Environmental stresses affect the nitrate distribution between roots and shoots, and transporters that remove nitrate from the xylem sap are essential for long-distance nitrate transport. This study shows that the nitrate transporter NRT1.8 is induced by cadmium and removes nitrate from xylem vessels and furthermore establishes a correlation between nitrate allocation and cadmium tolerance.
Abstract
Long-distance transport of nitrate requires xylem loading and unloading, a successive process that determines nitrate distribution and subsequent assimilation efficiency. Here, we report the functional characterization of NRT1.8, a member of the nitrate transporter (NRT1) family in Arabidopsis thaliana. NRT1.8 is upregulated by nitrate. Histochemical analysis using promoter-β-glucuronidase fusions, as well as in situ hybridization, showed that NRT1.8 is expressed predominantly in xylem parenchyma cells within the vasculature. Transient expression of the NRT1.8:enhanced green fluorescent protein fusion in onion epidermal cells and Arabidopsis protoplasts indicated that NRT1.8 is plasma membrane localized. Electrophysiological and nitrate uptake analyses using Xenopus laevis oocytes showed that NRT1.8 mediates low-affinity nitrate uptake. Functional disruption of NRT1.8 significantly increased the nitrate concentration in xylem sap. These data together suggest that NRT1.8 functions to remove nitrate from xylem vessels. Interestingly, NRT1.8 was the only nitrate assimilatory pathway gene that was strongly upregulated by cadmium (Cd2+) stress in roots, and the nrt1.8-1 mutant showed a nitrate-dependent Cd2+-sensitive phenotype. Further analyses showed that Cd2+ stress increases the proportion of nitrate allocated to wild-type roots compared with the nrt1.8-1 mutant. These data suggest that NRT1.8-regulated nitrate distribution plays an important role in Cd2+ tolerance.
INTRODUCTION
The nitrate assimilation pathway has been extensively studied. It consists of several steps, beginning with uptake into roots. Nitrate concentrations in soil vary considerably, mainly as a result of two microbial processes, mineralization and nitrification, that are highly sensitive to environmental conditions (Marschner, 1995). To cope with highly variable nitrate concentrations in soil, plants have developed both a high-affinity transport system (HATS) and a low-affinity transport system (LATS) (Glass et al., 1992; Crawford, 1995; Crawford and Glass, 1998; Forde, 2000). When the external nitrate concentration is high (>1 mM), LATS is preferentially used. When nitrate availability is limited, HATS is activated and takes over the nitrate uptake process (Glass et al., 1992; Crawford and Glass, 1998).
To date, two nitrate transporter gene families, NRT1 and NRT2, were identified as responsible for LATS and HATS, respectively (Glass et al., 1992; Orsel et al., 2002b; Tsay et al., 2007). In the Arabidopsis thaliana NRT1 family, the 53 NRT1 (PTR, peptide transporter) members include both nitrate and oligopeptide transporters. Among the characterized nitrate transporters, CHLorate resistant 1 (CHL1/NRT1.1) is the most studied and represents a major low-affinity uptake mechanism for nitrate (Tsay et al., 1993, 2007). Furthermore, CHL1/NRT1.1 functions as a dual-affinity transporter with regulation by phosphorylation (Wang et al., 1998; Liu et al., 1999). The NRT2 family consists of seven members in the Arabidopsis genome. NRT2.1 and NRT2.2 are involved in inducible high-affinity nitrate uptake (Cerezo et al., 2001; Li et al., 2007). Though functionally and phylogenetically distinct, the nitrate transport functions of both NRT1 and NRT2 are believed to be proton dependent (Paulsen and Skurray, 1994; Orsel et al., 2002a).
Once taken up into the root cytoplasm, nitrate is either translocated across the tonoplast and stored in the vacuoles, or it is reduced to nitrite and then partitioned to plastids where it is further assimilated to organic nitrogen (Orsel et al., 2002a). Alternatively, nitrate can be loaded into xylem vessels and subsequently unloaded (moved from the xylem sap into xylem parenchyma cells) in plant aerial tissues where it undergoes processes similar to those in roots (Marschner et al., 1997). So far, both the low-affinity nitrate transporter NRT2.7 and the chloride channel, CLCa, were identified as responsible for short-distance nitrate translocation into vacuoles (De Angeli et al., 2006; Chopin et al., 2007). For long-distance transport, recent studies found that NRT1.5 represents a major mechanism for loading nitrate into xylem vessels (Lin et al., 2008) and NRT1.7 for loading nitrate into the phloem (Fan et al., 2009).
For herbaceous species, most of the nitrate undergoes long-distance transport to leaves where its assimilation can be directly coupled to photosynthesis in chloroplasts (Smirnoff and Stewart, 1985; Andrews, 1986). However, the proportion of nitrate distributed to and assimilated in roots and shoots can vary substantially depending on the ambient environment. Under high light conditions, leaf assimilation is more energy efficient than root assimilation. However, when light intensity is limited, nitrate assimilation competes with CO2 fixation for photochemical energy and reductants; thus, leaf assimilation is disadvantageous (Smirnoff and Stewart, 1985). Prevalent root assimilation has also been reported when external nitrate availability is limited (Beevers and Hageman, 1980). These observations indicate that nitrate distribution between roots and shoots is of physiological importance in response to changing environments, though the regulating mechanisms remain largely unidentified.
In this study, we report the identification and characterization of Arabidopsis NRT1.8, which belongs to the 53-member NRT1 family (Crawford and Glass, 1998; Tsay et al., 2007). We show that NRT1.8 is an inducible low-affinity transporter for nitrate that is localized in the plasma membrane of xylem parenchyma cells. Our results suggest that NRT1.8 functions in taking up nitrate into xylem parenchyma cells, thus removing nitrate from the xylem sap, and that NRT1.8-mediated nitrate distribution plays a role in plant tolerance to Cd2+ stress.
