Abstract
In biological and ecological statistical inference, it is practically useful to provide a lower bound for species richness in a community. Chao (1984, 1989) derived a nonparametric lower bound for species richness in a single community. However, there have been no lower bounds proposed in the literature for the number of species shared by multiple communities. Based on sample species abundance or replicated incidence records from each of the N communities, we derive in this article a nonparametric approach to constructing a lower bound for the number of species shared by N (N ≥ 2) communities. The approach is valid for all types of species abundance distributions (for abundance data) or species detection probabilities (for replicated incidence data). Variance estimators for the proposed lower bounds are obtained by using typical asymptotic theory. Simulation results are reported to examine the performance of the lower bounds. Replicated incidence data of ciliate species collected in three areas from Namibia, southwest Africa, are used for illustration. We also briefly discuss the application of the proposed method to estimate the size of a shared population (i.e., the number of individuals in the intersection of multiple populations) based on capture-recapture data from each population.
Keywords: Abundance data, Community overlap, Diversity indices, Replicated incidence data, Shared species, Similarity
1. INTRODUCTION
Species richness in a single community (alpha diversity) is a classic concept for characterizing community diversity. The estimation of species richness has been extensively discussed in the literature; see Seber (1982), Bunge and Fitzpatrick (1993), Colwell and Coddington (1994), and Chao (2005) for reviews. For multiple communities, the number of shared species plays an important role for describing community overlap and forms a basis to construct various types of similarity indices or beta diversity. When compared with species richness in one community, the estimation of shared species richness in multiple communities has received relatively little attention. Although estimators for shared species richness in two communities were proposed (e.g., Chao et al. 2000; Chao, Shen, and Hwang 2006), these methods have not been extended to more than two communities.
It is intuitively understood that, if there are many undetectable or “invisible” species in a hyper-diverse community, then it is impossible to obtain a good estimate of species richness. Therefore, it is practically useful to provide a lower bound for species richness. A nonparametric lower bound in a single community was derived by Chao (1984, 1989) for abundance-based data and for replicated incidence (i.e., presence or absence) data. The Chao (1984, 1989) lower bound has been applied in various disciplines. For example, microbiologists used it to infer species richness in microbial hyper-diverse communities (Hughes et al. 2001; Bohannan and Hughes 2003; Stach et al. 2003; Schloss and Handelsman 2005). However, there have been no lower bounds proposed for the number of species shared by multiple communities.
Part of this research was initiated by analyzing soil ciliate species data collected in three areas of Namibia, southwest Africa by Foissner and colleagues (Foissner, Agatha, and Berger 2002). See Table 1 and Section 5 for data and detailed analysis. Questions concerning the alpha, beta, and gamma diversity of microorganisms and their biogeographical distribution (ubiquity or endemicity) have generated extensive discussion in the literature. However, previous diversity analysis for soil ciliate species (Chao et al. 2006) was limited to alpha diversity in a single community (or area). In order to investigate the community overlap or beta diversity based on multiple-community data in Namibia, we were motivated to estimate the number of species shared by at least two communities (or areas).
Table 1.
Data summary for three areas of Namibia (original data are given in Foissner, Agatha, and Berger 2002)
| Data |
||||
|---|---|---|---|---|
| Area | Number of soil samples |
Number of observed species |
Number of unique species |
Number of duplicate species |
| Area 1: Southern Namib Desert | 15 | 154 | 85 | 29 |
| Area 2: Central Namib Desert | 17 | 136 | 69 | 28 |
| Area 3: Etosha Pan | 19 | 234 | 125 | 44 |
| Total | 51 | 331 | ||
When sample abundance or replicated incidence records are available from each of the N (N ≥ 2) communities, we propose in this article a unified approach to constructing a nonparametric lower bound for the number of species shared by N communities. The proposed method is nonparametric in the sense that they are not dependent on the assumptions about the species abundance distribution (for abundance data) or the species detection probability (for replicated incidence data). Variance estimators for the proposed lower bounds are also obtained.
Since a review of the details on deriving the lower bound of species richness in a single community (Chao 1984, 1989) would greatly help to extend the framework to multiple communities, we provide such a review in Section 2 separately for abundance data (in Section 2.1) and replicated incidence data (in Section 2.2). In Section 3, we develop a lower bound for the number of species shared by two communities. In Section 4, a unified approach is described for the case of more than two communities. In Section 5, the replicated incidence data for ciliate species that motivated this research are analyzed as an illustrative example. Section 6 reports a simulation study in order to examine the performance of the proposed method. Some concluding remarks and relevant discussion are provided in Section 7.
2. ONE COMMUNITY
2.1 Abundance Data
We first review the lower bound of species richness (Chao 1984, 1989) in a single community. Assume that there are S species indexed from 1 to S and a fixed number of n individuals are independently observed in the community. Denote the species probabilities by (θ1, θ2, … , θS), where . That is, θi the probability that any randomly selected individual is classified to the ith species. Each probability is a combination of species abundance and individual detectability. If all individuals in the community have the same probabilities of being detected, then the species probabilities represent the true relative abundances.
Let Xi (species frequency) be the number of times, or individuals, that the ith species is observed in the sample, i = 1, 2, … , S. Only those species with Xi > 0 are observable in the sample. The species frequencies (X1, X2, … , XS) are assumed to follow a multinomial distribution with cell total n and probabilities (θ1, θ2, … , θS).
