Abstract
A pivotal gluconeogenic enzyme in Saccharomyces cerevisuae, fructose-1, 6-bisphosphatase (FBPase) was selectively turned over in vacuole via Vid (vacuole import and degradation) dependent pathway in response to the fresh glucose after chronic glucose starvation. TCO89, a novel and unique component of Tor Complex I (TORCI), was found to physically associate with FBPase and significantly affect FBPase degradation via Vid pathway. Further investigation indicated that Δtco89 mutant strongly impaired FBPase's importing into Vid vesicles and Vid24's association with Vid vesicles. Inactivation of TORCI by rapamycin treatment strongly blocked FBPase degradation. Other components of TORCI were also found to physically associate with FBPase. The P1S mutation of FBPase, reported to block its degradation, was observed to impair the association of FBPase with TORCI components. These results implicated an important regulatory role of TCO89 and TORCI in this pathway.
Keywords: FBPase, Vid, Tor Complex 1, TCO89, Protein degradation
Introduction
The fructose-1, 6-bisphosphatase (FBPase), a key enzyme of gluconeogenic pathway, is rapidly inactivated and selectively turned over when ample glucose is supplied after glucose starvation 1-3. The catabolite degradation of FBPase effectively prevents the energy futile cycle between glycolysis and gluconeogenesis 1- 4. Depending on the culture conditions, there are two pathways for FBPase degradation: proteasome-dependent degradation and vacuole import and degradation (Vid) dependent pathway 3- 6. It is reported that in one day starvation, FBPase was conjugated with multi-ubiquitin chain at Lys 48 during its degradation; the 26S proteasome mutant cim3-1blocked FBPase degradation at non-permissive temperature 4-6. In two-day or longer starvation condition, FPBase is degraded in vacuole, the homologue of lysosome of mammals 1,5.
The FBPase degradation via Vid pathway is one of an autophagic process in which long-lived proteins or cell organelles are engulfed in the membrane organelles and then targeted to vacuole for degradation 2, 7. Multiple proteins are involved in Vid pathway of FBPase degradation: this pathway shares components with endocytotic pathway such as V-ATPase complex and epsilon cop coatomer complex1 (COPI) subunits 8-9. When mutated in Stv1 or Vph1, the subunit of V-ATPase complex, FBPase degradation was blocked 8. When mutated in Sec28, the subunit of COPI, FBPase's association with Vid vesicles and its degradation were impaired 9. This pathway also required a unique protein Vid24, the protein marker of Vid vesicle, which functions in targeting FBPase containing vesicles to the vacuole 10.
Target of rapamycin (Tor), a conserved phosphatidylinositol 3-related protein kinase, plays pivotal roles in cell growth and cell metabolism 11-13. As a nutrition sensor, Tor proteins are highly conserved from yeast to mammals. In yeast, two Tor proteins form two functional distinct protein complexes: Tor Complex I (TORC1) and Tor Complex II (TORCII). TORCI contains unique components (Tor1 and TCO89) and shares components (Tor2, Lst8 and Kog1) with TORCII 14-15. Each component of TORC1 plays distinct but related functions 16-18. TORCI regulates protein synthesis, ribosome biogenesis, transcription and autophagy in response to the nutrition availability and energy status of cells, which are sensitive to rapamycin treatment 19-22.
To further investigate the mechanisms of Vid pathway, experiments have been performed to screen the yeast deletion library and GST fusion library 23. It has been identified a list of proteins that interact with and regulate FBPase degradation in this pathway. This study reported the role of TCO89, the subunit of TORCI, as a new component in FBPase degradation of Vid pathway. TCO89 was found to associate with FBPase and regulate FBPase degradation. The results showed that all other TORCI components associated with FBPase. Thus, this study also implicated a possible role for TORC1 in this pathway.
