Abstract
Although smallpox was eradicated as a global illness more than 30 years ago, variola virus and other related pathogenic poxviruses, such as monkeypox, remain potential bioterrorist weapons or could re-emerge as natural infections. Poxviruses express virulence factors that down-modulate the host’s immune system. We previously compared functional profiles of the poxviral complement inhibitors of smallpox, vaccinia, and monkeypox known as SPICE, VCP (or VICE), and MOPICE, respectively. SPICE was the most potent regulator of human complement and attached to cells via glycosaminoglycans. The major goals of the present study were to further characterize the complement regulatory and heparin binding sites of SPICE and to evaluate a mAb that abrogates its function. Using substitution mutagenesis, we established that (1) elimination of the three heparin binding sites severely decreases but does not eliminate glycosaminoglycan binding, (2) there is a hierarchy of activity for heparin binding among the three sites, and (3) complement regulatory sites overlap with each of the three heparin binding motifs. By creating chimeras with interchanges of SPICE and VCP residues, a combination of two SPICE amino acids (H77 plus K120) enhances VCP activity ~200-fold. Also, SPICE residue L131 is critical for both complement regulatory function and accounts for the electrophoretic differences between SPICE and VCP. An evolutionary history for these structure-function adaptations of SPICE is proposed. Finally, we identified and characterized a mAb that inhibits the complement regulatory activity of SPICE, MOPICE, and VCP and thus could be used as a therapeutic agent.
Many authorities suggest that smallpox and the emerging natural infection of monkeypox represent two of the most important agents that could be used in a bioterrorist attack (1–4). For example, smallpox produced as a stable aerosol has high infectivity and mortality. Given that ~80% of the population no longer has immunity to variola, mathematical models of smallpox attack scenarios estimate ~55,000 deaths in the context with a high-impact airport exposure (5). Furthermore, even if given preventatively, the current smallpox vaccine can produce life-threatening complications (6). The epidemic of monkeypox that occurred in the United States in 2003 (7) caused great concern and may have been larger than previously realized with a possible spread to rodents (8). The monkeypox virus that caused this infection was, fortunately, a less virulent strain (from West Africa) that did not contain the complement inhibitor (9, 10). New threats have also arisen regarding other poxviruses such as those infecting sheep and goats that could be used in economic bioterrorism (11). Our studies are part of the national biomedical research agenda, Project BioShield, which aims not only to enhance our understanding of the pathogenesis of poxviral and other potentially emerging infections but also to develop improved diagnostics/therapeutics (2, 12).
Poxviruses target the complement system by expressing virulence factors called poxviral inhibitors of complement enzymes, or PICES3 (13–19). The complement system has the capability to recognize, bind, and lyse pathogens including viruses and virally infected cells. The concomitant release of anaphylatoxins activates immune cells. The generation of opsonins coats Ags for their subsequent phagocytosis and for their presentation to the adaptive immune system. The PICES likely were highjacked from a mammalian source and then employed to down-modulate the host’s complement system (20–22).
Variola virus, the causative agent of smallpox, encodes a secreted complement regulatory protein called SPICE (19). The vaccine strain (vaccinia), encodes vaccinia complement protein (VCP) (23, 24). For monkeypox, two strains have been identified in which a major difference is the presence or absence of the complement regulatory protein MOPICE (9, 10). The less virulent strain from West Africa does not contain the gene for MOPICE, whereas the more virulent strain from the Congo basin (Central Africa) expresses MOPICE (9, 10). That PICES serve as virulence factors is further evidenced by studies demonstrating that vaccinia virus mutants not expressing VCP are attenuated in vivo (25) and that surviving ectromelia virus infection (mousepox) requires the complement system (26).
SPICE and VCP are secreted proteins consisting of 244 aa with only 11 differences between them (Fig. 1). These residues are functionally important, however, since SPICE is ~100-fold more potent than VCP in regulating C3b and 4- to 6-fold more efficient at inactivating C4b (13, 18, 19, 27). SPICE and VCP inhibit the complement system analogous to the host’s own regulators via cofactor activity and decay-accelerating activity. Cofactor activity (CA) refers to the limited proteolytic degradation of C3b and C4b that requires a cofactor protein working in concert with the plasma serine protease factor I. Decay-accelerating activity (DAA) refers to the ability to dissociate the catalytic serine protease domain from the C3- and C5-activating enzyme complexes known as convertases. Thus, PICES function similarly to human complement regulatory proteins and indeed are 30 – 40% identical to their human counterparts (15, 16, 20, 28). Each possesses four complement control repeats (CCPs), as does membrane cofactor protein (MCP; CD46) and decay-accelerating factor (DAF; CD55) (29, 30).
FIGURE 1.
Amino acid alignment of the four CCP repeats of SPICE and VCP. The 11 aa differences between the two secreted proteins are indicated. The overlined areas denote the putative heparin binding sites that consist of three amino acids. SPICE contains three domains (HS1, HS2, HS3), while VCP adds a fourth (HS4). Amino acid numbering conforms to the mature protein (does not include signal peptide).
Poxviral and human regulators also contain putative motifs for binding to glycosaminoglycans (GAGs), such as heparin and related sulfated molecules (31–33). SPICE binds to heparin similarly to VCP and MOPICE, but with a higher affinity than human complement regulator factor H (13). SPICE binds to multiple cell types via heparin and related sulfated GAGs and inhibits complement regulation similarly to related host regulators (34). Heparin binding is likely an important functional feature by providing a means for a secreted regulator to anchor to host cells, viruses, or virally infected cells where it can then down-regulate complement (33, 35).
Our overall goals are to better understand the virulence factors that underlie poxviral pathogenesis. Herein, we further characterize the heparin binding and complement regulatory sites of SPICE and evaluate a function-blocking mAb to SPICE with therapeutic potential.
Materials and Methods
Molecular engineering and expression of SPICE, VCP, chimeras, and mutants
As previously described, the gene for VCP was amplified by PCR using genomic DNA from vaccinia virus (Western Reserve) and cloned into the plasmid pSG5 (Stratagene) (13). SPICE was constructed from VCP by mutation of the 11 aa using nine primers (Integrated DNA Technologies) and the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene).
Point mutations in SPICE were produced utilizing the QuikChange Site-Directed Mutagenesis kit (Stratagene). The fidelity of clones was verified by DNA sequencing. Proteins were expressed transiently in either Chinese hamster ovary (CHO) cells using Fugene-6 (Roche Molecular Biochemicals) or 293T cells using Lipofectamine (Invitrogen). To disrupt the three putative heparin binding sites in CCP1, the first and third amino acids of the heparin binding sites (see Figs. 1 and 2A) were changed to alanine.
FIGURE 2.
SPICE mutations evaluated in this report. A, Substitution of an alanine for the first and third residues of the three amino acid heparin binding sites in CCP1 of SPICE. B, Substitution of alanines into SPICE at three locations in CCP2, namely, Y98, Y103, and K108. The corresponding peptide sequence of VCP is also shown (see text). C, SPICE-VCP chimeric mutants: in the first, SPICE residues 77H and 120K were substituted into VCP (VCP Q77H-E120K) or VCP residue 131S was substituted into SPICE (SPICE-L131S).
ELISA for poxviral complement inhibitors
Poxviral proteins were quantitated by ELISA (13). Briefly, the capture mAb, 5A10 (13) or mAb KL5.1 (see below) (36), was coated (5 μg/ml) on the plate overnight at 4°C and then blocked for 1 h at 37°C (1% BSA and 0.1% Tween 20 in PBS). Samples and standards (VCP) (13) were incubated for 1 h at 37°C and then washed with PBS containing 0.05% Tween 20. Next, a rabbit anti-VCP antiserum that cross-reacts with SPICE (13) was applied for 1 h at 37°C. After washing, HRP-coupled donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories) was added and incubated for 1 h at 37°C. After washing, 3,3′,5,5′-tetramethylbenzidine substrate (Pierce Biotechnology) was added and absorbance (630 nm) measured in an ELISA reader.
Functional assessment
Ligand binding, cofactor, and decay-accelerating assays have been described (13). Briefly, for C3b and C4b binding, an ELISA format was used in which human C3b or C4b (Complement Technologies) was coated overnight on a well and then blocked (13). Samples diluted in ELISA buffer (10 mM Tris (pH 7.4), 25 mM NaCl, 0.05% Tween 20, 4% BSA) were incubated for 1 h at 37°C and then washed (10 mM Tris (pH 7.4), 25 mM NaCl, 0.05% Tween 20). Rabbit anti-VCP antiserum was applied for 1 h at 37°C and then HRP-coupled donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories) was added for 1 h at 37°C. After washing, 3,3′,5,5′-tetramethylbenzidine substrate was added and absorbance (630 nm) measured. Ligand binding assays were routinely performed on serially diluted samples in three separate experiments.
