane of the embedded cross section. Fig. 2E shows the TEM of the “cross section of the cross section” of the IMAC bead loaded with copper. This result showed that the cross section in Fig. 2C was 25 nm thick. Nanoparticle density was calculated by dividing the surface area calculated by the length and width dimensions (250 and 25 nm, respectively) by the number of nanoparticles counted by visual inspection. Therefore, there is approximately 1 nanoparticle per 150 nm2.
Chromatographic separations
To demonstrate that the stationary phase is stable, we carried out multiple injections of myoglobin without reloading the column with metal between injections. The data from these runs can be seen in Fig. 3. These data show that immobilized copper remains on the stationary phase even after 60 injections and that the run time of myoglobin remains constant. This feature is advantageous for use of the column in multiple analyses.
Fig. 3.
Multiple injects of myoglobin. Elution was achieved by running a linear gradient from 20 mM Hepes/Mes and 1.0 M NaCl (pH 7.0) to 20 mM Hepes/Mes, 1.0 M NaCl, and 5 mM imidazole in 20 min. The eluent was adjusted with HCl or NaOH to achieve the proper eluent pH.
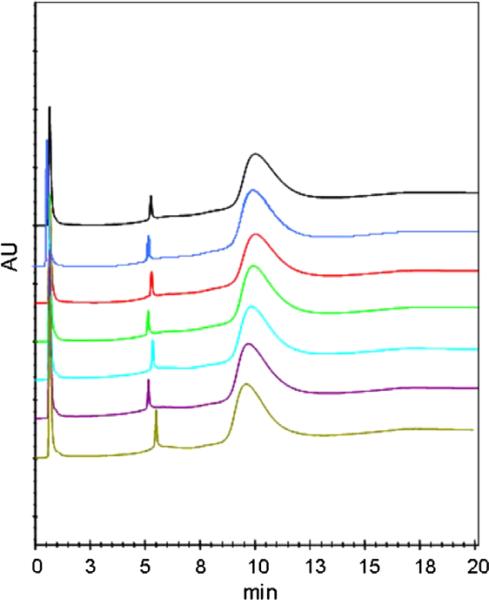
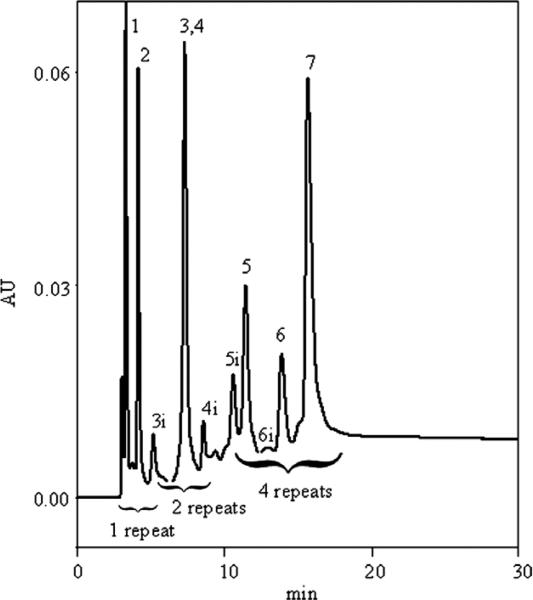
To demonstrate the selectivity of this IMAC column, we injected a mixture of synthetic peptides derived from the Cu(II) binding sites of the prion protein. The chromatographic trace can be seen in Fig. 4. All major peaks and impurity peaks were identified by injecting components individually. Because each octarepeat binds a single copper ion [27], we expect resolution of octarepeat number variants. The columns resolving power for octarepeat number variants can be seen in the separation of (PHGGGWGQ), KKRPKP(PHGGGWGQ)2, and KKRPKPWGQ(PHGGGWGQ)4 (peptides 1, 3, and 6, respectively) in Fig. 4. Because humans [28] with genes coding for extra octarepeat units are predisposed to CJD, there is interest in understanding the relationships between octarepeat number, structure of the copper-bound species, and predisposition to inherent CJD. The peptide library used in this study originally was constructed to facilitate spectroscopic studies of the Cu(II)–peptide complex structure for peptides with varying octarepeat numbers under varying conditions [29,30]. The hydrophilic extension (KKRPKP) was added to some of the peptides to increase the solubility, thereby enhancing the signal/noise ratio in spectroscopic measurements, and was found to have no effect on copper binding structure. The resolving power for octarepeat number variants here suggests that this column could be useful in resolving native PrP octarepeat number variants.
