Abstract
RNA interference (RNAi) technology has recently emerged as an important biological strategy for gene silencing. Previously, the efficacies of RNAi in cultured nucleus pulposus cells in vitro have been reported. However, RNAi in the disc in vivo has never been reported. Therefore, the aims of the present study were to establish a method for RNAi in the disc in vivo and to evaluate the applicability of this technique for endogenous genes in the intervertebral discs using Fas Ligand (FasL) as a representative endogenous gene. To evaluate the efficacy of RNAi in vivo, two reporter luciferase plasmids (Firefly and Renilla) were used. These plasmids and unmodified short interference RNA (siRNA) duplex for targeting Firefly luciferase were co-transfected into coccygeal intervertebral disc of Sprague-Dawley rats in vivo using the ultrasound gene transfer technique. To evaluate the RNAi of the endogenous gene in vivo, siRNAs targeting rat FasL were transfected with the same technique. Non-specific siRNA was used as the negative control. The discs receiving no siRNAs were used as the control. The inhibitory effect of Firefly luciferase against Renilla luciferase was obtained using the results of dual-luciferase assay. Down-regulation of endogenous FasL was calculated by the data from real-time PCR. Our results showed that siRNA for Firefly luciferase can dramatically down-regulate the Firefly luciferase gene expression in vivo compared with Renilla luciferase. The inhibitory effects were maintained for at least 24 weeks and at 24 weeks post transfection, the inhibitory rate was 80% compared with the control group. Furthermore, the siRNA co-transfection group inhibited endogenous FasL expression by 53% compared with the control group. The present study demonstrates long-term down-regulation mediated by unmodified siRNA is possible not only for the exogenous reporter gene, but also for endogenous FasL expression in rat discs in vivo. This application of RNAi might be promising as a local therapy for disc degeneration and associated disorders by down-regulating some of the genes that are harmful for the normal physiology of the disc and may cause disc degeneration.
Keywords: RNA interference (RNAi), Unmodified siRNA, Intervertebral disc, Animal model, Ultrasound gene therapy
Introduction
Degeneration of the intervertebral disc is an underlying etiology in many spinal disorders and is known to be a significant source of patient pain and morbidity [3, 8, 30]. Effective treatment or prevention of degenerative disc disease is not easily achieved because the molecular events that drive these processes are poorly understood. Recent studies have shown potential applications of the growth factor injection [22] and cell-mediated therapies [31] for regenerating degenerated discs by stimulating matrix synthesis. However, improvements are being made not only in the areas of protein delivery or cell therapy, but also in the regulation of gene therapy, which it has come to be realized, is a potentially powerful therapeutic tool for stimulating matrix synthesis [26, 27].
In gene therapy, vehicles called vectors are required to deliver exogenous genes of interest to the target cells and both viral and non-viral vectors are being developed to do this. Because of their natural ability to transfect human cells and influence transcription and translation, viruses have been the most commonly used gene therapy vectors to date. Use of less pathogenic viral constructs such as the adeno-associated viral vector (AAV) [4, 19] are among recent improvements aimed at increasing the safety of gene therapy. However, recent studies demonstrate strong efforts to develop efficient non-viral gene delivery systems that avoid the risk of transmission of infectious agents and limit the host immune response against viral products [13]. Of all methods reported, microbubble-enhanced ultrasound gene therapy is currently an available non-virus mediated, feasible gene transfection method to the disc cells in vivo [28].
In contrast, in the field of intradiscal gene therapy, new approaches such as decreasing catabolic factors of the matrix have been recently reported due to the limited physiology of the disc [35]. It is well known that the lumbar intervertebral discs are the largest avascular organ in the body and due to this unique property the inside of the disc allows little nutrition and oxygen [10], suggesting energy consuming approaches such as stimulating matrix synthesis are theoretically less likely to result in successful regeneration of the degenerated disc. RNA interference (RNAi) is one of the candidates for this new approach as it provides a powerful means of sequence-specific gene silencing [11]. RNAi was first reported in 1998 by Fire et al. who demonstrated that double-stranded RNA induced sequence-specific silencing of gene expression in nematode cells [6]. Since then, molecular mechanisms of RNAi have been intensively investigated in a variety of organisms, indicating that RNAi provides a quite useful means of functional analyses of genes as well as epigenetic engineering of cells of various species [11, 36]. Elbashir et al. [5] demonstrated that RNAi can be achieved in the mammalian cells using oligoribonucleotide duplex 21 or 22 bases in length (small interfering RNA; siRNA).
