Abstract
Why we sleep remains one of the enduring unanswered questions in biology. At its core, sleep can be defined behaviorally as a homeostatically regulated state of reduced movement and sensory responsiveness. The cornerstone of sleep studies in terrestrial mammals, including humans, has been the measurement of coordinated changes in brain activity during sleep measured using the electroencephalogram (EEG). Yet among a diverse set of animals, these EEG sleep traits can vary widely and, in some cases, are absent, raising questions as to whether they define a universal, or even essential, feature of sleep. Over the past decade, behaviorally defined sleep-like states have been identified in a series of genetic model organisms, including fish, flies and worms. Genetic analyses in these systems are revealing a remarkable conservation in the underlying mechanisms controlling sleep behavior. Taken together, these studies suggest an ancient origin for sleep and raise the possibility that model organism genetics may reveal the molecular mechanisms that guide sleep and wake.
Introduction
“Nothing in biology makes sense except in light of evolution”
—Theodosius Dobzhansky
Most carefully studied animals have been found to sleep or exhibit a sleep-like state [1,2]. Yet for the most part, while we sleep we cannot eat, mate, or protect ourselves from predation. Sleep deprivation can cause an irresistible drive to sleep. Rats chronically deprived of sleep by the ‘disk over water’ method die in about the same amount of time they would die in the absence of food [3]. Flies have also been shown to die when deprived of sleep [4]. While we understand the need to eat, we still do not understand how sleep contributes to survival.
To provide a framework for discussing sleep, we will first discuss the criteria that are widely accepted and appear to define features of sleep that are emblematic and/or indicate its functional significance [1,2,5–8]. The first set of criteria is behavioral and, thus, is most easily assessed and widely observed. One of the major criteria for sleep is behavioral quiescence, typically characterized by reduced motor activity. Second, elevated arousal thresholds accompany sleep. A higher intensity stimulus is required to elicit a response from a sleeping animal, compared to that required in the same animal when it is awake. The reduction in spontaneous movement as well as in arousability suggests that the sensory and motor systems of the brain are less active [9]. Third, sleep is homeostatically regulated [10]: if we lose a night of sleep, we experience an intense drive to sleep even during times when we would typically be awake. This increase in the amount or intensity of sleep is often termed a sleep rebound. This homeostatic regulation, like other homeostatic behaviors such as feeding, indicates that sleep is regulated and serves an important function. The homeostatic regulation has led to the suggestion that wakefulness may cause the accumulation of adverse changes in the brain or body, or depletion of a fuel needed to maintain wakefulness, which is sensed by a sleep homeostat that then triggers a restorative period of sleep reversing wake induced modifications.
In addition to behavioral criteria, there are electrical, pharmacological, and molecular criteria for defining sleep. These features have not yet been examined in a wide range of animals, and some show species-specific attributes. The cornerstone of sleep studies in mammals over the past century is the electroencephalogram (EEG). Several electrodes are attached to the scalp and voltage changes between electrodes are measured. These voltage changes are caused by the synchronized activity of thousands of neurons in the cerebral cortex. The more synchronous the activity, the larger the voltage change, reflecting the summed postsynaptic potentials impinging on neurons near the recording electrodes [11]. During deep slow-wave sleep, synchronous hyperpolarization of cortical neurons can occur repeatedly with a defined frequency (less than 4 Hz in humans), resulting in high amplitude ‘slow waves’. The amplitude of these slow (or delta) waves is thought to be a reflection of homeostatic drive [12].
Deep or slow-wave (non-rapid eye movement, non-REM) sleep is typically followed by rapid eye movement (REM) sleep in a repeating cycle which lasts approximately 90 minutes in humans, shorter periods in smaller animals and longer periods in larger animals [13]. The EEG in REM sleep closely resembles the waking EEG in most mammalian species, with generally low voltage activity in the neocortex. A large amplitude theta rhythm is seen in the hippocampus during REM sleep and also during certain waking states [9]. In many species, REM sleep is accompanied by rapid eye movements, and in humans vivid dream mentation is frequently reported [7]. At the neuronal level, REM sleep is characterized by high, irregular, waking-like rates of unit discharge in most brain neuronal groups. Conspicuous exceptions are the noradrenergic, serotonergic and histaminergic cell groups, which are tonically active in waking, but almost completely inactive in REM sleep. The reduced activity of these cell groups and the increased activity of certain cells containing the neuroransmitters γ-amino-butyric acid (GABA) and glycine may be responsible for the major differences between REM sleep and waking, specifically the loss of consciousness and the profound loss of muscle tone in most somatic muscles in REM sleep [9,14–20]. This loss of muscle tone prevents the expression of centrally commanded motor activity [21].
The complexity of sleep has contributed to the difficulty of its study. At least in mammals, sleep is not a unidimensional brain state. Rather, in terrestrial mammals and birds, it consists of at least two distinctly different brain states: the REM and slow wave (or non-REM) sleep states mentioned above, the latter being further subdivided into different stages. An array of brain loci, circuits and their neurotransmitters drive these various forms of sleep. Accompanying these changes in neural function are changes in hundreds, if not thousands, of genes encompassing not only parts of the brain that regulate sleep and wake behavior, but other regions as well [22–25]. Sleep changes during development, with more total sleep, especially REM sleep, being required early in life in land mammals (particularly altricial mammals [26–32]). Finally, sleep is manifest differently between species, even those that are closely related. For example, among carnivores, the domestic cat sleeps for 12.5 hours a day, while the closely related Genet sleeps for just 6.3 hours [13]. Among rodents, the Golden-mantled ground squirrel sleeps for 15.9 hours and the Degu sleeps for only 7.7 hours [13]. Among primates, the Owl monkey sleeps for 17 hours, while humans sleep for 7–8 hours [1] (Figure 1).
Figure 1.

Mammalian phylogenetic order is not strongly correlated with sleep parameters. Despite similar genetics and physiology, sleep times within mammalian orders overlap extensively. On the left are three pairs of animals that are in the same order but have very different sleep parameters. On the right are three pairs of animals from different orders with similar sleep amounts. Mammalian sleep times are not strongly correlated with phylogenetic order [1,13,33]. Photo of Eastern American Mole, courtesy Barbara L. Clauson and Robert M. Timm; the ajacent photo of J.M.S., courtesy of N.Y. Times. Photo of R.A., courtesy of Northwestern University. Photos of other animals courtesy of Wikipedia commons site.
Studies correlating sleep time with various behavioral and physiological parameters have found some small correlations between single measured or hypothesized conditions (such as sleep site safety), but have not been able to explain a substantial portion of the variance in sleep times between species [1,13,33] Of more concern, these studies have reached diametrically opposite conclusions as to the nature of correlations between sleep time and brain size or metabolic rate [13,33,34]. These differences are largely a consequence of post-hoc decisions made about whether closely related animals should be treated as individuals or as a group, which of the published sleep studies are adequate and should therefore be included in the data set, and how the data should best be handled mathematically. Despite these different assumptions and results, what all these studies have in common is that all significant correlations between physiological variables and sleep time explain only a small percentage of the variance [13,34,35]. Such correlations do not necessarily identify causal relations. Sleep in non-mammalian vertebrates differs from that in mammals [5]. Indeed, these different manifestations of sleep in different organisms have led to controversies concerning the most fundamental question in the field: what exactly is sleep and how should it be defined?
Here, we take a comparative phylogenetic approach to sleep. It is widely acknowledged that sleep is likely to be accompanied by restorative neural processes required for optimal brain function. Thus, organisms that have brains may have used sleep processes to deal with the unique requirements of neural circuits [36,37]. If true, then the puzzle of sleep might be solved by approaching simple model organisms that display sleep behaviors. Given the conservation of genomes across animal species, organisms with sequenced genomes and facile genetics present important advantages for studying the genetic underpinnings of sleep. Indeed, a number of laboratories have analysed sleep in a range of genetic model systems from zebrafish to fruit flies to nematodes. We suggest that the common elements of sleep present from worms to humans represent those properties present in their common ancestor. This common ancestry may be reflected in the shared deployment of genetic pathways important for control of sleep.