Fig. 4.
Separation of prion-derived peptide test mix. Peaks 1 to 7 correspond to the peptide structures in Table 1. Impurities separated from the major peak in the peptide samples are labeled with an “i”. The injection volume was 50 μl, which contained a total of 2.8 μg of each synthetic peptide. Elution was achieved by running a curve 7 gradient from 20 mM Hepes, 0.5 M NaCl, and 1 mM imidazole (pH 7.5) to 20 mM Hepes, 0.5 M NaCl, and 500 mM imidazole (pH 7.5) in 20 min.
Unexpectedly, peptides with small differences in sequence unrelated to the copper coordination sites were also separated. For example, titration and electron paramagnetic resonance (EPR) experiments on peptides 6 and 7 showed that they have the same number of Cu(II) binding sites and similar Cu(II) binding behavior (data not shown). These two peptides, which differ only by substitution of every glutamine residue in peptide 6 with an alanine residue (peptide 7), were well resolved. Similarly, peptides 5 and 6, which differ by the presence of a noncoordinating hydro-philic tail, are resolved. These data suggest that, in addition to the sum of the binding affnities from each coordination interaction site, the elution time for an analyte in an imidazole gradient is effected by other factors that may include local hydrophobicity, ligand environment, and ligand binding frame.
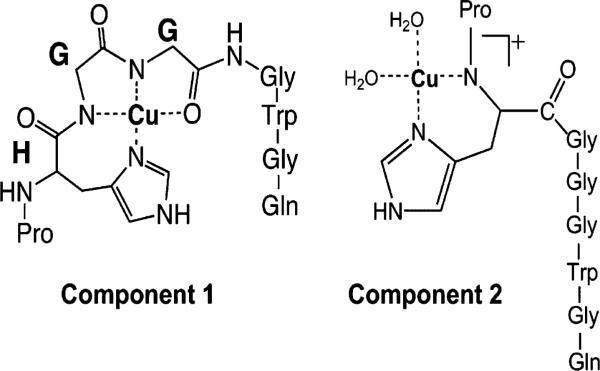
Three distinct Cu(II) binding modes, referred to as components 1, 2, and 3, have been observed for the synthetic prion peptide (PHGGGWGQ)n [29], where n ≥ 3. Component 1 and 2 binding modes can be seen in Fig. 5 [30]. The component 1 Cu(II) binding mode has a structure where the ligands for Cu(II) are the imidazole ring of histidine, the deprotonated amide nitrogens of the following two glycines, and the carbonyl oxygen of the second glycine [29]. The component 2 Cu(II) binding mode has a structure where the ligands for Cu(II) are the exocyclic amide nitrogen of the histidine and the imidazole ring nitrogen of the histidine [30]. Multiple histidines coordinate Cu(II) through their imidazole ring nitrogens in the component 3 binding mode. Because the peptides in the subsequent experiment contain only two octarepeats, and component 3 is observed only in synthetic prion peptide constructs with three or more octarepeats, we do not consider this binding as a contributor to retention or selectivity [30].
Fig. 5.
Structure of component 1 and component 2 copper binding modes.