Results and Discussion
TCO89 is required for FBPase degradation in Vid dependent pathway
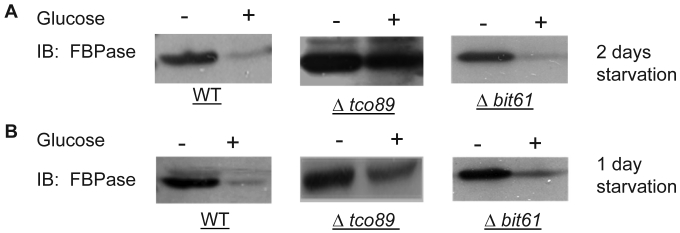
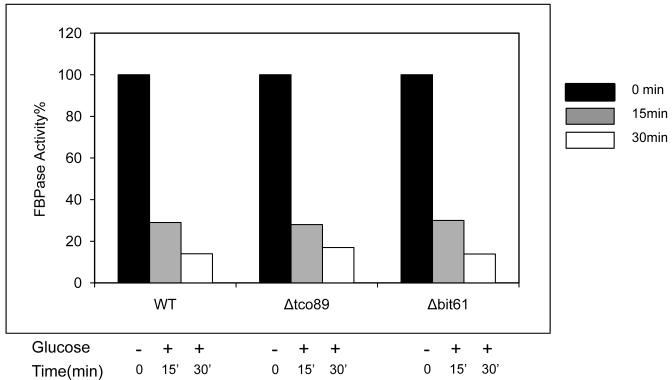
A list of potential FBPase interacting proteins were identified through HIS beads pull down assay and the MALDI Mass Spectra in the preliminary experiments. Among them, TCO89 was identified as one of the FBPase binding proteins. Besides, when screening the yeast deletion library, TCO89 was identified as a potential component of FBPase degradation pathway. It is reported that FBPase can be degraded via either Vid-dependent pathway or ubiquitin-proteasome pathway depending on the starvation conditions 3-6. In this study, two types of FBPase degradations were induced (Fig 1A, 1B). In two-day starvation, Vid-dependent degradation was induced; most of FBPase (90%) kept intact in Δtco89 mutant (Fig 1A); while there was only less than 5% of FBPase left in wild type strain. Similar to the wild type, mutant Δbit61 (one of unique components of Tor Complex II) did not affect FBPase degradations in Vid -dependent pathway (Fig 1A). However, in one-day starvation, ubiquitin-proteasome degradation was induced; as in wild type strain and Δbit61 mutant, little FBPase (15%) was left in the Δtco89 mutant, (Fig1B). Based on Figure 1, loss function of TCO89 exhibited great different effect on two FBPase degradation pathways, strong suggesting TCO89 played a more exclusive role in Vid-dependent pathway for FBPase degradation.
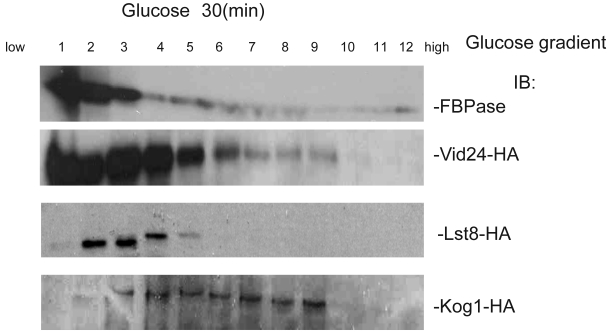
Fig 1.
Regulation of FBPase degradation by TCO89. All strains were glucose starved for indicated time (1day or 2 days). In the experiments, cell cultures were equally divided and two treatments were given as indicated. Same amount of total protein was loaded in the gel for every sample. A. Deletion of TCO89 in Vid-dependent FBPase degradation pathway. IB, Immuno-blotting; WT, w303; Δtco89 and Δbit61, deletion mutants. B. Deletion of TCO89 in proteasome-dependent FBPase degradation. C. The importation of FBPase into Vid vesicle in Δtco89 mutant. Pellet fraction (P) containing Vid vesicles while supernatant fraction (S) containing soluble proteins. D. GST pull-down assay showed the physical association of TCO89 with FBPase. IP, immuno-precipitation. E. Sucrose gradient analysis showed co-localization of TCO89, FBPase and Vid24 during FBPase degradation.
TCO89 is required for the FBPase import into Vid-vesicles
FBPase degradation via Vid pathway can be further divided into sequential events: inactivation of FBPase, importing of FBPase to Vid vesicles, docketing of FBPase vesicles to vacuole for degradation 27, 28. To further investigate the role of TCO89 in this pathway, several experiments were performed. We first checked the localization of FBPase by differential centrifugation (Fig1C). In 30 min FBPase degradation, it was found in wild type strain that 1/3 FBPase was targeted into vesicles (P) and 2/3 of FBPase was still left in cytosol (S); while it was found in Δtco89 mutant strain that only less than 1/10 FBPase was targeted into vesicles (P) and most of FBPase (9/10) was still left in cytosol (S). The results suggested that FBPase was severe blocked in the cytosol (S) and was unable to target into vesicles (P) properly due to loss function of TCO89 (Fig1C).The localization of specific molecular marker of Vid vesicles--Vid24-- was examined by differential centrifugation as well. Interestingly, Vid24 was found to exclusively reside in the cytosol (S) in the Δtco89 mutant (Fig1C); while as reported, the Vid24 resided mostly in the vesicles (P) in wild type strain (Fig1C) 10, 29. Moreover, comparing to the wild type strain, Vid24 expression level wasn't impaired much in the Δtco89 mutant. Since FBPase was accumulated in the Vid vesicles and could not target to vacuole for degradation in Δvid24 mutant 10, 29, our results suggested that TCO89 most likely regulated Vid24 localization. In further examining the activity of FBPase, FBPase wasn't found any change in its enzyme activity at both starvation condition and degradation condition in the Δtco89 mutant comparing to wild type control (S1), so TCO89 did not affect FBPase activity in either situations.