RESULTS
Isolation and Characterization of NRT1.8
The NRT1 gene family contains 53 members, with nitrate transporters having been identified in several divergent subfamilies (Tsay et al., 2007). Microarray data indicated that the At4g21680 gene in the NRT1 family is regulated by nitrate (Wang et al., 2003, 2004), suggesting that it might encode a nitrate transporter. Thus, we assigned the name NRT1.8 to the At4g21680 gene (Figure 1A). A full-length cDNA of NRT1.8 was cloned using high-fidelity RT-PCR based on EST information from the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/plant/jsf/athal/searchjsp/index.jsp). The sequence was further confirmed by a BLAST search against The Arabidopsis Information Resource database (TAIR; http://www.Arabidopsis.org/Blast/index.jsp). NRT1.8 is predicted to contain four exons and three introns (see Supplemental Figure 1A online), encoding a transmembrane protein with 589 amino acids and the typical 12 transmembrane domains observed in all other NRT1 family members. The deduced protein sequence of NRT1.8 is similar to all identified NRT1-type nitrate transporters and showed 64% identity with NRT1.5 (Lin et al., 2008), its closest family member (Figure 1A).
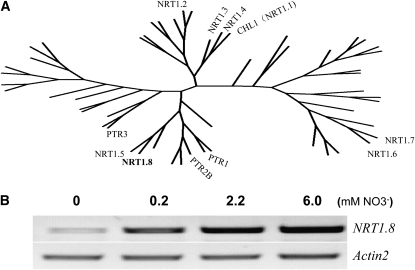
Figure 1.
NRT1.8 in the Arabidopsis NRT1 Family Is Nitrate Responsive.
(A) Amino acid sequences of all 53 members in the NRT1 family (Tsay et al., 2007) were downloaded from the TAIR website. Multiple alignment was performed using ClustalX 1.83 (Thompson et al., 1997) with default settings (protein gap open penalty, 10; protein gap extension penalty, 0.2; delay divergent sequences, 30%; use negative matrix, off; protein weight matrix, Gonnet series). The radial tree was drawn using PhyloDraw (Choi et al., 2000) based on the prior alignment result (see Supplemental Table 2 and Supplemental Data Set 1 online). Only characterized members are indicated.
(B) RT-PCR analysis of NRT1.8 expression in roots under different external nitrate concentrations. The amplification cycles were 30 and 20 for NRT1.8 and the loading control gene Actin2, respectively.
We further performed RT-PCR to characterize the expression pattern of NRT1.8 under various nitrate treatments. As shown in Figure 1B, after 3 d of nitrate starvation, a low level of NRT1.8 mRNA was detected in roots. NRT1.8 expression was induced by 0.2 mM NO3−, and stronger induction was observed when 2.2 and 6 mM NO3− were added (Figure 1B). These results showed that NRT1.8 is responsive to external nitrate in a concentration-dependent manner. In situ hybridization further confirmed this result (see Supplemental Figure 2 online).
NRT1.8 Is Preferentially Expressed in Arabidopsis Vascular Tissues
To determine the tissue-specific expression pattern of NRT1.8, histochemical analyses were performed using transgenic lines harboring the β-glucuronidase (GUS) gene driven by the NRT1.8 promoter (NRT1.8p:GUS). As shown in Figure 2A, GUS activity was maximally and constitutively detected in the vasculature of Arabidopsis seedlings (Figure 2A). Cross-sectioned young seedling roots showed NRT1.8p:GUS expression within the stele (Figure 2B). Longitudinal sectioning further demonstrated expression in parenchyma cells abutting xylem vessels (Figures 2C and 2D). These results suggested that NRT1.8 may be involved in xylem loading or unloading.
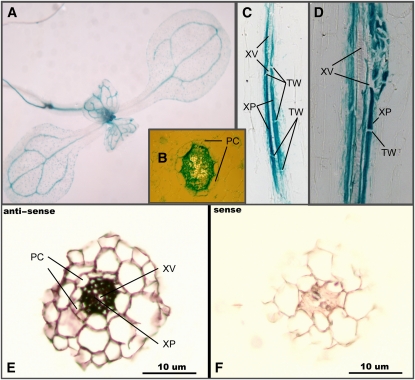
Figure 2.
NRT1.8 Is Expressed in Xylem Parenchyma Cells.
(A) to (D) Histochemical localization of GUS activity in transgenic plants expressing the GUS reporter gene under the control of the NRT1.8 promoter.
(A) Whole-mount seedlings.
(B) Cross-sectioned seedling roots.
(C) and (D) Longitudinally sectioned seedling roots.
(E) In situ hybridization of the antisense NRT1.8 probe to a section of Arabidopsis root tissue.
(F) In situ hybridization of the sense NRT1.8 probe to a section of Arabidopsis root tissue.
PC, pericycle cells; XV, xylem vessels; XP, xylem parenchyma; TW, transverse cell wall.
We further performed mRNA in situ hybridization to exclude the possibility that GUS activity leakage was responsible for these results. Consistent with the GUS analysis, in situ hybridization also localized NRT1.8 accumulation to xylem parenchyma cells in the stele (Figure 2E). In the control experiment using the sense NRT1.8 probe, no signal was detected (Figure 2F). These results confirmed that the GUS activity analysis was reliable, and NRT1.8 was expressed in xylem parenchyma cells within the vasculature.