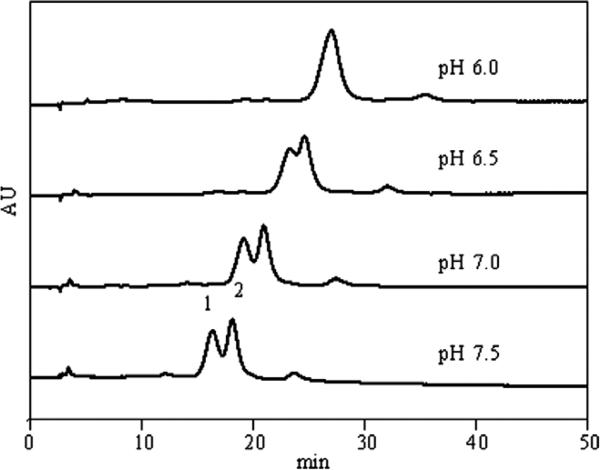
At pH 7.5 (PHGGGWGQ)2 binds predominantly in the component 1 binding mode. EPR data showed that substitution of the second glycine in each repeat unit of (PHGGGWGQ)2 with N-methylated glycine (sarcosine) (PHGXGWGQ)2 resulted in predominantly component 2 binding at pH 7.4 [29]. To test whether the column could resolve peptides that differed by copper binding mode, we carried out a gradient separation of the mixture of KKRPKP(PHGGGWGQ)2 and KKRP(PHGXGWGQ)2 at pH 7.5. Fig. 6 shows that the two peptides nearly baseline resolved at pH 7.5.
Fig. 6.
Separation of two-peptide mixture. Elution was achieved by running a linear gradient from 20 mM Nem/Hepes/Mes/acetic acid, 0.5 M NaCl, and 1 mM imidazole to 20 mM Nem/Hepes/Mes/acetic acid, 0.5 M NaCl, and 100 mM imidazole in 50 min. The eluent was adjusted with HCl or NaOH to achieve the proper eluent pH.
In synthetic peptides containing the octapeptide region, there are substantial changes in the ligand environment of copper with changes in pH [29]. At pH 7.5 and full copper occupancy, (PHGGGWGQ)4 binds predominantly in the component 1 binding mode. As the pH becomes more acidic (pH 6.5), component 2 becomes the predominant mode of Cu(II) binding [29,30]. If the IMAC column is resolving the peptides based on copper binding mode, we expect that the column would not be able to resolve peptides 3 and 4 when the eluent pH was adjusted to pH 6.5. Fig. 6 shows that peptides 3 and 4 are only partially resolved at pH 6.5 and coelute at pH 6.0. Recent studies have found that the octapeptide repeat binding mode is also dependent on octarepeat to Cu(II) stoichiometry [3,30]. Although the binding stoichiometry of the peptide interacting with Cu(II) on the IMAC column is not known, the chromatographic evidence strongly supports chromatographic resolution of peptides differing by mode of copper binding. Overall, these results suggest that the column has a high selectivity for not only the number of copper binding sites but also the affnity of the coordination interaction, including differences in copper binding structure. The column can be used over a broad pH range, and pH can be used as a lever to optimize resolution (Fig. 6).
Conclusions
A unique synthetic approach has been employed to nanoengineer the surface of PS–DVB beads for analytical IMAC applications. This approach allowed for the control of column capacity and nanoparticle size. TEM of bead cross sections showed nanoscale architectures, including an approximately 50-nm thick hydrophilic layer and approximately 3-nm diameter copper–polymer nanoparticles with 1 nanoparticle per 150 nm2. A single ferritin molecule was visualized interacting with a single nanoparticle, suggesting an optimal phase design for protein purification and on-column refolding experiments.
The column was able to resolve prion-derived peptides that differed by octarepeat number, Q → A substitution, presence of hydrophilic tail, and copper binding mode. The column was operated over a wide pH range. Resolution of species with different modes of copper binding was optimized by adjusting eluent pH, suggesting that pH can be used as a lever for optimizing separations.
We expect that this analytical IMAC column will provide a much-needed analytical tool not only for increasing the purity of PrPC, PrPSc, and glycosylation variants of PrP fractions but also for identifying new PrP isoforms or variants. Because this column is not limited to prion applications, these data suggest that the column will increase purity of His-tag protein purifications, intact protein (e.g., monoclonal antibody) purifications, copper binding peptide fractionation, and phosphopeptide enrichment applications.