Above all, it seems that TCO89 may mediate FBPase import and also regulate Vid24 association with Vid vesicles, and thus control the formation of FBPase-containing vesicles 29.
TCO89 associated with FBPase during starvation and degradation
To investigate the mechanism of TCO89 in regulating FBPase degradation, GST pull-down was performed at different conditions (starvation condition, degradation condition). FBPase was found to bind to TCO89 at both starvation and degradation conditions, but with less binding affinity at the degradation condition; as a control, UBC8 interacted with FBPase (Fig 1D) as reported 30. The results suggested that TCO89 associated with FBPase during degradation, but it seems to associate with FBPase at the initial steps of the degradation, later it may release FBPase into Vesicle or vacuole so less TCO89 binding was found in the degradation condition. To further investigate if the localizations of TCO89, FBPase and Vid24 were in the Vid vesicle, sucrose gradient analysis was performed. The sucrose gradient fraction showed TCO89 overlapped with four fractions of FBPase and three fractions of Vid24 during FBPase degradation in distribution (Fig 1E). As reported 10, FBPase also overlapped with Vid24 very well during FBPase degradation in distribution (Fig 1E). These results suggested that TCO89 partially interacted with FBPase in the Vid vesicle fraction in the degradation (Fig1D, 1E), thus it is possible that TCO89 regulated vesicle importation of FBPase through protein-protein interaction.
TOR Complex 1(TORCI) played a role in FBPase degradation
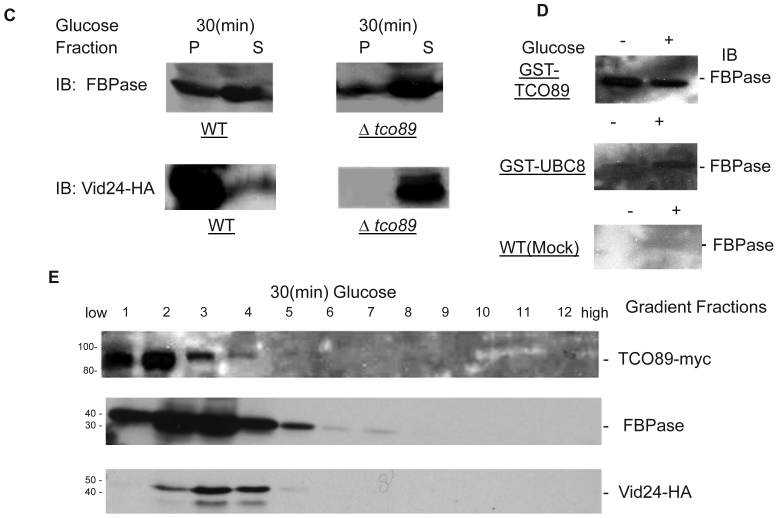
Although TCO89 is a component of TOR Complex 115, it is not known if TORC1 functions in Vid degradation pathway of FBPase. It is reported that TORC1's function is sensitive to rapamycin, the inhibitor of TOR kinases 15. Comparing to control group, rapamycin treatment blocked FBPase degradation in wild type (Fig. 2A). So the function of TORCI is critical for Vid degradation of FBPase. In confirming TORC1's role in FBPase degradation, we over-expressed TOR1 and found it caused seriously blockage of FBPase degradation (Fig 2B). However, the rapamycin treatment did not reverse the defect of FBPase degradation in TOR1overexpressing cells (Fig 2C), which suggested that the impairment of FBPase degradation in TOR1 over-expressing cells was not due to enhanced TOR1 kinase activity. In Drosophila, there is only one TOR protein, but it forms two TOR complexes 35-36. It was reported that over-expression of dTOR in wing showed phenotypes remarkably similar to the loss of dTOR function mutant, such as decreased cell size and cell-proliferation rate, as well as arrested cell cycle in the G1 phase during development 37. Since the N-terminal half of the TOR protein is comprised of a series of repeated HEAT motifs, alpha-helical domains which mediate protein-protein interactions 38, it was thus proposed that abnormally high levels of dTOR can reduce dTOR signaling output by binding and titrating away factors essential for normal TOR function 37. Recently, it was reported again that over-expression of dTOR can hypo- activate TORC1 activity and result in small eye Drosophila 39. It is known that maintaining of a proper molecular ratio of components in a complex is critical to the function of the complex and disturbance of the complex organization by over-expression of one component may cause dominant negative effect 40. In the preliminary experiments, we found that FBPase couldn't be phosphorylated in the TOR1 over-expressing cells, suggesting that TORCI kinase activity was impaired in the cells. It is possible that TOR1 ectopic expression disturbed the stoichiometry of TORCI components and impaired the function of TORC1 so may cause the dominant-negative effect on FBPase degradation.
