Abstract
The HAMP domain is a conserved motif widely distributed in prokaryotic and lower eukaryotic organisms, where it is often found in transmembrane receptors that regulate two-component signaling pathways. The motif links receptor input and output modules and is essential to receptor structure and signal transduction. Recently, a structure was determined for a HAMP domain isolated from an unusual archeal membrane protein of unknown function [Hulko, M., et al. (2006) Cell 126, 929–940]. This study uses cysteine and disulfide chemistry to test this archeal HAMP model in the full-length, membrane-bound aspartate receptor of bacterial chemotaxis. The chemical reactivities of engineered Cys residues scanned throughout the aspartate receptor HAMP region are highly correlated with the degrees of solvent exposure of corresponding positions in the archeal HAMP structure. Both domains are homodimeric, and the individual subunits of both domains share the same helix–connector–helix organization with the same helical packing faces. Moreover, disulfide mapping reveals that the four helices of the aspartate receptor HAMP domain are arranged in the same parallel, four-helix bundle architecture observed in the archeal HAMP structure. One detectable difference is the packing of the extended connector between helices, which is not conserved. Finally, activity studies of the aspartate receptor indicate that contacts between HAMP helices 1 and 2′ at the subunit interface play a critical role in modulating receptor on–off switching. Disulfide bonds linking this interface trap the receptor in its kinase-activating on-state, or its kinase inactivating off-state, depending on their location. Overall, the evidence suggests that the archeal HAMP structure accurately depicts the architecture of the conserved HAMP motif in transmembrane chemoreceptors. Both the on- and off-states of the aspartate receptor HAMP domain closely resemble the archeal HAMP structure, and only a small structural rearrangement occurs upon on–off switching. A model incorporating HAMP into the full receptor structure is proposed.
Two-component signaling pathways are widespread in lower eukaryotes and are ubiquitous in prokaryotes, where they are the most common type of signaling pathway and control a wide array of cell functions (1). Typically, these pathways are regulated by a transmembrane receptor possessing a conserved HAMP1 domain that links the receptor input and output modules (2–6). This HAMP motif is an essential signal transduction element that interconverts different types of mechanical signals passing between modules (3). Each subunit of the homodimeric HAMP is known to possess a helix–connector–helix organization, originally revealed by cysteine and disulfide scanning studies of the aspartate receptor of bacterial chemotaxis (4) and later discovered to be the signature of the HAMP motif (5), yet despite its broad importance, the three-dimensional fold of HAMP remained elusive until the recent report of a NMR structure for a thermophillic HAMP domain isolated from an archeal transmembrane protein (6). The atypical parent protein lacks the cytoplasmic output domain typically coupled to HAMP; moreover, the isolated HAMP domain lacks the structural constraints normally provided by transmembrane helices. Thus, it is important to test whether the parallel, four-helix bundle observed for this archeal HAMP accurately reflects HAMP architecture in a conventional, full-length, membrane-associated receptor.
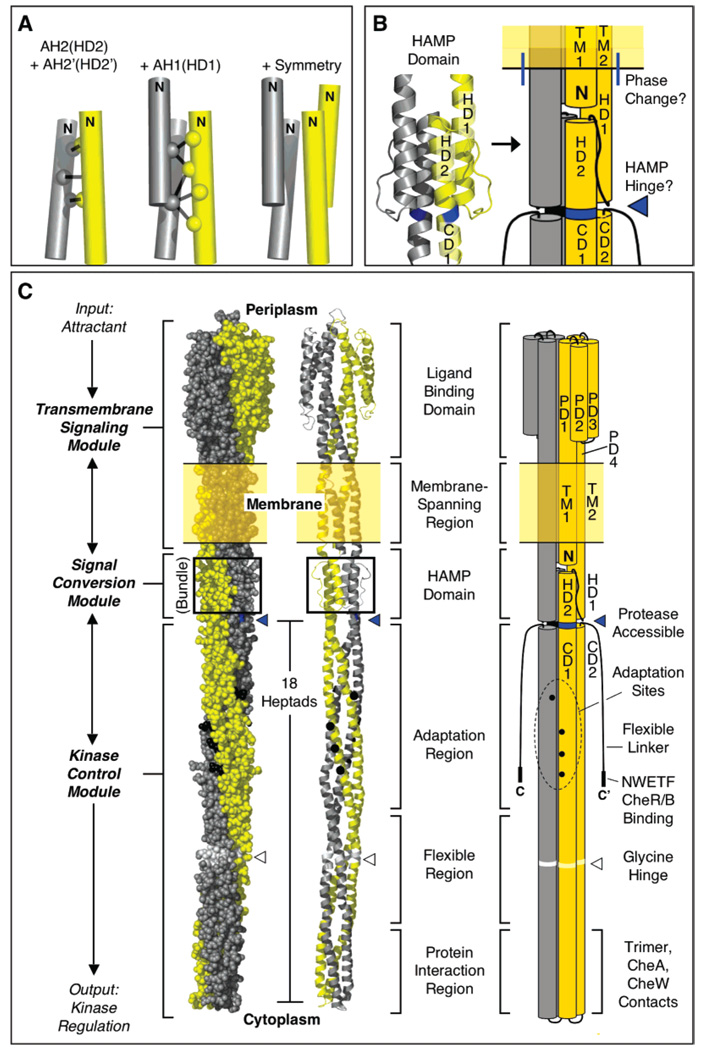
The prototypical chemoreceptors of Escherichia coli and Salmonella typhimurium, including the aspartate receptor, are the best-characterized receptors that regulate two-component pathways (2, 3). The fundamental unit of receptor structure and transmembrane signaling is the homodimer. Such dimers assemble to form trimers of dimers, which in turn are found in large patches of thousands of receptors at the cell pole. The homodimer can be divided into three modules: (i) the transmembrane signaling module comprised of the periplasmic ligand binding domain and the transmembrane helices, (ii) the cytoplasmic HAMP domain which serves as a signal conversion module, and (iii) the cytoplasmic kinase control module possessing the adaptation sites and a kinase docking region. Attractant binding signals are transmitted through the transmembrane signaling module via a piston displacement of the second transmembrane helix (TM2) (2, 7–9) and then are converted by HAMP into an interfacial signal and transmitted down the long subunit interface of the kinase control module to the His kinase CheA (10, 11). The prototypical nature of the aspartate receptor and its HAMP domain make this an ideal system in which to test the generality of the archeal HAMP model.
This study begins by comparing the previously determined cysteine and disulfide scanning data obtained for the aspartate receptor HAMP domain (4) to the NMR structure of the archeal HAMP domain (6). Then a new disulfide mapping analysis is carried out: proximal and distal pairs of positions are selected in the NMR model, and cysteine pairs are substituted at the corresponding positions in the aspartate receptor HAMP domain. The resulting di-Cys mutants are oxidized, and cysteine pairs that form disulfide bonds rapidly and slowly are identified. Finally, the functional effects of the cysteine pairs, and, where appropriate, the disulfide bonds produced by oxidation, are determined. The findings indicate that the aspartate receptor HAMP domain closely resembles the NMR structure of the archeal HAMP domain and also identify a helix–helix interface within HAMP that is crucial for on–off switching.
MATERIALS AND METHODS
Materials
Reagents were obtained from the following sources: [γ-32P]ATP from Perkin-Elmer, QuickChange site-directed mutagenesis kit from Stratagene, mutagenic oligonucleotides from Integrated DNA Technologies, sulfhydryl-specific probe 5-fluoresceinmaleimide (5FM) from Molecular Probes Invitrogen, and all other analytical grade chemicals from Sigma unless noted otherwise. E. coli strains were graciously provided by J. S. Parkinson (University of Utah, Salt Lake City, UT).