Table 1.
Synthetic prion peptides
| Peak number | Sequence |
|---|---|
| 1 | PHGGGWGQ |
| 2 | PHGXGWGQ |
| 3 | KKRPKP(PHGGGWGQ)2 |
| 4 | KKRP(PHGXGWGQ)2 |
| 5 | KKRPKPWGQ(PHGGGWGQ)4 |
| 6 | WGQ(PHGGGWGQ)4 |
| 7 | WGA(PHGGGWGA)4 |
Note. X, sarcosine (N-methyglycine).
Acknowledgments
We thank Dionex Corporation and the University of California, Santa Cruz, for support. This work was supported, in part, by National Institutes of Health grant GM 65790 (to G.L.M.) and National Science Foundation grant DMR 0090409 (to K.M.).
Footnotes
Abbreviations used: PrPC, cellular form of the prion protein; GPI, glycosylphosphatidylinositol; TSE, transmissible spongiform encephalopathy; CJD, Creutzfeldt–Jakob disease; IMAC, immobilized metal affnity chromatography; RP, reversed phase; HIC, hydrophobic interaction chromatography; IEC, ion exchange chromatography; TEM, transmission electron microscopy; IDA, iminodiacetic acid; Fmoc, N-(9-fluorenyl)methoxycarbonyl; HOBt, 1-hydroxybenzotriazole; HBTU, O-benzotriazolyl-N,N,N′,N′-tetramethyluronium hexafluorophosphate; DIEA, diisopropylethylamine; DBU, 1,8-diazabicyclo[5.4.0]undec-7-ene; DMF, dimethyl formamide; DIC, diisopropylcarbodiimide; THF, tetrahydrofuran; TFA, trifluoroacetic acid; PS–DVB, polystyrene divinylbenzene; EPR, electron paramagnetic resonance.
References
- 1.Prusiner SB. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 2.Donne DG, Viles JH, Groth D, Melhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Structure of the recombinant full-length hamster prion protein PrP (29–231): The N terminus is highly flexible. Proc. Natl. Acad. Sci. USA. 1997;94:13452–13456. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, Gerfen GJ, Peisach J, Millhauser G. Copper coordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–6803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Millhauser G. Copper binding in the prion protein. Acc. Chem. Res. 2004;37:79–85. doi: 10.1021/ar0301678. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Stockel J, Safar J, Wallace AC, Cohen FE, Prusiner SB. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 5.a Helfferich FG. “Ligand exchange”: A novel separation technique. Nature. 1961;189:1001–1002. [Google Scholar]; b Davankov VA, Semechkin AV. Ligand-exchange chromatography. J. Chromatogr. 1977;141:313–353. doi: 10.1016/s0021-9673(01)97930-0. [DOI] [PubMed] [Google Scholar]
- 6.a Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography: A new approach to protein fractionation. Nature. 1975;258:598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]; b Chaga GS. Twenty-five years of immobilized metal ion affinity chromatography: Past, present, and future. J. Biochem. Biophys. Methods. 2001;49:313–334. doi: 10.1016/s0165-022x(01)00206-8. [DOI] [PubMed] [Google Scholar]; c Hemdan ES, Zhao YJ, Sulkowski E, Porath J. Surface topography of histidine residues: A facile probe by immobilized metal ion affinity chromatography. Proc. Natl. Acad. Sci. USA. 1989;86:1811–1815. doi: 10.1073/pnas.86.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Hochuli E, Dobeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighboring histidine residues. J. Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]; b Hochuli C, Bannwarth W, Dobeli H, Gentz R, Stuber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Bio/Technology. 1988;6:1321–1325. [Google Scholar]