Many reviews cataloging the sleep behaviors of a variety of species have been published [2,5,6,13]. We will not repeat these analyses, but rather we will focus initially on mammalian sleep. The remarkable diversity in electroencephalographically defined sleep among mammals suggests that these aspects of sleep may have evolved relatively recently. We will then focus on three genetically tractable and widely used model organisms: zebrafish, flies and worms and what they may reveal about the ancient origins of sleep.
Diversity of Sleep among Animals
Many animals that exhibit clear sleep behaviors do not display the characteristic EEG signatures of sleep seen in mammals. For example, reptiles and amphibians have higher amplitude cortical activity during waking states than they do in quiescent states [38–40]. These findings suggest that EEG signatures are linked to the structure and function of mammalian neocortex, but are not a universal characteristic of sleep. Although a few older studies saw signs of activation during sleep, more recent and thorough studies have generally concluded that reptiles and amphibians do not have REM sleep [38–43]. A study in the turtle of neuronal activity in the brainstem regions known to generate REM sleep did not show the periodic activation pattern that underlies all of the phasic phenomena of REM sleep [44]. However, birds, which like mammals are homeotherms, have both REM and non-REM sleep [45–49].
The phenomena of sleep vary even within mammals. A consistent correlate of slow-wave sleep in humans is the release of growth hormone, particularly in younger individuals [50]; but growth hormone is not normally released during sleep in dogs [51]. In humans, arousal threshold is lowest during REM sleep, but in rats it is highest in this state [52–54]. Erections have been shown to be present during REM sleep in humans and rats [55], but the armadillo has erections only in non-REM sleep [56]. The physiological signs of REM sleep in both the platypus [57] and the related monotreme, the short nosed echidna [58], are largely restricted to the brainstem, in contrast to their propagation to the forebrain in adult placental and marsupial mammals.
The standard criteria defining sleep have even been modified for descriptions of ‘sleep’ in marine mammals. Unlike land mammals, marine mammals can sleep with one half of the brain at a time, and it has been said that they can swim while sleeping. Two unusual forms of this behavior have been described: sleep in otariids, such as the fur seal and the harbor seal; and sleep in cetaceans such as the bottlenose dolphin and the beluga whale.
On land, sleep in the fur seal resembles that in most terrestrial mammals: the EEG is bilaterally synchronized, and the animal closes both eyes, appears unresponsive and cycles between REM and slow wave sleep. In contrast, when the fur seal is in the water, it shows slow waves in one hemisphere, with the contralateral eye frequently being closed and the contralateral flipper immobile. The other eye is generally open or partially open and the other flipper is active in maintaining the animal’s position in the water [59,60]. So it appears that half of the brain and body are ‘asleep’ and the other half ‘awake’ by both EEG and behavioral criteria. REM sleep time is greatly reduced in the water and no rebound of lost REM sleep is seen when the fur seal returns to land, even after several weeks in the water [61].
The situation in the dolphin and other cetaceans is quite different [62,63]. They never show high voltage waves bilaterally for more than a few seconds. Rather, extended periods of slow waves appear only in one hemisphere at a time. Sometimes they float at the surface while showing unihemispheric slow waves; but often they swim with unihemispheric slow waves, and when they do there is no asymmetry in their motor activity, in contrast to the behavior seen in the fur seal. Regardless of which hemisphere is showing slow wave activity, they tend to circle in a counterclockwise direction. Mukhametov states that “the sleep behavior of these animals is indistinguishable from that of quiet waking” [64]. No evidence has been presented for elevated sensory response thresholds contralateral to the hemisphere that has slow waves. Indeed it seems that a substantial elevation of sensory threshold on one side of the body would be quite maladaptive given the danger of collisions while moving. Similarly, brain motor systems must be bilaterally active to maintain the bilaterally coordinated movement. Therefore forebrain and brainstem activity must differ radically from that seen in terrestrial mammals during sleep. The one study of unihemispheric slow wave rebound after unihemispheric slow wave deprivation in dolphins produced variable results, with little or no relation (and no significant difference) between the amount of slow waves lost in each hemisphere and the amount of slow waves recovered in each hemisphere when the animals were subsequently left undisturbed [64]. In another study it was shown that dolphins are able to maintain continuous vigilance for 5 days with no decline in accuracy. At the end of this period there was no detectable decrease of activity or evidence of inattention such as would be expected of a sleep deprived animal [65].
In some smaller cetaceans, such as the harbor porpoise [63] and Commerson’s dolphin [66], motor activity is essentially continuous from birth to death: they never float quietly at the surface or rest on the bottom. It is evident that they must have accurate sensory and motor performance and associated brain activation 24 hours a day to avoid collisions. Thus this behavior differs from the criteria normally used to define sleep.
A remarkable behavior is seen in newborn dolphins, killer whales and their mothers. All land mammals show maximal sleep and maximal quiescent immobility at birth, behaviors which have been assumed to be required for brain and body development. Newborn killer whales and dolphins, however, are continuously active, in the manner seen in adults of small dolphin species, for at least four weeks after birth. Although some unihemispheric slow waves might be present at these times, the eyes are open bilaterally when they surface at average intervals of less than one minute, indicating that any slow wave pattern could not last longer than this period [67]. Sleep is not restorative if interrupted on such a schedule in humans [68] or rats [69]. The cetacean mothers also cease extended periods of eye closure and floating behavior during the postpartum period. No rebound of lost immobility is seen. Rather the neonate and mother gradually transition to the adult pattern of periodic immobility over a 1–2 month period. In many cetacean species, migration occurs during the postpartum period. In all cetaceans, this period is the time of greatest danger from predation because of the small size of the calf, necessitating the mother and calf to be maximally alert [70–72].
The remarkable diversity of sleep traits among mammals raises the question of which, if any, physiological changes are a consistent accompaniment of sleep. When sleep-like states are compared between mammals, non-mammalian vertebrates and invertebrates, it is even more difficult to identify physiological commonalities. This diversity suggests that these physiological sleep traits may have evolved more recently and may serve species-specific adaptive functions [36,37]. Conversely, certain molecular commonalities do exist as indicated below. Bridging the gap between these molecular mechanisms and physiological functions is a major opportunity and challenge.
Genetic Model Organisms
One approach to analyzing the complexity and diversity of sleep and sleep-like states is to use simpler, more genetically tractable organisms to understand the core properties of sleep. Ideally, so-called genetic model organisms are small, produce large numbers of offspring, have short generation times, and have sequenced genomes. Studies of simpler organisms also have implications for the evolution of sleep. These organisms often reflect more ancient branches from the tree of animal lineages. Shared properties of sleep between simpler model organisms and more complex mammals may then reflect the biology of the ancient common ancestor of these diverged species.
The freshwater zebrafish is one such organism that has been established recently as a model for sleep studies (Figure 2A). The zebrafish develops outside the mother from embryo to early larval stages in just three days. Much of the early work on zebrafish sleep has therefore focused on larval sleep behavior, which is observed in as young as five day old larvae [73]. The advantage of zebrafish, especially relative to invertebrate models, is the conservation of neurotransmitter systems and neuroanatomy with mammalian models [74], and its diurnal activity pattern [75,76], resembling that of humans. Unlike mice and humans, the zebrafish is cold blooded, and thus, like other cold-blooded animals, is not likely to exhibit REM or slow-wave sleep. Nonetheless, it has been studied as a potential model for hypothalamic and brainstem sleep regulation.
Figure 2. Fish, flies and worms.