Creation and Isolation of Mutant Aspartate Receptors
Site-directed mutagenesis was performed as previously described (12) using the PCR-based QuickChange mutagenesis kit to engineer point mutations into the S. typhimurium aspartate receptor gene in plasmid pSCF6 (13). Mutated plasmids were transformed into E. coli strain RP3808 lacking the major endogenous receptors and other pathway components and expressed, and then receptor-containing membranes were isolated, quantitated, and stored as previously described (11). The final membranes were resuspended at a receptor concentration of ~50 µM in final buffer [20 mM sodium phosphate (pH 7.0 with NaOH), 10% glycerol, 0.1 mM EDTA, and 0.5 mM phenylmethanesulfonyl fluoride].
Oxidation of Receptors
Disulfide formation reactions were carried out for 2.5 µM receptor dimer in buffer A [50 mM Tris (pH 7.2 with HCl), 5 mM MgCl2, and 160 mM KCl] as previously described (4). Reactions were initiated by adding the redox catalyst Cu(II)(1,10-phenanthroline)3 in the presence of ambient-dissolved oxygen (200 µM). Reaction conditions were tuned for specific applications. Typically, EDTA was added to buffer the Cu(II) and thereby moderate the strength of the oxidation reaction. Specific concentrations of catalyst and EDTA as well as specific incubation times and temperatures are indicated for each reaction in the appropriate figure legend. Reactions were quenched by mixing the samples with 4× Laemmli nonreducing sample buffer containing 10 mM EDTA and 40 mM N-ethylmaleimide (NEM), and the mixtures were immediately heated to 95 °C for 1 min to promote unfolding; then products were resolved by Laemmli SDS–PAGE and quantitated by densitometry of the Coomassie-stained protein bands as previously described (11).
To assign the types and numbers of disulfide bonds present in disulfide-linked dimers, different oxidation reactions were carried out to trap the dimers formed during the early, intermediate, and late phases of the oxidation reactions, as specified in the appropriate figure legend. Following oxidation, two identical aliquots were quenched by adding 5-fluoresceinmaleimide (5FM) or N-ethylmaleimide to a final concentration of 500 µM and then incubating the sample for 10 min in the dark at 22 °C. Subsequently, the samples were mixed with 4× Laemmli nonreducing sample buffer containing 10 mM EDTA, and products were resolved by Laemmli SDS–PAGE. Fluorescently labeled products were imaged on a UV transilluminator using a 470 nm long-pass filter, prior to Coomassie staining and visualization of all product bands as noted above.
Receptor Activity Assays
Mutant receptors were assessed for their ability to regulate chemotaxis in vivo by expressing each receptor in E. coli strain RP8611, a strain lacking the aspartate receptor, and then assessing cellular chemotaxis at 30 °C in a standard soft agar swim plate assay, using minimal medium containing 100 µM aspartate (11, 14). Mutant receptors were assessed for their ability to bind, activate, and regulate CheA kinase by reconstituting receptor-containing, isolated membranes with purified CheA, CheW, and CheY [all Cys-less constructs as previously described (12)] and then quantitating receptor-regulated kinase activity in the standard in vitro assay under conditions where the kinase activity of CheA is rate-determining (12). Finally, mutant receptors were assessed for the ability of their adaptation sites to serve as substrates for methyl esterification by reconstituting receptor-containing membranes with purified CheR in the standard in vitro receptor methylation assay (12). For in vivo chemotaxis assays, all receptors were in their reduced state due to the reducing environment of the cytoplasm. For in vitro kinase and methylation assays, receptor-containing membranes were reduced or oxidized to completion by incubation for 20 min at 37 °C with 75 mM dithiothreitol (DTT) or 2 mM Cu(II)(1,10-phenanthroline)3 prior to reconstitution, respectively.
Standard Deviation
Error ranges represent the standard deviation of the mean (n ≥ 3).
RESULTS
Comparison of the NMR Model with Existing Cysteine and Disulfide Scanning Data
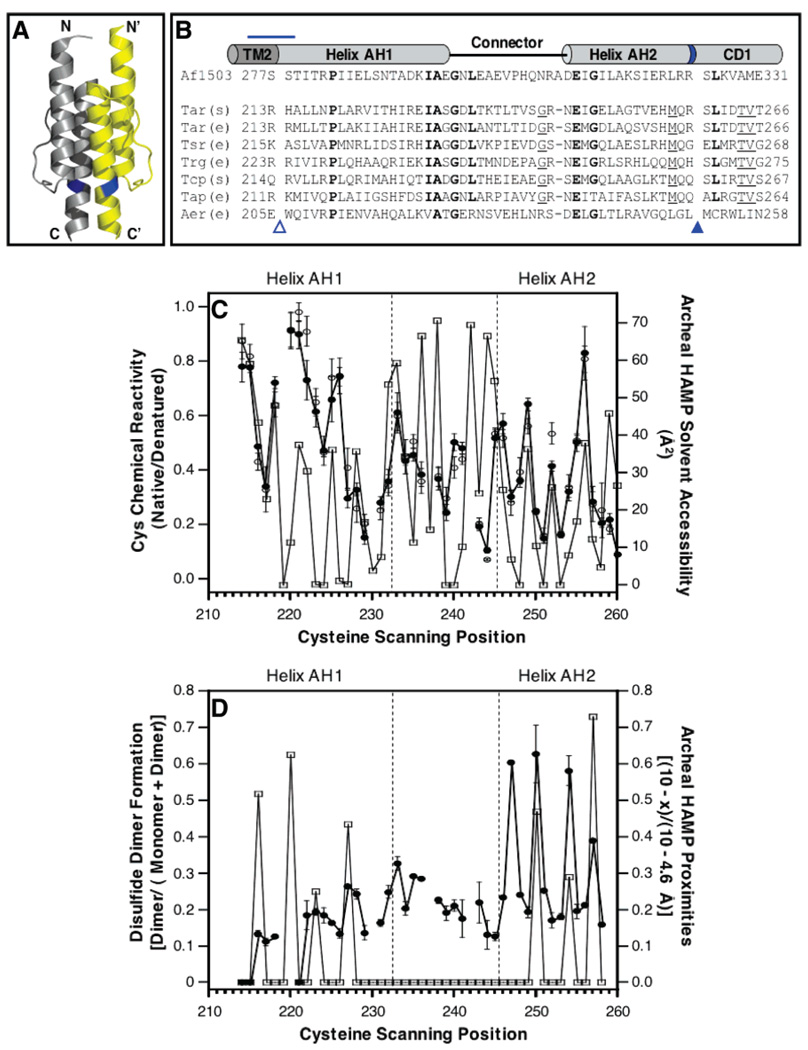
Panels A and B of Figure 1 present the NMR structure of the archeal HAMP domain and its sequence aligned to representative chemoreceptor HAMP domains, respectively. Our previous Cys scanning study of the HAMP region in full-length, membrane-bound aspartate receptor included both a chemical reactivity scan and a symmetric disulfide scan of the entire HAMP region (4). These chemical approaches revealed two amphiphilic helices, AH1 and AH2, each with an exposed face and a buried face. Connecting the two helices was a region lacking a defined secondary structure. If the NMR structure of the archeal HAMP domain (6) is an appropriate model for the aspartate receptor HAMP domain, then the same helical patterns observed in the previous Cys scanning data should be present in the NMR model.
Figure 1.