Fig 2.
The involvement of TORC1 in FBPase degradation. All strains were glucose starved for two days. In the experiments, the 2ml cell cultures were equally divided and different treatments were given as indicated. Same amount of total protein was loaded in the gel for every sample. A. FBPase degradation was rapamycin sensitive. Two-day starved cells (-) and starved cells supplied with glucose for 2hrs (+) were treated with ethanol or rapamycin (20nM) during FBPase degradation. B. Tor1 overexpression blocked FBPase degradation. WT: w303; Tor1 overexpression: ectopic expression of Tor1-V5; Δtor1, Δbit61, Δavo2, and Δavo3: deletion mutants for components of Tor Complex I and Tor complex II. C. Rapamycin did not relieve the impairment of FBPase degradation by Tor1 overexpression.
It is reported that TOR1 and TOR2 proteins share some similar functions and the TOR2 can complement the function of TOR1 in vivo 14, 31, so it is not surprising to find that loss function of TOR1 did not affect FBPase degradation (Fig. 2B). However, loss function of the unique components of TOR Complex II, such as Bit61, Avo2 or Avo3 did not affect the FBPase degradation in this study (Fig. 2B).Therefore, our results strongly implicated a possible role of TORCI, not TORCII in Vid-dependent pathway for FBPase degradation.
Similar to TCO89, other components of TORCI physically associated with FBPase
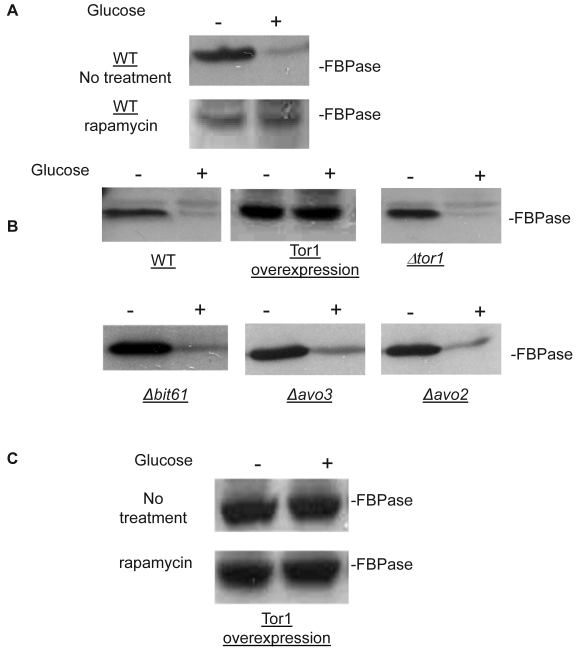
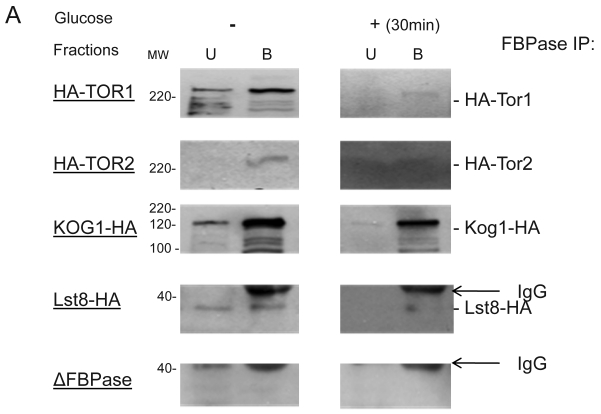
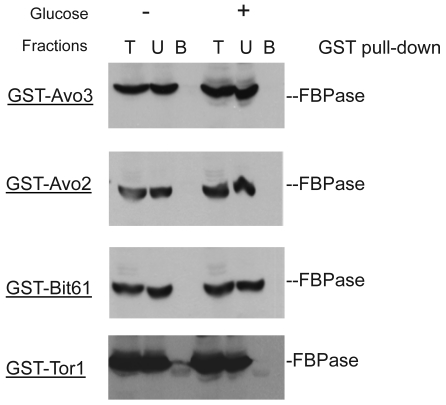
Though the MALDI mass spectra results have suggested the components of Tor Complex I associated with FBPase, immune-precipitation was also performed to confirm if TORC1 associated with FBPase. Consistent with TCO89, TOR1, TOR2, Kog1 and Lst8 all were found to interact with FBPase in the starvation status, and also interact with FBPase in degradation condition with lower affinity (Fig3A). However, the unique components of Tor Complex II, Bit61, Avo2 and Avo3 did not interact with FBPase in this study (S2).