(A) The adult zebrafish, Danio rerio. (B) The adult fruit fly, Drosophila melanogaster. (C) The adult nematode, Caenorhabdities elegans. Photos courtesy of Wikipedia Commons website.
Larval and adult zebrafish demonstrate many of the core properties of behavioral sleep (Table 1). Larvae exhibit prolonged periods of immobility lasting many minutes [73,77]. These immobile fish display specific place preference for sleep, often moving to the bottom of the chamber (larvae and adults) [77,78], or staying near the water surface (adults) [78]. They also exhibit specific postures, floating with their head pointed down (larvae) [77], or with a ‘drooping’ caudal fin (adult) [78]. This immobility is also accompanied by increases in arousal threshold. In larvae, this has been assessed by a mechanical tap [77] or by exposure to sudden darkness [73] and assessments of subsequent behavioral responses. In larvae, increased arousal threshold is evident after one minute of immobility, but does not increase significantly over longer periods of immobility, leading to the definition of 1 minute of immobility as sleep [73]. In adults, a variety of stimuli, including mechanical, acoustic, and electrical, have been applied, with the last the most effective [78]. In the adult, it was determined that just six seconds of inactivity was deemed as a minimal epoch of sleep [78]. It is not clear whether the difference (6 seconds in adults, 1 minute in larvae) reflects biological or analytical differences.
Table 1.
Model organisms and sleep traits.
| Caenorhabditis elegans (nematode) | Drosophila melanogaster (fruit fly) | Danio rerio (zebrafish) | |
|---|---|---|---|
| Posture | ? | Prone, supported | “drooping” fin |
| Reduced activity | + | + | + |
| Arousal threshold | + | + | + |
| Homeostasis | + | + | +1 |
Exception during constant light.
In both larval and adult zebrafish, there is evidence of homeostatic regulation. In larvae, deprivation by tapping the tank during the typical ‘sleep’ period, but not during the wake period, results in a compensatory sleep rebound during the following day [77]. In adults, sleep deprivation has been more difficult to accomplish, with electrical stimulation required to persistently disrupt sleep. Nonetheless, this stimulus also resulted in sleep rebound [78]. At least in adults, light can persistently deprive zebrafish of behavioral sleep. Yet no apparent sleep rebound is evident once animals are put into darkness [78]. It has been proposed that light can somehow bypass the homeostatic regulation of sleep. Nonetheless, the findings that zebrafish display some of the core behavioral properties of sleep suggests that it will be a valuable sleep model.
The invertebrate whose sleep has been most intensively studied is the fruit fly Drosophila melanogaster (Figure 2B). The fact that the fruit fly displays all of the core behavioral properties of sleep (Table 1) has engendered great enthusiasm and implies that sleep may have been present in the common ancestor of arthropods and vertebrates. The fruit fly central nervous system has over 200,000 neurons (the human brain has 1011 neurons) and does not have anatomic structures that clearly correspond to their vertebrate counterparts [79]. But the fly genome has about 14,000 genes, many of which are highly conserved between flies and humans at the level of sequence and even function [80–82]. Flies use many of the same or similar neurotransmitters, receptors and ion channels [83] as mammals, although some transmitters (for example, octopamine) are used more commonly in flies than in mammals [84] and other transmitters (for example, hypocretin/orexin) present in mammals have not been observed in flies.
Fly sleep is not typically monitored using video-based approaches but rather using the ‘Drosophila activity monitoring system’ (Figure 3) [85]. Here, a single fly is placed into a small glass tube with agar food at one end. The tube is place into a monitor containing 32 infrared emitter/detector pairs, one for each tube. Infrared beam breaks are counted as activity. Using this assay, the fly exhibits long periods of immobility, sometimes lasting for hours. A close examination of these immobile flies reveals a typical posture and place preference (near food when solitary) [86]. This behavioral quiescence is accompanied by increases in arousal threshold saturating at five minutes of immobility during the dark period, leading to the five-minute criterion for sleep that is commonly used [87,88]. Sleep depriving flies leads to a compensatory homeostatic rebound [86–88]. Sleep is usually regulated by a circadian clock, which (at least) times sleep and wake to occur at particular times of day in most organisms. The fly also demonstrates robust circadian regulation of its sleep state and has been one of the best models for understanding the molecular basis of circadian clocks [89–91].
Figure 3. The Drosophila activity monitoring system.
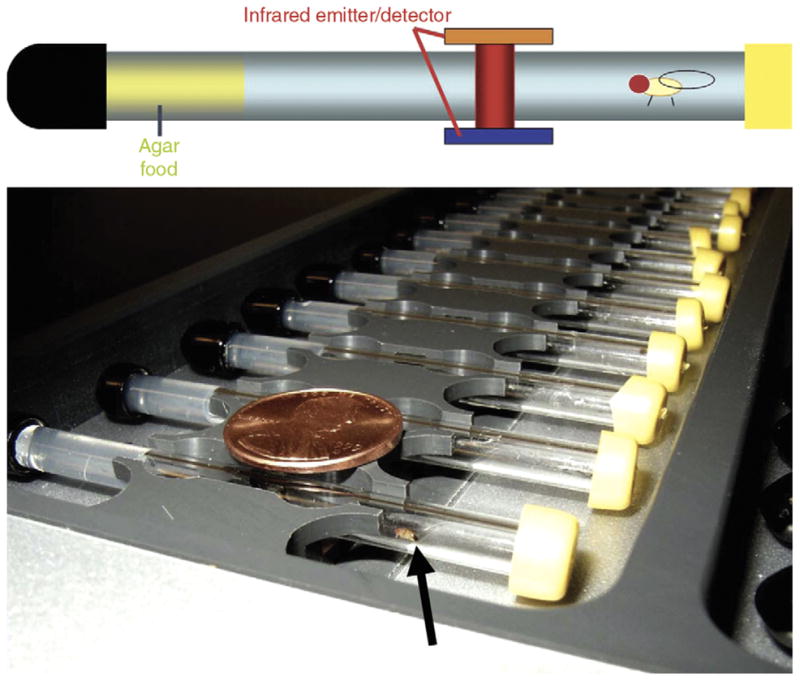
The top panel is a schematic of a behavioral assay tube with an agar food plug at one end. The tube is crossed by an infrared beam. The bottom panel is a photograph of an activity monitor. A U.S. penny (19 mm) is indicated for scale. A fruit fly is indicated by an arrow.
The most recent entrant, and perhaps the simplest organism that has been shown to have a sleep-like state, is the tiny (less than 1 mm long) roundworm, Caenorhabditis elegans (Figure 2C). The worm is notable for its remarkably simple and well-understood anatomy with just 959 cells (not including sperm and eggs) of which precisely 302 are neurons connected through 700 electrical and 5000 chemical synapses [92]. The presence of sleep-like states in C. elegans suggests that the common ancestor of bilaterally symmetric animals may have slept.
The study of sleep in worms has focused on a developmental behavior termed ‘lethargus’ — a period of behavioral quiescence that occurs before each of its four larval molts, the final molt leading to the adult worm [93]. Lethargus occurs about every 10–16 hours and lethargus periods last about 2–4 hours each. These periods of relative immobility are accompanied by reductions in responsiveness to mechanical and olfactory stimuli, such as a tap (Table 1) [94]. Depriving worms of these quiescent periods results in a compensatory rebound of quiescence, indicating homeostatic control [94].
Unlike the sleep-like states of zebrafish and flies, lethargus of C. elegans is not under circadian control, but rather under the control of a developmental program that precisely controls the timing of molts [95]. Remarkably, the worm ortholog of the fly and mammalian circadian clock gene period, lin-42, oscillates with the timing of the molts [96]. Thus, sleep in fish, flies and worms is linked to the oscillation of the per gene whether circadian or developmentally controlled. While sleep in worms has only been claimed for the larval stages, adult worms do display circadian rhythms of moving speed [97] and satiety-induced behavioral quiescence [98].