Previous studies of the conserved HAMP motif. (A) Recent NMR structure of a HAMP domain isolated from an archeal transmembrane protein of unknown function (6), illustrating in cartoon form the parallel, four-helix bundle architecture generated by the association of two identical subunits (yellow and gray) in a symmetric homodimer. (B) Sequence alignment of representative HAMP domains (5, 6), indicating their putative helix–loop–helix regions. Bold residues are most widely conserved in these sequences; underlined residues are conserved except in the archeal and Aer sequences. The blue horizontal line highlights a change of helix register in the aspartate receptor (4); the empty triangle indicates the location of the membrane–water interface (4); and blue band and filled triangle denote the location of an arginine side chain, conserved in some but not all receptors, which is the major site of proteolysis in the S. typhimurium aspartate receptor (25). The latter position may represent the border between HAMP amphiphilic helix 2 (AH2) and cytoplasmic domain helix 1 (CD1) (see the text). (C) Comparison of cysteine chemical reactivities, previously measured for the aspartate receptor HAMP [thick line, ●, (−) Asp; ○, (+) Asp] (4), to the solvent accessibilities calculated for the corresponding positions in the archeal HAMP NMR structure (thin line, □) (6). The correlation is strong in the helical regions, but not in the connector region. (D) Comparison of symmetric disulfide dimer formation, previously measured for the aspartate receptor HAMP (thick line, ●) (4), to spatial proximities calculated for the corresponding cysteine pairs in the archeal HAMP NMR structure (thin line, □) (6). The correlation is strong only in the region of the second helix. Spatial proximities were calculated using the indicated equation where x is the distance in Å between β-carbons.
Figure 1C compares the chemical reactivity data previously published for the aspartate receptor HAMP domain (residues 213–259) (4) to the corresponding solvent accessibilities calculated using the standard rolling water-sized sphere method from the NMR model of the archeal HAMP domain (4, 6). The chemical reactivities, which were originally measured to provide a qualitative estimate of solvent exposure, display an oscillating pattern with α-helical periodicity in the AH1 (residues 213–232) and AH2 (245–259) regions, where high and low reactivities correspond to exposed and buried positions, respectively. Strikingly, these reactivities are highly correlated with the solvent accessibilities calculated from the NMR model, so the same positions are found at the peaks and troughs of both data sets in the AH1 and AH2 regions. By contrast, the correlation between reactivities and accessibilities is poor in the connector region. The simplest interpretation is that helices AH1 and AH2 of the aspartate receptor HAMP correspond to helices 1 and 2 of the NMR model, with the same exposed and buried faces as in the NMR model. However, the packing of the connector region is not the same.
Figure 1D compares the symmetric disulfide scan previously carried out for the aspartate receptor HAMP domain (4) to the spatial proximities calculated from the NMR model (6). For symmetric Cys pairs on the AH1 and AH1′ helices in the aspartate receptor, mild oxidation did not generate high extents of disulfide bond formation, even for the Cys pairs predicted by the NMR model to be most proximal. By contrast, for symmetric Cys pairs on the AH2 and AH2′ helices, the same mild oxidation did yield high extents of disulfide bond formation among three symmetric pairs of buried positions predicted by the NMR model to be buried with high AH2–AH2′ proximities. The findings suggest that the AH2–AH2′ helix pair may collide more rapidly than the AH1–AH1′ helix pair, since disulfide formation rates are often correlated with sulfhydryl collision frequencies (other factors can contribute as well; see below). The NMR model alone cannot account for the significant differences between the AH1–AH1′ and AH2–AH2′ disulfide formation rates, since the distances between their helix pairs are similar (6). However, in the context of the full-length, membrane-bound receptor, the slower AH1–AH1′ collision rate could arise from additional constraints provided by the coupling of helices AH1 and AH1′ to transmembrane helices TM2 and TM2′ (see Discussion). Thus, the NMR model adequately explains the previous chemical reactivity and disulfide scans.
Designing a Rigorous Disulfide Mapping Test of the NMR Model
The disulfide mapping approach is based on the assumption that, under identical oxidizing conditions, proximal Cys pairs in a stable protein structure will generally collide and form disulfide bonds more rapidly than distal Cys pairs (15,16). To form a disulfide bond under oxidizing conditions, two Cys residues must collide, requiring a β-carbon–β-carbon distance of ≤4.6 Å, and furthermore must exhibit angular orientations within an allowed range of geometries. The sulfhydryl moieties of the Cys side chains must be accessible to the oxidation agent, and the local environment must allow deprotonation of the sulfhydryls to yield their redox-active sulfanion states. If all these conditions are met, the two sulfhydryl residues can collide with a favorable geometry, be oxidized, and form a disulfide bond.
The previous cysteine and disulfide scanning study revealed important features of HAMP structure but could not fully define the helix packing because only symmetric interactions between the AH1–AH1′ and AH2–AH2′ helix pairs were examined (4). In the NMR HAMP model, the four helices of the dimer are arranged in a parallel, four-helix bundle (6). To test this model, the present targeted disulfide mapping analysis asks whether the asymmetric AH1–AH2′ interface observed in the NMR structure is present in the aspartate receptor HAMP domain. In the NMR model, the AH1 and AH2′ helices form extensive close contacts. Moreover, since these two helices are in different subunits, the formation of a disulfide bond between them will yield an easily detected covalent dimer. Such characteristics make the AH1–AH2′ interface ideal for disulfide mapping. Furthermore, if the AH1–AH2′ packing arrangement can be determined, this constraint, together with constraints provided by the previous AH2–AH2′ disulfide scan (4) and homodimer symmetry (17), should be sufficient to define the general architecture of helix packing in the native aspartate receptor.
The disulfide mapping approach was applied to the AH1–AH2′ interface by designing two types of engineered Cys pairs: one type predicted by the NMR HAMP structure to be proximal and the other type predicted to be distal (6). Multiple examples of both proximal and distal Cys pairs were employed to check for self-consistency. Suitable proximal and distal pairs were selected by examining all possible AH1–AH2′ pairs of positions in the NMR model (6). For each pair, four β-carbon–β-carbon distances were measured since the introduction of two engineered Cys residues (X and Y) into both subunits of the symmetric dimer can yield four different types of disulfide bonds: two symmetric, intersubunit disulfides (X–X′ and Y–Y′); one asymmetric, intersubunit disulfide (X–Y′); and one asymmetric, intra-subunit disulfide (X–Y). This study operationally defined unique proximal pairs of positions as those for which the intersubunit, asymmetric distance (X–Y′) was less than 7 Å, while the three other distances (X–X′, Y–Y′, and X–Y) all exceeded 10 Å. Distal pairs were defined as those for which all four distances exceeded 10 Å.