Cofactor assays utilized biotinylated human C3b or C4b, human factor I (Complement Technologies), and varying concentrations of the cofactor proteins in a low salt (25 mM NaCl) buffer (13). Cleavage fragments were analyzed on 10% reducing gels followed by Western blotting. Detection was with ExtrAvidin peroxidase conjugate (Sigma-Aldrich). Analysis was performed by densitometric scanning. Assays were performed in duplicate on two to four separate occasions.
DAAs were performed in microtiter dishes as described (13). Briefly, for classical pathway convertase assembly, Ab-coated sheep erythrocytes (Complement Technologies) were incubated with human C1 (Complement Technologies) (1 μg/ml) for 15 min at 30°C and washed with dextrose gelatin veronal buffer (DGVB2+; Complement Technologies). Human C4 (Complement Technologies) (2.2 μg/ml) was added for 15 min at 30°C and erythrocytes were washed. Human C2 (Complement Technologies) (0.25 μg/ml) was added for 4 min at room temperature and then the cells were washed. For DAA assessment, 50 μl of inhibitor (30 ng) was added to an equal volume of prepared cells for 10 min at 30°C. For lytic development, guinea pig serum (0.5 ml) (Colorado Serum Company) diluted 1/20 in 40 mM EDTA-gelatin veronal buffer for 30 min 37°C was added. Following centrifugation at 2000 × g, the A414 of the supernatant was determined. Assays were performed at least three times with each condition in duplicate.
Heparin-affinity chromatography
Heparin binding was assessed by loading serum-free supernatants from transient transfections of SPICE or its heparin mutants on a 1-ml HiTrap HP heparin column (GE Healthcare) previously equilibrated with 10 mM phosphate (pH 7.2). The column was washed with 5 ml of equilibration buffer at a flow rate of 1 ml/min, and bound proteins were eluted with a linear gradient from 0 to 2 M NaCl. Fractions were assessed using Western blots with a polyclonal Ab to VCP/SPICE (13).
Generation of murine anti-SPICE hybridoma
Mice were immunized with recombinantly produced VCP (13) (rVCP). A mouse with high titer Ab was sacrificed for fusion at the Washington University Hybridoma Center. An ELISA was developed to select function-blocking mAbs based on competitive inhibition of SPICE binding to human C3b by hybridoma supernatants. In this assay, human C3b-coated plates (5 μg/ml) were blocked (PBS, 0.05% Tween 20, 1.0% BSA for 1 h at 37°C), and rVCP (1500 ng/ml) added to hybridoma supernatant was mixed and preincubated for 30 min at 37°C in low salt buffer (10 mM Tris (pH 7.4) and 25 mM NaCl, 0.05% Tween 20, 4% BSA). This mixture was added to the C3b-coated wells and incubated for 1 h at 37°C. Washes were performed in low salt buffer (10 mM Tris (pH 7.4), 25 mM NaCl, and 0.05% Tween 20). Detection of binding was by the addition of 1/5000 dilution of a rabbit polyclonal Ab to VCP (1 h at 37°C) that cross-reacts with SPICE and MOPICE (13). After washing, a secondary HRP-labeled secondary Ab was incubated for 1 h at 37°C followed by addition of substrate and measurement of absorbance. Loss of binding was taken as an indication of the presence of a function-blocking mAb. One hybridoma, KL5.1, was isolated and subcloned. Ascites fluid was prepared and the mAb was purified (performed at Harlan Laboratories). The studies employing animals complied with all required federal guidelines and institutional practices.
Vaccinia immune globulin
Reference Vaccinia Immune Globulin (VIGIV; lot 1) was obtained from the Standard and Testing Section of Center for Biologics Evaluation and Research/Food and Drug Administration courtesy of Dr. C. Anderson (Food and Drug Administration, Department of Health and Human Services). Control IgG was obtained from the serum of unvaccinated individuals via purification over protein A affinity media (GE Healthcare) per the manufacturer’s directions.
Results
Hierarchy of activity of heparin binding sites
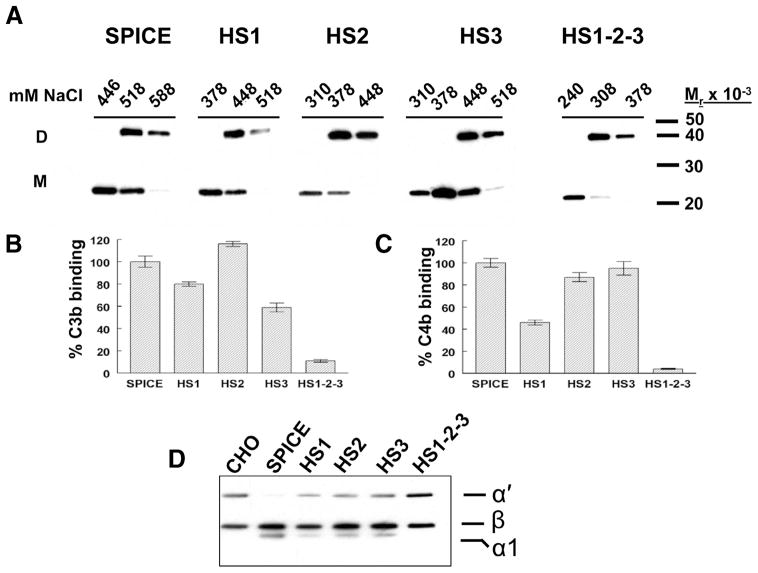
To examine the influence of three putative heparin binding sites in SPICE that are also common to other PICES, we altered each in SPICE to create mutant proteins HS1, HS2, and HS3 (Figs. 1 and 2A). We also produced a construct that eliminated all three sites (HS1-2-3). These proteins were transiently expressed in CHO or 293T cells and supernatants chromatographed over a heparin column. Proteins were eluted by increasing the salt concentration, and fractions were monitored via Western blotting. As compared with SPICE, mutation of each site reduced binding to heparin (Fig. 3A and Table I) and mutation of all three sites (HS1-2-3) severely reduced heparin binding (Fig. 3A and Table I). Mutation of the second site decreased heparin binding the most (Table I, 328 ± 3 mM sodium chloride peak monomer elution) as compared with the other two mutants. These results establish a hierarchy of activity for the three sites, with the second site being the most important for heparin interactions. They further demonstrate that some heparin binding is retained, despite mutation of the three sites, suggesting that overall positive charge may be involved.
FIGURE 3.
Functional profile of mutants in heparin binding sites of SPICE. A, SPICE and four heparin mutants (see Fig. 2) were transiently expressed in 293T cells. Supernatants were chromatographed over heparin affinity columns, eluted with increasing salt (NaCl) concentrations, and fractions were monitored in Western blots with a polyclonal Ab to SPICE. “D” and “M” designations indicate dimer and monomer forms of the proteins. B and C, Supernatants were assessed for C3b (B) and C4b (C) binding in an ELISA format in which human C3b or C4b was adsorbed to microtiter plates. SPICE and mutant supernatants were applied and then detected with a polyclonal Ab. SPICE binding was set at 100%. Data represent the means ± SEM for three experiments. D, Biotinylated C3b was incubated with human factor I and supernatants from the transfectants. Methods per Table I.
Table I.
Heparin binding and complement regulatory profile of SPICE HS mutantsa
| Protein | Peak Elutionb (mM NaCl) | C3b Bindingc (% Wild type) | C4b Bindingc (% wild type) | C3b Cofactord (% wild type) | C4b Cofactord (% wild type) |
|---|---|---|---|---|---|
| SPICE | 447 ± 5 | 100 ± 5 | 100 ± 4 | 100 ± 7 | 100 ± 8 |
| HS1 | 384 ± 3 | 80 ± 2 | 46 ± 2 | 42 ± 4 | 48 ± 3 |
| HS2 | 328 ± 3 | 116 ± 3 | 87 ± 4 | 54 ± 5 | 88 ± 6 |
| HS3 | 388 ± 4 | 59 ± 4 | 95 ± 6 | 54 ± 2 | 76 ± 8 |
| HS1-2-3 | 239 ± 6 | 11 ± 1 | 4 ± 1 | 8 ± 5 | None detected |
SPICE with mutated heparin binding sites (see Figs. 1 and 2A) were expressed in 293T cells and the supernatants were evaluated.
SPICE supernatants were chromatographed over a heparin column, eluted with increasing salt concentrations, and fractions were monitored via Western blot with a polyclonal Ab (see Fig. 3A). Peak elution of monomer represents the mean ± SEM of three or four experiments.
C3b and C4b binding were performed in an ELISA format in which human C3b or C4b was adsorbed to microtiter plates. SPICE and mutant supernatants (10 ng/ml) were applied and then detected with a polyclonal Ab. SPICE binding was set at 100%. Data are percentages of wild-type SPICE and represent means ± SEM for four experiments.