Plasma Membrane Localization of NRT1.8
To investigate the subcellular localization of the NRT1.8 protein, NRT1.8 was fused in frame with enhanced green fluorescent protein (EGFP) and subcloned into the binary vector pGreenII. Transient expression of NRT1.8:EGFP in onion epidermal cells showed that green fluorescence was localized to the plasma membrane of plasmolyzed cells (Figures 3A and 3B) compared with the diffuse nucleocytoplasmic localization of the EGFP control (Figures 3C and 3D). To further confirm this result, we expressed NRT1.8:EGFP in Arabidopsis protoplasts. Consistently, green fluorescence was also observed at the protoplast plasma membrane (Figures 3E and 3F). These data indicated that NRT1.8 is plasma membrane localized.
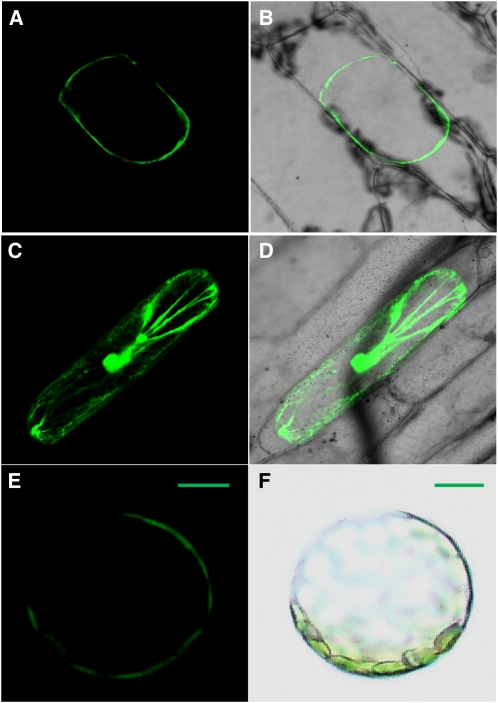
Figure 3.
Subcellular Localization of NRT1.8.
Onion epidermal cells transiently transformed with either the NRT1.8:EGFP fusion or unfused EGFP were incubated in 0.8 M mannitol to induce plasmolysis and then imaged by confocal microscopy.
(A) Fluorescence image of epidermal cell expressing the NRT1.8:EGFP fusion protein.
(B) Merged EGFP fluorescence and bright-field image.
(C) Fluorescence image of epidermal cell expressing EGFP as a control.
(D) Merged control EGFP fluorescence and bright-field image.
(E) and (F) Arabidopsis protoplasts expressing the NRT1.8:EGFP protein (E) and bright-field image (F).
Bars = 20 μM.
NRT1.8 Can Transport Nitrate and Regulates Nitrate Removal from Xylem
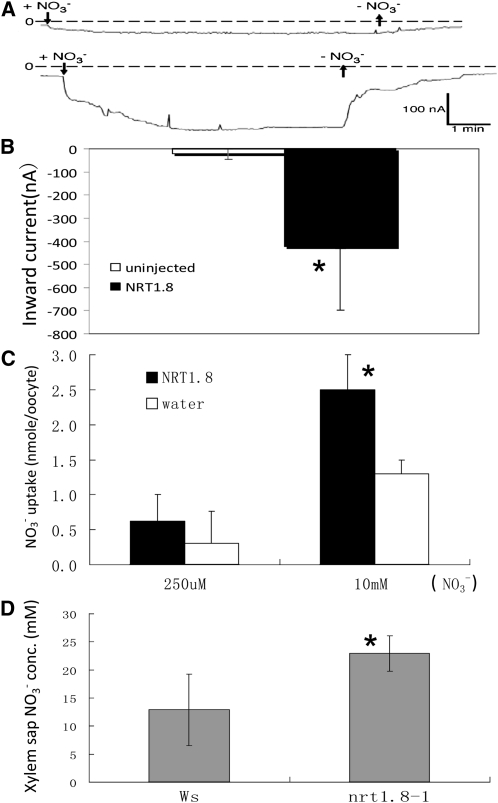
Electrophysiological analyses were performed using Xenopus laevis oocytes injected with NRT1.8 cRNA to determine whether NRT1.8 is a nitrate transporter. In uninjected oocytes, exposure to 10 mM nitrate at pH 5.5 induced a small inward current when oocytes were clamped to −40 mV (Figure 4A, top). On average, this current was approximately −20 nA (Figure 4B). By contrast, 11 out of 11 NRT1.8-injected oocytes from four separate batches showed large inward currents (Figure 4A, bottom), which averaged approximately −420 nA (Figure 4B). Uptake analyses in oocytes showed that NRT1.8 mediated nitrate uptake at 10 mM external nitrate, but not 250 μM nitrate (Figure 4C), which is comparable to NRT1.5 (Tsay et al., 1993, 2007; Lin et al., 2008). To determine the apparent affinity of NRT1.8 for nitrate, we exposed oocytes to 1, 2, 5, and 10 mM HNO3 at pH 5.5. In NRT1.8-injected oocytes, currents began to saturate at 10 mM nitrate (see Supplemental Figure 3 online). Higher nitrate concentrations induced leak currents also in uninjected control oocytes. Based on data from five oocytes from two batches, we calculated a Km of NRT1.8 for nitrate of 12.0 ± 4.8 mM (error denotes sd). Currents at 10 mM nitrate averaged −340 ± 48 nA, whereas in uninjected oocytes, they averaged −38 ± 40 nA (average current from five oocytes from the same batches as NRT1.8-injected oocytes). We concluded that NRT1.8 is a low-affinity nitrate transporter.
Figure 4.
NRT1.8 Transports Nitrate.