Let fk, k = 0, 1, … , n, (frequency counts) be the number of species represented by k times, or individuals, in the sample. That is, , where I(A) is the usual indicator function, i.e., I(A) = 1 if the event A occurs, and 0 otherwise. Here, f0 denotes the number of undetected species in the sample. Thus, we have . Let D denote the number of distinct species observed in the sample, that is, .
A parametric approach to estimating species richness is to assume that (θ1, θ2, … , θS) follows some types of distributions characterized by a few parameters. For example, Fisher, Corbet, and Williams (1943) assumed that , where (λ1, λ2, … , λS) are a random sample from a gamma distribution. MacArthur’s (1957) broken-stick model assumed that (λ1, λ2, … , λS) are a random sample from an exponential distribution. There are other types of abundance distributions; see Magurran (2004) for a review.
Since S = D + f0, our estimating target becomes E(f0). Under the assumptions that (θ1, θ2, … , θS) are fixed unknown parameters, we have the following expectation, respectively, for the expected number of undetected species, singletons, and doubletons:
| (2.1a) |
| (2.1b) |
| (2.1c) |
Based on Equations (2.1a) to (2.1c), Chao (1989) used the following Cauchy–Schwarz inequality
| (2.2) |
to obtain a theoretical bound for E(f0):
| (2.3) |
The inequality becomes an equality if and only if all probabilities are equal (a homogeneous case). If f2 > 0, we can replace the expected values by the observed data in Equation (2.3) and a lower bound for species richness becomes:
| (2.4) |
with the bound being achieved under a homogeneous community. We remark that instead of treating (θ1, θ2, … , θS) as fixed parameters, they can be modeled as random effects selected from an unknown distribution. Under a random-effect model, parallel derivation results in the same estimator. A bias-corrected estimator in a homogeneous case turns out to be
| (2.4a) |
The lower bound in Equation (2.4) was proposed by Chao (1984) using an alternative derivation. For abundance data, the sample size n is often large so that the term (n − 1)/n in the bound can be dropped and the estimator in Equation (2.4) is reduced to . This simplified estimator has been referred to as the Chao1 estimator in the biological and ecological literature (e.g., Colwell and Coddington 1994; Walther and Morand 1998; Hughes et al. 2001). It is also featured in several computer software packages including EstimateS (Colwell 2004), DOTUR (Schloss and Handelsman 2005), SPADE (Chao and Shen 2003), and WS2m (Turner, Leitner, and Rosenzweig 1999).
An estimated variance formula derived in Chao and Shen (2003) for the estimator in Equation (2.4) is
| (2.5) |
where K = (n − 1)/n. When f2 = 0, it is suggested using the bias-corrected form and the lower bound becomes . In this instance, the variance formula is modified to
| (2.6) |
One advantage of using the Chao1 estimator is that the estimated number of undetected species depends only on the first two frequency counts, i.e., the numbers of singletons and doubletons. This implies that ecologists do not need to obtain the exact frequency of any species that has at least three individuals in the sample. The estimator is especially useful if counting the exact number of individuals for each species appearing in the sample requires substantial effort.
2.2 Replicated Incidence Data
In many microorganism surveys, only species presence/absence data can be collected because there are too many individuals to be counted. For example, in the ciliate species data (Section 5) and other microbial data, it is not possible to count exactly the number of individuals and thus only the presence/absence of each observed species was recorded. Accordingly, only replicated incidence data were available.
Assume that there are t samples and they are indexed 1, 2, … , t. We use the general term “sample” which could also refer to a team, occasion, transect line, a fixed period of time, or an investigator. The presence or absence of any species for these t samples is recorded to form a species-by-sample incidence matrix. In most applications, sufficient statistics from the species-by-sample incidence matrix are the incidence-based frequency counts (Q1, Q2, … , Qt), where Qk denotes the number of species that are detected in exactly k samples, k = 1, 2, … , t. Hence, Q1 represents the number of “unique” species (those that are detected in only one sample) and Q2 represents the number of “duplicate” species (those that are detected in only two samples).
Assume that the species detection probabilities, defined as the chance of encountering at least one individual of a given species in any sample, are (θ1, θ2, … , θS) and these probabilities are kept constant across the samples. We remark that, unlike the constraint in abundance data, may be greater than 1 for incidence-based data.
Parallel derivations to those in Section 2.1 can be made with n being replaced by t, and the counts (f1, f2, … , fn) replaced by (Q1, Q2, … , Qt). Therefore, an estimator based on t replicated incidence records for multiple samples has the form , which is referred to in the literature as the Chao2 estimator. The number of samples t for incidence data may not be large, so we suggest retaining the term (t − 1)/t in the estimator. This estimator was originally derived by Chao (1987) for capture-recapture data as a lower bound. A bias-corrected form is . See Chao and Shen (2003) for an approximate variance formula.
3. TWO COMMUNITIES
3.1 Abundance Data
This section extends our approach to the estimation of the number of species shared by two communities. Assume that there are S1 species in community I and there are S2 species in community II. The species probabilities in communities I and II are denoted (θ11, θ21, … , θS1,1) and (θ12, θ22, … , θS2,2), respectively. . Let of the number shared species be S12. Without loss of generality, we assume that the first S12 species are the shared species.