Conservation of the Sleep Mechanism
The presence of behaviorally defined sleep-like states in such a diverse array of creatures, even in animals as simple as C. elegans, raises the question of whether sleep evolved independently in each of these animals or whether sleep was present in a primitive form in the common ancestor of worms, flies, fish, and even humans. While studies of the genetics and pharmacology of sleep are still in their infancy, particularly for the genetic model species, the remarkable similarity in the genetic and pharmacological control of sleep provides compelling support for the latter hypothesis. This suggests that sleep was present in the common ancestor of all bilaterally symmetric organisms over 600 million years ago. Indeed, even the cnidarian jellyfish, which represents an even more ancient branch of the animal kingdom, has been reported to show sleep-like states [99,100]. We shall focus on genes with functions that affect sleep, discussing three areas of conservation: circadian clock genes, signaling pathways, and neurotransmission. There are many excellent gene expression profiling studies [22–25,87,101,102] that also suggest conservation of sleep mechanism but we do not have space to discuss them here.
One of the most well conserved pathways for sleep regulation is the circadian clock, or more precisely, the circadian clock genes (Table 2). Circadian clocks are composed of transcriptional feedback loops that are highly conserved between flies and mammals. In Drosophila, the CLOCK (CLK) transcription factor along with its heterodimeric partner CYCLE (CYC), activates the period (per) and timeless (tim) genes (reviewed in [103]). The PER and TIM proteins feedback and repress CLK/CYC. PER and TIM are also modified by phosphorylation by kinases such as DOUBLETIME (DBT), leading to their degradation and allowing the cycle to proceed. Remarkably, mutations in the human orthologs of the per and Dbt genes have been shown to be responsible for familial advanced sleep phase syndrome [104,105]. Individuals affected with this dominantly inherited syndrome sleep and wake 3–4 hours before their unaffected siblings [106]. Not surprisingly, zebrafish uses similar genes to those identified in flies and mammals to regulate its circadian clocks [75,107]. As mentioned above, in C. elegans lethargus is also linked to the oscillation of its per gene, lin-42, but this oscillation is controlled by a developmental, not a circadian clock. Taken together, these observations suggest that sleep is linked to oscillation of ‘clock’ genes, and that this may even have preceded the evolution of fully-fledged circadian clocks, at least in animals. At least some unicellular organisms show circadian or ultradian rhythms [108,109], but evidence for sleep in unicellular organisms is lacking (see below).
Table 2.
Conserved sleep mechanisms.
| Worms | Flies | Fish | Mammals | |
|---|---|---|---|---|
| Clock Genes | +1 | + | + | + |
| Cyclic AMP | + | + | ? | + |
| Cyclic GMP | + | + | ? | ? |
| EGF | + | + | ? | + |
| GABA | ? | + | + | + |
| Adenosine | ? | + | + | + |
| Dopamine | ? | + | ? | + |
| Histamine | ? | + | + | + |
| Melatonin | ? | ? | + | + |
| Hypocretin/orexin | ? | ? | +2 | + |
| Potassium channels | ? | + | ? | + |
Clock gene expression is associated with developmentally timed sleep.
The precise direction of hypocretin sleep regulation is under debate.
Interestingly, disruption of clock function can affect not only the timing of sleep but also the amount of sleep. Lesion of the mammalian circadian pacemaker, the suprachiasmatic nucleus (SCN), can increase sleep time in some primates [110] and in mice [111] (although not in rats) [112]. Nonetheless, mutations of the Clock gene not only disrupt circadian aspects of sleep but also result in reduced sleep in both flies [113] and mice [114]. Mutations of the heterodimeric partner of CLK, CYC, also result in reduced sleep levels [4,113]: male cyc mutant flies display reduced sleep rebound [113] and female cyc mutant flies are hypersensitive to the lethal effects of sleep deprivation [4]. Knockouts of the mouse CYC ortholog, Bmal1, actually exhibit an increase in sleep time [115]; however, they display reduced sleep rebound similar to male flies [115].
A number of signal transduction pathways also appear to be commonly deployed in the regulation of sleep (Table 2). The cyclicAMP(cAMP) pathway, including the cAMP-dependent protein kinase A and cAMP activated transcription factor CREB, plays a role in promoting wakefulness in worms, flies, and mice. In Drosophila, a series of mutations in cAMP pathway components or overexpression of components that would increase cAMP levels or activity of downstream components, increases wake behavior, whereas mutations that result in the converse increase sleep [116,117]. Similarly, in C. elegans, mutations that increase cAMP lead to increase responsiveness to sensory stimuli [94]. In mice, knockout of two CREB isoforms results in reduced wakefulness [118]. Interestingly, this pathway has been implicated as a central player in long term memory formation in both flies and mice, suggesting potential genetic links between sleep regulation and memory consolidation [119]. Consistent with this hypothesis, a key neural locus for learning and memory, the mushroom bodies, is also an important neural substrate for sleep regulation [117,120].
Cyclic GMP signaling may also play a conserved role in sleep regulation. Gain- and loss-of-function mutants in egl-4, which encodes the worm ortholog of cGMP-dependent protein kinase (PKG), result in increased and decreased behavioral quiescence, respectively [94]. Similarly, a mutation in the Drosophila PKG foraging (for) locus, which lowers PKG activity, is associated with reduced sleep, suggesting potential conservation of sleep mechanisms between flies and worms [94].
A third signaling pathway that is conserved for sleep control is that of the epidermal growth factor receptor (EGF receptor), with increases in EGF resulting in increased sleep/quiescence, while reductions in EGF receptor signaling result in increased wake/activity. Remarkably, the EGF receptor has been shown to control sleep behavior in worms, flies, and mice (Table 2). The EGF receptor is a transmembrane receptor tyrosine kinase that is activated by secreted growth factor ligands such as EGF and transforming growth factor-alpha (TGFα). EGF infusion enhances slow-wave sleep in rabbits [121]. The EGF receptor ligand, TGFα, is rhythmically expressed and secreted by the mammalian SCN [122]. TGFα infusion into the third ventricle substantially inhibited wheel-running activity while loss of function EGF receptor mutants exhibited increased daytime activity [122], although effects on light induced behavior have been questioned [123]. In flies, induction of EGF ligand secretion by overexpression of EGF processing proteins, Rho and Star, results in increased sleep, while targeted rho loss of function by RNA interference (RNAi) results in reduced sleep [124]. In worms, transient induction of Egf (lin-3) expression results in behavioral quiescence even in adult animals, while egfr (let-23) loss-of-function mutations result in increased activity during lethargus periods [125]. The remarkable conservation of EGF signaling across evolution suggests it is a component of an ancient sleep pathway.
Sleep is largely regarded as a neurally driven phenomenon and a number of neurotransmitters appear to play conserved roles in sleep (Table 2). We will comment only on the system-level effect of manipulation of gene/neurotransmitter function and compare the effects between organisms. More precise manipulations particularly in mammalian systems suggest more complex regulatory functions within discrete circuits.
The most commonly used drugs that induce sleep and treat sleep disorders function by activating GABA receptors. GABAergic compounds such as benzodiazepines and barbiturates also induce sleep in fish as they do in ‘higher’ organisms [77,126]. In Drosophila, GABA receptor mutants that reduce desensitization of the GABA receptor (resulting in maintained GABA signaling) fall asleep more quickly (show reduced sleep latency) [127]; elegant genetic analyses revealed that these GABA receptor mutants block desensitization and sleep latency increases induced by drugs [127].