We previously reported the efficacies of RNAi in cultured nucleus pulposus cells in vitro, using unmodified siRNA duplexes targeting exogenous reporter gene for the first time [15]. However, RNAi in the disc in vivo has never been reported. Therefore, the objectives of this study were: (1) to investigate the efficacies and the longevity of RNAi using exogenous reporter gene by means of microbubble-enhanced ultrasound gene transfer technique for intervertebral disc cells in vivo and (2) to estimate the applicability of this method for endogenous genes of the intervertebral disc in vivo using Fas Ligand (FasL) as a representative endogenous gene. FasL was chosen as it has previously been reported that nucleus pulposus cells constitutively express FasL and it is recognized as one of the marker gene of nucleus pulposus cells [14, 33].
Materials and methods (Table 1)
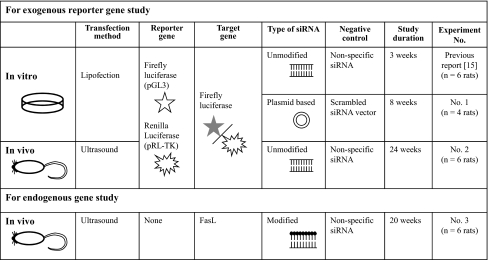
Table 1.
Overview of each experiment
Preparation of reporter gene and siRNAs
Reporter gene expression plasmids
For the efficacy and longevity analyses of exogenous gene silencing, two luciferase plasmids, pGL3 Control Vector (encoding Firefly luciferase gene) and pRL-TK Vector (encoding Renilla luciferase gene) (Promega, Madison, USA), were used for Experiments no. 1 and 2. Renilla luciferase gene expression was used as an internal control.
siRNA and siRNA expression vectors targeting for exogenous reporter gene
We used unmodified siRNA duplex for targeting Firefly luciferase (Catalog no. D-001400-01-20 Dharmacon/20 μM) in the Experiment no. 2, and non-specific siRNA (Dharmacon/20 μM) in the negative control group for the Experiments no. 2 and 3.
The siRNA expression vectors were used for Experiments no. 1. Oligo 1 was targeting for Firefly luciferase (Table 2) and ligated into siRNA expression vector (p-silencer; Ambion, Austin, USA). As a negative control against siRNA expression vector, we used scrambled siRNA vector, which oligo 2 was ligated into siRNA expression vector (p-silenecer; Ambion) (Table 2).
Table 2.
Sequences of siRNAs used in Experiments no. 1 and 3
| Oligo no. 1 | |
| Sense | 5′-GATCCCGCTTACGCTGAGTACTTCGATTCAAGAGATCGAAGTACTCAGCGTAAGTTTTTTGGAAA-3′ |
| Anti-sense | 5′-AGCTTTTCCAAAAAACTTACGCTGAGTACTTCGATCTCTTGAATCGAAGTACTCAGCGTAAGCGG-3′ |
| Oligo no. 2 (scrambled) | |
| Sense | 5′-GATCCCGTAATATTCCGTGGGCCTACTTCAAGAGAGTAGGCCCACGGAATATTATTTTTTGGAAA-3′ |
| Anti-sense | 5′-AGCTTTTCCAAAAAATAATATTCCGTGGGCCTACTCTCTTGAAGTAGGCCCACGGAATATTACGG-3′ |
| FasL siRNA no. 1 | |
| Sense | 5′-GUAUGAGAGUAGAGCGAUUUU-3′ |
| Anti-sense | 5′-PAAUCGCUCUACUCUCAUACUU-3′ |
| FasL siRNA no. 2 | |
| Sense | 5′-UCACAUGAGUACAACGCGUUU-3′ |
| Anti-sense | 5′-PACGCGUUGUACUCAUGUGAUU-3′ |
| FasL siRNA no. 3 | |
| Sense | 5′-GGUUAGGAAUGUAUCAACUUU-3′ |
| Anti-sense | 5′-PAGUUGAUACAUUCCUAACCUU-3′ |
| FasL siRNA no. 4 | |
| Sense | 5′-GGGCAGUAUUUAAUCUUACUU-3′ |
| Anti-sense | 5′-PGUAAGAUUAAAUACUGCCCUU-3′ |
siRNAs targeting for endogenous FasL
To establish the down-regulation of the endogenous gene, a mixture of four siRNAs targeting rat FasL (20 μM) and one non-specific siRNA (used for the negative control/20 μM) with the following sense and antisense sequences were used (Table 2). All siRNAs to target rat FasL were designed by Dharmacon, Inc. (Chicago, USA). Dharmacon SMART pool® uses an algorithm to combine four SMART selected siRNA duplexes in a single pool (product number QS-004997-00-70).