Two random samples (sample I with size n1 and sample II with size n2) are taken from communities I and II, respectively. Assume that D12 shared species are observed. Denote the observed frequencies in the two communities, respectively, by (X11, X21, … , XS1,1) and (X12, X22, … , XS2,2). Define for any two nonnegative integers j and k,
| (3.1a) |
| (3.1b) |
| (3.1c) |
That is, fjk denotes the number of shared species that are observed j times in sample I and k times in sample II. In particular, f11 denotes the number of shared species that are singletons in both samples, and f00 denotes the number of shared species that are undetected in both samples. Also, f+0 denotes the number of shared species that are observed in sample I but not observed in sample II, and a similar interpretation for f0+.
Since S12 = D12 + f+0 + f0+ + f00 and only D12 is observable, our approach is to find a lower bound for each of the expected values of the other three terms, i.e., E(f+0), E(f0+), and E(f00). Assuming a multinomial model for each of the two sets of frequencies, we have
| (3.2a) |
| (3.2b) |
| (3.2c) |
- A lower bound for E(f+0): Note we have
The following Cauchy–Schwarz inequality
leads to
The equality holds if and only if community 2 is homogenous in species probabilities.(3.3) - Similarly, a lower bound for E(f0+) is
The equality holds if and only if community 1 is homogenous in species probabilities.(3.4) - A lower bound for E(f00) is obtained by noting
Again, a similar Cauchy-Schwarz inequality
gives
Combining the above three lower bounds, we thus have a lower bound for the shared species richness (let Ki = (ni − 1)/ni)(3.5)
In many cases, the sample sizes n1 and n2 are large for abundance data; thus, the terms (n1 − 1)/n1 and (n2 − 1)/n2 can be dropped in the above formula. The estimator in Equation (3.6) can be regarded as an extension of the Chao1 estimator to two communities. When f2+ = 0 or f+2 = 0, a bias-corrected estimator is(3.6)
Note that only observed shared species are involved in the formulas of Equations (3.6) and (3.7), thus observed nonshared species play no role in our estimation, although any observed nonshared species could actually be a shared species. Because the proposed estimator can be regarded as a function of the statistics (D12, f11, f22, f1+, f2+, f+1, f+2), we obtain a variance estimator by using a standard asymptotic approach under a multinomial model. Then the estimated variance can be used to construct a confidence interval for the true parameter using a log-transformation (Chao 1987).(3.7)
3.2 Replicated Incidence Data
The method developed for abundance data can be directly adapted to deal with the replicated incidence case. All notation and model formulation are similar to those in Section 3.1. Assume that there are t1 samples randomly taken from community I and t2 samples from community II. In each sample, only presence/absence data are recorded. The two sets of probabilities (θ11, θ21, … , θS1,1) and (θ12, θ22, … , θS2,2) in the incidence case represent species detection probabilities in any sample from communities I and II, respectively.
Let Xi1 and Xi2 denote the number of samples that the ith species is detected in communities I and II, respectively. Let denote the number of shared species that are detected in j samples in community I and k samples in community II. Similarly, we can define Qj+ and Q+k. By applying a method analogous to that in Section 3.1, it can be shown that the lower bound and the bias-corrected version for the number of shared species based on incidence counts have the same forms as in Equations (3.6) and (3.7), except that the samples sizes n1 and n2 should be, respectively, by t1 and t2, and abundance counts replaced by incidence counts. We remark that an approximate estimator of shared species richness was derived in Chao, Shen, and Hwang (2006) for both types of data based on the Laplace approximation formula, but that estimator cannot be theoretically verified to be a lower bound.
4. MORE THAN TWO COMMUNITIES
The approach proposed in Section 3 has an obvious extension to the case of more than two communities. We first describe the derivation for three communities. Extension to more than three communities is direct. Here a “shared” species is defined as that the species belongs to all communities. Assume that there are S123 species shared by three communities I, II, and III and a random sample is taken from each of the three communities. The three samples are called samples I, II, and III with sizes n1, n2, and n3, respectively. Let D123 denote the observed shared species richness in the three samples. Then
| (4.1) |
where f++0 denotes the number of shared species that are observed in samples I, II, but not observed in sample III, f000 denotes the number of shared species that are undetected in all three samples, and a similar interpretation for other terms in Equation (4.1).
Based on a similar type of inequality as in Equations (3.3) and (3.4), we can get a lower bound for the expected value of each term of f++0 + f+0+ + f0++ as shown in the second term to the fourth term in the right hand side of Equation (4.2).
Based on a similar type of inequality as in Equation (3.5), we can get a lower bound for the expected value of each term of f00+ + f0+0 + f+00 as shown in the fifth term to the seventh term of Equation (4.2).
Extending Equation (3.5), we have a lower bound for E(f000) as showninthe last term of Equation (4.2).
Combining the above, we have a lower bound for S123 as follows:
| (4.2) |
An estimated variance can be obtained by an asymptotic method. Extending Equations (3.6) and (4.2) with self-explanatory notation generalization, we have a lower bound for four communities:
| (4.3) |
Thus, we have provided a unified approach to formulating lower bounds for any number of communities. However, the estimated variance estimator becomes quite complicated. Currently, we have variance estimators only up to five communities.
Based on Equations (4.2) and (4.3), similar lower bounds for replicated incidence data can be obtained by replacing frequency counts by incidence counts and each sample size by the number of samples. Variance estimators are derived in an analogous way.