One issue that has arisen is the precise role of the hypocretin/orexin system in zebrafish. Loss of hypocretin cells, or mutations in the hypocretin receptors and ligands, results in the sleep disorder narcolepsy in both humans and mice [128–133]. Narcolepsy is characterized by persistent sleepiness, rapid transitions from wake directly to REM sleep and sudden losses of muscle tone (cataplexy) [131,134,135]. The hypocretin/orexin system plays a pivotal role in maintaining the wakeful state [135]. Expression of a heat-shock inducible form of hypocretin/orexin in larval zebrafish disrupted sleep, supporting the view that hypocretin/orexin has a wake-promoting role [73]. Similarly, injection of hypocretin/orexin peptide into the brains of adult goldfish increased locomotor activity [136]. But adult fish bearing loss-of-function alleles of the single hypocretin/orexin receptor displayed modestly reduced sleep time with increased sleep fragmentation [78]. Furthermore, some anatomical studies suggested that the hypocretin/orexin receptor is expressed in different circuits in adult fish than in mammalian models, while others have suggested that the hypocretin/orexin circuit architecture is preserved [73,78,137]. The differences between the results of the two genetic studies may be attributed to many factors, including assessment at different developmental stages (larval versus adult) with potentially different neural circuitries, use of inducible versus non-inducible genetic manipulations with their potential of developmental and/or compensatory changes, gain-of-function with potentially ectopic effects versus loss-of-function approaches, and manipulation of ligand versus receptor genes. Nonetheless, these studies highlight a role for hypocretin/orexin in sleep in zebrafish, the precise nature of which awaits additional experimentation. Hypocretin/orexin has not been identified in invertebrates.
In many animals, melatonin is secreted at night and induces sleep in diurnal animals, including humans [138–141]. Zebrafish also rhythmically produces melatonin [142] and melatonin also induces sleep —in particular, changes in the activity and arousal threshold — in zebrafish [77]. The sleep inducing effects of melatonin have not been described in invertebrates.
A variety of other transmitters also may play conserved roles between flies and mammals. The ATP breakdown product adenosine increases in response to wakefulness and can, in turn, induce sleep by acting through specific G-protein coupled receptors [143]. Caffeine is thought to act as an adenosine receptor antagonist. In Drosophila, caffeine induces wakefulness, while the adenosine agonist cyclohexyladenosine induces sleep [86,87]. Antihistamines are also noted to induce sleep in mammals [144]. Similarly, antihistamines can induce sleep in both Drosophila [87] and zebrafish [126]. Drugs or mutants that enhance or inhibit dopaminergic transmission and activity result in reduced and increased sleep in Drosophila [145,146], as they do in mammals. Even the novel wake-promoting agent modafinil, which is thought to act via dopaminergic pathways [147], similarly promotes wakefulness in flies [148]. Conservation also extends to the potassium channel family [149,150].
Conclusions
The remarkable diversity of sleep behavior coupled to the apparent conservation of basic sleep mechanisms raises many important issues. The diversity of sleep traits in all animals, and even among mammals, suggests that these unique aspects of sleep serve specific functions. The underlying mechanistic basis, especially at the molecular level, will be of great interest. On the other hand, the conservation of certain molecular mechanisms linked to sleep control across diverse species suggests that simple model organisms such as the fly and worm can be effectively used to reveal the genetic basis of sleep in higher organisms. While it is unlikely that plants or even unicellular organisms manifest the neurochemical and physiological machinery that underlies sleep seen in more complex organisms, we cannot exclude the possibility that they may show biochemical precursors of sleep. As basic sleep mechanisms are uncovered in multicellular animals, it will be of interest to see if similar genetic pathways are operating together in plants or unicellular organisms, particularly those with circadian clocks. Finally, will uncovering ancient sleep mechanisms that tell us how we sleep explain why we sleep? Only time will tell, but the entrance of simple genetic models may bring us one step closer to determining core sleep functions.
Acknowledgments
We thank David Raizen, David Prober, and Jena Pitman for helpful comments. R.A. acknowledges the support of the National Institutes of Health (R01MH067870 and R01NS052903) and the March of Dimes. J.M.S. acknowledges the support of the Medical Research Service of the Department of Veteran’s Affairs, the National Science Foundation and the National Institutes of Health. (R01MH64109 and 1RO1-NS42947).
Contributor Information
Ravi Allada, Email: r-allada@northwestern.edu.
Jerome M. Siegel, Email: jsiegel@ucla.edu.
References
- 1.Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005;437:1264–1271. doi: 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 3.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–184. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 4.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 5.Siegel JM. Do all animals sleep? Trends Neurosci. 2008;31:208–213. doi: 10.1016/j.tins.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tobler I, Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Vol. 4. Philadelphia: Elsevier Saunders; 2005. Phylogeny of sleep regulation; pp. 77–90. [Google Scholar]
- 7.Rechtschaffen A, Siegel JM, Kandel ER, Schwartz JH, Jessel TM. Principles of Neuroscience. Vol. 4. New York: McGraw Hill; 2000. Sleep and Dreaming; pp. 936–947. [Google Scholar]
- 8.Tobler I. Is sleep fundamentally different between mammalian species? Behav Brain Res. 1995;69:35–41. doi: 10.1016/0166-4328(95)00025-o. [DOI] [PubMed] [Google Scholar]
- 9.Siegel JM, Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Vol. 4. Philadelphia: Elsevier Saunders; 2005. REM sleep; pp. 120–135. [Google Scholar]
- 10.Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- 11.Steriade M, Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Vol. 4. Philadelphia: Elsevier Saunders; 2005. Brain electrical activity and sensory processing during waking and sleep; pp. 101–119. [Google Scholar]
- 12.Borbely AA, Achermann P, Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Vol. 4. Philadelphia: Elsevier Saunders; 2005. Sleep homeostasis and models of sleep regulation; pp. 405–417. [Google Scholar]
- 13.Zepelin H, Siegel JM, Tobler I, Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Vol. 4. Philadelphia: Elsevier Saunders; 2005. Mammalian sleep; pp. 91–100. [Google Scholar]
- 14.Fenik VB, Davies RO, Kubin L. Noradrenergic, serotonergic and GABAergic antagonists injected together into the XII nucleus abolish the REM sleep-like depression of hypoglossal motoneuronal activity. J Sleep Res. 2005;14:419–429. doi: 10.1111/j.1365-2869.2005.00461.x. [DOI] [PubMed] [Google Scholar]