For the C3b and C4b cofactor assays, biotinylated C3b or C4b was incubated with human factor I and transfectant supernatants (1.7 ng/ml of SPICE or mutant for C3b assays and 6.7 ng/ml for C4b assays) and evaluated on Western blots using HRP-ExtrAvidin. For C3b cofactor activity, comparisons were made based on the loss of the α′-chain and development of the α1 cleavage fragment. For C4b cofactor activity, comparisons were made based on density of the α′ band relative to the β–chain. Data are percentages of wild-type SPICE and represent means ± SEM for four experiments.
Heparin sites impact complement regulation
We next assessed if altering heparin binding sites modulated the complement inhibitory profile of SPICE. CCP1 of SPICE and VCP have identical amino acid sequences. Previous studies have implicated CCP1 in complement inhibition (32, 33, 37, 38) and residues K12, K14, K41, K43, K64, R65, and R66 as being important for C3b/C4b interactions (33, 39) As shown in Fig. 3B–D and summarized in Table I, each of the heparin mutants had a reduced functional repertoire of ligand binding and/or CA. The mutant lacking all three sites failed to bind either C3b or C4b and lacked CA. Mutants HS1, HS2, and HS3 lost ~50% of their C3b CA. Mutants HS1 and HS3 showed decreased binding to C3b. Additionally, the HS1 mutation also showed diminished C4b binding and C4b CA (Table I, 46 ± 2% and 48 ± 3%, respectively). These data demonstrate that all three heparin binding sites overlap complement regulatory sites. Although in many cases ligand binding and CA parallel one another, several studies of human regulators have identified mutants in which binding and CA are separable functions (40, 41).
SPICE functional sites in CCP2
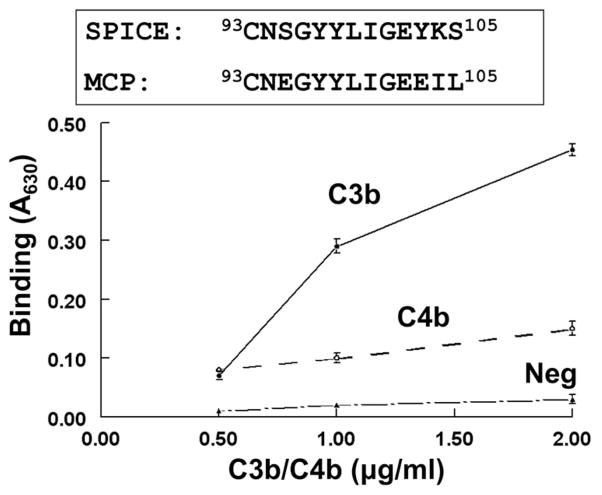
While there are only 11 aa that differ between SPICE and VCP, SPICE possesses 100- to 1000-fold enhanced regulatory activity (13, 19, 27, 39). We assessed a SPICE peptide (aa 93–105) in CCP2 that has 69% homology to a functionally important site of the human regulator MCP (40) (Fig. 4). Indeed, one stretch of the SPICE peptide (aa 93–102) is 90% identical to the corresponding aa of MCP. Moreover, the amino acid number positions of the peptides in this region are identical in the mature proteins of SPICE and MCP. An ELISA was utilized in which the SPICE peptide was attached to microtiter wells. C3b bound to the SPICE peptide, while C4b binding was minimal (Fig. 4). This result establishes that this MCP-homologous peptide in SPICE is a key region for C3b binding.
FIGURE 4.
Binding by SPICE peptide to C3b and C4b. The peptide is 69% homologous to human complement regulator MCP. After adsorption of the SPICE peptide to microtiter wells, C3b or C4b was incubated and binding detected with a goat polyclonal Ab to C3 or C4. Neg, a peptide to SPICE (aa 107–123) that showed no binding. A representative experiment of three is shown. Data represent the means ± SEM.
Since this same region bears 3 of the 11 aa differences between SPICE and VCP, point mutations of these 3 aa were next evaluated. Each was mutated to an alanine (Y98A, Y103A, and K108A) (see Figs. 1 and 2B). Following expression in CHO cells and quantitation, the mutants were functionally evaluated (Table II). Mutant Y98A had decreased C4b binding, C4b CA, and classical pathway DAA, but was equivalent to wild type relative to its C3 interactions. On the other hand, Y103A lost 90% of its C3b CA and DAA, while maintaining wild-type activity for C3b and C4b binding and for C4b CA. Surprisingly, K108A mutant had increased C3b and C4b binding and DAA for the classical pathway. These mutants again support the concept of ligand binding, CA, and DAA as being distinct and separable regulatory activities and further establish this peptide in SPICE as being involved in these functions.
Table II.
Complement regulatory profile of point mutations in CCP2 of SPICEa
| SPICE Mutant | C3b Binding | C4b Binding | C3b Cofactor Activity | C4b Cofactor Activity | DAA (CP) |
|---|---|---|---|---|---|
| Y98A | 98 ± 4 | 61 ± 3 | 100 ± 4 | 50 ± 2 | 60 ± 8 |
| Y103A | 96 ± 5 | 99 ± 3 | 10 ± 1 | 96 ± 3 | 10 ± 1 |
| K108A | 184 ± 18 | 180 ± 19 | 119 ± 12 | 112 ± 13 | 172 ± 17 |
Data are presented as percentage activity of wild-type SPICE (mutants and SPICE expressed in CHO cells). C3b/C4b binding and cofactor assay conditions are described in Table I. For DAA for the classical pathway C3 convertase, supernatants of SPICE or mutant (20 ng) were incubated 10 min with prepared sheep erythrocytes. Guinea pig serum was then added for 30 min. DAA is described relative to SPICE and represents percentage of inhibition as calculated from comparisons to conditions without added inhibitor. Data represent means ± SEM for three or four experiments.
Two SPICE amino acids substituted into VCP CCP2 enhance regulation of human complement
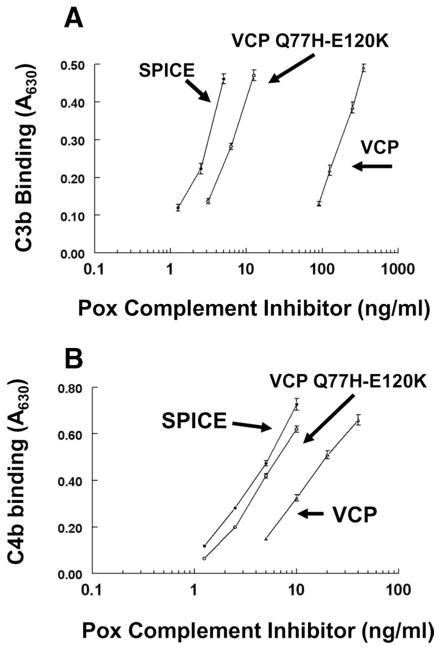
As noted, SPICE differs from VCP by 11 aa. We constructed SPICE by stepwise substitution of two to three residues at a time into VCP. As a result, chimeric constructs were developed bearing combinations of SPICE residues onto the backbone of VCP. The chimeras were transfected into CHO cells, and the supernatant was quantified and functionally evaluated. As described below, one chimeric construct in CCP2, with only two SPICE amino acids (VCP Q77H-E120K), was particularly informative relative to SPICE structure and function (Figs. 2C and 5).
FIGURE 5.
C3b and C4b binding by a VCP chimeric mutant. A VCP construct carrying two SPICE residues (VCP Q77H-E120K), as well as SPICE and VCP, were transiently expressed in CHO cells. Supernatants were quantified and then assessed for ligand binding by ELISA as per Table I. A, C3b binding and (B) C4b binding. Representative experiment of three. Data represent the means ± SEM. Also, see Table III.
C3b and C4b binding
As expected, SPICE bound C3b ~60-fold more efficiently than VCP (Fig. 5A) (13, 19, 27, 39). Unexpectedly, this chimeric mutant bearing only 2 of the 11 residues of SPICE enhanced C3b binding capability of VCP ~30-fold (Fig. 5A). This finding is consistent with a previous report that single amino acid substitutions into VCP of these residues (VCP-Q77H or VCP-E120K) augmented complement regulatory activity (27).
C4b binding by SPICE, VCP, and this mutant were also assessed by ELISA (Fig. 5B). In contrast to C3b binding, SPICE bound human C4b 4-fold more efficiently than VCP, consistent with previous results (13, 18, 19, 27, 39). Again, the functional profile of the chimeric construct was more similar to SPICE, showing ~4-fold enhanced activity over VCP. These ligand binding results suggest that evolutionary forces impacting SPICE, compared with VCP, primarily created a more efficient interaction with human C3b. These results also reinforce the key role of CCP2 in C3b binding.