5. EXAMPLE
A total of 51 soil samples were taken from three areas of Namibia; see Table 1 for a description of relevant data information. Generally, collections were made from a variety of soil and vegetation types of the respective area. About 10 small subsamples were taken from an area of about 100 m2 and mixed to a composite soil sample. In each soil sample, presence/absence of ciliate species was recorded. Species were determined by combining live observation, silver impregnation, and scanning electron microscopy. Detailed sampling locations, procedures, and species identification were described in Foissner (1999, 2006) and Foissner, Agatha, and Berger (2002). After presence/absence of soil ciliate species was recorded for each sample, the replicated incidence data were merged by species identity and a total of 331 species were recorded in our data. All data in EXCEL spreadsheets are available from the authors upon request.
We illustrate one-community species richness estimation for each area and shared species richness estimation for any two areas (three combinations) and for all three areas; see Table 2. We provide for each case a lower bound for species richness or shared species richness in Table 2. The communities considered in our applications are highly heterogeneous and thus we adopt the original form of estimators. That is, our estimates are calculated from Equations (2.4), (3.6), and (4.2) with sample size there being replaced by the number of samples and frequency counts being replaced by incidence counts. The bias-corrected formulas which are derived under a homogeneous case are not reported here. For each estimate, its associated SE (standard error) as well as the 95% confidence interval based on a log-transformation are also shown in Table 2. The percentage of undetected shared species with respect to the estimated minimum is given in the last column.
Table 2.
Lower bounds for species richness and shared species richness (see Table 1 for area definition)
| Lower bounds |
|||||
|---|---|---|---|---|---|
| Area | Number of observed |
Estimate | SE | 95% CI | % of undetected |
| Species richness for one area: | |||||
| Area 1 | 154 | 270 | 34.9 | (219, 361) | 43% |
| Area 2 | 136 | 216 | 26.1 | (179, 285) | 37% |
| Area 3 | 234 | 402 | 41.4 | (339, 505) | 42% |
| (Average 41%) | |||||
| Shared species richness for two areas: | |||||
| Areas (1, 2) | 77 | 114 | 17.2 | (92, 165) | 32% |
| Areas (1, 3) | 97 | 189 | 33.6 | (143, 281) | 49% |
| Areas (2, 3) | 84 | 157 | 30.2 | (117, 243) | 46% |
| (Average 42%) | |||||
| Shared species richness for all three areas: | |||||
| (1, 2, 3) | 65 | 126 | 30.1 | (89, 217) | 48% |
All estimates indicate that there are still a substantial fraction of undetected species and shared species in the current data. For the alpha diversity, on average, about 41% of species diversity is still undetected. This is consistent with the finding in Chao et al. (2006). For estimating shared species richness, the observed number of shared species substantially underestimates the true number of shared species. Our approach reveals the extent of under-estimation for the observed number of shared (an average 42% for any two areas and 48% for three areas) and provides helpful information for understanding community overlap of micro-organisms.
6. SIMULATION
Since our data analysis was based on replicated incidence data, we carried out a simulation study to investigate the performance of the proposed lower bounds for such kinds of data. We examined the shared species richness estimation for two and three communities. In each community, five types of species detection probabilities were considered: one homogeneous and four heterogeneous communities with 200 species in each. The five sets of species detection probabilities along with their average (
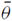 ) and coefficient of variation (CV) are given as follows:
) and coefficient of variation (CV) are given as follows:
Type I: θi = 0.10, i = 1, … , 200 (
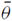 = 0.10, CV = 0.0),
= 0.10, CV = 0.0),Type II: θi ~ Uniform(0, 1), i = 1, … , 200 (
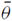 = 0.50, CV = 0.58),
= 0.50, CV = 0.58),Type III: θi ~ Beta(1, 2), i = 1, … , 200 (
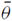 = 0.33, CV = 0.71),
= 0.33, CV = 0.71),Type IV: θi = 10/(i + 10), i = 1, … , 200 (
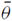 = 0.15, CV = 1.01),
= 0.15, CV = 1.01),Type V: θi = 3/(i + 3), i = 1, … , 200 (
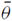 = 0.06, CV = 1.55).
= 0.06, CV = 1.55).
Type I denotes a homogeneous case, i.e., all species have the same probability of being detected in any sample; Types II and III assume that the probabilities represent a random sample, respectively, from a uniform or a beta density. (That is, for each simulation trial, we generated a sample of size of 200 as species probabilities.) Types IV and V are in a form of truncated logarithmic series, which is widely prevalent in modeling natural frequency data. It is also called Zipf’s law in linguistics and behavioral sciences. The value of CV characterizes the degree of heterogeneity among detection probabilities.
We considered all 15 possible combination cases of any two communities: (I versus I), (I versus II), … , (V versus V) as our target communities. We assume that the first 120 species are the shared species. Thus S1 = S2 = 200 and S12 = 120. Table 3 presents the simulation results for the case of two communities.
Table 3.