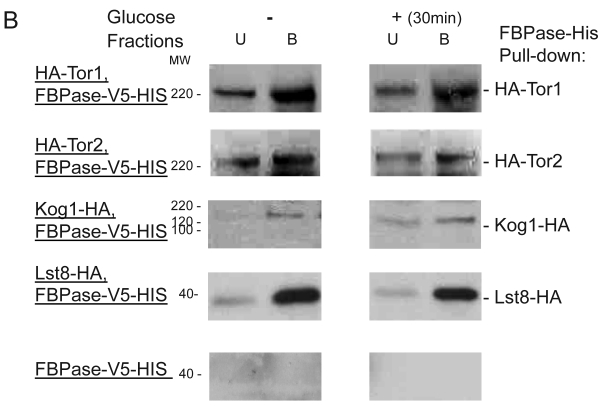
To further confirm the interaction of TORC1 with FBPase, HIS pull-down assay was performed. Similar results were found that TOR1, TOR2, Kog1 and Lst8 associated with FBPase in both starvation status and degradation status (Fig 3B). The sucrose gradient results also showed Kog1 and Lst8 co-localized with FBPase in Vid fractions: seven fractions of Kog1 and five fractions of Lst8 respectively overlapped with FBPase in distribution during FBPase degradation (S3), while Vid24 was localized with Kog1 and Lst8 at the vesicle fraction respectively (S3). The previous study reported that TORCI was found in the membrane fractions and also may locate in vesicles membranes 24, 32. So, it is likely that TORCI regulates FBPase degradation via its association with FBPase, and it seems that the components of TORCI gradually dissociated from FBPase during the degradation process. We proved that all components of TOCR1 physically associated with FBPase by three independent methods: sucrose gradient experiment, His Pull-down and immuno-precipitation. We will further test which component(s) of TORC1 binds to FBPase directly.
Fig 3.
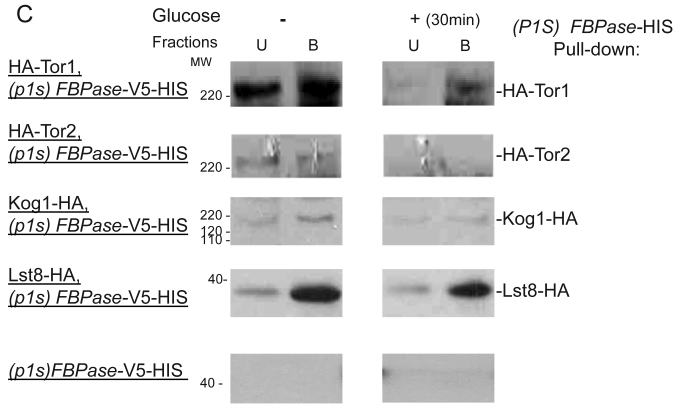
FBPase interacted with components of Tor Complex I. All strains were glucose starved for two days. In the experiment, the cell cultures were equally divided and different treatments were given as indicated. Immunoprecipitation and pull-down assay were performed as described in the material and methods. Equal total protein was used in the different treatment. A. FBPase associated with TOR Complex 1 components by immuno-precipitation. Proteins were precipitated with rabbit anti-FBPase antibody and protein A bead. The interaction proteins were blotted with HA antibody. Starved cells (-) and starved cells supplied with glucose for 30min (+) were used. U, unbound protein, B, bound protein. B. FBPase associated with TOR Complex 1 components by HIS pull-down assay. C.P1S FBPase mutant weakly interacted with TORCI in HIS pull-down. Immuno-blotting was with mouse anti-HA antibody.