In vitro study procedures
Cell isolation and culture
Male Sprague-Dawley rats (SD rats), approximately 20 weeks of age and 600 g in weight (CLEA Japan, Tokyo, Japan), were used in the current in vitro study. The rats were euthanized by an overdose of Nembutal (100 mg/kg). Caudal spines were obtained in aseptic conditions within 1 h of death. The surface of the intervertebral disc was exposed, a small incision was made, and the prolapsed gelatinous nucleus pulposus tissue was harvested. Those tissues were placed in a complete tissue culture medium consisting of Dulbecco’s modified Eagle’s medium (DMEM/Invitrogen, Carlsbad, USA), supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin. The pH of the medium was maintained between 7.0 and 7.2. Then the tissues were placed in a solution of Type I collagenase (0.25 mg/mL in PBS) for approximately 1 h. The cells were released from the matrix by a centrifuge (1,200 rpm × 10 min) and placed into 35-mm tissue culture dishes in six-well plates (2 ml medium/well) maintained in 5% CO2 and 95% air.
Transfection by lipofection
The cells were plated in six-well plates (2 mL medium/well) to reach about 80% confluence at transfection. The cells were co-transfected with two different (Firefly and Renilla) luciferase plasmids and siRNA expression vector for targeting Firefly luciferase (Dharmacon, Chicago, USA) as the experimental group, and scrambled siRNA vector (Dharmacon) as the negative control group (Experiment no. 1/Tables 1, 2) by a reported transfection protocol (Lipofectamine 2000; Invitrogen) with modification. Co-transfection by two different luciferase plasmids (Firefly and Renilla) in the absence of siRNA expression vector was used as a control group for normalization of the obtained data. Per well, 2.0 μg of pGL3 and 0.4 μg of pRL-TK plasmid (Promega, Madison, USA), 1 μg of siRNA expression vector or scrambled vector were used for Experiment no. 1 (Table 1). The transfection medium was changed to culture medium (2.5 mL) after 3 h. Each luciferase expression was subsequently monitored with dual-luciferase reporter assay system (Promega).
In vivo study procedures
All animal procedures were performed under the guidance of the Animal Research Committee at the author’s institution.
Animals, surgical procedures, and in vivo gene transfer
A 12-week-old male SD rats were used in this in vivo study (weight: 409–498 g/CLEA). Rats were anesthetized by an intraperitoneal injection of Nembutal (50 mg/kg) before sedation by inhalation of diethyl ether (Wako Chemicals, Osaka, Japan). Under general anesthesia, the coccygeal intervertebral discs were surgically exposed. For the exogenous reporter gene study, a 2 μl solution containing luciferase plasmids (pGL3/2.5 μg and pRL-TK/0.5 μg) and siRNA targeting Firefly luciferase (20 pmol) mixed with 20% ultrasonography contrast agent (microbubbles; Optison/Nepagene, Chiba, Japan) was used (Experiment no. 2). A 2 μl solution containing the luciferase plasmids group in the absence of siRNAs was used as the control group to normalize the obtained data. For the endogenous FasL gene study, a 2 μl solution containing siRNAs targeting rat FasL (20 pmol) mixed with 20% ultrasonography contrast agent was used (Experiment no. 3). A 2 μl solution in the absence of siRNAs was used as the control group.
Each solution was injected into three coccygeal intervertebral discs of rats (usually C7-8, 8–9, 9–10) to triplicate the results using a microsyringe with 31G needle. Different rats were used for each solution (n = 6 animals/each group). Usually, once the needle penetrated the annulus fibrosus, resistance to insertion suddenly decreased and the depth of insertion into the disc was approximately 2 mm. After injection, a gel for ultrasound transmission (Rich-mar, Inola, USA) was applied on the surface of the target discs, and at the optimized intensity (2.0 W/cm2) and for optimized durations (60 s), therapeutic ultrasound was irradiated (Nepagene) on the surface of the injected discs via ultrasound gel. These ultrasound gels were removed after irradiation and all wounds were routinely closed [28]. Antibiotics were supplied to the animals immediately after the operation. Animals were sacrificed at various intervals after operation.