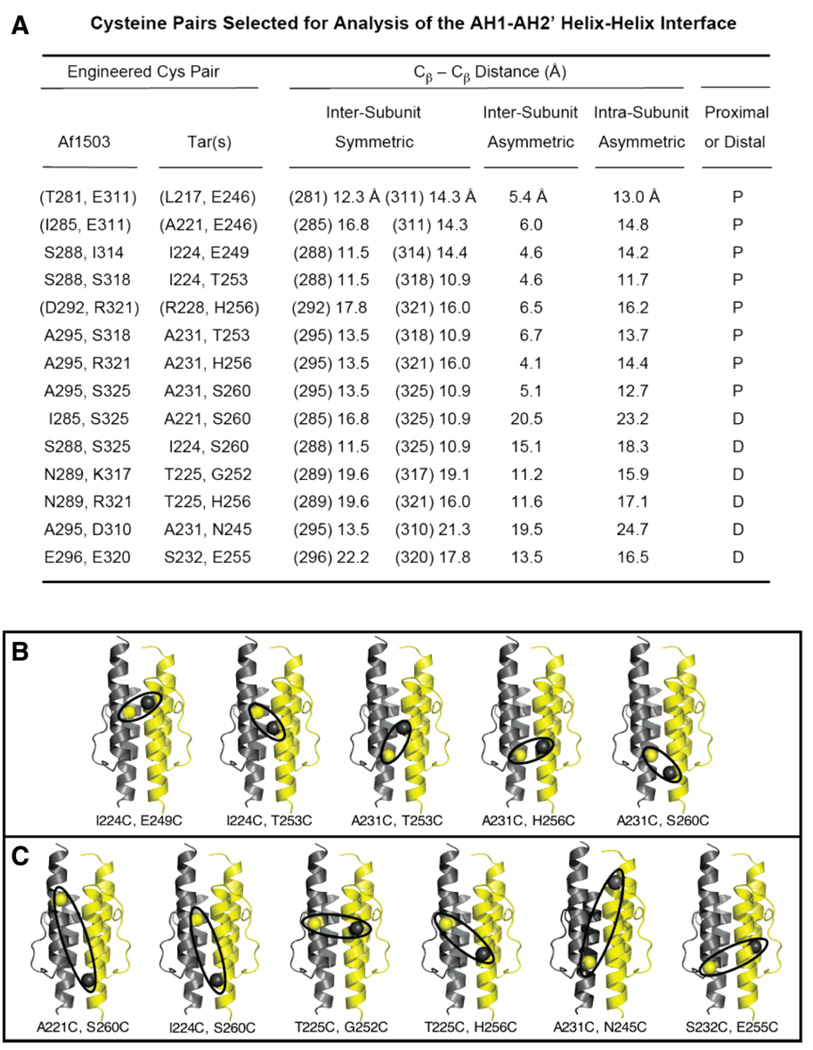
Figure 2A lists the eight unique, proximal pairs of positions targeted for proximal Cys substitutions, as well as six pairs of positions targeted for distal Cys substitutions. Also summarized are the four β-carbon–β-carbon distances observed for each Cys pair in the NMR HAMP model (6). The eight proximal Cys pairs represent a complete set: no other AH1–AH2′ pairs satisfy the operational definition of a unique proximal pair. The six distal Cys pairs represent a subset of the potential distal pairs but were selected because their Cys substitutions lie at or near the positions chosen for Cys substitution in the proximal pairs, to ensure that the proximal and distal pairs experienced similar chemical and motional environments.
Figure 2.
Cys pairs selected for analysis of the HAMP AH1–AH2′ helical interface. (A) Summary of pairs of positions selected in the archeal HAMP structure (6), and the corresponding pairs in the S. typhimurium aspartate receptor. Also shown are the four β-carbon–β-carbon distances measured for each pair in the homodimeric, archeal HAMP structure. Each proximal pair (P) possesses an asymmetric, intersubunit distance of < 7 Å, while the other three distances simultaneously exceed 10 Å. In each distal pair (D), all four distances exceed 10 Å. Parentheses indicate low-yield di-Cys mutants not suitable for further analysis (see the text). (B and C) Locations of the proximal (B) and distal (C) Cys pairs employed for disulfide mapping. Ovals highlight the asymmetric, intersubunit separation for each pair.
Construction and Expression of Di-Cys Receptors
To create aspartate receptors containing the selected proximal and distal Cys pairs, the HAMP sequence alignment of Figure 1B (5, 6) was used to identify the aspartate receptor positions that correspond to the selected positions in the NMR model of the archeal HAMP domain. Subsequently, PCR site-directed mutagenesis of the aspartate receptor gene tar was used to construct the 14 di-Cys mutants as confirmed by DNA sequencing. The resulting mutants were expressed by plasmid pSCF6 under control of the native tar promoter in an E. coli strain lacking all major components of the chemotaxis pathway, including the aspartate receptor and adaptation enzymes (CheB and CheR, respectively). The absence of adaptation enzymes ensured that each mutant receptor population possessed identical adaptation sites and thus was homogeneous.
When expressed, 11 of the 14 di-Cys mutants were stable, membrane-incorporated receptors accounting for 8–17% of the total membrane protein (TMP). The other three mutants exhibited very low yields (<1% TMP) insufficient for further study. These low-yield mutants, indicated by parentheses in Figure 2A, were A221C/E246C, L217C/E246C, and R228C/H256C, all members of the proximal class. Two of these three mutants possessed the E246C substitution, which neutralizes a conserved glutamate side chain located at the N-terminus of helix AH2 where its negative charge may be needed to stabilize the helix dipole. Panels B and C of Figure 2 illustrate the locations of the remaining Cys pairs in five proximal and six distal di-Cys mutants, each of which is a stable, active receptor as indicated by its normal expression level and its measurable ability to bind and stimulate CheA kinase (see below).
Disulfide Formation Reactions of Di-Cys Mutants
For each of the five proximal Cys pairs, the NMR model (6) predicts that little or no backbone motion (0–2.4 Å) would be required to form the asymmetric, intersubunit (X–Y′) disulfide bond. By contrast, for all remaining disulfide bonds possible for proximal Cys pairs, and for all disulfides possible for distal Cys pairs, the NMR model predicts that significantly larger backbone motions (>5.4 Å) would be required for bond formation. Thus, the five proximal Cys pairs are predicted to more rapidly collide and form dimer-linking disulfide bonds than the six distal pairs under identical oxidizing conditions.
To quantitatively compare the propensities of the proximal and distal Cys pairs to form disulfide bonds, it was necessary to tune the oxidation chemistry so that both rapid and slow disulfide formation reactions could be monitored under the same reaction conditions. Various mild, intermediate, and strong oxidation chemistries were tested, and an intermediate chemistry was found to generate the best discrimination between rapid and slow reactions. The optimized chemistry utilized ambient molecular oxygen (~200 µM) as the oxidation agent, and the oxidation reaction was triggered by addition of a redox catalyst, 1.0 mM Cu(II)(1,10-phenanthroline)3, together with 1.5 mM EDTA to moderate the oxidation strength by buffering the Cu(II). The reaction was allowed to proceed for 1.0 min at 30 °C before being quenched with a large excess of NEM (40 mM) to block the remaining free Cys residues, and EDTA (10 mM) to chelate the catalytic Cu(II). For comparison, parallel reactions were carried out for each di-Cys mutant and its two corresponding single-Cys mutants. Following the reaction, monomeric and disulfide-linked dimer products were resolved by SDS–PAGE and quantitated by densitometry. Fractional disulfide formation was calculated as the density ratio (dimer)/(monomer + dimer).