(A) Representative inward currents elicited by 10 mM NO3− at pH 5.5 and a holding potential of −40 mV were recorded for control (top) and NRT1.8 cRNA injected oocytes (bottom).
(B) Quantification of the currents recorded; *P < 0.001, n = 11 for both injected and uninjected oocytes from four separate batches.
(C) High- and low-affinity nitrate uptake activity at pH 5.5. Oocytes were incubated for 3 h with 250 μM or 10 mM nitrate. The amount of nitrate removed from the medium (high-affinity activity) or retained in the oocytes (low-affinity activity) was analyzed by HPLC as described by Huang et al. (1999). *P <0.01, n = 5 samples for both high- and low-affinity uptake assays. Each sample consisted of four oocytes.
(D) NO3− concentration in xylem sap; *P < 0.01, n = 6 separate samples for both Ws and the nrt1.8-1 mutant; each sample pooled sap from approximately nine independent plants. Error bars denote sd in (B) to (D).
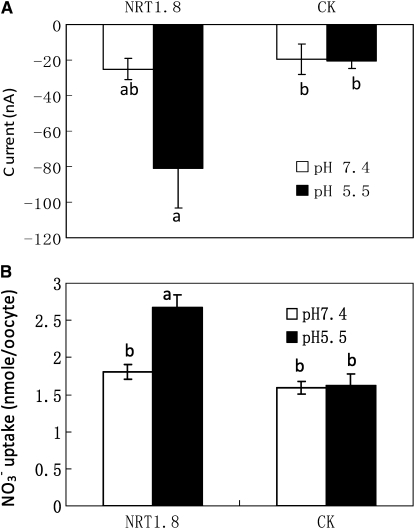
In oocyte experiments, nitrate was added as HNO3. No additional monovalent mineral cations were present in the bath solution that contained 0.15 mM CaCl2 (Huang et al., 1999). In addition, increasing external HNO3 induced larger inward currents and shifted the reversal potential to more positive values. Together, these data indicated that the nitrate-induced inward current in NRT1.8-injected oocytes represents proton-coupled nitrate uptake. This is typical for most of the other characterized nitrate transporters in the NRT1 family (Tsay et al., 1993, 2007; Lin et al., 2008). Consistent with this, NRT1.8-mediated nitrate currents (Figure 5A) or uptake (Figure 5B) were larger than background at pH 5.5, but not at pH 7.4.
Figure 5.
Nitrate Transport by NRT1.8 Is pH Dependent.
(A) Oocytes injected with NRT1.8 cRNA (NRT1.8) or uninjected oocytes (CK) were clamped at −20 mV and exposed to 5 mM HNO3 at pH 7.4 (open bars) and subsequently at pH 5.5 (closed bars). Current differences before and after addition of 5 mM HNO3 at each pH were averaged from four NRT1.8-injected and four uninjected oocytes from three batches. For NRT1.8, currents at pH 7.4 and 5.5 were statistically different at P < 0.06.
(B) Oocytes injected with NRT1.8 cRNA or uninjected oocytes were incubated with 10 mM nitrate for 3 h at pH 7.4 and 5.5, respectively. The amount of nitrate retained in oocytes was determined by HPLC. n = 10 samples from two batches, and each sample consisted of four oocytes. For NRT1.8, nitrate uptake at pH 5.5 and 7.4 was significantly different at P < 0.001.
Error bars denote se. Lowercase letters indicate whether current averages were statistically different at P < 0.05 by t tests.
Considering that NRT1.8 is expressed in the plasma membranes of xylem parenchyma cells (Figures 2 and 3) and is a functional nitrate uptake transporter (Figures 4A to 4C), we suspected that NRT1.8 might function to take up nitrate into xylem parenchyma cells from the xylem. Disruption of NRT1.8 should thus result in nitrate accumulation in the xylem sap. To test this hypothesis, xylem sap was collected from wild-type Wassilewskija (Ws) plants and a T-DNA insertion mutant nrt1.8-1, in which full-length NRT1.8 transcript accumulation was not detected (see Supplemental Figure 1B online). As expected, the nitrate concentration in nrt1.8-1 xylem sap was significantly higher than in that from Ws plants (Figure 4D, P < 0.01). These results suggest that NRT1.8 functions in transporting nitrate across xylem parenchyma cell membrane to unload nitrate from the xylem sap.
NRT1.8 Is Upregulated by Cd2+ Stress, and the nrt1.8-1 Mutant Is Cd2+ Sensitive
NRT1.8 was initially identified based on its dramatic upregulation by Cd2+ stress in microarray experiments (see Supplemental Table 1 online) and hypothesized to provide an oligopeptide-mediated Cd2+ transport mechanism (Gong et al., 2003), since PTR-type oligopeptide transporters belong to the same family as NRT1 transporters (Tsay et al., 2007). However, our data demonstrate that NRT1.8 does not transport tripeptide glutathione (see Supplemental Figure 6 online) and is unlikely to transport the much larger oligopeptide phytochelatins (PCs/PCn, n = 2 to 11), the most studied Cd2+ chelators, but is a nitrate transporter (Figure 4). Subsequently, we analyzed why Cd2+ strongly induces NRT1.8. The nonessential heavy metal Cd2+ has been observed to severely inhibit nitrate assimilation (Sanita di Toppi and Gabbrielli, 1999). Microarray analyses showed that NRT1.8 was the only gene in the nitrate assimilation pathway that was dramatically upregulated by Cd2+ stress (see Supplemental Table 1 online), suggesting that NRT1.8 might be an essential component in the interaction between nitrate assimilation and tolerance to Cd2+ stress.