Evaluation of gene silencing
Exogenous reporter gene expression
Harvested cultured cells (in vitro study) or tissues of nucleus pulposus (in vivo study) were lysed using a passive lysis buffer (50 μl per disc) according to modified instructions in the dual-luciferase reporter assay system (Promega). Each luciferase activity in the samples was measured by Luminometer (ATP-3010; Advantec, Japan) with a delay time of 2 s and an integration of 10 s. The averages from triplicates were used for evaluation. To evaluate the inhibitory effects of Firefly luciferase gene expression generated by siRNA, the ratio of Firefly luciferase to Renilla luciferase (internal control) activity was calculated to compensate for the potential differences in transfection efficiencies between cells or animals. The ratios were then normalized to those observed in cells or animals receiving no siRNA (control).
Endogenous gene expression
Rats were sacrificed 4 and 20 weeks after gene transduction. The nucleus pulposus tissues from three discs in each rat were harvested and all specimens were immediately frozen in liquid nitrogen. The frozen tissues were homogenized, and total RNA was isolated using an RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Reverse transcription to cDNA was performed using 1 μg total RNA in the presence of murine leukemia virus RT and oligo d(T) primer (Perkin Elmer, Boston, USA). The mixture was incubated at 75°C for 5 min and 37°C for 120 min.
The relative levels of mRNA encoding FasL in disc cells were compared by Perfect Real-Time SYBR® Premix Ex Taq™ (TAKARA BIO INC., Tokyo, Japan) using an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA, USA). Predesigned primers for rat FasL and rat GAPDH (50 pmol/μl) as control were purchased from TAKARA BIO INC. (Tokyo, Japan) (Fas ligand sense: 5′-CACCAACCACAGCCTTAGAGTATCA-3′, anti-sense: 5′-ACTCCAGAGATCAAAGCAGTTCCA-3′, GAPDH sense: 5′-GACAACTTTGGCATCGTGGA-3′, anti-sense: 5′-ATGCAGGGATGATGTTCTGG-3′). This procedure used 50 PCR cycles at 95°C for 15 s, at 60°C for 34 s. Reaction volume was 20 μl per tube. The normalized amount of each sample was then determined by dividing the copy numbers of each mRNA by those of GAPDH mRNA. The threshold was set at the mid-point of the log-liner phase amplification plot and defined as Ct, which represents the number of cycles at this point. For each experimental sample, the relative quantity of mRNA was determined from the standard curve, which was obtained from healthy intervertebral disc cells. Each sample was normalized by GAPDH and the relative quantity of FasL mRNA expression was determined.
Statistical analysis
All values were expressed as mean ± SD. Statistical analysis using the Mann-Whitney’s U-test was conducted between the negative control group and the experimental group. The P value was set at 0.01.
Results
DNA vector-based siRNA-mediated RNAi in vitro (Experiment no. 1)
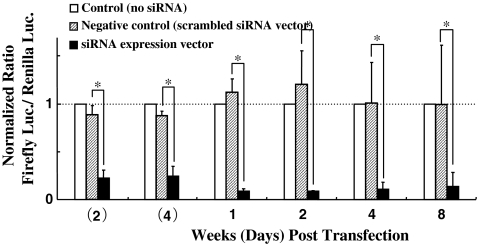
The siRNA expression vector groups showed significant inhibition of exogenous reporter gene expression (Firefly luciferase) at all time points, and the inhibitory effect of DNA vector was sustained for 8 weeks (56 days). The negative control groups did not inhibit Firefly luciferase activity (Fig. 1).
Fig. 1.
The longevity of RNAi effect mediated by DNA vector-based siRNA in nucleus pulposus cells in vitro. In all cases, the ratio of Firefly luciferase to Renilla luciferase activity was calculated to compensate for the potential differences in transfection efficiencies. The ratios were then normalized to those observed in cells receiving no siRNA expression vectors (control). The scrambled siRNA expression vector was used as the negative control. The plotted data are averages from triplicates ±SD. n = 4 animals. *P < 0.01
Unmodified siRNA-mediated RNAi in vivo (Experiment no. 2)
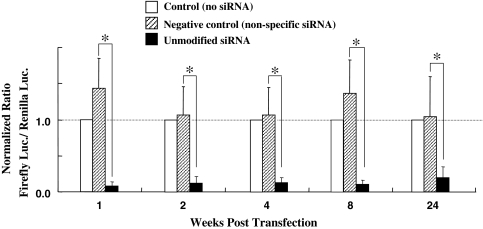
Our results showed that siRNA for Firefly luciferase can dramatically down-regulate the Firefly luciferase gene expression compared with Renilla luciferase. The average inhibitory effect of siRNA group was about 90% compared with the control group (Fig. 2). In contrast, both luciferase (Firefly and Renilla) gene expressions in the negative control group remained unaffected. An inhibitory effect of 80% was observed after 24 weeks (168 days) compared with the control group.