Simulation results for two communities (the true parameter S12 = 120; simulation trials = 2000), : the original lower bound; : the bias-corrected form
| Cases | Average of observed shared |
Estimator | Average estimate |
Sample SE |
Average estimated SE |
Sample RMSE |
Coverage of 95% CI |
|---|---|---|---|---|---|---|---|
| I vs. I | 92.5 | 122.1 | 11.32 | 11.14 | 11.50 | 0.93 | |
| 119.8 | 10.63 | 11.16 | 10.63 | 0.96 | |||
| I vs. II | 100.5 | 119.0 | 8.47 | 8.85 | 8.53 | 0.96 | |
| 117.1 | 7.84 | 7.95 | 8.36 | 0.96 | |||
| I vs. III | 95.8 | 116.9 | 10.02 | 9.84 | 10.51 | 0.94 | |
| 114.3 | 9.01 | 8.84 | 10.66 | 0.91 | |||
| I vs. IV | 98.5 | 121.6 | 9.61 | 9.56 | 9.74 | 0.94 | |
| 119.5 | 8.87 | 9.20 | 8.88 | 0.96 | |||
| I vs. V | 69.9 | 115.7 | 20.65 | 19.36 | 21.09 | 0.93 | |
| 110.4 | 17.66 | 18.24 | 20.10 | 0.93 | |||
| II vs. II | 111.5 | 116.6 | 5.56 | 5.32 | 6.50 | 0.89 | |
| 115.2 | 4.62 | 4.14 | 6.67 | 0.83 | |||
| II vs. III | 103.7 | 112.1 | 6.73 | 6.08 | 10.39 | 0.84 | |
| 111.2 | 6.35 | 5.49 | 10.85 | 0.80 | |||
| II vs. IV | 106.5 | 117.6 | 6.59 | 6.69 | 7.02 | 0.95 | |
| 116.2 | 6.06 | 5.92 | 7.13 | 0.94 | |||
| II vs. V | 75.8 | 110.4 | 16.26 | 15.81 | 18.88 | 0.90 | |
| 107.9 | 15.45 | 14.63 | 19.64 | 0.87 | |||
| III vs. III | 104.0 | 113.4 | 7.81 | 7.44 | 10.20 | 0.85 | |
| 111.2 | 6.47 | 6.17 | 10.94 | 0.79 | |||
| III vs. IV | 101.8 | 115.5 | 7.94 | 7.66 | 9.14 | 0.93 | |
| 113.6 | 7.15 | 6.77 | 9.61 | 0.90 | |||
| III vs. V | 72.2 | 108.6 | 16.74 | 16.94 | 20.23 | 0.90 | |
| 104.7 | 15.23 | 15.19 | 21.60 | 0.85 | |||
| IV vs. IV | 105.0 | 120.5 | 7.94 | 7.94 | 7.95 | 0.95 | |
| 118.5 | 7.08 | 7.49 | 7.25 | 0.96 | |||
| IV vs. V | 75.7 | 114.1 | 17.83 | 18.04 | 18.78 | 0.94 | |
| 109.1 | 15.68 | 16.31 | 19.08 | 0.92 | |||
| V vs. V | 57.5 | 106.9 | 27.90 | 26.21 | 30.81 | 0.88 | |
| 98.0 | 21.95 | 23.22 | 31.06 | 0.87 |
For three communities, we considered 35 possible combination cases of any three communities: (I, I, I), (I, I, II), … , (V, V, V). We assumed that S1 = S2 = S3 = 200, S12 = S13 = S23 = 120, and S123 = 80. The overlap structure is described as follows: (a) the first 80 species in each community are shared by all three communities, (b) the last 40 species in each community are unique species, and (c) the species shared by communities I and II are the 81 ~ 120th species in community I and the 81 ~ 120th species in community II; the species shared by communities I and III are the 121 ~ 160th species in community I and the 81 ~ 120th species in community III; and the species shared by communities II and III are the 121 ~ 160th species in community II and the 121 ~ 160th species in community III. Table 4 presents the simulation results for the case of three communities.
Table 4.
Simulation results for three communities (the true parameter S123 = 80; simulation trials = 2000), : the original lower bound; the bias-corrected form
| Cases | Average of observed shared |
Estimator | Average estimate |
Sample SE |
Average estimated SE |
Sample RMSE |
Coverage of 95% CI |
|---|---|---|---|---|---|---|---|
| (I, I, I) | 54.3 | 84.3 | 13.29 | 13.20 | 13.97 | 0.94 | |
| 79.8 | 11.31 | 11.15 | 11.31 | 0.95 | |||
| (I, I, II) | 58.7 | 81.1 | 10.33 | 10.88 | 10.40 | 0.95 | |
| 77.7 | 9.11 | 9.08 | 9.40 | 0.94 | |||
| (I, I, III) | 56.2 | 80.4 | 11.99 | 11.93 | 12.00 | 0.95 | |
| 76.2 | 10.07 | 9.66 | 10.76 | 0.94 | |||
| (I, I, IV) | 60.0 | 83.4 | 10.57 | 10.72 | 11.10 | 0.93 | |
| 79.9 | 9.32 | 9.21 | 9.31 | 0.95 | |||
| (I, I, V) | 47.7 | 83.5 | 16.86 | 17.23 | 17.22 | 0.95 | |
| 76.8 | 13.74 | 13.57 | 14.10 | 0.94 | |||
| (I, II, II) | 65.3 | 79.4 | 8.17 | 8.55 | 8.19 | 0.96 | |