- 8.a Pergami P, Jaffe H, Safar J. Semipreparative chromatographic method to purify the normal cellular isoform of the prion protein in nondenatured form. Anal. Biochem. 1996;236:63–73. doi: 10.1006/abio.1996.0132. [DOI] [PubMed] [Google Scholar]; b Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Melhorn I, Huang Z, Fletterick RJ, Cohen FE, Prusiner SB. Conversion of a-helices b-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Safar J, Roller PP, Gajdusek DC, Gibbs CJ. Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J. Biol. Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 9.Yin S-M, Zheng Y, Tien P. On-column purification and refolding of recombinant bovine prion protein: Using its octarepeat sequences as a natural affinity tag. Protein Express. Purif. 2003;32:104–109. doi: 10.1016/S1046-5928(03)00195-5. [DOI] [PubMed] [Google Scholar]
- 10.Shaked Y, Rosenmann H, Hijazi N, Halimi M, Gabizon R. Copper binding to the PrP isoforms: A putative marker of their conformation and function. J. Virol. 2001;75:7872–7874. doi: 10.1128/JVI.75.17.7872-7874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller H, Strom A, Hunsman G, Stuke A. Biochem. J. submitted. [Google Scholar]
- 12.Leonard M. New packaging materials for protein chromatography. J. Chromatogr. B. 1997;699:3–27. doi: 10.1016/s0378-4347(97)00160-6. [DOI] [PubMed] [Google Scholar]
- 13.a Frechet JMJ. Science. Vol. 263. 1994. Functional polymers and dendrimers: Reactivity, molecular architecture, and interfacial energy; pp. 1710–1715. [DOI] [PubMed] [Google Scholar]; b Tomalia DA, Durst HD. In: Topics in Current Chemistry. Weber E, editor. Vol. 165. Springer-Verlag; Berlin: 1993. pp. 193–319. [Google Scholar]; c Hawker C, Frechet JMJ. Preparation of polymers with controlled molecular architecture: A new convergent approach to dendritic macromolecules. J. Am. Chem. Soc. 1990;112:7638–7647. [Google Scholar]
- 14.a Szwarc M, Levy R, Milkovich J. Polymerization initiated by electron transfer to monomer: A new method of formation of block polymers. J. Am. Chem. Soc. 1956;78:2656. [Google Scholar]; b Grubbs RH, Gilliom LR. Titanacyclobutanes derived from strained, cyclic olefins: The living polymerization of norbornene. J. Am. Chem. Soc. 1986;108:733–742. [Google Scholar]; c Webster O. Living polymerization methods. Science. 1991;251:887–893. doi: 10.1126/science.251.4996.887. [DOI] [PubMed] [Google Scholar]; d Wang JS, Matyjaszewski K. Controlled living radical polymerization–atom-transfer radical polymerization in the presence of transition–metal complexes. J. Am. Chem. Soc. 1995;117:5614–5615. [Google Scholar]
- 15.Harth E, Van Horne B, Lee VY, Germack DS, Gonzales CP, Miller RD, Hawker CJ. A facile approach to architecturally defined nanoparticles via intramolecular chain collapse. J. Am. Chem. Soc. 2002;124:8653–8660. doi: 10.1021/ja026208x. [DOI] [PubMed] [Google Scholar]
- 16.Jung HM, Price KE, McQuade DT. Synthesis and characterization of cross-linked reverse micelles. J. Am. Chem. Soc. 2003;125:5351–5355. doi: 10.1021/ja0271983. [DOI] [PubMed] [Google Scholar]
- 17.Huang H, Remsen EE, Kowalewski T, Wooley KL. Nanocages derived from shell cross-linked micelle templates. J. Am. Chem. Soc. 1999;121:3805–3806. [Google Scholar]
- 18.McCarthy PA, Huang J, Yang S, Wang HS. Synthesis and characterization of water-soluble chiral conducting polymer nano-composites. Langmuir. 2002;18:259–263. [Google Scholar]
- 19.a Matyjaszewski K, Xia J. J. Chem. Rev. Vol. 101. 2001. Atom transfer radical polymerization; pp. 2921–2990. [DOI] [PubMed] [Google Scholar]; b Matyjaszewski K, Davis TP, editors. Handbook of Radical Polymerization. John Wiley; Hoboken, NJ: 2002. [Google Scholar]