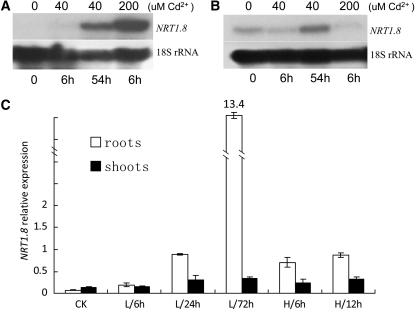
To test our hypothesis, we performed RNA gel blots to confirm and further characterize the NRT1.8 expression pattern under Cd2+ stress. As shown in Figure 6A, NRT1.8 was undetectable in roots without Cd2+ or with 40 μM Cd2+ for 6 h, while at 54 h, NRT1.8 expression was significantly induced by 40 μM Cd2+. By contrast, 200 μM Cd2+ strongly induced NRT1.8 within 6 h (Figure 6A). In shoots, in addition to a constitutively low expression, NRT1.8 was also induced by long-term exposure to 40 μM Cd2+ (Figure 6B). Quantitative RT-PCR analyses further confirmed these results (Figure 6C). In roots, the NRT1.8 mRNA level under the control condition was about 7% of that for the Actin2 gene. When exposed to 20 μM Cd2+, the expression level gradually increased with prolonged exposure and reached ∼13-fold of that for Actin2 after 72 h (Figure 6C). Treatment with 200 μM Cd2+ rapidly and significantly induced NRT1.8 expression in roots (Figure 6C; P < 0.001). By contrast, only prolonged Cd2+ treatment could significantly induce NRT1.8 expression in shoots (Figure 6C, closed bars), most likely because of the time needed for Cd2+ transport and accumulation in shoots. These results indicated that Cd2+ regulates NRT1.8 expression in a dose- and time-dependent manner, consistent with the expression pattern under NO3− induction (Figure 1B; see Supplemental Figure 2 online). Note that 200 μM Cd2+ is very toxic to plants; thus, long-term experiments were not performed at this Cd2+ concentration.
Figure 6.
NRT1.8 Expression under Cd2+ Stress.
(A) and (B) NRT1.8 expression was determined in roots (A) and shoots (B) by RNA gel blotting after plants were exposed to 0, 40, and 200 μM Cd2+ treatment for the indicated times. 18S rRNA was used as a loading control.
(C) Quantitative RT-PCR analysis of NRT1.8 expression in plants exposed to 20 μM (L) and 200 μM (H) Cd2+ for the indicated times. The y axis shows NRT1.8 RNA levels normalized to that of Actin2. n = 3, and values are mean ± sd. CK, control condition.
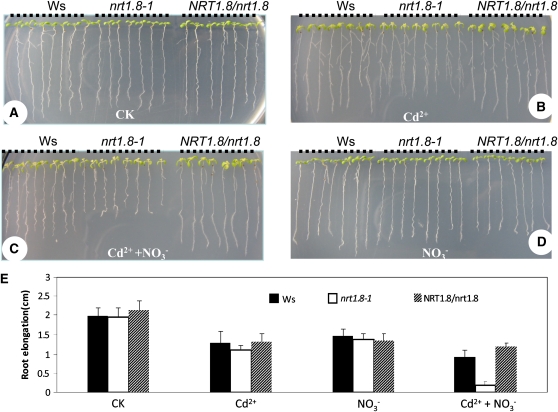
Subsequently, in vivo functional analyses were also performed. When grown on minimal medium supplemented with 50 μM Cd2+, both nrt1.8-1 and Ws showed similar reduced growth (Figures 7B and 7E) compared with the control (Figures 7A and 7E). Considering that NRT1.8 is a nitrate transporter, we then treated with various combinations of nitrate and Cd2+. As shown in Figure 7C, when both 25 mM NO3− and 50 μM Cd2+ were applied, root elongation of nrt1.8-1 was significantly reduced compared with that of the wild type (Figures 7C and 7E; P < 0.001), while no significant difference was observed in rosettes (Figures 7B and 7C). When exposed to 25 mM NO3− only, no obvious difference was observed between nrt1.8-1 and Ws (Figures 7D and 7E; P > 0.19). Further elevating NO3− to 50 mM (the nitrogen level in 1× Murashige and Skoog medium is 60 mM) in combination with 50 μM Cd2+ resulted in a more apparent growth reduction in mutant aerial parts as well as secondary root formation (see Supplemental Figure 4C online). Complementation lines (see Supplemental Figure 1C online) harboring the genomic sequence of NRT1.8 exhibited a less severe Cd2+ sensitivity phenotype comparable to the wild type (Figures 7A to 7D). These results indicated that the wild-type NRT1.8 transporter increases Cd2+ tolerance in a nitrate-dependent manner in Arabidopsis.
Figure 7.
The nrt1.8-1 Mutant is Cd2+ Sensitive.
(A) to (D) Plant growth under control condition (A), 50 μM Cd2+ (B), 25 mM NO3− plus 50 μM Cd2+ (C), or 25 mM NO3− (D). Black dotted lines indicate Ws, nrt1.8-1, and complementation line NRT1.8/nrt1.8, respectively. Images are not at same scale.
(E) Root elongation between days 2 and 10 after transfer to plates with treatments from (A) to (D). n = 7 to 8 plants, and values are mean ± sd. CK, control condition.
[See online article for color version of this figure.]