CA for C3b and C4b
We next assessed C3b and C4b CA of this mutant (Table III). Similar to our previous data, SPICE was ~400-fold more potent as a cofactor for C3b than was VCP (13). Consistent with the C3b binding data, the chimera showed ~200-fold increased activity over parental VCP, indicating again the key role of these two amino acids in CCP2 for enhancing SPICE regulatory activity for C3b. These data agree with and extend the observations of Sfyroera et al. (27) and Yadav et al. (39), who individually added these SPICE residues to VCP and both mutants enhanced C3b binding and decreased C3b deposition in ELISA-based assays. These data again illustrate that ligand binding and CA are separable functions. Also, despite only a 4-fold increase in C4b binding (Fig. 5B), C4b CA was enhanced ~200-fold. Overall, these results point to an increase in CA as being key to the increased virulence of SPICE vs VCP for regulating human complement.
Table III.
Cofactor activity and DAA of VCP-SPICE chimera (VCP-77H/120K)a
| Protein | C3b Cofactor Activity (ng)b | C4b Cofactor Activity (ng)b | DAA (CP) (% SPICE)c |
|---|---|---|---|
| SPICE | 25 ± 4 | 25 ± 6 | 100 ± 3 |
| VCP | 10,000 ± 100 | 10,000 ± 110 | 14 ± 2 |
| VCP Q77H-E120Kd | 50 ± 7 | 50 ± 8 | 79 ± 8 |
SPICE, VCP, and VCP Q77H-E120K (see Figs. 2C and 5) were expressed in CHO cells. Mean ± SEM of three or four experiments.
C3b/C4b cofactor method per Table I, except these data represent nanograms needed to cleave 40–60% of the biotinylated ligand in the presence of factor I.
DAA methods per Table II (% SPICE activity).
VCP Q77H-E120K is VCP carrying SPICE residues 77H and 120K.
DAA for the classical pathway
Consistent with the more SPICE-like activity seen for both binding and cofactor assays, DAA (Table III) for the C3 convertase of the classical pathway was also enhanced. The two SPICE amino acids demonstrated ~5-fold greater activity relative to VCP and approached wild-type SPICE activity. These data suggest that the two SPICE amino acids share overlapping regulatory activity.
Electrophoretic mobility of VCP and SPICE influenced by amino acid residue 131
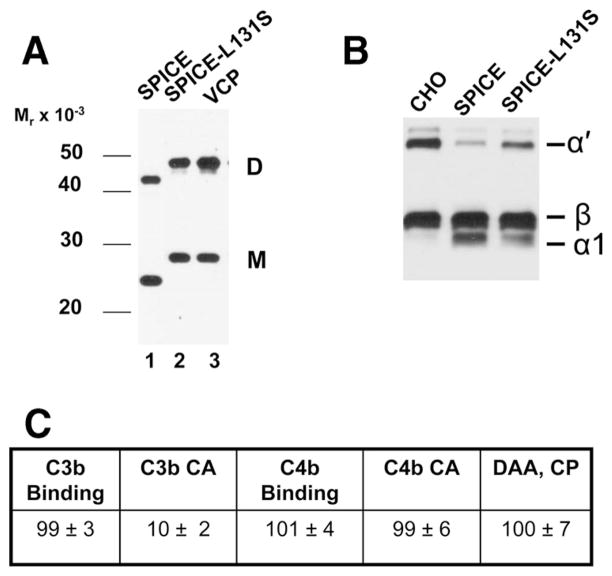
Although VCP and SPICE each contain 244 residues with a similar mass (VCP molecular mass 26,745 Da; SPICE molecular mass 26,863 Da), SPICE migrates with a faster Mr on gels (nonreducing SDS-PAGE) (13, 19). Both monomers and dimers show this pattern (13). In the process of constructing SPICE from VCP, we noted that SPICE-VCP chimeras with the VCP residue at 131 (serine) migrated with a Mr similar to VCP, while those containing the SPICE residue (leucine) migrated with a Mr similar to SPICE (not shown). Consequently, we substituted the VCP residue into SPICE (mutant SPICE L131S, Fig. 2C). Western blot analysis of the transfectant media revealed that monomers or dimers of SPICE L131S now migrated with a Mr similar to VCP (Fig. 6A). Thus, the serine to leucine change at residue 131 is responsible for the Mr difference between these two proteins.
FIGURE 6.
SPICE residue L131 influences electrophoretic mobility and function. A, Western blot of SPICE substituted with a serine residue of VCP (i.e., L131S) and expressed in CHO cells. Ten percent nonreducing SDS-PAGE of supernatants. SPICE substituted with the single VCP residue (i.e., L131S) (lane 2) now has an identical Mr to that of VCP. B and C, Functional analyses of SPICE-L131S. CHO in B represents control CHO media. Assays (as described in Table I) were performed for CA for C3b (B and C) and C4b (C), ligand binding (C), and classical pathway decay acceleration for the classical pathway C3 convertase (C). Data in C represent means ± SEM of three to four experiments.
We next assessed the functional impact of this mutation (Fig. 6, B and C). SPICE L131S demonstrated a specific loss (~90%) of C3b CA (Fig. 6, B and C). In contrast, it retained an activity profile similar to wild-type SPICE with respect to C3b and C4b binding, C4b CA, and classical pathway DAA (Fig. 6C). We conclude that the SPICE residue L131 produces a conformational change that specifically enhances CA against human C3b. This is consistent with the greater CA of SPICE as compared with VCP (13, 19, 27, 39) and that single amino acids can modulate distinct complement inhibitory activities (Table II). Thus, alteration of a single amino acid in SPICE nearly abrogates its ability to serve as an efficient cofactor while not interfering with its binding to C3b. This finding suggests that L131 in SPICE is a site for interaction with factor I.
mAb to SPICE blocks regulatory activity
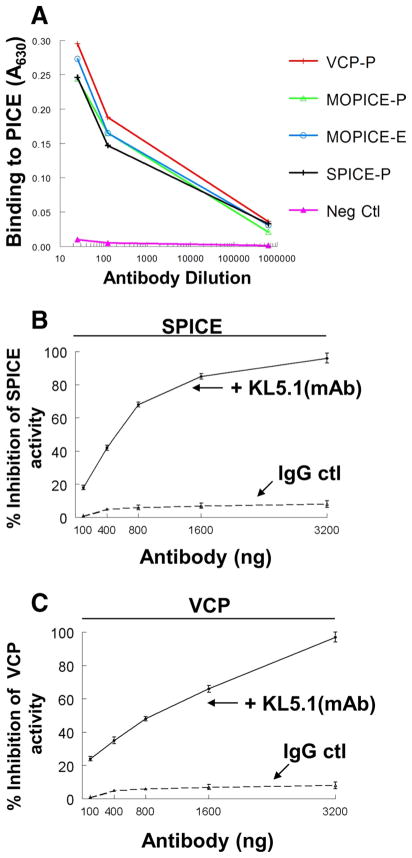
In addition to further defining the active sites in SPICE, we desired to develop mAbs that block SPICE activity. To obtain a function-blocking mAb, we employed an ELISA to select hybridoma supernatants that could competitively inhibit SPICE binding to human C3b (see Materials and Methods). We cloned a mAb KL5.1 that bound SPICE, MOPICE, and VCP equivalently across a range of dilutions (Fig. 7A). Additionally, the Ab bound similarly if the poxviral protein was produced in mammalian, yeast, or an Escherichia coli expression system (Fig. 7A and not shown).
FIGURE 7.
mAb KL5.1 binds to SPICE, MOPICE, and VCP and blocks complement regulatory activity. A, The PICE proteins were adsorbed to a microtiter plate and dilutions of Ab added. Detection was with an HRP-linked polyclonal anti-mouse gamma globulin. “P” or “E” designations refer to whether the protein was produced in Pichia pastoris or E. coli expression systems, respectively. Shown is a representative experiment from three. B, mAb KL5.1 blocks classical pathway C3 convertase DAA of SPICE. SPICE activity was set at 100%. C, KL5.1 blocks classical pathway C3 convertase DAA of VCP. Assay conditions as in Table I using 20 ng of SPICE or 200 ng of VCP. Representative experiments of three for each set of conditions. Data represent the means ± SEM.
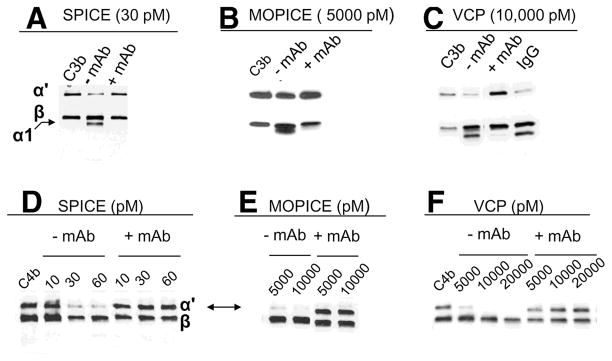
We evaluated the ability of mAb KL5.1 to inhibit DAA of SPICE and VCP (MOPICE lacks this function) (13). KL5.1 inhibited DAA of SPICE and VCP in a dose-dependent manner (Fig. 7, B and C). We next determined if the mAb could block cofactor function of SPICE, VCP, and MOPICE. In C3b and C4b cofactor assays, mAb KL5.1 blocked cleavage mediated by each of the three PICES (Fig. 8). Thus, mAb KL5.1 inhibits both CA and DAA.