Fig. 2.
The longevity of RNAi effect mediated by unmodified siRNA in nucleus pulposus in vivo. The inhibition effect was sustained for at least 24 weeks. The negative control groups did not inhibit Firefly luciferase activity. In all cases, the ratio of Firefly luciferase to Renilla luciferase activity was calculated to compensate for the potential differences in transfection efficiencies between animals. The ratios were then normalized to those observed in animals receiving no siRNA (control). The plotted data are averages from triplicates ±SD. n = 6 animals/each group.*P < 0.01
Efficacies of RNAi effect for endogenous FasL gene (Experiment no. 3)
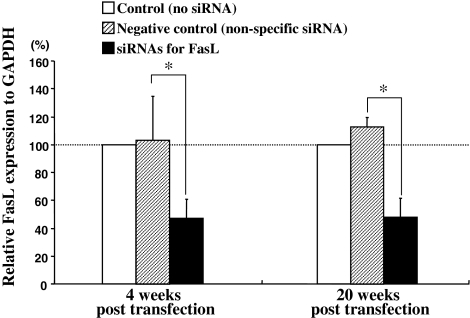
The siRNA co-transfection targeting rat FasL gene group was inhibited by 53% in endogenous FasL gene expression at mRNA level compared with the control group at both 4- and 20-week post transfection (Fig. 4). The negative control groups did not show any inhibition effect.
Fig. 4.
Efficacies of down-regulation for endogenous FasL gene in vivo. Transfection of the siRNA targeting rat FasL gene was inhibited by 53% in endogenous FasL gene expression compared with the control group (receiving no siRNA) at both 4 and 20 weeks. The plotted data are averages from triplicates ±SD. n = 6 animals/each group. *P < 0.01
Discussion
Gene array studies have revealed that the efficacy of RNAi depends on the efficacy of transduction, and that the longevity depends on the stability of the transduced gene in the cell [1]. First, for the efficacy of transduction, many kinds of transduction methods have been developed in vivo models. Systemic transduction, for example, hydrodynamic tail vein injection (or volume overload) of siRNAs is an alternative and also effective method for silencing (endogenous) genes, especially in the liver [12]. However, the disadvantage of this method is that the liver is specifically targeted and effects on gene expression in other organs are difficult to achieve. According to previous reports, systemic transduction shows major disadvantages such as needing very high amounts of siRNAs on which the unwanted interferon response seems to be dependent [12]. Other approaches for the delivery of siRNAs in vivo have also been described. Local delivery of siRNAs into the eye has been used to show that siRNAs targeting the vascular endothelial growth factor (VEGF) pathway could be therapeutically beneficial in neovascularization [16]. Electroporation has been used to deliver siRNAs into the kidney [32], brain [2], eyes [23], muscles [9, 18], and skin [37] of rodents. Intradermal administration of nucleic acids via gene gun has been used to deliver siRNA in vivo to enhance cancer vaccine potency [17]. Ultrasound could be also be potentially used to deliver siRNA [20, 34], although there have been no reports of this technique for delivery of siRNA and, as far as we know, our current paper is the first report of successful siRNA transduction in vivo using an ultrasound method.
Second, the instability of siRNA molecules in vivo remains an important issue and a major obstacle [1]. Usually, siRNA effects are transient, remaining only a few weeks at best without any vector-based RNAi [29]. Obviously, the stabilization protection of siRNAs to maintain a prolonged effect of RNAi is required. To overcome this problem, many other systems have been developed using chemically modified siRNA such as short hairpin RNA (shRNA) molecules, additional molecules for resistance to nuclease degradation [7], and various vectors such as plasmid DNA [1]. In principle, these DNA-based systems offer the advantage of constitutive expression of siRNAs with a potentially higher level and unlimited duration of gene silencing. We previously reported that the inhibitory effect of unmodified siRNA transfection disappeared completely by 3 weeks in nucleus pulposus cells in vitro [15]. In contrast, in this research, we discovered that the DNA vector-based RNAi in cultured nucleus pulposus cells showed significant inhibition at all time points, and the inhibitory effect of the DNA vector was sustained up to 8 weeks (Figs. 1, 3). These results indicate the usefulness of DNA vector-based RNAi in the intervertebral disc in terms of longevity of RNAi effects.