Nitrate Distribution under Cd2+ Stress Is Altered by NRT1.8
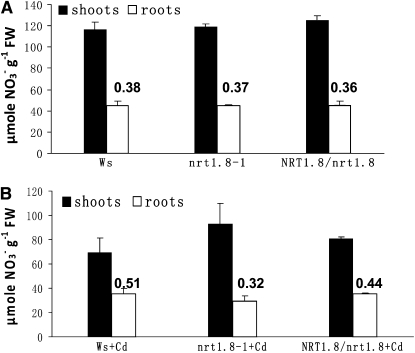
To investigate the underlying mechanism of nitrate-dependent Cd2+ sensitivity, we determined the nitrate concentration in Arabidopsis shoots and roots, normalized by fresh weight to compare plant lines with differing growth rates. Under normal conditions, the nitrate level in both shoots and roots was similar in Ws, nrt1.8-1, and NRT1.8/nrt1.8 plants. The root/shoot nitrate ratio was 0.38 in Ws, 0.37 in mutant nrt1.8-1, and 0.36 in the complementation line NRT1.8/nrt1.8 (Figure 8A; P > 0.55). But when NRT1.8 expression is strongly induced in roots by Cd2+ (Figures 6A and 6C), the proportion of nitrate concentration in Ws roots increased to 51% of that in shoots (a root/shoot nitrate ratio of 0.51; Figure 8B). By contrast, in the nrt1.8-1 mutant, Cd2+ stress decreased the root/shoot nitrate ratio from 0.37 (Figure 8A) to 0.32 (Figure 8B; P < 0.05). In the complementation lines, the Cd2+-induced root/shoot nitrate ratio alteration (an increase from 0.36 under control condition to 0.44 under Cd2+ stress) was comparable to that observed in the wild type. These data demonstrate that under Cd2+ stress, a greater proportion of nitrate accumulates in the wild-type Ws roots compared with nrt1.8-1 (Figure 8B; P < 0.01) and suggest that the induction of NRT1.8 in roots (Figures 6A and 6C) permits Ws to continue removing nitrate from the xylem when Cd2+ toxicity limits nitrate reductase activity. Note that under Cd2+ stress, the overall nitrate concentration was decreased in both Ws and nrt1.8-1 mutant (Figures 8A and 8B), as would be expected due to Cd2+ toxicity (Hernandez et al., 1997; Sanita di Toppi and Gabbrielli, 1999).
Figure 8.
Nitrate Distribution Is Altered in the nrt1.8-1 Mutant.
(A) NO3− concentration in wild-type (Ws), nrt1.8-1 mutant, and NRT1.8/nrt1.8 complementation plants under control conditions.
(B) NO3− concentration in wild-type (Ws), nrt1.8-1 mutant, and NRT1.8/nrt1.8 complementation plants under Cd2+ treatment.
The numbers above each bar represent the root/shoot nitrate ratio. Data were normalized to fresh weights (FW). Experiments were performed for six replicate samples, and each sample contained more than nine independent plants. Values are mean ± sd.
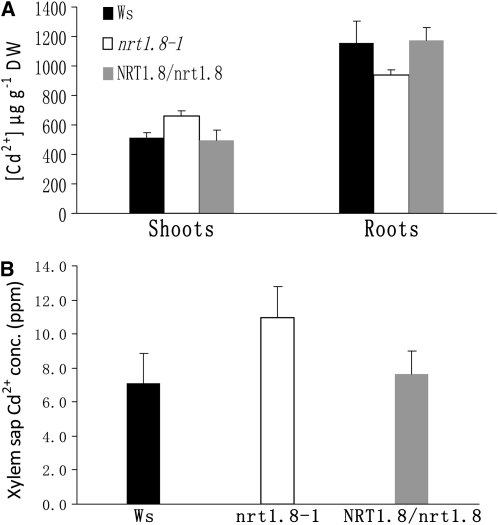
The Cd2+ distribution within plants was also determined. As shown in Figure 9A, more Cd2+ accumulated in nrt1.8-1 shoots when compared with both Ws and NRT1.8/nrt1.8, while in roots, less Cd2+ was detected in the mutant than in the wild type and the complementation lines (P < 0.05). Interestingly, more Cd2+ accumulation was also observed in xylem sap from nrt1.8-1 than in sap from the wild-type control and the complementation lines (Figure 9B; P < 0.05).
Figure 9.
Cd2+ Distribution Is Altered in the nrt1.8-1 Mutant.
(A) Cd2+ concentration in shoot and root tissues. Plants were grown hydroponically for 3 weeks and then exposed to 20 μM Cd2+ for 3 d. Shoots and roots were harvested and subjected to determination of Cd2+ concentration in the wild type (Ws), nrt1.8-1 mutant, and complementation line NRT1.8/nrt1.8. Data were normalized to dry weight (DW). Experiments were performed for six replicates, and more than nine independent plants were pooled per replicate.
(B) Cd2+ concentration in xylem sap. Plants were treated with 5 μM Cd2+ for 3 d, xylem sap was collected as described in Methods, and Cd2+ concentration (conc.) was determined using inductively coupled plasma–mass spectrometry (n = 6).
Values are mean ± sd.