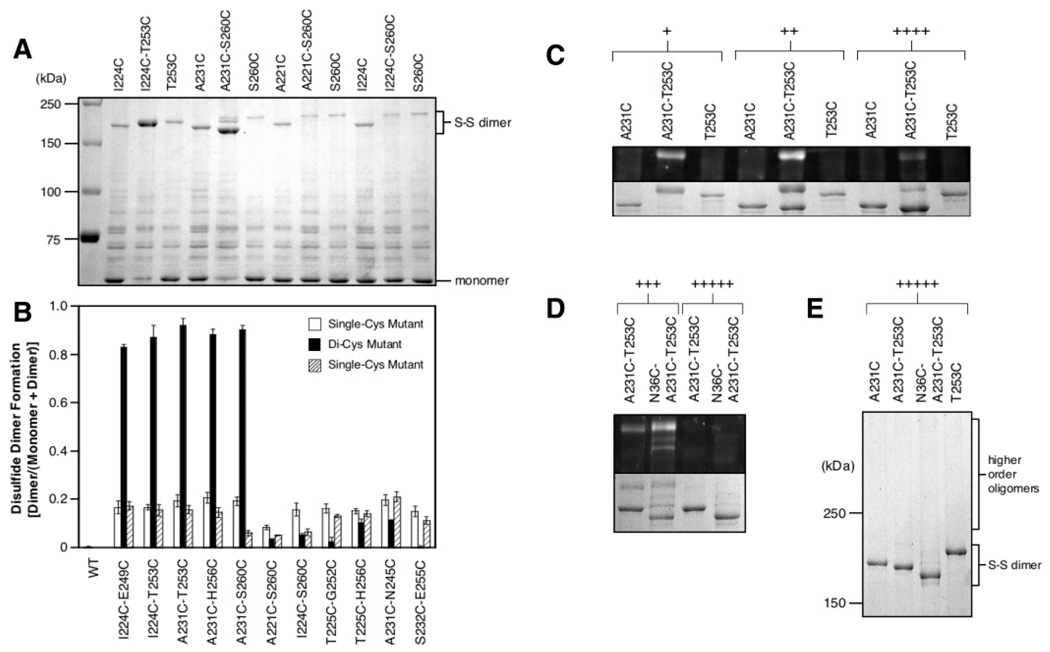
Figure 3A illustrates the reaction products observed for representative proximal and distal di-Cys receptors, as well as for the corresponding single-Cys receptors. Notably, for the proximal di-Cys receptors, nearly all of the receptor monomer is converted to dimer during the reaction. By contrast, for the distal di-Cys receptors, most of the product remains in the monomeric state following the reaction. Similarly, for the single-Cys receptors of both classes, most product is monomeric. In all cases, the dimeric products observed upon oxidation were reduced back to monomers by treatment with 50 mM DTT (1 min, 95 °C) before SDS–PAGE (data not shown). As noted previously (18), these dimers exhibit slower migration on SDS–PAGE than expected for their molecular weight because the disulfide bond cross-linking the two denatured polypeptide chains yields an X shape that experiences a high level of friction when moving through a molecular mesh. Moreover, the number and positions of cross-links along the chains modulate the migration rate (18), enabling resolution of different dimeric products.
Figure 3.
Comparison of disulfide-linked dimer formation for proximal and distal Cys pairs, and assignment of products. (A) Oxidation of four representative di-Cys mutant aspartate receptors, and the corresponding single-Cys mutants, in isolated E. coli membranes. Reactions were initiated by addition of the redox catalyst Cu(II)(1,10-phenanthroline)3 (1 mM final) to receptor (2.5 µM dimer) in 160 mM KCl and 50 mM Tris (pH 7.2 with HCl), 5 mM MgCl2, and 1.5 mM EDTA, followed by incubation for 1 min at 30 °C, quenching, and analysis by Coomassie SDS–PAGE. The I224C/T253C and A231C/S260C Cys pairs are predicted to be proximal, while the A221C/S260C and I224C/S260C pairs are predicted to be distal (Figure 2A). (B) Extent of disulfide-linked dimer formation for all 11 di-Cys mutants and the corresponding single-Cys mutants. Oxidation reactions were carried out and analyzed as described for panel A. The I224C/E249C, I224C/T253C, A231C/T253C, A231C/H256C, and A231C/S260C pairs are predicted to be proximal, while the A221C/S260C, I224C/S260C, T225C/G252C, T225C/H256C, A231C/N245C, and S232C/E255C pairs are predicted to be distal. (C) Kinetic product analysis for a representative di-Cys mutant, A231C/T253C, and the corresponding single-Cys mutants. Oxidation at 30 °C as in panel A, except that oxidation strength and time were varied to examine early (+), middle (++), and late (++++) products as follows: (+) 0.5 mM redox catalyst, 5 mM EDTA, 1 min; (++) 1 mM redox catalyst, 0.8 mM EDTA, 5 min; (++++) 1 mM redox catalyst, no EDTA, 10 min. Reactions were quenched with 5-fluoresceinmaleimide (5FM, 500 µM) to visualize the unreacted, free Cys residues remaining after oxidation. The expanded view of the dimer region compares fluorescent (top) and all (bottom) dimeric products. The most fluorescent band is an early oxidation product containing one asymmetric disulfide and two free cysteines modified with 5FM during quenching. (D) Oxidized product analysis for a representative tri-Cys mutant, N36C/A231C/T253C, and the corresponding di-Cys mutant. Oxidation as in panel A, except that oxidation strength and time were varied to examine middle (+++) and late (+++++) products as follows: (+++) 1 mM redox catalyst, no EDTA, 5 min at 30 °C; (+++++) 2 mM redox catalyst, no EDTA, 20 min at 37 °C. Reactions were quenched with 5FM as in panel C. Images show the expanded dimer region comparing fluorescent (top) and all (bottom) dimeric products. Following oxidation, the uppermost fluorescent dimer contained one asymmetric disulfide and either two free cysteines for the di-Cys mutant, or four free cysteines for the tri-Cys mutant. The latter product was labeled by as many as four 5FM molecules during quenching and thus exhibits the highest ratio of fluorescence/Coomassie stain. (E) Same as panel D but showing the entire oligomer region for the Coomassie gel. No oligomers larger than dimers are detected. In panels C and E, single-Cys mutants were overloaded relative to di-Cys mutants to ensure visibility of their relatively rare dimeric products.
Figure 3B summarizes the fractional extents of disulfide dimer formation measured for all 11 di-Cys receptors, and for the corresponding single-Cys receptors. Strikingly, the five di-Cys receptors predicted by the NMR structure of the archeal HAMP domain to possess unique, proximal Cys pairs all yielded rapid disulfide-linked dimer formation. By contrast, the distal di-Cys receptors, and all single-Cys receptors, formed disulfide dimers at least 4-fold more slowly (Figure 3B). (The observed 4-fold differences must underestimate the true differences, since the proximal di-Cys reactions were approaching completion.) Without exception, the observed pattern of disulfide formation rates was fully consistent with the structural predictions of the NMR HAMP model (6). While the pattern was highly correlated with Cys pair proximity in the NMR model, it was poorly correlated with the chemical reactivities and solvent exposures of Cys residues (compare Figure 1C and Figure 3B), indicating that sulfhydryl proximities, not their intrinsic reactivities or accessibilities, dominated the rates. To further test this conclusion, the mechanisms of disulfide bond formation that led to dimeric products were determined.
Determination of Reaction Mechanism
To directly assign SDS–PAGE bands to specific products, a kinetic approach was developed to ascertain whether the rapidly formed dimer bands contained (a) one or two disulfide bonds formed between (b) symmetric or asymmetric Cys pairs. Each of the five unique, proximal di-Cys mutants was oxidized to three different extents by different reaction conditions, and then the reactions were quenched with 5-fluoresceinmaleimide (5FM) to label any remaining free Cys residues. The resulting 5FM fluorescence and Coomassie-stained SDS–PAGE images are illustrated in Figure 3C for the representative A231C/T253C di-Cys receptor. At low levels of oxidation, the major early dimeric product was labeled with the 5FM probe and exhibited a mobility slower than that of dimers of the corresponding single-Cys mutants. A similar mobility shift was observed for representative early dimers even in the absence of 5FM labeling; thus, the mobility change was not simply an artifact of the fluorescent probe (data not shown). It follows that the early dimer was formed by a single asymmetric disulfide bond that yielded a migration rate different from that of either of the corresponding symmetric disulfide bonds and also retained two free Cys residues that were labeled with 5FM. At the highest level of oxidation, the early dimer containing a single, asymmetric disulfide bond largely disappeared. In its place arose a faster migrating dimeric product which no longer labeled with 5FM. The simplest explanation is that a second asymmetric disulfide bond was formed within the same dimer, protecting the remaining Cys pair from 5FM labeling and increasing the gel mobility by making the molecule more compact (as typically observed for intrachain disulfide bonds (15)). Analogous results were observed for all five proximal di-Cys mutants, indicating that in each case the major disulfide-linked dimer observed upon terminal oxidation contained two asymmetric bonds (X–Y′ and X′–Y).