It was reported that an N-terminal mutation of FBPase (p1s fbpase) blocked FBPase degradation 5. In this study, this mutation was also found to severely weaken the association of FBPase with TOR1, TOR2, Kog1 and Lst8 respectively comparing to the wild type FBPase (Fig 3B &3C), therefore suggesting that TORC1's association with FBPase is required for FBPase degradation. As reported, FBPase can be phosphorylated by cAMP dependent pathway prior to its degradation and cAMP pathway cross-talked with TORC1 pathway in regulating autophagy 5, 33-34. We found that loss of function of TORCI also decreased the phosphorylation level of FBPase and thus failed to inactivate FBPase activity in response to glucose (preliminary data not shown). Therefore, with the Thr /Ser protein kinase activity, TORC1 most likely regulates FBPase degradation via phosphorylation as it regulates other proteins. In summary, FBPase degradation was analyzed in the mutants of Tor Complex I. The results suggested that TCO89 was essential for FBPase to be degraded properly in Vid dependent pathway —TCO89 was especially required for FBPase to be targeted to the Vid vesicles through protein-protein interaction. Besides, the function of TORCI was required for this pathway as well.
Material and methods
Yeast strains
Yeast strains used in this study are listed in Table 1.
Table 1.
yeast strains in this study
| Strain | Genotype |
|---|---|
| W303 | MATa leu2,3-112 ade2 his3-Δ200 trp1-1 ura3-52 |
| HLY650 | MATαhisΔ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| BY4742 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (Euroscarf, Germany) |
| Δtco89 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 tco89::kanmx4 (Euroscarf, Germany) |
| Δarp8 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 arp8::kanmx4 (Euroscarf, Germany) |
| Δbit61 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 bit61::kanmx4 (Euroscarf, Germany) |
| Δavo2 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 avo2::kanmx4 (Euroscarf, Germany) |
| Δavo3 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 avo3::kanmx4 (Euroscarf, Germany) |
| Δtor1 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 tor1::kanmx4 (Euroscarf, Germany) |
| GST-TCO89 | Matα his3-Δ200,leu2-3, 112, ura3-52, pep4::URA, GST-TCO89::LEU 23 |
| GST-ARP8 | Matα his3-Δ200,leu2-3,112, ura3-52,pep4::URA, GST-ARP8::LEU 23 |
| Tor1-V5-HIS | Matα his3-Δ200 ura3-52 leu2, 3-112 trp1-1, Tor1 -V5-HIS URA( this study) |
| FBPase -V5-HIS | Mata his3-Δ200 ura3-52 leu2,3-112 trp1-1, FBPase -V5-HIS URA( this study) |
| PLY298 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+3HA-TOR1: His3MX6 22 |
| PLY122 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ 3HA-TOR2: His3MX6 22 |
| PLY307 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ LST8-HA:TRP1 22 |
| PLY306 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ KOG1-HA: TRP1 22 |
| PLY336 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+TCO89-MYC:TRP1 22 |
| HLY2100 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+3HA- TOR1: His3MX6, FBPase -V5-HIS LEU( this study) |
| HLY2101 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ 3HA-TOR2:His3MX6, FBPase -V5-HIS LEU( this study) |
| HLY2102 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ LST8-HA: TRP1 FBPase -V5-HIS LEU( this study) |
| HLY2103 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ KOG1-HA: TRP1 FBPase -V5-HIS LEU( this study) |
| HLY2104 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ 3HA-TOR1:His3MX6, P1S fbpase -V5-HIS LEU( this study) |
| HLY2105 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ 3HA-TOR2:His3MX6, P1S fbpase -V5-HIS LEU( this study) |
| HLY2106 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ LST8-HA: TRP1, P1S fbpase -V5-HIS LEU( this study) |
| HLY2107 | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ KOG1-HA: TRP1, P1S fbpase -V5-HIS LEU( this study) |
Yeast starvation and rapamycin treatment
Yeast cells (1:250 ratios) were inoculated in YPKG (1% yeast extract, 2% peptone,1% KCH3COOH, 0.5% dextrose) or YPKGal (1% yeast extract, 2% peptone,1% KCH3COOH, 0.5% Galactose) medium for either one day or two days to perform glucose starvation. To induce FBPase degradation, YPKG or YPKGal medium was removed and fresh YPD was added for indicated time. Without specific indication, the yeast starvation was 2 days and FBPase degradation was induced for 30 min. For rapamycin treatment, yeast cultures were grown in 20nM rapamycin (Sigma) YPD medium, control group was treated with YPD containing same amount of ethanol 24.
Western blotting and antibodies
Western blotting was performed with standard protocol 10. Primary antibodies included monoclonal mouse anti-HA antibody (1:5000) (Roche), monoclonal mouse anti-V5 antibody (1:5000) (Life Technologies) and polyclonal rabbit anti-FBPase antibody (1:10000) 25. Secondary antibody included HRP conjugated anti-rabbit antibody (1:5,000) and HRP conjugated anti-mouse antibody (1:5,000) (Amersham Biosciences).