| 76.6 | 6.93 | 6.68 | 7.71 | 0.94 | |||
| (I, II, III) | 60.9 | 77.8 | 9.61 | 9.85 | 9.86 | 0.95 | |
| 74.1 | 7.70 | 7.42 | 9.73 | 0.91 | |||
| (I, II, IV) | 65.1 | 80.7 | 7.99 | 8.59 | 8.02 | 0.95 | |
| 77.8 | 6.92 | 6.93 | 7.26 | 0.95 | |||
| (I, II, V) | 51.5 | 78.4 | 12.89 | 13.42 | 12.99 | 0.96 | |
| 74.2 | 11.18 | 11.28 | 12.60 | 0.93 | |||
| (I, III, III) | 60.7 | 77.5 | 9.90 | 9.92 | 10.21 | 0.95 | |
| 73.8 | 8.22 | 7.61 | 10.27 | 0.90 | |||
| (I, III, IV) | 62.2 | 79.0 | 9.13 | 9.17 | 9.18 | 0.95 | |
| 75.7 | 7.59 | 7.33 | 8.71 | 0.93 | |||
| (I, III, V) | 49.3 | 77.9 | 13.97 | 14.62 | 14.13 | 0.96 | |
| 72.6 | 11.74 | 11.68 | 13.89 | 0.92 | |||
| (I, IV, IV) | 66.7 | 82.4 | 7.92 | 8.12 | 8.27 | 0.93 | |
| 79.7 | 6.80 | 6.85 | 6.81 | 0.96 | |||
| (I, IV, V) | 53.1 | 81.5 | 13.71 | 14.09 | 13.78 | 0.95 | |
| 76.7 | 11.72 | 11.60 | 12.17 | 0.95 | |||
| (I, V, V) | 43.5 | 80.4 | 19.43 | 20.00 | 19.42 | 0.94 | |
| 72.2 | 15.30 | 14.80 | 17.18 | 0.91 | |||
| (II, II, II) | 73.0 | 78.0 | 5.63 | 5.76 | 5.98 | 0.89 | |
| 75.6 | 4.12 | 3.37 | 6.01 | 0.80 | |||
| (II, II, III) | 67.8 | 76.1 | 6.81 | 7.36 | 7.84 | 0.93 | |
| 73.0 | 5.24 | 4.78 | 8.70 | 0.83 | |||
| (II, II, IV) | 72.4 | 79.1 | 5.98 | 6.11 | 6.05 | 0.94 | |
| 76.6 | 4.47 | 3.94 | 5.62 | 0.89 | |||
| (II, II, V) | 57.3 | 77.0 | 11.38 | 11.25 | 11.77 | 0.95 | |
| 73.5 | 9.82 | 9.24 | 11.76 | 0.91 | |||
| (II, III, III) | 66.1 | 75.5 | 8.22 | 8.15 | 9.36 | 0.90 | |
| 72.0 | 5.89 | 5.17 | 9.94 | 0.80 | |||
| (II, III, IV) | 67.4 | 77.4 | 7.58 | 7.62 | 8.03 | 0.93 | |
| 74.1 | 5.63 | 5.12 | 8.15 | 0.86 | |||
| (II, III, V) | 53.4 | 75.2 | 12.11 | 12.41 | 13.04 | 0.94 | |
| 70.7 | 10.01 | 9.73 | 13.65 | 0.88 | |||
| (II, IV, IV) | 72.4 | 80.1 | 5.66 | 6.16 | 5.66 | 0.96 | |
| 77.6 | 4.23 | 4.24 | 4.84 | 0.95 | |||
| (II, IV, V) | 57.6 | 78.5 | 11.68 | 11.70 | 11.77 | 0.95 | |
| 74.8 | 10.08 | 9.55 | 11.36 | 0.92 | |||
| (II, V, V) | 47.1 | 74.6 | 15.61 | 15.22 | 16.52 | 0.93 | |
| 69.9 | 13.55 | 12.63 | 16.92 | 0.87 | |||
| (III, III, III) | 67.1 | 75.5 | 7.87 | 7.58 | 9.09 | 0.90 | |
| 72.3 | 5.98 | 5.00 | 9.76 | 0.78 | |||
| (III, III, IV) | 67.5 | 76.6 | 7.34 | 7.23 | 8.09 | 0.92 | |
| 73.7 | 5.66 | 5.06 | 8.49 | 0.83 | |||
| (III, III, V) | 53.6 | 75.4 | 12.22 | 12.46 | 13.07 | 0.93 | |
| 70.8 | 10.28 | 9.80 | 13.78 | 0.88 | |||
| (III, IV, IV) | 69.2 | 78.5 | 7.21 | 6.69 | 7.35 | 0.95 | |
| 75.9 | 5.36 | 4.88 | 6.77 | 0.91 | |||
| (III, IV, V) | 55.0 | 77.3 | 12.32 | 12.48 | 12.61 | 0.94 | |
| 72.9 | 10.33 | 9.93 | 12.54 | 0.90 | |||
| (III, V, V) | 45.2 | 74.4 | 16.31 | 16.49 | 17.22 | 0.93 | |
| 68.4 | 13.54 | 12.93 | 17.79 | 0.86 | |||
| (IV, IV, IV) | 74.2 | 81.6 | 4.88 | 5.26 | 5.13 | 0.91 | |
| 79.6 | 3.94 | 4.08 | 3.96 | 0.95 | |||
| (IV, IV, V) | 59.1 | 79.8 | 11.61 | 11.50 | 11.61 | 0.95 | |
| 76.0 | 9.88 | 9.44 | 10.66 | 0.93 | |||
| (IV, V, V) | 48.9 | 77.4 | 16.15 | 16.07 | 16.35 | 0.94 | |
| 71.9 | 13.39 | 12.77 | 15.67 | 0.90 | |||
| (V, V, V) | 41.7 | 77.7 | 20.65 | 21.91 | 20.77 | 0.94 | |
| 68.6 | 16.08 | 15.63 | 19.71 | 0.87 |
For any fixed combination of communities, we generated 20 replicated incidence samples from each community according to a specified type of detection probabilities. Then for each generated dataset, the observed number of shared species was recorded; the original lower bound (or ) and the bias-corrected version (or ) as well as their SE estimates and associated 95% confidence intervals were obtained. The resulting averages in Tables 3 and 4 were based on 2000 simulation trials. The percentage of 2000 simulated data sets in which the 95% confidence intervals covered the true parameter was recorded and shown in the last column in each table.