- 15.John J, Wu MF, Boehmer LN, Siegel JM. Cataplexy-active neurons in the posterior hypothalamus: implications for the role of histamine in sleep and waking behavior. Neuron. 2004;42:619–634. doi: 10.1016/s0896-6273(04)00247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kodama T, Lai YY, Siegel JM. Changes in inhibitory amino acid release linked to pontine-induced atonia: an in vivo microdialysis study. J Neurosci. 2003;23:1548–1554. doi: 10.1523/JNEUROSCI.23-04-01548.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai YY, Kodama T, Siegel JM. Changes in monoamine release linked to pontine inhibition of muscle tone: An in vivo microdialysis study. J Neurosci. 2001;21:7384–7391. doi: 10.1523/JNEUROSCI.21-18-07384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodama T, Lai YY, Siegel JM. Enhancement of acetylcholine release during REM sleep in the caudomedial medulla as measured by in vivo microdialysis. Brain Res. 1992;580:348–350. doi: 10.1016/0006-8993(92)90967-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel JM. Mechanisms of sleep control. J Clin Neurophysiol. 1990;7:49–65. doi: 10.1097/00004691-199001000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai YY, Siegel JM. Cardiovascular and muscle tone changes produced by microinjection of cholinergic and glutamatergic agonists in dorsolateral pons and medial medulla. Brain Res. 1990;514:27–36. doi: 10.1016/0006-8993(90)90432-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahowald MW, Schenck CH. Insights from studying human sleep disorders. Nature. 2005;437:1279–1285. doi: 10.1038/nature04287. [DOI] [PubMed] [Google Scholar]
- 22.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 23.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 24.Cirelli C, Faraguna U, Tononi G. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98:1632–1645. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- 25.Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–457. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 26.Hoppenbrouwers T, Sterman MB. Development of Sleep State Patterns in the Kitten. Exp Neurol. 1975;49:822–838. doi: 10.1016/0014-4886(75)90062-x. [DOI] [PubMed] [Google Scholar]
- 27.Jouvet-Mounier D, Astic L, Lacote D. Ontogenesis of the states of sleep in rat, cat, and guinea pig during the first postnatal month. Dev Psychobiol. 1970;2:216–239. doi: 10.1002/dev.420020407. [DOI] [PubMed] [Google Scholar]
- 28.McGinty DJ, Stevenson M, Hoppenbrouwers T, Harper RM, Sterman MB, Hodgman J. Polygraphic studies of kitten development: sleep state patterns. Dev Psychobiol. 1977;10:455–469. doi: 10.1002/dev.420100506. [DOI] [PubMed] [Google Scholar]
- 29.Roffwarg HP, Muzio JN, Dement WC. Ontogenetic development of the human sleep-dream cycle. Science. 1966;152:604–619. doi: 10.1126/science.152.3722.604. [DOI] [PubMed] [Google Scholar]
- 30.Seelke AM, Karlsson KA, Gall AJ, Blumberg MS. Extraocular muscle activity, rapid eye movements and the development of active and quiet sleep. Eur J Neurosci. 2005;22:911–920. doi: 10.1111/j.1460-9568.2005.04322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegel JM. Functional implications of sleep development. PLoS Biol. 2005;3:e178. doi: 10.1371/journal.pbio.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamasy V, Koranyi L, Lissak K. Early postnatal development of wakefulness-sleep cycle and neuronal responsiveness: A multiunit activity study on freely moving newborn rat. Electroenceph Clin Neurophysiol. 1980;49:102–111. doi: 10.1016/0013-4694(80)90356-9. [DOI] [PubMed] [Google Scholar]
- 33.Lesku JA, Roth TC, Amlaner CJ, Lima SL. A phylogenetic analysis of sleep architecture in mammals: the integration of anatomy, physiology, and ecology. Am Nat. 2006;168:441–453. doi: 10.1086/506973. [DOI] [PubMed] [Google Scholar]
- 34.Capellini I, Barton RA, McNamara P, Preston BT, Nunn CL. Phylogenetic analysis of the ecology and evolution of mammalian sleep. Evolution. 2008 May 14; doi: 10.1111/j.1558-5646.2008.00392.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lesku JA, Roth TC, Rattenborg NC, Amlaner CJ, Lima SL. Phylogenetics and the correlates of mammalian sleep: A reappraisal. Sleep Med Rev. 2008;12:229–244. doi: 10.1016/j.smrv.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Huber R, Ghilardi MF, Massimini M, Tononi G. Local sleep and learning. Nature. 2004;430:78–81. doi: 10.1038/nature02663. [DOI] [PubMed] [Google Scholar]
- 37.Vyazovskiy VV, Tobler I. Handedness leads to interhemispheric EEG asymmetry during sleep in the rat. J Neurophysiol. 2008;99:969–975. doi: 10.1152/jn.01154.2007. [DOI] [PubMed] [Google Scholar]
- 38.Hobson JA. Electrographic correlates of behavior in the frog with special reference to sleep. Electroenceph Clin Neurophysiol. 1967;22:113–121. doi: 10.1016/0013-4694(67)90150-2. [DOI] [PubMed] [Google Scholar]
- 39.Hobson JA, Goin OB, Goin CJ. Electrographic correlates of behavior in tree frogs. Nature. 1968;220:386–387. doi: 10.1038/220386a0. [DOI] [PubMed] [Google Scholar]
- 40.Tauber ES, Rojas-Ramirez J, Hernandez-Peon R. Electrophysiological and behavioral correlates of wakefulness and sleep in the lizard (Ctenosaura pectinata) Electroenceph Clin Neurophysiol. 1968;24:424–443. doi: 10.1016/0013-4694(68)90102-8. [DOI] [PubMed] [Google Scholar]
- 41.Ayala-Guerrero F, Huitron-Resendiz S. Sleep patterns in the lizard Ctenosaura pectinata. Physiol Behav. 1991;49:1305–1307. doi: 10.1016/0031-9384(91)90369-y. [DOI] [PubMed] [Google Scholar]
- 42.De Vera L, Gonzalez J, Rial RV. Reptilian waking EEG: slow waves, spindles and evoked potentials. Electroenceph Clin Neurophysiol. 1994;90:298–303. doi: 10.1016/0013-4694(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 43.Flanigan WF. Sleep and wakefulness in iguanid lizards, Ctenosaura pectinata and Iguana iguana. Brain Behav Evol. 1973;8:401–436. doi: 10.1159/000124366. [DOI] [PubMed] [Google Scholar]
- 44.Eiland MM, Lyamin OI, Siegel JM. State-related discharge of neurons in the brainstem of freely moving box turtles, Terrapene carolina major. Arch Ital Biol. 2001;139:23–36. [PMC free article] [PubMed] [Google Scholar]
- 45.Amlaner CJ, Ball NJ, Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Philadelphia: W.B. Saunders Company; 1994. Avian sleep; pp. 81–94. [Google Scholar]
- 46.Newman S, Rattenborg N, Obermeyer WH, Benca RM. Effects of sleep deprivation by the disk over water method in pigeons. Sleep. 2004;27:A163–A164. doi: 10.1016/j.physbeh.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Rattenborg NC, Obermeyer WH, Vacha E, Benca RM. Acute effects of light and darkness on sleep in the pigeon (Columba livia) Physiol Behav. 2005;84:635–640. doi: 10.1016/j.physbeh.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Gonzalez D, Lesku JA, Rattenborg NC. Increased EEG spectral power density during sleep following short-term sleep deprivation in pigeons (Columba livia): evidence for avian sleep homeostasis. J Sleep Res. 2008;17:140–153. doi: 10.1111/j.1365-2869.2008.00636.x. [DOI] [PubMed] [Google Scholar]
- 49.Rattenborg NC. Response to commentary on evolution of slow-wave sleep and palliopallial connectivity in mammals and birds: a hypothesis. Brain Res Bull. 2007;72:187–193. doi: 10.1016/j.brainresbull.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 50.Van Cauter E, Copinschi G. Interrelationships between growth hormone and sleep. Growth Horm IGF Res. 2000;10(Suppl B):S57–S62. doi: 10.1016/s1096-6374(00)80011-8. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi Y, Ebihara S, Nakamura Y, Takahashi K. A model of human sleep-related growth hormone secretion in dogs: effects of 3, 6, and 12 hours of forced wakefulness on plasma growth hormone, cortisol, and sleep stages. Endocrinology. 1981;109:262–272. doi: 10.1210/endo-109-1-262. [DOI] [PubMed] [Google Scholar]
- 52.Read PA, Horne RS, Cranage SM, Walker AM, Walker DW, Adamson TM. Dynamic changes in arousal threshold during sleep in the human infant. Pediatr Res. 1998;43:697–703. doi: 10.1203/00006450-199805000-00020. [DOI] [PubMed] [Google Scholar]
- 53.Piallat B, Gottesmann C. The reticular arousal threshold during the transition from slow wave sleep to paradoxical sleep in the rat. Physiol Behav. 1995;58:199–202. doi: 10.1016/0031-9384(94)00351-5. [DOI] [PubMed] [Google Scholar]