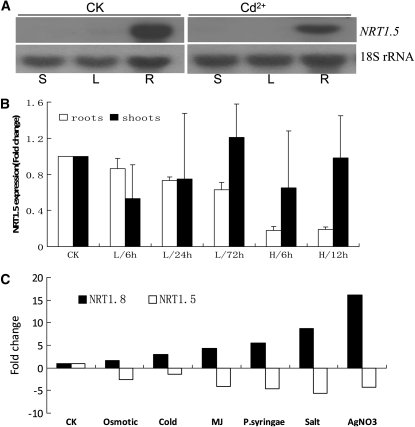
NRT1.5 and NRT1.8 Expression Is Oppositely Regulated under Stresses
NRT1.5 was shown to load nitrate into the xylem (Lin et al., 2008), and our data suggest that NRT1.8 functions to unload nitrate from the xylem (Figures 2 to 4). Furthermore, our microarray data showed that NRT1.5 and NRT1.8 were oppositely regulated by Cd2+ stress (see Supplemental Table 1 online). These observations indicate that NRT1.8 and NRT1.5 may be fine-tuned to regulate nitrate distribution between shoots and roots under Cd2+ stress. To test our hypothesis, RNA gel blots were performed. As shown in Figure 10A, under control conditions, NRT1.5 showed constitutively high expression in roots, while in shoots, no NRT1.5 mRNA was detected. When treated with 200 μM Cd2+ for 6 h, NRT1.5 expression was reduced (Figure 10A), as is observed in microarray analyses (see Supplemental Table 1 online). Further quantitative RT-PCR showed that in roots, low-level Cd2+ treatments (20 μM) gradually reduced NRT1.5 mRNA levels with time, while high-level Cd2+ treatment rapidly and significantly reduced NRT1.5 expression (Figure 10B, open bars). This was in contrast to what was observed for NRT1.8 (Figure 6C, open bars). In shoots, no significant change was observed for NRT1.5 mRNA levels (Figure 10B, closed bars). Note that NRT1.5 expression levels are very low in aerial parts (Figure 10A). Consequently, the standard deviation of NRT1.5 expression levels in shoots measured by quantitative RT-PCR was large (Figure 10B, closed bars).
Figure 10.
Opposite Regulation of NRT1.8 and NRT1.5 Expression under Stress Conditions.
(A) Four-week-old plants were exposed to 200 μM Cd2+ for 6 h or were kept under control conditions (CK). NRT1.5 expression was determined in stems (S), rosette leaves (L), and roots (R) by RNA gel blots. 18S rRNA was used as a loading control.
(B) Quantitative RT-PCR of NRT1.5 expression in plants treated with 20 μM (L) or 200 μM (H) Cd2+ for the indicated times. n = 3, and values are mean ± sd. Fold change compared with control (CK) is shown.
(C) Expression data of NRT1.8/At4g21680 and NRT1.5/At1g32450 were obtained from Genevestigator (Zimmermann et al., 2004). Examples representing the following stresses were selected: osmotic (mannitol), cold (low temperature), MJ (methyl jasmonate), P.syringae (Pseudomonas syringae), salt (NaCl), and heavy metal (AgNO3). Fold change compared with control (CK) is shown. Detailed information and more data are available from Genevestigator (https://www.genevestigator.com/gv/index.jsp).
Further analyses revealed a similar opposite pattern between NRT1.8 and NRT1.5 mRNA level changes under various environmental stresses. As shown for some stress examples in Figure 10C, NRT1.8 was consistently upregulated, while NRT1.5 was downregulated, and more examples were found in the Genevestigator database (https://www.genevestigator.com/gv/index.jsp; Zimmermann et al., 2004). These observations suggest that the coordinated opposite regulation of NRT1.5 and NRT1.8 might be a universal mechanism in response to environmental stresses.
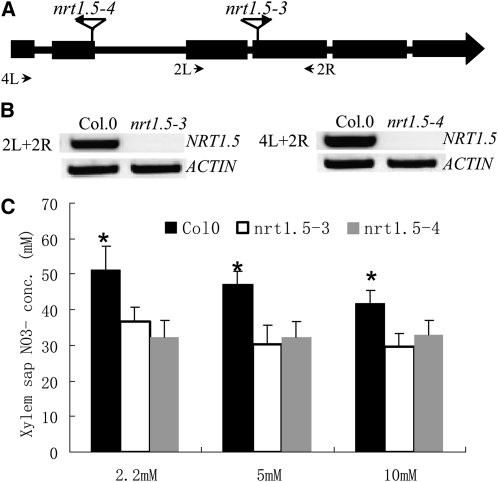
Lower Nitrate Accumulation in nrt1.5 Xylem Sap
Considering that NRT1.5 functions to load nitrate into the xylem, disruption of NRT1.5 is expected to reduce the nitrate concentration in the xylem. A previous study observed that the rate of nitrate secretion into the xylem sap was reduced in nrt1.5-1 (Lin et al., 2008). To complement our observation that the xylem sap nitrate concentration increased in the nrt1.8-1 mutant (Figure 4D), we measured xylem sap nitrate concentrations in mutant lines of a different NRT1 transporter, the allelic NRT1.5 mutants nrt1.5-3 and nrt1.5-4, in which intact NRT1.5 mRNA was undetectable (Figures 11A and 11B). Consistent with our hypothesis that NRT1.5 and NRT1.8 have opposite functions, less nitrate accumulated in xylem sap of nrt1.5 mutant plants compared with the wild-type control (Figure 11C), supporting that NRT1.5 functions to load nitrate into the xylem (Lin et al., 2008).
Figure 11.
Decreased Nitrate Concentration in nrt1.5 Xylem Sap.
(A) Schematic representation of two allelic T-DNA insertion lines nrt1.5-3 (SAIL_72_F04) and nrt1.5-4 (SALK_063393) were PCR screened and sequenced as described (Krysan et al., 1999) to identify homozygous mutants.
(B) RT-PCR did not detect intact NRT1.5 mRNA in nrt1.5-3 (left) and nrt1.5-4 (right) mutant plants.
(C) The homozygous mutants and wild-type Columbia-0 were grown hydroponically for 4 weeks, subjected to nitrate starvation for 7 d, and then transferred to medium with 2.2, 5, or 10 mM nitrate for 12 h. Xylem sap was collected for 6 h, and nitrate concentrations were determined as described in Methods. Values are mean ± sd, n = 5, and the asterisk indicates significant differences between wild-type Columbia-0 and the two mutant lines at P < 0.01.