FIGURE 8.
mAb KL5.1 blocks CA of SPICE, MOPICE, and VCP. Method per Table I and as indicated, except 30 μg/ml mAb or IgG control was preincubated 20 min before addition of C3b or C4b. C3b cofactor assays for (A) SPICE, (B) MOPICE, and (C) VCP. The C3b control (no added cofactor) is shown in the first lane. The α1 cleavage fragment is indicated. D–F, Dose-dependent blocking of C4b CA by the mAb is shown for SPICE (D), MOPICE (E), and VCP (F). Concentration (pM) of PICES is indicated. The C4b control (no added cofactor) is shown in the first lane (D and F).
We subsequently successfully employed KL5.1 as a capture mAb for quantification by ELISA and as a detection mAb in Western blots (not shown). The former allowed a detection range from 0.1 to 2 ng/ml for VCP and SPICE (not shown). In Western blotting analysis, the mAb reacted with the three PICES and mutants, except for the HS3 and HS1-2-3 mutants, suggesting that HS3 (K64-R65-R66) comprises part of the epitope for the mAb. Since this mAb was selected based on blocking of C3b binding to PICES, its ability to inhibit function thus is consistent with the selection strategy. Furthermore, these data indicate a contribution to C3b and C4b binding by the peptide in HS3 (K64-R65-R66), a linker area connecting CCPs 1 and 2.
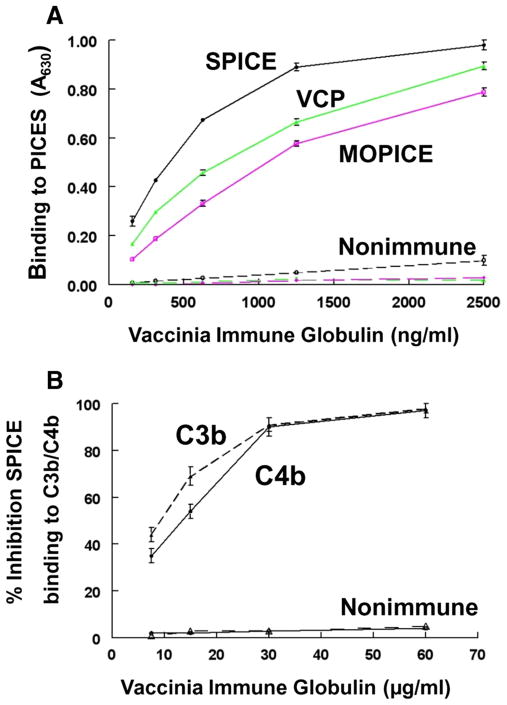
Since this function-blocking mAb to SPICE was generated in mice, we next asked if the human immune response that arises from smallpox vaccination generates Abs that bind to SPICE and inhibit SPICE binding to C3b and C4b. We utilized the Vaccinia Immune Globulin (VIG) produced for the Centers for Disease Control to support treatment of adverse events resulting from smallpox vaccination (42). SPICE (as well as VCP and MOPICE) were coated on microtiter wells and binding by VIG or nonimmune control IgG was assessed. VIG, but not control IgG, bound to all PICES in a dose-dependent fashion (Fig. 9A). This demonstrates the presence of reactive Abs not only to SPICE and VCP, but also a population of cross-reacting Abs to the monkeypox complement inhibitor. Next, a competitive inhibition assay was performed in an ELISA format in which SPICE was preincubated with VIG dilutions before addition to C3b- or C4b-coated wells (Fig. 9B). VIG, but not the IgG control, inhibited the binding by SPICE to both C3b and C4b in a dose-dependent manner. Thus, smallpox vaccination produces Abs that bind to SPICE and block interaction of SPICE with C3b and C4b.
FIGURE 9.
Presence of Abs to PICES and function-blocking Abs to SPICE in VIG. A, Recombinant SPICE, VCP, and MOPICE expressed in E. coli were coated onto microtiter plates (5 μg/ml) and dilutions of VIG or nonimmune IgG from unvaccinated individuals was assessed for binding. Detection with HRP-linked donkey anti-globulin. Data represent the means ± SEM for three experiments. B, SPICE supernatant (10 ng/ml) expressed by CHO cells was preincubated with varying concentrations of VIG (▲, ●) or control IgG (△, ○) for 30 min at 37°C then incubated on wells coated with C3b (△, ▲) or C4b (○, ●) for 1 h at 37°C. Detection of SPICE binding was by rabbit Ab and HRP anti-globulin. Percentage inhibition obtained by comparing binding to non-IgG control. Representative experiment of three. Data represent the means ± SEM.
Discussion
The present study represents a continuation of our investigations into the structure-function relationships of SPICE (13, 34). In this report we focus on heparin binding sites, amino acid substitutions that alter biologic activity, and a mAb that abrogates regulatory function.
Heparin binding sites
The binding to heparin and related GAGs by SPICE is a functional attribute that provides a mechanism for a secreted virulence factor to anchor to host tissues, the microbe itself, and infected cells (35). This scenario is analogous to that of factor H, a major regulator of the alternative pathway (reviewed in Ref. 43). Heparin binding allows factor H to modulate complement activation at sites of tissue damage and SPICE to inhibit complement activation at sites of infection (43).
Our studies point out that all three heparin binding sites in CCP1of SPICE impact its ability to attach to GAGs. Their elimination reduced, but did not abrogate, the ability of SPICE to bind to heparin. This suggests that the overall positive charge may be involved in SPICE binding to heparin or that there are other unidentified binding sites. Furthermore, there may be other mechanisms by which PICES bind to cells. For example, Girgis et al. demonstrated that VCP may covalently interact via disulfide bond formation with viral membrane protein A56 to attach to the surfaces of infected cells (44).
The crystal structure and models of SPICE’s homolog in vaccinia, VCP, indicates that it is an extended molecule that binds heparin via clusters of positive charges (27, 32, 33, 45, 46). The amino acid sequence of VCP predicts four putative heparin binding sites with the motif K/R-X-K/R (31). Three of these heparin binding sites are located in CCP1 of SPICE, VCP (Fig. 10A), and other PICES (13, 31). Additionally, the overall positive charge of VCP could contribute to heparin binding (31). Only two of these motifs, one in CCP4 (see Fig. 1) and a second in the linker between CCP1 and 2 (K64-R65-R66), were considered likely heparin binding sites (32). The crystallographic structure of VCP complexed with heparin suggested that only the CCP4 site bound heparin (33). SPICE and VCP have the identical putative heparin binding sequences in CCP1, as does MOPICE, the monkeypox homolog. However, SPICE and MOPICE do not have this site in CCP4, yet in our studies, VCP, SPICE, and MOPICE bound nearly equivalently to heparin (13). Taken together, these results suggest that the GAG binding site in CCP4 is unlikely to be the only one utilized.
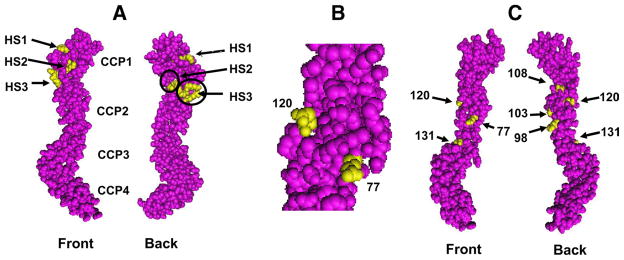
FIGURE 10.
Modeling of SPICE based on the crystal structure of VCP. A, Three heparin binding sites of VCP are highlighted in yellow. VCP and SPICE have identical CCP1 sequence. Front and back views and approximate areas for CCP1–4 are indicated. B, Side view of VCP residues 120 and 77, showing proximity of each. C, Additional mutations of SPICE prepared in this report are modeled onto VCP (yellow residues) (see Fig. 2 for residues).
The heparin binding and complement regulatory sites of SPICE appear to overlap. These data are consistent with structural studies of VCP in complex with heparin (33) and overlapping sites in human regulators (32, 40, 43, 47–49). Mutation of each heparin binding site decreased C3b CA to ~50% of the activity of parental SPICE. Likewise, alteration of the first heparin binding site reduced C4b binding and CA by ~50%. Complement binding and heparin binding sites also overlap in the plasma complement regulator, factor H. For example, in factor H CCP7 and the most carboxyl-terminal domain, CCP20, appear to carry heparin and C3d binding sites (43, 49). These data relate to clinical issues since polymorphisms of the heparin/C3b/C3d binding sites of factor H are associated with age-related macular degeneration (50–52) and with atypical hemolytic uremic syndrome (50, 53–55).