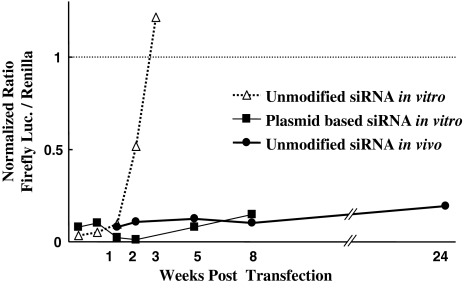
Fig. 3.
Comparison of longevity of RNAi effect among each method. According to our previous publication [15], the inhibitory effect of unmodified siRNA in vitro disappears completely by 21 days (3 weeks). In contrast, the RNAi effect of the vector-based siRNA groups in vitro was sustained up to 56 days (8 weeks). On the other hand, the inhibitory effect of unmodified siRNA in vivo continued for at least 168 days (24 weeks). *P < 0.01
On the other hand, unmodified siRNA in our in vivo model effectively inhibited exogenous reporter gene expression up to 24 weeks (Figs. 2, 3). Furthermore, although only mRNA level evaluation of endogenous FasL gene expression was conducted in this current study and protein level evaluation should be made in future studies, sustained down-regulation of endogenous FasL gene was possible up to 20 weeks (140 days) using the same technique (Fig. 4). The persistence of siRNA activity in mammalian cells varies with the proliferative status of the cells, such that siRNA activity lasts for 3–7 days in proliferating cells, but can persist for 3 weeks or more in terminally differentiated cells such as neurons [29]. Therefore, our unusual result showing the long-lasting duration of RNAi mediated by unmodified siRNA in the disc indicates that disc cells are very stable in terms of cell proliferation, especially in the in vivo situation. As the disc is a unique structure in the body known to be the largest avascular organ and the nucleus pulposus is well encapsulated by endplates and annulus fibrosus, it is potentially very suitable for local siRNA delivery. Additionally, due to being an avascular organ, oxygen or nutrition supply depend on the passive diffusion mainly via endplates, leading to little nutrition and oxygen tension especially in the center part of the disc [10]. Owing to the anaerobic metabolism, lactates are produced, resulting in low pH inside the disc [24]. Furthermore, the inside of the disc is usually maintained at a dynamic and changeable high-positive pressure [25]. These factors make the interior of the intervertebral disc a harsh biological environment and accordingly, the nucleus pulposus cells must be highly differentiated to survive in this special environment. The different duration of RNAi effects mediated by unmodified siRNA between nucleus pulposus cells in vitro (3 weeks) and in vivo (at least 24 weeks) is probably due to the different nucleus pulposus cell status. It is reasonable that the nucleus pulposus cells in culture, may be potentially undifferentiated in quite different environments from the in vivo situation, leading to a higher proliferation activity, resulting in a shorter RNAi effect in vitro compared with in vivo.
Usually, transfection efficiency used by direct transduction of 21-nt dsRNA (unmodified siRNA) is much higher than plasmid transduction [21]. Moreover, despite the advent of modification strategies to improve cellular uptake and biostability in vivo, there is still a necessity to test each DNA vector-based siRNA independently, which is time consuming and costly, because the inhibitory effects of the DNA vector are still uncertain compared with unmodified siRNA. Hence, the direct use of unmodified siRNA molecules may offer advantages and may be preferable to using DNA-based strategies or chemically modified siRNAs. Accordingly, our results showing a long-lasting period of RNAi mediated by unmodified siRNA in the disc are probably sufficient for studying the effect of down-regulational genes in several disease models as well as in therapeutic use.
Conclusions
In conclusion, the current study demonstrated the effectiveness of DNA vector-based RNAi technique in vitro in terms of prolonged RNAi effect. Additionally, our study revealed that simple unmodified siRNA-mediated RNAi effect in intervertebral discs in vivo resulted in a long-lasting period up to 24 weeks (168 days). Furthermore, the present study demonstrates long-term down-regulation is possible not only for exogenous reporter gene, but also for endogenous FasL expression (mRNA level) in rat discs in vivo. This unusual long period of RNAi in intervertebral disc in vivo may be due to stability of disc cells which should be highly differentiated. Because of the prolonged silencing effect after a single intradiscal transduction of unmodified siRNA, this gene delivery method is suitable for investigating the functions of endogenous genes in the disc. Moreover, this application of RNAi might be promising as a local therapy for disc degeneration and associated disorders by down-regulating some harmful genes for the normal physiology of the disc, which may cause disc degeneration.