- 54.Neckelmann D, Ursin R. Sleep stages and EEG power spectrum in relation to acoustical stimulus arousal threshold in the rat. Sleep. 1993;16:467–477. [PubMed] [Google Scholar]
- 55.Hirshkowitz M, Schmidt MH. Sleep-related erections: clinical perspectives and neural mechanisms. Sleep Med Rev. 2005;9:311–329. doi: 10.1016/j.smrv.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Affanni JM, Cervino CO, Marcos HJ. Absence of penile erections during paradoxical sleep. Peculiar penile events during wakefulness and slow wave sleep in the armadillo. J Sleep Res. 2001;10:219–228. doi: 10.1046/j.1365-2869.2001.00259.x. [DOI] [PubMed] [Google Scholar]
- 57.Siegel JM, Manger PR, Nienhuis R, Fahringer HM, Shalita T, Pettigrew JD. Sleep in the platypus. Neuroscience. 1999;91:391–400. doi: 10.1016/s0306-4522(98)00588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegel JM, Manger P, Nienhuis R, Fahringer HM, Pettigrew J. The echidna Tachyglossus aculeatus combines REM and nonREM aspects in a single sleep state: implications for the evolution of sleep. J Neurosci. 1996;16:3500–3506. doi: 10.1523/JNEUROSCI.16-10-03500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyamin OI, Mukhametov LM, Siegel JM. Relationship between sleep and eye state in Cetaceans and Pinnipeds. Arch Ital Biol. 2004;142:557–568. [PMC free article] [PubMed] [Google Scholar]
- 60.Lyamin OI, Chetyrbok IS. Unilateral EEG activation during sleep in the Cape fur seal, Arctocephalus pusillus. Neurosci Lett. 1992;143:263–266. doi: 10.1016/0304-3940(92)90279-g. [DOI] [PubMed] [Google Scholar]
- 61.Lyamin OI, Oleksenko AI, Polyakova IG, Mukhametov LM. Paradoxical sleep in northern fur seals in water and on land. J Sleep Res. 1996;5(suppl):130. [Google Scholar]
- 62.Mukhametov LM, Polyakova IG. EEG investigation of sleep in porpoises (Phocoena phocoena) J Higher Nervous Activity. 1981;31:333–339. [Google Scholar]
- 63.Mukhametov LM. Sleep in marine mammals. Exp Brain Res. 2007;8:227–238. [Google Scholar]
- 64.Oleksenko AI, Mukhametov LM, Polykova IG, Supin AY, Kovalzon VM. Unihemispheric sleep deprivation in bottlenose dolphins. J Sleep Res. 1992;1:40–44. doi: 10.1111/j.1365-2869.1992.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 65.Ridgway S, Carder D, Finneran J, Keogh M, Kamolnick T, Todd M, Goldblatt A. Dolphin continuous auditory vigilance for five days. J Exp Biol. 2006;209:3621–3628. doi: 10.1242/jeb.02405. [DOI] [PubMed] [Google Scholar]
- 66.Mukhametov LM, Lyamin OI, Shpak OV, Manger P, Siegel JM. Swimming styles and their relationship to rest and activity states in captive Commerson’s dolphins. Proceedings of the 14th Biennial Conference on the Biology of Marine Mammals; Vancouver. Nov.27–Dec.3; 2002. p. 152. [Google Scholar]
- 67.Lyamin O, Pryaslova J, Lance V, Siegel J. Animal behaviour: continuous activity in cetaceans after birth. Nature. 2005;435:1177. doi: 10.1038/4351177a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bonnet MH, Kryger MH, Roth T, Dement WC. Sleep Deprivation. Vol. 3. Philadelphia: W.B. Saunders; 2000. pp. 53–71. [Google Scholar]
- 69.Rechtschaffen A, Bergmann BM. Sleep deprivation in the rat: an update of the 1989 paper. Sleep. 2002;25:18–24. doi: 10.1093/sleep/25.1.18. [DOI] [PubMed] [Google Scholar]
- 70.Sekiguchi Y, Arai K, Kohshima S. Sleep behaviour: sleep in continuously active dolphins. Nature. 2006;441:E9–E10. doi: 10.1038/nature04898. [DOI] [PubMed] [Google Scholar]
- 71.Gnone G, Moriconi T, Gambini G. Sleep behaviour: activity and sleep in dolphins. Nature. 2006;441:E10–E11. doi: 10.1038/nature04899. [DOI] [PubMed] [Google Scholar]
- 72.Lyamin OI, Pryaslova J, Lance V, Siegel JM. Sleep behaviour: Sleep in continuously active dolphins; Activity and sleep in dolphins (Reply) Nature. 2006;441:E11. [Google Scholar]
- 73.Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26:13400–13410. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wullimann MF, Rupp B, Reichter H. In Neuroanatomy of the Zebrafish Brain: A Topological Atlas. Boston: Birkhauser Verlag; 1996. p. 144. [Google Scholar]
- 75.Hurd MW, Debruyne J, Straume M, Cahill GM. Circadian rhythms of locomotor activity in zebrafish. Physiol Behav. 1998;65:465–472. doi: 10.1016/s0031-9384(98)00183-8. [DOI] [PubMed] [Google Scholar]
- 76.Cahill GM, Hurd MW, Batchelor MM. Circadian rhythmicity in the locomotor activity of larval zebrafish. Neuroreport. 1998;9:3445–3449. doi: 10.1097/00001756-199810260-00020. [DOI] [PubMed] [Google Scholar]
- 77.Zhdanova IV, Wang SY, Leclair OU, Danilova NP. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903:263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 78.Yokogawa T, Marin W, Faraco J, Pezeron G, Appelbaum L, Zhang J, Rosa F, Mourrain P, Mignot E. Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 2007;5:2379–2397. doi: 10.1371/journal.pbio.0050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nichols CD. Drosophila melanogaster neurobiology, neuropharmacology, and how the fly can inform central nervous system drug discovery. Pharmacol Ther. 2006;112:677–700. doi: 10.1016/j.pharmthera.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 80.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 81.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Celniker SE, Rubin GM. The Drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- 83.Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 84.Burchett SA, Hicks TP. The mysterious trace amines: protean neuromodulators of synaptic transmission in mammalian brain. Prog Neurobiol. 2006;79:223–246. doi: 10.1016/j.pneurobio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 85.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Meth Enzymol. 2005;393:759–772. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- 86.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 87.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 88.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 89.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 90.Helfrich-Forster C. Techniques that revealed the network of the circadian clock of Drosophila. Meth Enzymol. 2005;393:439–451. doi: 10.1016/S0076-6879(05)93021-8. [DOI] [PubMed] [Google Scholar]
- 91.Collins B, Blau J. Even a stopped clock tells the right time twice a day: circadian timekeeping in Drosophila. Pflugers Arch. 2007;454:857–867. doi: 10.1007/s00424-006-0188-9. [DOI] [PubMed] [Google Scholar]
- 92.White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Phil Trans R Soc Lond B. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- 93.Cassada RC, Russell RL. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- 94.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 95.Ambros V. Control of developmental timing in Caenorhabditis elegans. Curr Opin Genet Dev. 2000;10:428–433. doi: 10.1016/s0959-437x(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 96.Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 97.Saigusa T, Ishizaki S, Watabiki S, Ishii N, Tanakadate A, Tamai Y, Hasegawa K. Circadian behavioural rhythm in Caenorhabditis elegans. Curr Biol. 2002;12:R46–R47. doi: 10.1016/s0960-9822(01)00669-8. [DOI] [PubMed] [Google Scholar]
- 98.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kavanau JL. Is sleep’s ‘supreme mystery’ unraveling? An evolutionary analysis of sleep encounters no mystery; nor does life’s earliest sleep, recently discovered in jellyfish. Med Hypoth. 2006;66:3–9. doi: 10.1016/j.mehy.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 100.Seymour JE, Carrette TJ, Sutherland PA. Do box jellyfish sleep at night? Med J Aust. 2004;181:707. doi: 10.5694/j.1326-5377.2004.tb06529.x. [DOI] [PubMed] [Google Scholar]