From the two tables, the traditional approach of using the observed number of shared species as an estimator of shared species richness is clearly not appropriate. The observed number of shared species exhibits severely negative bias in all cases. When at least one set of detection probabilities is Type V (low average probability and high heterogeneity), the bias is substantial.
The performance of the lower bounds as estimators of shared species richness improves when more shared information are available. The magnitudes of bias, sample SE, and sample RMSE decrease as more shared species are observed. The bias-corrected bound is always lower than the original bound, but these two bounds are generally comparable with respect to RMSE. In terms of bias, the bias-corrected bound is useful when all communities are homogeneous as in the case (I versus I) in Table 3 or when there are at least two communities are homogeneous as in the three cases (I, I, I), (I, I, IV), and (I, I, V) in Table 4. This is expected because the bias-corrected form is derived under a homogeneous condition. Thus, unless in the special case that most communities are homogeneous, we suggest using the original lower bound. Since the CV of species detection probabilities measures the degree of heterogeneity, a CV estimator can be used to quantify the degree of heterogeneity present in data; see Chao et al. (2000).
When a sufficient amount of shared information is available (say, at least 70% of the shared species are observed), the lower bound in most cases is close to the true parameter. Thus it can be used as an estimator of shared species richness. When there are not sufficient shared data, our approach only provides a reliable lower bound. The magnitude of downwards bias mainly depends on the average and CV of the detection probabilities as well as the number of replicated samples. Further work is needed to determine more sophisticated guidelines about how large the samples should be to provide sufficient shared information.
Simulations also show that the estimated standard errors using the asymptotic method, although biased slightly downwards, are generally satisfactory when compared with the sample standard errors. The confidence interval based on the estimated SE for the original estimator performs reasonably well as most coverage probabilities are close to the anticipated nominal confidence coefficient of 95%.
7. CONCLUDING REMARKS AND DISCUSSION
Using the Cauchy–Schwarz inequality for the expected frequency counts based on abundance or replicated incidence data, we have developed a simple and useful lower bound for the number of species shared by multiple communities. The proposed lower bounds for abundance and replicated incidence data are natural extensions of the previous estimators used for a single community. Simulation results have shown that the performance of the lower bounds under several types of abundance distributions is generally satisfactory. The estimators discussed in this article will be featured in Program SPADE (Species Prediction And Diversity Estimation) following publication of this article (Chao and Shen 2003).
For estimating species richness in one community, we have discussed in Section 2 that our lower bound for the undetected species is in terms of the number of singletons and doubletons (for abundance data) or of uniques and duplicates (for replicated incidence data). Similar advantage holds for the case of two communities. For example, Equation (3.6) implies that the estimated number of undetected shared species for abundance data requires only information of the frequencies f1+, f+1, f2+, f+2, f 11, and f22. As a result, having the exact species frequency is not necessary for species that have at least three individuals in any of the two communities. Parallel conclusions are also valid for replicated incidence data and for more than two communities.
One critical assumption about our sampling model for abundance data is that we assume that individuals are randomly selected with replacement from each of the target community. Under this assumption, the species frequencies follow a multinomial distribution. However, in the case of sampling without replacement, the corresponding distribution becomes a generalized hyper-geometric distribution, which is less mathematically tractable. Besides, sampling fraction (i.e., the ratio of sample size and total population size) should be considered in the model framework. Research on the sampling without replacement is still undergoing. Also, for multiple incidence data, one restrictive assumption is that the species detection probability, although it is allowed to vary among species, is kept as a constant across all samples. This assumption may not be satisfied if samples are taken from areas where species occurrences are spatially aggregated.
In our proposed lower bounds, we did not consider relevant covariate information such as distance between communities and habitat types. Hillebrand et al. (2001) and Green et al. (2004) used species overlap information to assess the similarity of microbes as a function of geographic distance. These authors discovered the distance-decay relationship for microbial assemblages. Thus, the communities that are similar (close geographically and similar habitat) would generally have more overlap than one farther apart. How to incorporate covariate information in the estimation of shared species richness merits more research.
Boulinier et al. (1998) pointed out a simple analogy between the species replicated incidence data in a community and capture-recapture studies of a closed population. Thus, the estimation of species richness in a community based on replicated incidence data is equivalent to the estimation of the size of a population based on capture-recapture data. The analogy can be extended to the general case of multiple communities. That is, the estimation of shared species richness based on multiple incidence data from each community is equivalent to the estimation of the size of a shared population based on capture-recapture data from each population. Consequently, the proposed methodology for replicated incidence data can be directly applied to estimate the size of a shared population. This application and relevant topics are currently under investigation; see Chao, Pan, and Chiang (2008).
ACKNOWLEDGMENTS
This work was supported by the Taiwan National Science Council (Project 96-2118-M007-001) to HYP and AC, and by the Austrian Science Foundation (FWF project P19699-B17) to WF. Part of the material is based on the Ph.D. work of the first author under the supervision of the second author. The authors thank the Editor, Associate Editor, and two reviewers for carefully reading the manuscript and providing very thoughtful comments and suggestions, which significantly improved the article.