Two amino acid changes in VCP create SPICE-like activity
SPICE inactivates human complement more efficiently than does VCP or MOPICE (13, 18, 19, 27). Substitutions of two amino acids of SPICE (H77 and K120) were sufficient to convert VCP to SPICE-like inhibitory activity for human complement. This chimeric VCP construct substituted with only two SPICE amino acids enhanced the C3b and C4b CA of VCP ~200-fold. This finding is consistent with that of Sfyroera et al., who constructed two separate point mutations (VCP-Q77H and VCP-E120K) and each enhanced C3b binding and alternative pathway inhibition (27). Also, our prior studies on the corresponding amino acids in MCP (K119) and in CR1 (K1016), located in homologous positions to SPICE (K120), demonstrated the importance of this charged residue for C3b/C4b binding and/or CA (40, 46). Because VCP substituted with 77H and 120K enhanced C3b binding by ~30-fold, but demonstrated a 200-fold increase in CA, one or both of these amino acids likely comprise part of a site for an interaction with factor I. These two residues lie on the surface of the molecule in close proximity (Fig. 10B).
Homologous peptide in SPICE and MCP
A peptide in SPICE CCP2 is highly homologous to MCP with 7 identical amino acids from position 96 to 102 and 9 of 10 identical amino acids from position 93 to 102 (Fig. 4). In MCP, this is a site for C4b regulation as well as measles virus binding (40, 56). In SPICE this peptide houses 3 of the 11 aa differences compared with VCP (Y98, Y103, and K108). These three residues lie on the same face of VCP (Fig. 10C). An alanine substitution indicated that Y98A primarily altered C4b interactions and that Y103A impacted predominantly C3b CA. Surprisingly, K108A showed a gain of function for ligand binding and DAA. This finding, however, is consistent with those of Sfyroera et al. (27) and Yadav et al. (39), who both showed that VCP substituted with the corresponding SPICE amino acid at this residue (VCP-E108K) enhanced regulatory activity. These data also emphasize the critical role played by this peptide in mediating regulatory activity. Furthermore, unaltered C3b binding, but a loss of C3b CA, points to this being a site for a factor I interaction. Loss of DAA by this residue (Y103A) also suggests a role for this site in inhibiting the C3 convertase. Overall, these data further demonstrate how alteration of a single residue may cause a highly specific change in the functional repertoire of a complement regulatory protein. Furthermore, these mutants confirm that ligand binding, CA, and DAA are distinct and separable regulatory activities, a lesson also learned from human regulators (40, 41, 57, 58).
SPICE residue L131 accounts for its electrophoretic difference with VCP
The importance of individual SPICE residues for structure and function is further illustrated by SPICE residue L131. Mutation to a serine (homologous amino acid in VCP) changes the Mr of SPICE to that of VCP and selectively abrogates C3b CA. This alteration in Mr again demonstrates how a structural alteration in SPICE correlates with an effect on structure and function. Interestingly, L131 lies in a homologous region in MCP that is functionally important (40).
A function-blocking anti-SPICE mAb
The strategy of selecting hybridomas that competitively inhibit SPICE binding to human ligands C3b and C4b led to the identification of mAb KL5.1 that inhibits the complement regulatory activity of SPICE, VCP, and MOPICE. Its antigenic binding site maps to CCP1 and is influenced by residues in the third heparin binding motif (K64-R65-R66).
Poxviral complement regulators are attractive targets for therapeutic intervention. VCP inhibits Ab-dependent, complement-enhanced neutralization of vaccinia virus virions (25). Novel mAbs to VCP inhibited complement-enhanced neutralization of vaccinia virus virions (59). Animal studies demonstrate that viruses lacking VCP are attenuated (24, 25). Additionally, mice deficient in complement components (C3, C4, factor B) have increased mortality to ectromelia infection (26). Coupled with our demonstration of the presence of SPICE function-blocking Abs in VIG, PICES likely attenuate host complement activation and thus become a target in the treatment of poxviral infections (59–61). However, other mechanisms likely play important roles in controlling responses against smallpox. It was recently shown that unimmunized BALB/c mice depleted of CD4 and CD8 effector T cells did not survive infection by pathogenic vaccinia virus (62). In contrast, the same dose was sublethal in intact unimmunized mice. These data demonstrate that antigenic-specific CD4 and CD8 cells prevented mortality of infected immunized animals, and that T cell immunity was critical for natural resistance to vaccinia virus in unimmunized animals (62).
Evolutionary history of PICES
Human disease caused by variola virus has been reported in the human population for >2000 years. New studies suggest an even older origin, possibly stemming from African rodents 16,000–68,000 years ago (63). A proposal for the evolutionary history of SPICE and its homologous virulence factors are considered below. Initially, a prototype complement inhibitor, probably consisting of four CCP repeats, was hijacked from a mammalian source and inserted in the viral genome (20, 22, 64). Next, structural changes were introduced that enhanced function in general and for a specific host’s complement system (see below). The originally captured gene could have encoded a secreted protein, specifically a forerunner of factor H or C4b binding protein that possessed heparin binding sites. Another possibility is that an MCP/DAF-like membrane protein, which consisted of four CCP repeats, was captured and then modified to become a secreted protein. For example, a peptide (residues 93–102) in SPICE CCP2 bears a strong homology to MCP (90%) (40). A secreted protein has several advantages over a membrane-bound inhibitor. It, of course, would not require synthesis of a hydrophobic transmembrane or GPI anchor and would be able to function in solution within the local infectious environment. Furthermore, a secreted protein with GAG binding ability could attach to the virus and virally infected cells as well as cells at the local site of infection to protect them against complement attack. The outcome of this virulence activity profile is a less efficacious local inflammatory response and reduced destruction of the virus and virally infected cells; in other words, a means to delay and subvert the innate immune response of the host.
In addition to possibly being converted to a secreted factor, four types of structural modifications are hypothesized to have taken place to increase virulence (Fig. 1). The first is the addition of an “extra” cysteine at the amino terminus and a free cysteine at the carboxyl terminus in the case of the truncated MOPICE (13, 18). This “extra” cysteine allows for 5–25% of SPICE, VICE (vaccinia inhibitor of complement enzymes), MOPICE, and EMICE to be secreted as a disulfide-linked homodimer (Ref. 13 and J. P. Atkinson, unpublished observations). This dimeric protein is 10- to 20-fold more efficient than the monomeric species at inhibiting complement activation (13). The dimerization advantage also has been utilized by mammalian complement inhibitors. Thus, DAF-2 is a covalently linked homodimer (albeit nondisulfide) of DAF-1 (65). The orangutan expresses ~50% of its DAF on erythrocytes as a covalently linked homodimer (66). Complement receptor type 1 (CR1, CD35) consists of multiple highly homologous C3b/C4b binding sites, each with complement regulatory activity. Two such sites, tandemly arranged, are required to efficiently inhibit C5 convertases and are more efficient at inhibiting C3 convertases (41, 67).
A second modification is the introduction of amino acid changes in key peptides housing complement ligand binding and regulatory sites. These amino acid substitutions increase the efficiency of, for example, SPICE vs VCP in inhibiting human complement (13, 19, 27, 39). These studies demonstrate how changing a few amino acids between VCP and SPICE results in up to a 1000-fold greater activity of SPICE in inhibiting human complement.
Third, there appears to have been a refinement of the heparin (anionic binding sites) of the pox inhibitors. Particularly striking is the 100% homology of the three heparin interactive sites in CCP1 of SPICE, VICE, MOPICE, EMICE, and IMP (the cowpox complement inhibitor, CPV-GRI strain). This implies that the number of sites and their spacing within CCP1 are evolutionarily conserved to facilitate binding to mammalian GAGs and related structures. The desired functional outcome is more efficient binding to the virus itself, virally infected cells, and uninfected host cells.
Fourth, more specific regulatory activity is evolving; namely, more potent CA for C3b and C4b and DAA for the classical pathway is being enhanced, while DAA for the alternative pathway is decreasing (20). This “subspecialization” of inhibitory activity is somewhat puzzling because of the importance of the alternative pathway and its amplification loop in innate immunity. However, as demonstrated by human regulators (especially MCP), CA is capable by itself of inhibiting alternative pathway amplification, independent of DAA (68, 69). Furthermore, CA represents a “permanent fix” (i.e., inactivation of C3b or C4b), while DAA is only a “temporary fix”. Another issue that could be driving these functional changes might relate to inhibiting complement activation early in an infection, especially if the host expresses complement-activating natural Abs or lectins that recognize viral particles (26).