In principle, the rapidly formed, disulfide-linked dimers observed upon oxidation of the five unique, proximal di-Cys receptors could be produced by intra- or interdimer collisions. To resolve these possibilities, each of the five unique, proximal Cys pairs was combined with a third Cys residue at the N36 position to yield a tri-Cys mutant. The periplasmic N36C mutation has previously been shown to rapidly and efficiently form an intradimer disulfide bond which retains normal receptor structure and activity (7, 18, 19), and the periplasmic location of the N36C–N36C′ bond helps ensure that it will not perturb disulfide formation reactions involving the cytoplasmic HAMP domain. When the five tri-Cys mutants were oxidized under conditions designed to trap early and late products, the earliest dimeric products were highly fluorescent since rapid formation of a single asymmetric disulfide bond in HAMP left four free Cys residues that were labeled with 5FM during quenching, as illustrated for the representative A231C/T253C/N36C tri-Cys receptor in Figure 3D. Strong oxidation of this tri-Cys mutant yielded a terminal dimeric product that migrated slightly more rapidly than the corresponding terminal di-Cys product and which was nonfluorescent, indicating that all six Cys residues were incorporated into three disulfide bonds, including the N36C–N36C′ bond. Most importantly, strong oxidation yielded no detectable higher-order oligomers (Figure 3E), demonstrating that virtually all collisions leading to disulfide bond formation occurred within the same dimer. Analogous results were also observed for the other four proximal di-Cys receptors (data not shown). Thus, the five proximal di-Cys mutants each formed a disulfide-linked dimer via an intradimer reaction, since in each case introduction of the N36C–N36C′ disulfide bond failed to yield higher-order disulfide-linked products.
Overall, the disulfide mapping evidence is fully consistent with the NMR model of the archeal HAMP domain (6). The asymmetric Cys pairs predicted by the model to lie in the proximity of the AH1–AH2′ helix–helix interface were found to rapidly form disulfide bonds via collisions within the same dimer. Moreover, the symmetric and asymmetric Cys pairs predicted to be distal formed disulfide bonds much more slowly.
Effects of Di-Cys Mutations on Receptor Function in Vivo and in Vitro
To a first approximation, the receptor, and thus the HAMP domain, is a two-state switch with “on” and “off” signaling conformations (3). An important question is whether the HAMP domain structure defined by the NMR model resembles only one of these states or is similar to both states. To address this question, the functional effects of di-Cys mutations and disulfide bonds were examined.
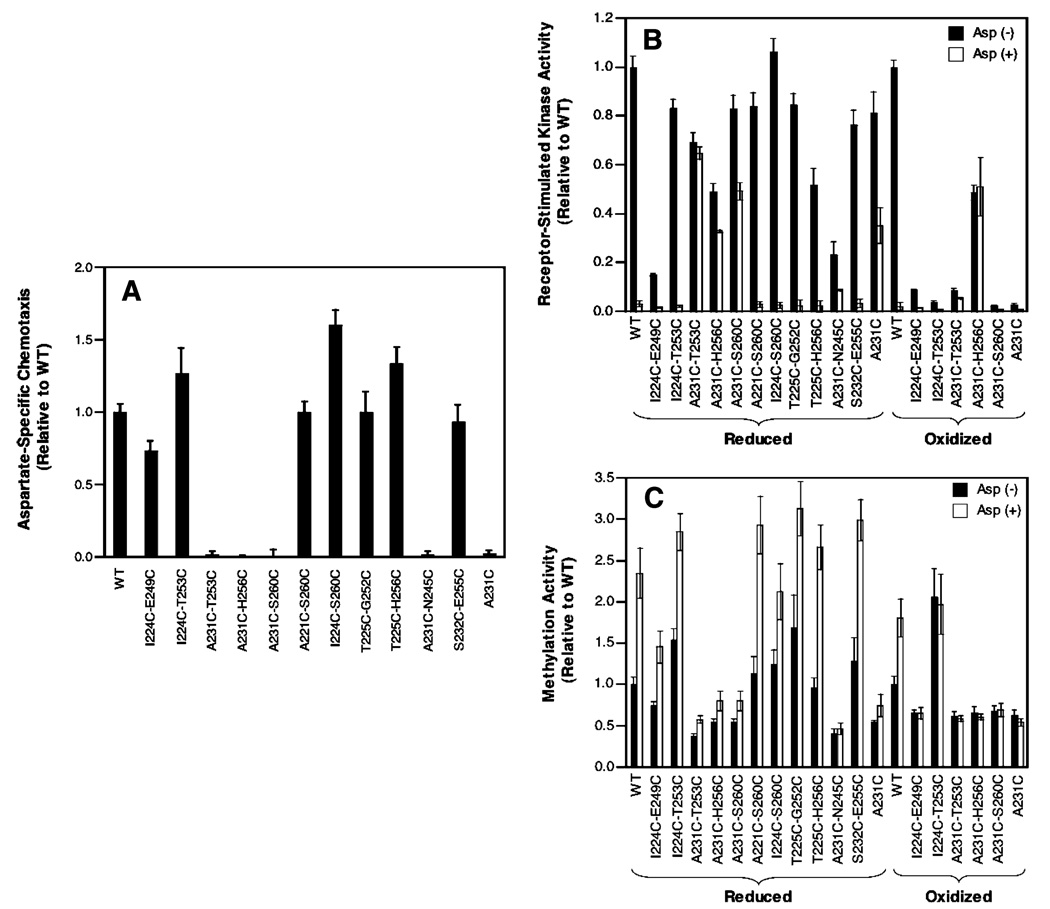
The ability of the di-Cys receptors to restore chemotaxis to cells in vivo was assessed by the standard swim plate assay (14). Figure 4A shows that 7 of the 11 di-Cys mutant receptors retain function similar to that of the wild-type receptor in the chemotactic swarm assay. All 7 exhibit the ability to restore cellular chemotaxis to a level within 70% of that observed for the wild-type receptor. The four remaining di-Cys mutants (A231C/T253C, A231C/H256C, A231C/S260C, and A231C/N245C) share the A231C substitution and completely block chemotaxis, as does A231C alone in a single-Cys mutant. These mutant receptors, which are known to express at levels similar to that of the wildtype receptor (see above), must be significantly damaged in their ability to bind or regulate CheA kinase, or in their adaptation properties. To resolve these possibilities, the effect of di-Cys mutations on receptor function in vitro was analyzed.
Figure 4.
Effect of Cys substitutions and disulfide bonds on receptor activity in vivo and in vitro. (A) Comparison of reduced WT and mutant receptor activities in the standard in vivo swim plate assay of bacterial chemotaxis in soft agar (11, 14). (B) Comparison of wildtype and mutant receptor activites following receptor reduction (left) or oxidation (right) in the standard in vitro assay measuring receptor-regulated CheA kinase activity in the reconstituted signaling complex (12). Shown are kinase activities in the absence and presence of attractant aspartate. (C) Comparison of wild-type and mutant receptor activities following reduction (left) or oxidation (right) in the standard in vitro assay measuring the extent of receptor methylation by CheR (12), in the absence and presence of aspartate.