Differential fractionation
Differential fractionation was performed as described 10. Cells were lysed with lysis buffer (50 mM Hepes-NaOH, pH 7.4, 5 mM MgSO4, 40 mM (NH4)2SO4, 0.1mM EDTA, 1mM PMSF). Total soluble proteins were subjected to a series of centrifugations. The high speed supernatant and pellet were analyzed with Western blotting.
GST pull-down
Yeast strains containing GST fused with the amino-terminus of TCO89 or UBC8 open reading frame respectively were used. GST pull-down was performed as described 10. Cells were lysed in GST buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 1mM PMSF). Total lysates were spun down at 13,000g for 20min. The supernatants were incubated with equilibrated glutathione-Sepharose 4B beads (Amersham Biosciences) for 2 hours at 4°C. GST beads were collected by spinning down at 500g for 1min after incubation. The incubated GST beads were washed with lysis buffer for 3 times. And GST bounded proteins were analyzed with Western blotting.
FBPase Immunoprecipitation
Immunoprecipitation was performed as described 10. 10 ml yeast cells were lysed in IP buffer (50 mM Tris, pH 7.4, 200 mM NaCl, 5 mM EDTA, 1 mM PMSF). Total lysates were spun down at 200,000g for 2 hours to enrich vesicle proteins. The pellets were suspended and incubated with 1ul of FBPase antibody to form antigen- antibody complex at 4°C for 2 hour. Then equilibrated protein A beads (Amersham Biosciences) were added and followed with 1 hour rotating at 4°C. FBPase interacting proteins were precipitated by collecting protein A beads at 500g for 1min. Incubated Protein A beads were washed for 3 times. The FBPase interaction proteins were analyzed by Western blotting.
HIS pull-down
FBPase open reading frame fused with V5 and HIS tag at the carboxyl-terminus was transformed into respective yeast strains. HIS pull-down was performed according to the product manual (Qiagen). 10 ml yeast cells were lysed in HIS lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.05% Tween20, pH 8.0, 1mM PMSF and 1x cocktail proteinase inhibitors (Roche). Total lysates were spun down at 200,000g for 2 hours at 4°C. Pellets were resuspended and incubated with equilibrated Ni-NTA agarose for 2-3 hours at 4°C. Incubated Ni-NTA agarose was collected and washed 3 times with 10x volume of washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 0.05% Tween 20, pH 8.0, 1mM PMSF). Elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 0.05% Tween20, pH8.0, 1mM PMSF) was added and incubated for 5min at 4°C. Elute was collected for Western blotting analysis.
Sucrose-gradient
Sucrose gradient was performed as described 26. 10 ml of cell were starved from glucose for 2 days. Total cell lysates were subjected to differential fractionation. High-speed pellets (100,000x g) were loaded to the top of sucrose gradient and were centrifuged at 100,000g for 14-16 hours at 4°C. Protein samples from gradients were fractionated into 1mls. And the 1ml protein sample fractions were precipitated with 10% of TCA. The precipitations of the proteins samples were washed and suspended with SDS loading buffer. All protein fraction samples were subjected to Western blotting analysis.
Determination of protein concentration
Protein concentration in different yeast extracts were determined by the Bradford method using BSA as the protein standard 41.
Fig S1.
Loss function of TCO89 did not affect FBPase activity in enzyme activity assay. Yeast strains were starved as described. Samples of no glucose added (-) and glucose added for 15 min and 30 min (+) were subjected to enzyme activity assay as reported 1.
Fig S2.
GST pull-down showed that unique components of Tor complex 2 did not associate with FBPase.
Fig S3.
Sucrose gradient showed that Lst8 and Kog1 co-subcellular localized with FBPase and Vid24.
Acknowledgments
The author is grateful to Dr Chiang Hui-Ling and her lab for the support. The author also thanks Prof. Ted Powers (University of UC Davis) for his generous gift strains. The author also thanks Dr Li Lei for his critical reading.
Abbreviations
- FBPase
fructose-1, 6-bisphosphatase
- Vid
vacuole import and degradation.