In summary, dimerization, multiple cellular attachment schemes, function-enhancing amino acid substitutions, and refinement of the heparin binding sites have transformed SPICE into an exceptionally efficacious inhibitor of the human complement system.
Acknowledgments
We thank Elizabeth Moulton and Steven Lawrence for helpful discussions, Madonna Bogacki for administrative assistance, and Christine Anderson for providing Vaccinia Immune Globulin.
Footnotes
This work was supported by National Institutes of Health Grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research.
Abbreviations used in this paper: PICES, poxviral inhibitors of complement enzymes; CA, cofactor activity; CCP, complement control protein; CHO, Chinese hamster ovary (cells); DAA, decay-accelerating activity; DAF, decay-accelerating factor; GAG, glycosaminoglycan; MCP, membrane cofactor protein (CD46); MOPICE, monkeypox inhibitor of complement enzymes; SPICE, smallpox inhibitor of complement enzymes; VICE, vaccinia inhibitor of complement enzymes; VIG, Vaccinia Immune Globulin; VCP, vaccinia complement control protein.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Parker S, Nuara A, Buller RM, Schultz DA. Human monkeypox: an emerging zoonotic disease. Future Microbiol. 2007;2:17–34. doi: 10.2217/17460913.2.1.17. [DOI] [PubMed] [Google Scholar]
- 2.Fauci AS, Challberg MD. Host-based antipoxvirus therapeutic strategies: turning the tables. J Clin Invest. 2005;115:231–233. doi: 10.1172/JCI24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson DA. The looming threat of bioterrorism. Science. 1999;283:1279–1282. doi: 10.1126/science.283.5406.1279. [DOI] [PubMed] [Google Scholar]
- 4.Weiner DB. Keeping the lock on smallpox. Nat Med. 2003;9:1115–1116. doi: 10.1038/nm0903-1115. [DOI] [PubMed] [Google Scholar]
- 5.Bozzette SA, Boer R, Bhatnagar V, Brower JL, Keeler EB, Morton SC, Stoto MA. A model for a smallpox-vaccination policy. N Engl J Med. 2003;348:416–425. doi: 10.1056/NEJMsa025075. [DOI] [PubMed] [Google Scholar]
- 6.Lane JM, Goldstein J. Adverse events occurring after smallpox vaccination. Semin Pediatr Infect Dis. 2003;14:189–195. doi: 10.1016/s1045-1870(03)00032-3. [DOI] [PubMed] [Google Scholar]
- 7.Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, Wegner MV, Kazmierczak JJ, Stratman EJ, Li Y, Fairley JA, et al. The detection of monkeypox in humans in the Western Hemisphere (see comment) N Engl J Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- 8.Hammarlund E, Lewis MW, Carter SV, Amanna I, Hansen SG, Strelow LI, Wong SW, Yoshihara P, Hanifin JM, Slifka MK. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat Med. 2005;11:1005–1011. doi: 10.1038/nm1273. [DOI] [PubMed] [Google Scholar]
- 9.Chen N, Li G, Liszewski MK, Atkinson JP, Jahrling PB, Feng Z, Schriewer J, Buck C, Wang C, Lefkowitz EJ, et al. Virulence differences between monkeypox virus isolates from West Africa and the Congo Basin. J Virol. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Likos AM, Sammons SA, Olson VA, Frace AM, Li Y, Olsen-Rasmussen M, Davidson W, Galloway R, Khristova ML, Reynolds MG, et al. A tale of two clades: monkeypox viruses. J Gen Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- 11.Babiuk S, Bowden TR, Boyle DB, Wallace DB, Kitching RP. Capripoxviruses: an emerging worldwide threat to sheep, goats and cattle. Transbound Emerg Dis. 2008;55:263–272. doi: 10.1111/j.1865-1682.2008.01043.x. [DOI] [PubMed] [Google Scholar]
- 12.Harrison SC, Alberts B, Ehrenfeld E, Enquist L, Fineberg H, McKnight SL, Moss B, O’Donnell M, Ploegh H, Schmid SL, et al. Discovery of antivirals against smallpox. Proc Natl Acad Sci USA. 2004;101:11178–11192. doi: 10.1073/pnas.0403600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liszewski MK, Leung MK, Hauhart R, Buller RM, Bertram P, Wang X, Rosengard AM, Kotwal GJ, Atkinson JP. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J Immunol. 2006;176:3725–3734. doi: 10.4049/jimmunol.176.6.3725. [DOI] [PubMed] [Google Scholar]
- 14.Kotwal GJ. Poxviral mimicry of complement and chemokine system components: what’s the end game? Immunol Today. 2000;21:242–248. doi: 10.1016/s0167-5699(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 15.Mullick J, Kadam A, Sahu A. Herpes and pox viral complement control proteins: “the mask of self”. Trends Immunol. 2003;24:500–507. doi: 10.1016/s1471-4906(03)00207-2. [DOI] [PubMed] [Google Scholar]
- 16.Dunlop LR, Oehlberg KA, Reid JJ, Avci D, Rosengard AM. Variola virus immune evasion proteins. Microbes Infect. 2003;5:1049–1056. doi: 10.1016/s1286-4579(03)00194-1. [DOI] [PubMed] [Google Scholar]
- 17.Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 18.Uvarova EA, Shchelkunov SN. Species-specific differences in the structure of orthopoxvirus complement-binding protein. Virus Res. 2001;81:39–45. doi: 10.1016/S0168-1702(01)00332-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosengard AM, Liu Y, Nie Z, Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc Natl Acad Sci USA. 2002;99:8808–8813. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciulla E, Emery A, Konz D, Krushkal J. Evolutionary history of orthopoxvirus proteins similar to human complement regulators. Gene. 2005;355:40–47. doi: 10.1016/j.gene.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarian SH, McFadden G. Immune evasion by poxviruses. Future Virol. 2006;1:123–132. [Google Scholar]
- 22.Babkin IV, Shchelkunov SN. Molecular evolution of poxviruses (in Russian) Russ J Genet. 2008;44:895–908. [Google Scholar]
- 23.Kotwal GJ, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 24.Kotwal GJ, Isaacs SN, McKenzie R, Frank MM, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs SN, Kotwal GJ, Moss B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc Natl Acad Sci USA. 1992;89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulton EA, Atkinson JP, Buller RM. Surviving mousepox infection requires the complement system. PLoS Pathog. 2008;4:1–14. doi: 10.1371/journal.ppat.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sfyroera G, Katragadda M, Morikis D, Isaacs SN, Lambris JD. Electrostatic modeling predicts the activities of orthopoxvirus complement control proteins. J Immunol. 2005;174:2143–2151. doi: 10.4049/jimmunol.174.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Favoreel HW, Van de Walle GR, Nauwynck HJ, Pensaert MB. Virus complement evasion strategies. J Gen Virol. 2003;84:1–15. doi: 10.1099/vir.0.18709-0. [DOI] [PubMed] [Google Scholar]
- 29.Liszewski MK, Kemper C, Price JD, Atkinson JP. Emerging roles and new functions of CD46. Springer Semin Immunopathol. 2005;27:345–358. doi: 10.1007/s00281-005-0002-3. [DOI] [PubMed] [Google Scholar]
- 30.Liszewski MK, Atkinson JP. Membrane cofactor protein (CD46) and decay accelerating factor (CD55) In: Rother K, Till GO, Hansch GM, editors. The Complement System. 2. Springer; Berlin: 1998. pp. 146–162. [Google Scholar]
- 31.Smith SA, Mullin NP, Parkinson J, Shchelkunov SN, Totmenin AV, Loparev VN, Srisatjaluk R, Reynolds DN, Keeling KL, Justus DE, et al. Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J Virol. 2000;74:5659–5666. doi: 10.1128/jvi.74.12.5659-5666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murthy KH, Smith SA, Ganesh VK, Judge KW, Mullin N, Barlow PN, Ogata CM, Kotwal GJ. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell. 2001;104:301–311. doi: 10.1016/s0092-8674(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 33.Ganesh VK, Smith SA, Kotwal GJ, Murthy KHM. Structure of vaccinia complement protein in complex with heparin and potential implications for complement regulation. Proc Natl Acad Sci USA. 2004;101:8924–8929. doi: 10.1073/pnas.0400744101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Liszewski MK, Bertram P, Leung MK, Hauhart R, Zhang L, Atkinson JP. Smallpox inhibitor of complement enzymes (SPICE): regulation of complement activation on cells and mechanisms of its cellular attachment. J Immunol. 2008;181:4199–4207. doi: 10.4049/jimmunol.181.6.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blom AM, Mark L, Spiller OB. Viral heparin-binding complement inhibitors: a recurring theme. Adv Exp Med Biol. 2007;598:105–125. doi: 10.1007/978-0-387-71767-8_9. [DOI] [PubMed] [Google Scholar]