- 101.Mackiewicz M, Pack AI. Functional genomics of sleep. Respir Physiol Neurobiol. 2003;135:207–220. doi: 10.1016/s1569-9048(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 102.Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA, Pack AI. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27:337–350. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]
- 103.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 105.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 106.Jones CR, Campbell SS, Zone SE, Cooper F, DeSano A, Murphy PJ, Jones B, Czajkowski L, Ptacek LJ. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 107.Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock [see comments] Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 108.Johnson CH, Golden SS. Circadian programs in cyanobacteria: adaptiveness and mechanism. Annu Rev Microbiol. 1999;53:389–409. doi: 10.1146/annurev.micro.53.1.389. [DOI] [PubMed] [Google Scholar]
- 109.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 110.Edgar DM, Dement WC, Fuller CA. Effect of SCN lesions on sleep in squirrel monkeys: evidence for opponent processes in sleep-wake regulation. J Neurosci. 1993;13:1065–1079. doi: 10.1523/JNEUROSCI.13-03-01065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Easton A, Meerlo P, Bergmann B, Turek FW. The suprachiasmatic nucleus regulates sleep timing and amount in mice. Sleep. 2004;27:1307–1318. doi: 10.1093/sleep/27.7.1307. [DOI] [PubMed] [Google Scholar]
- 112.Eastman CI, Mistlberger RE, Rechtschaffen A. Suprachiasmatic nuclei lesions eliminate circadian temperature and sleep rhythms in the rat. Physiol Behav. 1984;32:357–368. doi: 10.1016/0031-9384(84)90248-8. [DOI] [PubMed] [Google Scholar]
- 113.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 114.Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 116.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 117.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 118.Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol. 2003;90:1152–1159. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- 119.Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr Drug Targets CNS Neurol Disord. 2005;4:481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- 120.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 121.Kushikata T, Fang J, Chen Z, Wang Y, Krueger JM. Epidermal growth factor enhances spontaneous sleep in rabbits. Am J Physiol. 1998;275:R509–R514. doi: 10.1152/ajpregu.1998.275.2.R509. [DOI] [PubMed] [Google Scholar]
- 122.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 123.Mrosovsky N, Redlin U, Roberts RB, Threadgill DW. Masking in waved-2 mice: EGF receptor control of locomotion questioned. Chronobiol Int. 2005;22:963–974. doi: 10.1080/07420520500395086. [DOI] [PubMed] [Google Scholar]
- 124.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGF receptor and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 125.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 126.Renier C, Faraco JH, Bourgin P, Motley T, Bonaventure P, Rosa F, Mignot E. Genomic and functional conservation of sedative-hypnotic targets in the zebrafish. Pharmacogenet Genomics. 2007;17:237–253. doi: 10.1097/FPC.0b013e3280119d62. [DOI] [PubMed] [Google Scholar]
- 127.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABA(A) receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–451. doi: 10.1016/s0092-8674(00)81973-x. [DOI] [PubMed] [Google Scholar]
- 129.Willie JT, Chemelli RM, Sinton CM, Tokita S, Williams SC, Kisanuki YY, Marcus JN, Lee C, Elmquist JK, Kohlmeier KA, et al. Distinct narcolepsy syndromes in Orexin receptor-2 and Orexin null mice: molecular genetic dissection of Non-REM and REM sleep regulatory processes. Neuron. 2003;38:715–730. doi: 10.1016/s0896-6273(03)00330-1. [DOI] [PubMed] [Google Scholar]
- 130.Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M, Cornford M, Siegel JM. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27:469–474. doi: 10.1016/s0896-6273(00)00058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Siegel JM, Boehmer LN. Narcolepsy and the hypocretin system- where motion meets emotion. Nat Clin Pract Neurol. 2006;2:548–556. doi: 10.1038/ncpneuro0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nishino S, Ripley B, Overeem S, Lammers GJ, Mignot E. Hypocretin (orexin) deficiency in human narcolepsy. The Lancet. 2000;355:39–41. doi: 10.1016/S0140-6736(99)05582-8. [DOI] [PubMed] [Google Scholar]
- 133.Peyron C, Faraco J, Rogers W, Ripley B, Overeem S, Charnay Y, Nevsimalova S, Aldrich M, Reynolds D, Albin R, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–997. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 134.Siegel JM. Narcolepsy: A key role for hypocretins (orexins) Cell. 1999;98:409–412. doi: 10.1016/s0092-8674(00)81969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Siegel JM. Hypocretin (orexin): role in normal behavior and neuropathology. Annu Rev Psychol. 2004;55:125–148. doi: 10.1146/annurev.psych.55.090902.141545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakamachi T, Matsuda K, Maruyama K, Miura T, Uchiyama M, Funahashi H, Sakurai T, Shioda S. Regulation by orexin of feeding behaviour and locomotor activity in the goldfish. J Neuroendocrinol. 2006;18:290–297. doi: 10.1111/j.1365-2826.2006.01415.x. [DOI] [PubMed] [Google Scholar]
- 137.Kaslin J, Nystedt JM, Ostergard M, Peitsaro N, Panula P. The orexin/hypocretin system in zebrafish is connected to the aminergic and cholinergic systems. J Neurosci. 2004;24:2678–2689. doi: 10.1523/JNEUROSCI.4908-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhdanova IV, Wurtman RJ, Morabito C, Piotrovska VR, Lynch HJ. Effects of low oral doses of melatonin, given 2–4 hours before habitual bedtime, on sleep in normal young humans. Sleep. 1996;19:423–431. doi: 10.1093/sleep/19.5.423. [DOI] [PubMed] [Google Scholar]
- 139.Zhdanova IV, Cantor ML, Leclair OU, Kartashov AI, Wurtman RJ. Behavioral effects of melatonin treatment in non-human primates. Sleep Res Online. 1998;1:114–118. [PubMed] [Google Scholar]
- 140.Stone BM, Turner C, Mills SL, Nicholson AN. Hypnotic activity of melatonin. Sleep. 2000;23:663–669. [PubMed] [Google Scholar]
- 141.Dollins AB, Zhdanova IV, Wurtman RJ, Lynch HJ, Deng MH. Effect of inducing nocturnal serum melatonin concentrations in daytime on sleep, mood, body temperature, and performance. Proc Natl Acad Sci USA. 1994;91:1824–1828. doi: 10.1073/pnas.91.5.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kazimi N, Cahill GM. Development of a circadian melatonin rhythm in embryonic zebrafish. Brain Res Dev Brain Res. 1999;117:47–52. doi: 10.1016/s0165-3806(99)00096-6. [DOI] [PubMed] [Google Scholar]
- 143.Basheer R, Strecker RE, Thakkar MM, McCarley RW. Adenosine and sleep-wake regulation. Prog Neurobiol. 2004;73:379–396. doi: 10.1016/j.pneurobio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 144.Mignot E, Taheri S, Nishino S. Sleeping with the hypothalamus: emerging therapeutic targets for sleep disorders. Nat Neurosci Suppl. 2002;5:1071–1075. doi: 10.1038/nn944. [DOI] [PubMed] [Google Scholar]
- 145.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 146.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nishino S, Mao J, Sampathkumaran R, Shelton J. Increased dopaminergic transmission mediates the wake-promoting effects of CNS stimulants. Sleep Res Online. 1998;1:49–61. [PubMed] [Google Scholar]
- 148.Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. Modafinil maintains waking in the fruit fly drosophila melanogaster. Sleep. 2003;26:139–146. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- 149.Espinosa F, Marks G, Heintz N, Joho RH. Increased motor drive and sleep loss in mice lacking Kv3-type potassium channels. Genes Brain Behav. 2004;3:90–100. doi: 10.1046/j.1601-183x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 150.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]