References
- 1.Chiang HL, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991;350:313–8. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- 2.Chiang HL, Schekman R, Hamamoto S. Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J Biol Chem. 1996;271:9934–41. doi: 10.1074/jbc.271.17.9934. [DOI] [PubMed] [Google Scholar]
- 3.Horak J, Regelmann J, Wolf DH. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J Biol Chem. 2002;277:8248–54. doi: 10.1074/jbc.M107255200. [DOI] [PubMed] [Google Scholar]
- 4.Regelmann J, Schüle T, Josupeit FS. et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell. 2003;14:1652–63. doi: 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung GC, Brown CR, Wolfe AB. et al. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–50. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 6.Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995;270:26446–50. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 7.Stromhaug PE, Klionsky DJ. Approaching the Molecular Mechanism of Autophagy. Traffic. 2001;2:524. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Brown CR, Chiang HL. Degradation of the gluconeogenic enzyme fructose-1, 6-bisphosphatase is dependent on the vacuolar ATPase. Autophagy. 2005;1:146–56. doi: 10.4161/auto.1.3.2036. [DOI] [PubMed] [Google Scholar]
- 9.Brown CR, Wolfe AB, Cui D. et al. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem. 2008;283:26116–27. doi: 10.1074/jbc.M709922200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang MC, Chiang HL. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol. 1998;140:1347–56. doi: 10.1083/jcb.140.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–9. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 12.Kunz J, Henriquez R, Schneider U. et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–96. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 13.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471– 84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Loewith R, Jacinto E, Wullschleger S. et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457– 68. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 15.Reinke A, Anderson S, McCaffery JM. et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–62. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 16.Roberg KJ, Bickel S, Rowley N. et al. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics. 1997;147:1569–84. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335– 48. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 18.Martin DE, Hall MN. The expanding TOR signaling network. Curr Opin Cell Biol. 2005;17:158– 66. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Beretta L, Gingras AC, Svitkin YV. et al. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–64. [PMC free article] [PubMed] [Google Scholar]
- 20.Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–38. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beck T, Hall MN. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–92. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 22.Urban J, Soulard A, Huber A. et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–74. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Cui DY, Brown CR, Chiang HL. The type 1 phosphatase Reg1p-Glc7p is required for the glucose-induced degradation of fructose-1,6-bisphosphatase in the vacuole. J Biol Chem. 2004;279:9713–24. doi: 10.1074/jbc.M310793200. [DOI] [PubMed] [Google Scholar]
- 24.Kunz J, Schneider U, Howald I. et al. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem. 2000;275:37011–20. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman M, Chiang HL. Isolation of degradation-deficient mutants defective in the targeting of fructose-1, 6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996;143:1555–66. doi: 10.1093/genetics/143.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang PH, Chiang HL. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–10. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown CR, Liu J, Hung GC. et al. The Vid vesicle to vacuole trafficking event requires components of the SNARE membrane fusion machinery. J Biol Chem. 2003;278:25688–99. doi: 10.1074/jbc.M210549200. [DOI] [PubMed] [Google Scholar]
- 28.Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J Cell Biol. 2000;150:65–76. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh HL, Chen Y, Brown CR. et al. Biochemical analysis of fructose-1,6-bisphosphatase import into vacuole import and degradation vesicles reveals a role for UBC1 in vesicle biogenesis. J Biol Chem. 2001;276:10398–406. doi: 10.1074/jbc.M001767200. [DOI] [PubMed] [Google Scholar]
- 30.Santt O, Pfirrmann T, Braun B. et al. The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell. 2008;19:3323–33. doi: 10.1091/mbc.E08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci U S A. 1996;93:13780–5. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturgill TW, Cohen A, Diefenbacher M. et al. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7:1819–30. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacinto E, Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem J. 2008;410:19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- 34.Yorimitsu T, Zaman S, Broach JR. et al. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180– 9. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanfel MN, Shamieh LS, Kaeberlein M. et al. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–74. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Billington CJJr, Pan DJ. et al. Drosophila Target of Rapamycin Kinase Functions as a Multimer. Genetics. 2006;172:355–62. doi: 10.1534/genetics.105.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hennig KM, Neufeld TP. Inhibition of cellular growth and proliferation by dTOR overexpression in Drosophila. Genesis. 2002;34:107–10. doi: 10.1002/gene.10139. [DOI] [PubMed] [Google Scholar]
- 38.Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–6. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 39.Wang YH, Huang ML. Reduction of Lobe leads to TORC1 hypoactivation that induces ectopic Jak/STAT signaling to impair Drosophila eye development. Mech Dev. 2009;126:781–90. doi: 10.1016/j.mod.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Nishikawa T, Hagihara K, Serada S. et al. Transcriptional complex formation of c-Fos, STAT3, and hepatocyte NF-1 alpha is essential for cytokine-driven C-reactive protein gene expression. J Immunol. 2008;180:3492–501. doi: 10.4049/jimmunol.180.5.3492. [DOI] [PubMed] [Google Scholar]
- 41.Bradford M. A rapid and sensitive Method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248– 54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]