- 20.a Matyjaszewski K, Miller PJ, Shukla N, Immaraporn B, Gelman A, Luokala BB, Siclovan TM, Kickelbick G, Vallent T, Hoffmann H, Pakula T. Polymers at interfaces: using atom transfer radical polymerization in the controlled growth of homo- and block copolymers from silicon surfaces in the absence of untethered sacrificial initiator. Macromolecules. 1999;32:8716–8724. [Google Scholar]; b de Boer B, Simon HK, Werts MPL, van der Vegte EW, Hadziioannou G. “Living” free radical photopolymerization initiated from surface-grafted iniferter monolayers. Macromolecules. 2000;33:349–356. [Google Scholar]; c Brittain W. Polymer brushes: Surface immobilized polymers. Surface Sci. 2004;570:1–12. [Google Scholar]
- 21.Xiao D, Wirth MJ. Surface initiated radical polymerization of acrylamide on silica at room temperature. Macromolecules. 2002;35:2919–2925. [Google Scholar]
- 22.a Zheng G, Stover HDH. Grafting of polystyrene from narrow disperse polymer particles by surface-initiated atom transfer radical polymerization. Macromolecules. 2002;35:6828–6834. [Google Scholar]; b Perruchot C, Khan MA, Kamitsi A, Armes SP, von Werne T, Patten T. Synthesis of well-defined, polymer-grafted silica particles by aqueous ATRP. Langmuir. 2001;17:4479–4481. [Google Scholar]; c Pyun J, Matyjaszewski K. Synthesis of nanocomposite organic/inorganic hybrid materials using controlled/“living” radical polymerization. Chem. Mater. 2001;13:3436–3448. [Google Scholar]
- 23.Meyer U, Svec F, Frechet JMJ, Hawker CJ, Irgum K. Use of stable free radicals for the sequential preparation and surface grafting of functionalized macroporous monoliths. Macromolecules. 2000;33:7769–7775. [Google Scholar]
- 24.Zhao B, Brittain WJ. Polymer brushes: Surface-immobilized macromolecules. Prog. Polym. Sci. 2000;25:677–710. [Google Scholar]
- 25.a Ejaz M, Yamamoto S, Ohno K, Tsujii Y, Fukuda T. Controlled graft polymerization of methyl methacrylate on silicon substrate by the combined use of the Langmuir–Blodgett and atom transfer radical polymerization techniques. Macromolecules. 1998;31:5934–5936. [Google Scholar]; b Husseman M, Malmstrom EE, McNamara M, Mate M, Mecerreyes O, Benoit DG, Hedrick JL, Mansky P, Huang E, Russell TP, Hawker CJ. Controlled synthesis of polymer brushes by “living” free radical polymerization techniques. Macromolecules. 1999;32:1424–1431. [Google Scholar]
- 26.Massover WH. UItrastructure of ferritin and apoferritin: A review. Micron. 1993;24:389–437. [Google Scholar]
- 27.Miura T, Hori A, Mototani H, Takeuchi H. Raman spectroscopy of pH dependence of copper binding to prion octapeptide. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 28.Goldfarb LG, Brown P, McCombie WR, Goldgaber D, Swergold GD, Willis PR, Cervenakova L, Baron H, Gibbs CJ, Gajdusek DC. Transmissible familial Creutzfeldt–Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proc. Natl. Acad. Sci. USA. 1991;88:10926–10930. doi: 10.1073/pnas.88.23.10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, Antholine WE, Ball HL, Cohen FE, Prusiner SB, Millhauser GL. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chattopadhyay M, Walter E, Newell D, Jackson P, Aronoff-Spencer E, Peisach J, Gerfen GJ, Bennett B, Antholine WE, Millhauser G. The octarepeat domain of the prion protein binds Cu(II) with three distinct coordination modes at pH 7.4. J. Am. Chem. Soc. 2005;127:12647–12656. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]