Acknowledgments
The authors thank Ms.Tubby for their help in preparing the manuscript. We also thank Ms. Yasuda and Ms. Tanaka for their sophisticated technical assistance. This work was supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Akaneya Y, Jiang B, Tsumoto T. RNAi-induced gene silencing by local electroporation in targeting brain region. J Neurophysiol. 2005;93:594–602. doi: 10.1152/jn.00161.2004. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GB. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 4.Carter PJ, Samulski RJ. Adeno-associated viral vectors as gene delivery vehicles. Int J Mol Med. 2000;6:17–27. doi: 10.3892/ijmm.6.1.17. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Fougerolles A, Manoharan M, Meyers R, Vornlocher HP. RNA interference in vivo: toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278–296. doi: 10.1016/S0076-6879(04)92016-2. [DOI] [PubMed] [Google Scholar]
- 8.Freemont AJ, Peacock TE, Goupille P, Hoyland JA, O’Brien J, Jayson MI. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet. 1997;350:178–181. doi: 10.1016/S0140-6736(97)02135-1. [DOI] [PubMed] [Google Scholar]
- 9.Golzio M, Mazzolini L, Moller P, Rols MP, Teissié J. Inhibition of gene expression in mice muscle by in vivo electrically mediated siRNA delivery. Gene Ther. 2005;12:246–251. doi: 10.1038/sj.gt.3302405. [DOI] [PubMed] [Google Scholar]
- 10.Grunhagen T, Wilde G, Soukane DM, Shirazi-Adl SA, Urban JP. Nutrient supply and intervertebral disc metabolism. J Bone Joint Surg. 2006;88-A:30–35. doi: 10.2106/JBJS.E.01290. [DOI] [PubMed] [Google Scholar]
- 11.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 12.Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol. 2004;22:1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- 13.Herweijer H, Wolff JA. Progress and prospects: naked DNA gene transfer and therapy. Gene Ther. 2003;10:453–458. doi: 10.1038/sj.gt.3301983. [DOI] [PubMed] [Google Scholar]
- 14.Inui Y, Nishida K, Doita M, Takada T, Miyamoto H, Yoshiya S, et al. Fas-ligand expression on nucleus pulposus begins in developing embryo. Spine. 2004;29:2365–2369. doi: 10.1097/01.brs.0000143172.07771.fc. [DOI] [PubMed] [Google Scholar]
- 15.Kakutani K, Nishida K, Uno K, Takada T, Shimomura T, Maeno K, et al. Prolonged down regulation of specific gene expression in nucleus pulposus cell mediated by RNA interference in vitro. J Orthop Res. 2006;24:1271–1278. doi: 10.1002/jor.20171. [DOI] [PubMed] [Google Scholar]
- 16.Kim B, Tang Q, Biswas PS, Xu J, Schiffelers RM, Xie FY, et al. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes: therapeutic strategy for herpetic stromal keratitis. Am J Pathol. 2004;165:2177–2185. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TW, Lee JH, He L, Boyd DA, Hardwick JM, Hung CF, et al. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer Res. 2005;65:309–316. [PubMed] [Google Scholar]
- 18.Kishida T, Asada H, Gojo S, Ohashi S, Shin-Ya M, Yasutomi K, et al. Sequence-specific gene silencing in murine muscle induced by electroporation-mediated transfer of short interfering RNA. J Gene Med. 2004;6:105–110. doi: 10.1002/jgm.456. [DOI] [PubMed] [Google Scholar]
- 19.Lattermann C, Oxner WM, Xiao X, Li J, Gilbertson LG, Robbins PD, et al. The adeno associated viral vector as a strategy for intradiscal gene transfer in immune competent and pre-exposed rabbits. Spine. 2005;30:497–504. doi: 10.1097/01.brs.0000154764.62072.44. [DOI] [PubMed] [Google Scholar]
- 20.Lawrie A, Brisken AF, Francis SE, Cumberland DC, Crossman DC, Newman CM. Microbubble-enhanced ultrasound for vascular gene delivery. Gene Ther. 2000;7:2023–2027. doi: 10.1038/sj.gt.3301339. [DOI] [PubMed] [Google Scholar]