REFERENCES
- Bohannan BJM, Hughes J. New Approaches to Analyzing Microbial Biodiversity Data. Current Opinion in Microbiology. 2003;6:182–187. doi: 10.1016/s1369-5274(03)00055-9. [DOI] [PubMed] [Google Scholar]
- Boulinier T, Nichols JD, Sauer JR, Hines JE, Pollock KH. Estimating Species Richness: The Importance of Heterogeneity in Species Detectability. Ecology. 1998;79:1018–1028. [Google Scholar]
- Bunge J, Fitzpatrick M. Estimating the Number of Species: A Review. Journal of the American Statistical Association. 1993;88:364–373. [Google Scholar]
- Chao A. Nonparametric Estimation of the Number of Classes in a Population. Scandinavian Journal of Statistics. 1984;11:265–270. [Google Scholar]
- Chao A. Estimating the Population Size for Capture-Recapture Data With Unequal Catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- Chao A. Estimating Population Size for Sparse Data in Capture-Recapture Experiments. Biometrics. 1989;45:427–438. [Google Scholar]
- Chao A. Species Estimation and Applications. In: Balakrishnan N, Read CB, Vidakovic B, editors. Encyclopedia of Statistical Sciences. 2nd ed. Vol. 12. Wiley; New York: 2005. pp. 7907–7916. [Google Scholar]
- Chao A, Shen TJ. Program SPADE (Species Prediction And Diversity Estimation) 2003. program and user’s guide at http://chao.stat.nthu.edu.tw.
- Chao A, Hwang W-H, Chen Y-C, Kuo C-Y. Estimating the Number of Shared Species in Two Communities. Statistica Sinica. 2000;10:227–246. [Google Scholar]
- Chao A, Li PC, Agatha S, Foissner W. A Statistical Approach to Estimate Soil Ciliate Diversity and Distribution Based on Data From Five Continents. Oikos. 2006;114:479–493. [Google Scholar]
- Chao A, Pan HY, Chiang SC. The Petersen–Lincoln Estimator and Its Extension to Estimate the Size of a Shared Population. Biometrical Journal. 2008;50:957–970. doi: 10.1002/bimj.200810482. [DOI] [PubMed] [Google Scholar]
- Chao A, Shen T-J, Hwang W-H. Application of Laplace’s Boundary-Mode Approximations to Estimate Species and Shared Species Richness. Australian and New Zealand Journal of Statistics. 2006;48:117–128. [Google Scholar]
- Colwell RK. EstimateS: Statistical Estimation of Species Richness and Shared Species From Samples. 2004. Version 7.5, user’s guide and application published at http://viceroy.eeb.uconn.edu/estimates.
- Colwell RK, Coddington JA. Estimating Terrestrial Biodiversity Through Extrapolation. Philosophical Transactions of the Royal Society of London B—Biological Sciences. 1994;345:101–118. doi: 10.1098/rstb.1994.0091. [DOI] [PubMed] [Google Scholar]
- Fisher RA, Corbet AS, Williams CB. The Relation Between the Number of Species and the Number of Individuals in a Random Sample of an Animal Population. Journal of Animal Ecology. 1943;12:42–58. [Google Scholar]
- Foissner W. Protist Diversity: Estimates of the Near Imponderable. Protist. 1999;150:363–368. doi: 10.1016/S1434-4610(99)70037-4. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and Dispersal of Micro-Organisms: A Review Emphasizing Protists. Acta Protozoologica. 2006;45:111–136. [Google Scholar]
- Foissner W, Agatha S, Berger H. Soil Ciliates (Protozoa, Ciliophora) From Namibia (Southwest Africa), With Emphasis on Two Contrasting Environments, the Etosha Region and the Namib Desert. Denisia. 2002;5:1–1459. [Google Scholar]
- Green J, Holmes AJ, Westoby M, Oliver I, Brlscoe D, Dangerfield M, Gillings M, Beattle AJ. Spatial Scaling of Microbial Eukaryote Diversity. Nature. 2004;432:747–753. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Hillebrand H, Watermann F, Karez R, Berninger U-G. Differences in Species Richness Patterns Between Unicellular and Multicellular Organisms. Oecologia. 2001;126:114–124. doi: 10.1007/s004420000492. [DOI] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the Uncountable: Statistical Approaches to Estimating Microbial Diversity. Applied and Environmental Microbiology. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH. On the Relative Abundances of Bird Species. Proceedings of the National Academy of Sciences. 1957;43:193–295. doi: 10.1073/pnas.43.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran AE. Measuring Biological Diversity. Blackwell; Oxford, U.K.: 2004. [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a Computer Program for Defining Operational Taxonomic Units and Estimating Species Richness. Applied and Environmental Microbiology. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seber GAF. The Estimation of Animal Abundance. 2nd ed. Griffin; London: 1982. [Google Scholar]
- Stach JEM, Maldonado LA, Masson DG, Ward AC, Goodfellow WM, Bull AT. Statistical Approaches for Estimating Actinobacterial Diversity in Marine Sediments. Applied Environmental Microbiology. 2003;69:6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner W, Leitner W, Rosenzweig ML. WS2m: Software for Estimating Diversity. 1999. program and user’s guide at http://eebweb.arizona.edu/diversity.
- Walther BA, Morand S. Comparative Performance of Species Richness Estimation Methods. Parasitology. 1998;116:395–405. doi: 10.1017/s0031182097002230. [DOI] [PubMed] [Google Scholar]