- 36.Liszewski MK, Bertram P, Leung MK, Hauhart R, Zhang L, Atkinson JP. Smallpox inhibitor of complement enzymes (SPICE): regulation of complement activation on cells and mechanism of its cellular attachment. J Immunol. 2008;181:4199–4207. doi: 10.4049/jimmunol.181.6.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosengard AM, Alonso LC, Korb LC, Baldwin WM, 3rd, Sanfilippo F, Turka LA, Ahearn JM. Functional characterization of soluble and membrane-bound forms of vaccinia virus complement control protein (VCP) Mol Immunol. 1999;36:685–697. doi: 10.1016/s0161-5890(99)00081-4. [DOI] [PubMed] [Google Scholar]
- 38.Mullick J, Bernet J, Panse Y, Hallihosur S, Singh AK, Sahu A. Identification of complement regulatory domains in vaccinia virus complement control protein. J Virol. 2005;79:12382–12393. doi: 10.1128/JVI.79.19.12382-12393.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadav VN, Pyaram K, Mullick J, Sahu A. Identification of hot spots in the variola virus complement inhibitor (SPICE) for human complement regulation. J Virol. 2008;82:3283–3294. doi: 10.1128/JVI.01935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liszewski MK, Leung M, Cui W, Bala Subramanian V, Parkinson J, Barlow PN, Manchester M, Atkinson JP. Dissecting sites important for complement regulatory activity in membrane cofactor protein (MCP; CD46) J Biol Chem. 2000;275:37692–37701. doi: 10.1074/jbc.M004650200. [DOI] [PubMed] [Google Scholar]
- 41.Krych-Goldberg M, Atkinson JP. Structure function relationships of complement receptor type 1. Immunol Rev. 2002;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- 42.Hopkins RJ, Lane JM. Clinical efficacy of intramuscular vaccinia immune globulin: a literature review. Clin Infect Dis. 2004;39:819–826. doi: 10.1086/422999. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt CQ, Herbert AP, Hocking HG, Uhrin D, Barlow PN. Translational mini-review series on complement factor H: structural and functional correlations for factor H. Clin Exp Immunol. 2008;151:14–24. doi: 10.1111/j.1365-2249.2007.03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girgis NM, Dehaven BC, Fan X, Viner KM, Shamim M, Isaacs SN. Cell surface expression of the vaccinia virus complement control protein is mediated by interaction with the viral A56 protein and protects infected cells from complement attack. J Virol. 2008;82:4205–4214. doi: 10.1128/JVI.02426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henderson CE, Bromek K, Mullin NP, Smith BO, Uhrin D, Barlow PN. Solution structure and dynamics of the central CCP module pair of a poxvirus complement control protein. J Mol Biol. 2001;307:323–339. doi: 10.1006/jmbi.2000.4477. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Morikis D. Immunophysical properties and prediction of activities for vaccinia virus complement control protein and smallpox inhibitor of complement enzymes using molecular dynamics and electrostatics. Biophys J. 2006;90:3106–3119. doi: 10.1529/biophysj.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hessing M, Vlooswijk RA, Hackeng TM, Kanters D, Bouma BN. The localization of heparin-binding fragments on human C4b-binding protein. J Immunol. 1990;144:204–208. [PubMed] [Google Scholar]
- 48.Krych M, Clemenza L, Howdeshell D, Hauhart R, Hourcade D, Atkinson JP. Analysis of the functional domains of complement receptor type 1 (C3b/C4b receptor; CD35) by substitution mutagenesis. J Biol Chem. 1994;269:13273–13278. [PubMed] [Google Scholar]
- 49.Hellwage J, Jokiranta TS, Friese MA, Wolk TU, Kampen E, Zipfel PF, Meri S. Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J Immunol. 2002;169:6935–6944. doi: 10.4049/jimmunol.169.12.6935. [DOI] [PubMed] [Google Scholar]
- 50.Prosser BE, Johnson S, Roversi P, Herbert AP, Blaum BS, Tyrrell J, Jowitt TA, Clark SJ, Tarelli E, Uhrin D, et al. Structural basis for complement factor H linked age-related macular degeneration. J Exp Med. 2007;204:2277–2283. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, Abecasis GR. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manuelin T, Hellwage J, Meri S, Capriolo J, Noris M, Heinen S, Jozsi M, Neumann HPH, Remuzzi G, Zipfel PF. Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in haemolytic uraemic syndrome. J Clin Invest. 2003;111:1181–1190. doi: 10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jozsi M, Heinen S, Hartmann A, Ostrowicz CW, Halbich S, Richter H, Kunert A, Licht C, Saunders RE, Perkins SJ, et al. Factor H and atypical hemolytic uremic syndrome: mutations in the C-terminus cause structural changes and defective recognition functions. J Am Soc Nephrol. 2006;17:170–177. doi: 10.1681/ASN.2005080868. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez-Corral P, Gonzalez-Rubio C, Rodriguez de Cordoba S, Lopez-Trascasa M. Functional analysis in serum from atypical hemolytic uremic syndrome patients reveals impaired protection of host cells associated with mutations in factor H. Mol Immunol. 2004;41:81–84. doi: 10.1016/j.molimm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Hsu EC, Dorig RE, Sarangi F, Marcil A, Iorio C, Richardson CD. Artificial mutations and natural variations in the CD46 molecules from human and monkey cells define regions important for measles virus binding. J Virol. 1997;71:6144–6154. doi: 10.1128/jvi.71.8.6144-6154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krych M, Hauhart R, Atkinson JP. Structure-function analysis of the active sites of complement receptor type 1. J Biol Chem. 1998;273:8623–8629. doi: 10.1074/jbc.273.15.8623. [DOI] [PubMed] [Google Scholar]
- 58.Richards A, Liszewski MK, Kavanagh D, Fang CJ, Moulton EA, Fremeaux-Bacchi V, Remuzzi G, Noris M, Goodship THJ, Atkinson JP. Implications of the initial mutations in membrane cofactor protein (MCP; CD46) leading to atypical hemolytic uremic syndrome. Mol Immunol. 2007;44:111–122. doi: 10.1016/j.molimm.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Isaacs SN, Argyropoulos E, Sfyroera G, Mohammad S, Lambris JD. Restoration of complement-enhanced neutralization of vaccinia virus virions by novel monoclonal antibodies raised against the vaccinia virus complement control protein. J Virol. 2003;77:8256–8262. doi: 10.1128/JVI.77.15.8256-8262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benhnia MR, McCausland MM, Moyron J, Laudenslager J, Granger S, Rickert S, Koriazova L, Kubo R, Kato S, Crotty S. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J Virol. 2009;83:1201–1215. doi: 10.1128/JVI.01797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benhnia MR, McCausland MM, Su HP, Singh K, Hoffmann J, Davies DH, Felgner PL, Head S, Sette A, Garboczi DN, Crotty S. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J Virol. 2008;82:3751–3768. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belyakov IM, Earl P, Dzutsev A, Kuznetsov VA, Lemon M, Wyatt LS, Snyder JT, Ahlers JD, Franchini G, Moss B, Berzofsky JA. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc Natl Acad Sci USA. 2003;100:9458–9463. doi: 10.1073/pnas.1233578100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci USA. 2007;104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nickells MW, Alvarez JI, Lublin DM, Atkinson JP. Characterization of DAF-2, a high molecular weight form of decay-accelerating factor (DAF; CD55), as a covalently cross-linked dimer of DAF-1. J Immunol. 1994;152:676–685. [PubMed] [Google Scholar]
- 66.Nickells MW, Atkinson JP. Characterization of CR1- and membrane cofactor protein-like proteins of two primates. J Immunol. 1990;144:4262–4268. [PubMed] [Google Scholar]
- 67.Krych-Goldberg M, Hauhart RE, Porzukowiak T, Atkinson JP. Synergy between two active sites of human complement receptor type 1 (CR1; CD35) in complement regulation: implications for the structure of the classical pathway C3 convertase and generation of more potent inhibitors. J Immunol. 2005;175:4528–4535. doi: 10.4049/jimmunol.175.7.4528. [DOI] [PubMed] [Google Scholar]
- 68.Barilla-LaBarca ML, Liszewski MK, Lambris JD, Hourcade D, Atkinson JP. Role of membrane cofactor protein (CD46) in regulation of C4b and C3b deposited on cells. J Immunol. 2002;168:6298–6304. doi: 10.4049/jimmunol.168.12.6298. [DOI] [PubMed] [Google Scholar]
- 69.Liszewski MK, Fang CJ, Atkinson JP. Inhibiting complement activation on cells at the step of C3 cleavage. Vaccine. 2008;26(Suppl 8):122–127. doi: 10.1016/j.vaccine.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]