- 21.Lentz G. The RNA interference revolution. Braz J Med Biol Res. 2005;38:1749–1757. doi: 10.1590/s0100-879x2005001200003. [DOI] [PubMed] [Google Scholar]
- 22.Masuda K, An HS. Prevention of disc degeneration with growth factors. Eur Spine Suppl. 2006;3:S422–S432. doi: 10.1007/s00586-006-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachemson A. Intradiscal measurements of pH in patients with lumbar rhizopathies. Acta Orthop Scand. 1969;40:23–42. doi: 10.3109/17453676908989482. [DOI] [PubMed] [Google Scholar]
- 25.Nachemson A, Elfstrom G. Intravital dynamic pressure measurements in lumbar discs. A study of common movements, maneuvers and exercises. Scand J Rehabil Med. 1970;Suppl 1:1–40. [PubMed] [Google Scholar]
- 26.Nishida K, Kang JD, Suh JK, Robbins PD, Evans CH, Gilbertson LG. Adenovirus-mediated gene transfer to nucleus pulposus cells. Implications for the treatment of intervertebral disc degeneration. Spine. 1998;23:2437–2442. doi: 10.1097/00007632-199811150-00016. [DOI] [PubMed] [Google Scholar]
- 27.Nishida K, Kang JD, Gilbertson LG, Moon SH, Suh JK, Vogt MT, et al. Modulation of the biologic activity of the rabbit intervertebral disc by gene therapy: an in vivo study of adenovirus-mediated transfer of the human transforming growth factor beta 1 encoding gene. Spine. 1999;24:2419–2425. doi: 10.1097/00007632-199912010-00002. [DOI] [PubMed] [Google Scholar]
- 28.Nishida K, Doita M, Takada T, Kakutani K, Miyamoto H, Shimomura T, et al. Sustained transgene expression in intervertebral disc cells in vivo mediated by microbubble-enhanced ultrasound gene therapy. Spine. 2006;31:1415–1419. doi: 10.1097/01.brs.0000219945.70675.dd. [DOI] [PubMed] [Google Scholar]
- 29.Omi K, Tokunaga K, Hohjoh H. Long-lasting RNAi activity in mammalian neurons. FEBS Lett. 2004;558:89–95. doi: 10.1016/S0014-5793(04)00017-1. [DOI] [PubMed] [Google Scholar]
- 30.Peng B, Hao J, Hou S, Wu W, Jiang D, Fu X, et al. Possible pathogenesis of painful intervertebral disc degeneration. Spine. 2006;31:560–566. doi: 10.1097/01.brs.0000201324.45537.46. [DOI] [PubMed] [Google Scholar]
- 31.Sakai D, Mochida J, Iwashima T, Watanabe T, Nakai T, Ando K, et al. Differentiation of mesenchymal stem cells transplanted to a rabbit degenerative disc model: potential and limitations for stem cell therapy in disc regeneration. Spine. 2005;30:2379–2387. doi: 10.1097/01.brs.0000184365.28481.e3. [DOI] [PubMed] [Google Scholar]
- 32.Takabatake Y, Isaka Y, Mizui M, Kawachi H, Shimizu F, Ito T, et al. Exploring RNA interference as a therapeutic strategy for renal disease. Gene Ther. 2005;12:965–973. doi: 10.1038/sj.gt.3302480. [DOI] [PubMed] [Google Scholar]
- 33.Takada T, Nishida K, Doita M, Kurosaka M. Fas ligand exists on intervertebral disc cells: a potential molecular mechanism for immune privilege of the disc. Spine. 2002;27:1526–1530. doi: 10.1097/00007632-200207150-00009. [DOI] [PubMed] [Google Scholar]
- 34.Taniyama Y, Tachibana K, Hiraoka K, Aoki M, Yamamoto S, Matsumoto K, et al. Development of safe and efficient novel nonviral gene transfer using ultrasound: enhancement of transfection efficiency of naked plasmid DNA in skeletal muscle. Gene Ther. 2002;9:372–380. doi: 10.1038/sj.gt.3301678. [DOI] [PubMed] [Google Scholar]
- 35.Wallach CJ, Sobajima S, Watanabe Y, Kim JS, Georgescu HI, Robbins P, et al. Gene transfer of the catabolic inhibitor TIMP-1 increases measured proteoglycans in cells from degenerated human intervertebral discs. Spine. 2003;28:2331–2337. doi: 10.1097/01.BRS.0000085303.67942.94. [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Nolan E, Kreitschitz S, Rabussay DP. Enhanced delivery of naked DNA to the skin by non-invasive in vivo electroporation. Biochim Biophys Acta. 2002;1572:1–9. doi: 10.1016/s0304-4165(02)00270-2. [DOI] [PubMed] [Google Scholar]