The standard in vitro receptor-coupled kinase assay (12, 20, 21) measures the ability of the receptor to regulate the activity of CheA kinase in the reconstituted receptor–CheA–CheW signaling complex. Since the adaptation enzymes are absent, this assay is well-suited for detecting even subtle effects of mutations on receptor-mediated kinase regulation. Isolated E. coli membranes containing a given overexpressed receptor in its fully reduced state were reconstituted with purified CheA and CheW to assemble the functional signaling complex, and then the activity of the complex was measured in the absence and presence of aspartate by monitoring the steady-state level of phospho-CheY. Figure 4B indicates that, in the absence of attractant, all 11 di-Cys mutants retained the ability to bind CheA and stimulate measurable levels of kinase activity. Moreover, 9 of the 11 di-Cys mutants stimulated kinase activity to within 2-fold of that observed for the wild-type receptor. Strikingly, however, all four of the di-Cys mutants containing A231C, as well as the A231C single-Cys mutant, failed to fully inactivate the kinase upon addition of attractant (compare to the wild type). It follows that the A231C substitution locks the receptor in the kinase-activating on-state, to a degree that depends on the specific mutant, thereby explaining the failure of A231C-containing mutants to function in the in vivo chemotaxis assay.
For the five proximal di-Cys mutants, strong oxidation conditions could be used to drive disulfide bond formation nearly to completion, thereby enabling analysis of their kinase regulation in the oxidized state as well as the reduced state. Figure 4B shows that for four of the five proximal Cys pairs, disulfide bond formation virtually eliminated kinase activation in the reconstituted signaling complex. By contrast, the A231C–H256C′ disulfide bond retained approximately 50% of the native activation and exhibited lock-on character. Notably, this disulfide is formed between the most proximal Cys pair, whose β-carbons are separated in the NMR model by only 4.1 Å and thus are predicted to be close enough to form a disulfide bond with little or no backbone motion.
The standard in vitro receptor methylation assay measures the ability of the receptor to regulate the methyl esterification of its adaptation sites by the adaptation enzyme CheR (12, 22). This assay, coupled with the in vitro kinase regulation assay, enables positive identification of mutations which drive the receptor toward the on- and off-states, respectively, since these two states yield opposite activities in the two assays. Figure 4C shows that the reduced receptors which exhibited normal aspartate-induced kinase inhibition also displayed aspartate-induced activation of receptor methylation, as observed for the wild-type receptor. By contrast, the locked-on receptors all failed to exhibit aspartate-induced stimulation of receptor methylation, as expected for mutations which covalently trap the receptor in the native on-state. Notably, when the five proximal disulfide bonds were formed by oxidation, all retained significant methylation rates, indicating that disulfide linkage did not dramatically perturb their structure, but the linkage did lock their conformations since they no longer responded to aspartate. Moreover, the I224C–T253C′ disulfide yielded high methylation activity indistinguishable from that of the asparate-occupied wildtype receptor in both the absence and presence of aspartate, indicating this disulfide linkage locks the receptor in its offstate. The corresponding Cys pair is separated by only 4.6 Å in the NMR model and thus is predicted to require little or no backbone motion to collide and form a disulfide bond.
Overall, the in vivo and in vitro functional assays revealed that all 11 of the reduced di-Cys receptors retained the ability to bind and activate CheA kinase to varying degrees. However, the four di-Cys receptors possessing the A231C substitution were locked in the kinase-activating on-state and thus failed to function in the in vivo chemotaxis assay. Formation of a disulfide bond between A231C and the closest position in the NMR model, H256C′, locked the receptor in the on-state, while formation of a disulfide between I224C and nearby T253C′ locked the off-state. It follows that the AH1–AH2′ interface is critical for on–off switching.
DISCUSSION
Implications for HAMP Domain Structure
Three lines of past and present evidence indicate that the symmetric, four-helix bundle architecture recently described in the NMR structure of an isolated archeal HAMP domain (6) is an accurate description of HAMP in the full-length, membrane-bound aspartate receptor. First, the chemical reactivities previously measured for Cys residues scanned through the aspartate receptor HAMP domain (4) exhibit the same α-helical patterns of solvent exposure and burial calculated for the archeal HAMP structure. It follows that the homologous AH1 and AH2 helices of the two HAMP domains share similar sequence locations, exposed faces, and buried faces. Second, previous disufide scanning studies (4) and our disulfide mapping studies of the AH2–AH2′ and AH1–AH2′ helix packing surfaces, respectively, reveal the same pattern of spatial proximities in the asparate receptor that were observed in the archeal HAMP structure. These findings confirm that the two HAMP domains share the same parallel arrangement of the AH1–AH2′ and AH2–AH2′ helix pairs and exhibit similar detailed contacts at the subunit interface. Third, as illustrated by the simple geometric analysis in Figure 5A, the parallel and proximal locations of the AH1–AH2′ and AH2–AH2′ helix pairs in the asparate receptor, together with the C2 symmetry of the homodimer, are sufficient to define this HAMP domain as a parallel, four-helix bundle like the archeal HAMP structure. In short, the packing and relative orientations of helices appear to the same in these two HAMP domains, within the resolution of Cys scanning and mapping methods. Since all studies of the aspartate receptor were carried out in the full-length, membrane-bound receptor, it follows that the HAMP domain of this native chemoreceptor possesses the same parallel, four-helix bundle architecture displayed by the archeal HAMP domain.
Figure 5.
Model for HAMP structure in the full-length aspartate receptor. (A) The model is built using disulfide mapping constraints (4) to define the geometry of the AH2–AH2′ helix pair, then building in the AH1–AH2′ interaction based on additional disulfide mapping constraints (Figure 3), and finally using the known C2 symmetry of the homodimer to complete the parallel, four-helix bundle (see the text). (B and C) Schematic model incorporating HAMP into the full-length receptor, illustrating a new helix nomenclature. For clarity, the model is shown in space-filling, ribbon, and cylinder formats which together portray the overall shape, subunit supercoiling, and helical structure of the homodimer. Highlighted is (i) the junction between the end of helix TM2 and the beginning of HAMP helix HD1, where there is a helical phase change (see the text), and (ii) the major proteolysis site, Arg259. The protease accessibility of the latter site suggests that position 259 lies at or near C-terminus of the distinct HAMP structural domain. The model of the full-length homodimer illustrates the three distinct modules (transmembrane signaling, signal conversion, and kinase control) of the receptor, as well as the three functional regions (adaptation, flexible, and protein interaction) of the latter module (3, 27).
The one significant difference between the asparate receptor and archeal HAMP domains is found in the connector region between the AH1 and AH2 helices. Chemical reactivity data indicate that the connector packs against these two helices in the aspartate receptor (4), but the observed reactivities yield a pattern of exposure and burial different from that seen in the connector region of the archeal HAMP structure (6). Thus, connector packing does not appear to be the same in the two HAMP domains, which is not surprising since their connector sequences and lengths are different.
How can the parallel, four-helix bundle architecture of the HAMP domain be incorporated into a structural model for the full-length chemoreceptor dimer? Panels B and C of Figure 5 illustrate the simplest model, making use of the known boundary conditions in which the N-terminus of HAMP is coupled to the C-terminus of transmembrane signaling helix TM2, while the C-terminus of HAMP is coupled to the N-terminus of long cytoplasmic domain helix CD1. Now that all the helical elements of the chemoreceptor are identified, panels B and C of Figure 5 also propose a new system of helix nomenclature designed to maximize the consistency and descriptive character of helix names. In this scheme, the four periplasmic domain helices are renamed PD1–4, the two transmembrane helices remain TM1 and TM2, the two HAMP domain helices are renamed HD1 and HD2, and the two cytoplasmic domain helices remain CD1 and CD2. The HAMP domain is proposed to begin where TM2 emerges from the membrane into the cytoplasm (residue H214 in the aspartate receptor) and end at the junction between the HAMP domain parallel four-helix bundle and the cytoplasmic domain antiparallel four-helix bundle.
Important features appear to be located at the junctions between the HAMP domain and the adjacent regions. The cytoplasmic, N-terminal end of TM1, where numerous mutations disrupt function, may directly contact HD1 of the HAMP domain. Moreover, the junction between TM2 and HD1 may not be a simple, continuous helix, since the faces of TM2 (23) and HD1 (4, 6) oriented toward the center of the dimer exhibit a different helical register. In contrast, the buried faces of HD2 (4, 6) and CD1 (24) do possess the same helical register, arguing that the junction between these two regions is a simple, continuous helix, at least in one signaling state. Such continuity would place Arg259 of the aspartate receptor at a position on HD2 where the four-helix bundles of the HAMP and cytoplasmic domains meet. In this region, the gap between the adjacent four-helix bundles would reduce the number of helix–helix interactions and could expose Arg259 to solvent. This picture helps explain the previous observation that Arg259 is the major proteolysis site of the aspartate receptor (25), thereby providing further support for the proposed model of HAMP architecture in the full-length receptor. Since proteases typically cannot hydrolyze an intact helix, the helical region containing the Arg259 region may well be unstable. It follows that the junction between the two four-helix bundles may serve as a hinge in receptor assembly, disassembly, or on–off switching. Our current working model proposes that the R259 proteolysis site at the junction between the two bundles defines the C-terminus of the HAMP domain (Figure 5B,C). However, a comparable proteolytic analysis has not yet been carried out for other receptors, and conserved residues C-terminal to this site have been used to argue that HAMP extends several helical turns further (5). Additional studies are needed to rigorously define the location of the HAMP C-terminus.
Implications for HAMP Domain On–Off Switching
Five lines of evidence indicate that the parallel, four-helix bundle HAMP architecture described above closely resembles the structure of the kinase-activating on-state and that this structure undergoes only a small rearrangement during switching to the off-state. First, the disulfide mapping studies of the HD1–HD2′ (AH1-AH2′) interface presented here were carried out for the apo receptor in the absence of aspartate to stabilize the on-state conformation. Second, the observation that A231C is a lock-on mutation suggests that this mutation might trap the on-state by increasing the strength of HD1–HD2′ packing. Indeed, in the NMR structure of the archeal HAMP domain (6), the conserved alanine at the homologous position serves as a knob in a socket at the HD1–HD2′ interface (26), but the short alanine side chain yields suboptimal packing contacts with its hole residues. Substitution of the larger cysteine would be expected to strengthen the socket, thereby stabilizing the HD1–HD2′ interaction and the on-state, as observed. Third, the A231C–H256C′ disulfide bond between the A231C knob and its closest hole residue forms rapidly and covalently stabilizes the HD1–HD2′ interface, thereby locking the receptor in its on-state. Fourth, the I224C–T253C′ disulfide bond also forms rapidly and stabilizes the HD1–HD2′ interface but locks the receptor in its off-state. Fifth, chemical reactivities previously measured for Cys residues scanned throughout the HAMP domain were similar in the absence and presence of attractant aspartate (Figure 2A) (4), suggesting that the HAMP conformational change induced by attractant-induced switching to the off-state is subtle and does not greatly alter helix packing or architecture of the four-helix bundle. Overall, the evidence suggests that the parallel, four-helix bundle architecture of the aspartate receptor HAMP domain is characteristic of both the on- and off-states, with only a minor rearrangement during on–off switching.
Although the structural changes that occur in the transmembrane signaling, HAMP, and kinase control modules are concerted, it is useful to separately consider the different type of structural rearrangement occurring in each module. In the current working model, binding of attractant to the transmembrane signaling module triggers a piston-type displacement of its PD4–TM2 signaling helix toward the cytoplasm (2, 3, 7–9). The HAMP domain, serving as a signal converter, transforms this piston displacement of TM2 into a different structural change that strains the subunit interface, presumably by altering CD1–CD1′ helix–helix packing (3, 10, 11). Finally, the extended kinase control module transmits this interfacial signal to CheA kinase (3, 10, 11). These findings suggest that the HD1–HD2′ interface is central to on–off switching. Interestingly, the geometries of the newly discovered lock-on and lock-off disulfides in the HAMP domain are consistent with the hypothesis that the HD1 helix undergoes a piston displacement similar in magnitude and direction to that described for TM2 (see Figure 2B). It is not yet known how HAMP converts the piston displacement into a strain of the CD1–CD1′ interface, but one can imagine several possibilities. In one, the piston displacement of the TM2–HD1 structure destabilizes the HAMP four-helix bundle, thereby facilitating concerted rotations of the four helices about their long axes to yield a different bundle conformation (6). At the same time, the concerted rotations of HD2 and HD2′ would strain the CD1–CD1′ interface. Such a picture combines elements of the published piston and rotational models for transmembrane and HAMP signaling, respectively (2, 6). Alternatively, if the HD2–HD2 interface possesses a stable pivot axis perpendicular to the helix axes, the piston displacement could pivot the two HAMP subunits relative to one another, thereby generating a scissors-type movement that strains the CD1–CD1′ interface. Further studies are needed to resolve these and other on–off switching models. More broadly, given that the parallel, four-helix bundle architecture of HAMP is conserved, it is likely that the on–off switching mechanism of this motif is conserved as well.
ACKNOWLEDGMENT
We gratefully acknowledge Drs. J. S. Parkinson, G. L. Hazelbauer, and M. D. Manson for helpful discussions; Dr. Parkinson for the kind gift of strains; and M. Gonzalez for advice on SOCKET (26).
Footnotes
Support provided by NIH Grant R01 GM-040731 (to J.J.F.).
Abbreviations: HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, and phosphatases; 5FM, 5-fluoresceinmaleimide; NEM, N-ethylmaleimide; DTT, dithiothreitol; EDTA, ethylenediamine-tetraacetic acid.
REFERENCES
- 1.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 2007 doi: 10.1016/j.tibs.2007.09.014. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 6.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 7.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: A model. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller AS, Falke JJ. Side Chains at the Membrane-Water Interface Modulate the Signaling State of a Transmembrane Receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 10.Bass RB, Falke JJ. The aspartate receptor cytoplasmic domain: In situ chemical analysis of structure, mechanism and dynamics. Struct. Folding Des. 1999;7:829–840. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: Electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA kinase of bacterial chemotaxis: Chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chervitz SA, Lin CM, Falke JJ. Transmembrane signaling by the aspartate receptor: Engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adler J. Chemoreceptors in bacteria. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- 15.Careaga CL, Falke JJ. Thermal motions of surface α-helices in the d-galactose chemosensory receptor. Detection by disulfide trapping. J. Mol. Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: Applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 18.Falke JJ, Dernburg AF, Sternberg DA, Zalkin N, Milligan DL, Koshland DE., Jr Structure of a bacterial sensory receptor: A site-directed sulfhydryl study. J. Biol. Chem. 1988;263:14850–14858. [PubMed] [Google Scholar]
- 19.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 20.Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ninfa EG, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 22.Chelsky D, Gutterson NI, Koshland DE., Jr A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal. Biochem. 1984;141:143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- 23.Chervitz SA, Falke JJ. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 1995;270:24043–24053. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danielson MA, Bass RB, Falke JJ. Cysteine and disulfide scanning reveals a regulatory α-helix in the cytoplasmic domain of the Asp receptor. J. Biol. Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mowbray SL, Foster DL, Koshland DE., Jr Proteolytic fragments identified with domains of the aspartate chemoreceptor. J. Biol. Chem. 1985;260:11711–11718. [PubMed] [Google Scholar]
- 26.Walshaw J, Woolfson DN. Socket: A program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 27.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]