DISCUSSION
Long-distance transport of nitrate determines nitrate distribution and subsequent assimilation between different tissues; thus, elucidation and regulation of the tightly controlled process would be of great importance to nutrient use efficiency (Marschner et al., 1997). In this study, we demonstrate that NRT1.8, a close homolog of NRT1.5, is a nitrate transporter and is expressed in xylem parenchyma cells. nrt1.8-1 mutant plants show increased levels of nitrate in the xylem sap, consistent with the model that NRT1.8 encodes a xylem unloading transporter.
NRT1.8 Functions to Remove Nitrate from Xylem Vessels
A transporter for nitrate loading into xylem sap was recently reported (Lin et al., 2008), and here we report a transporter for nitrate unloading from xylem sap. The NRT1.8 gene from the NRT1 family was nitrate inducible (Figure 1B; see Supplemental Figure 2 online), which suggested that it might encode a low-affinity nitrate transporter. Electrophysiological analyses and nitrate uptake assays showed that NRT1.8 mediates low-affinity nitrate uptake (Figures 4A to 4C). An inward current was elicited by millimolar nitrate in NRT1.8 cRNA-injected oocytes at pH 5.5 (Figures 4A, 4B, and 5), consistent with a proton-coupled nitrate uptake model that is used by other identified NRT1 family members (Tsay et al., 2007). Further analyses using GUS staining and in situ hybridization indicated that NRT1.8 is expressed predominantly in the vasculature and located in xylem parenchyma cells (Figure 2), suggesting a functional role for NRT1.8 in nitrate long-distance transport. Moreover, we show data demonstrating that NRT1.8 is localized to the plasma membrane (Figure 3) and that functional disruption of NRT1.8 led to nitrate accumulation in xylem sap (Figure 4D). Taken together, we conclude that NRT1.8 functions to remove nitrate from xylem vessels.
NRT1.8 Is an Essential Component Regulating Nitrate Distribution
Efficient long-distance transport of nitrate to aerial tissues of plants requires successive nitrate loading into the xylem and unloading to xylem parenchyma cells. Interestingly, consistent with the findings here that NRT1.8 is a nitrate unloading transporter (Figure 4) and findings that NRT1.5 is a nitrate loading transporter (Lin et al., 2008), an increased nitrate concentration was observed in nrt1.8-1 xylem sap (Figure 4D), while nrt1.5 mutants showed a decreased nitrate secretion rate into xylem (Lin et al., 2008) and reduced xylem sap nitrate concentrations (Figure 11C). Furthermore, NRT1.8 and NRT1.5 are closely related phylogenetically (Figure 1A). These data lead to the model that the distinct functions of NRT1.8 and NRT1.5 have evolved by subfunctionalization following an ancestral gene duplication event. Consistent with this postulation, when 4-week-old plants are under typical nonstress conditions, NRT1.8 expression in shoots (Figure 6B) is higher than in roots (Figure 6A), and NRT1.5 expression higher in roots than shoots (Figure 10A, CK). Under these conditions, NRT1.5 and NRT1.8 mainly effect the fine-tuning of long-distance nitrate transport from roots to shoots.
Subfunctionalization is also consistent with differential expression responses to environmental stresses. When under Cd2+ stress conditions, NRT1.8 is upregulated while NRT1.5 is downregulated in Arabidopsis roots (see Supplemental Table 1 online; Figures 6A, 6C, 10A, and 10B). Similar expression patterns were observed repeatedly for NRT1.8 and NRT1.5 under various stress conditions, including salt, cold, metal stresses, and biotic stress (Figure 10C) (www.genevestigator.com/gv/index.jsp; Zimmermann et al., 2004). These observations indicate that in addition to NRT1.8's role in nitrate transport to aerial tissues, a more important role of NRT1.8 might be to determine which tissues will accumulate nitrate in response to environmental cues. This hypothesis is supported by data showing that Cd2+ induces NRT1.8 expression in roots (Figures 6A and 6C) with a corresponding increase in wild-type root nitrate concentration (Figures 8A and 8B). A similar mineral distribution mechanism has been reported for the xylem parenchyma cell-localized HKT1 (high-affinity K+ transporter 1) and SKC1 (Shoot K+ content 1) sodium transporters (Maser et al., 2002; Gong et al., 2004; Ren et al., 2005; Sunarpi et al., 2005). Disruption of these Na+ transporters results in overaccumulation of Na+ in xylem sap and shoots (Maser et al., 2002; Gong et al., 2004; Ren et al., 2005; Sunarpi et al., 2005; Horie et al., 2006; Davenport et al., 2007; Moller et al., 2009). Here, we observed that in the mutant nrt1.8-1, a greater proportion of nitrate accumulated in shoots under cadmium stress (Figure 8B). It is worth noting that NRT1.8 expression is also induced in shoots under prolonged Cd2+ treatment (Figures 6B and 6C), which may contribute to recycling nitrate from shoots to roots. This would partially contribute to increasing the root/shoot nitrate ratio (Figure 8B), possibly via a similar mechanism proposed for HKT1/SKC1, in which Na+ is unloaded from xylem vessels to xylem parenchyma cells, then transported into the phloem and recycled to roots (Maser et al., 2002; Gong et al., 2004; Ren et al., 2005; Sunarpi et al., 2005; Horie et al., 2006; Davenport et al., 2007).
The Role of NRT1.8-Regulated Nitrate Distribution under Cadmium Stress
Cd2+, a nonessential toxic heavy metal, has long been observed to severely inhibit nitrate uptake, transport, and assimilation (Sanita di Toppi and Gabbrielli, 1999), resulting in biomass and protein content reduction (Chaffei et al., 2004). Furthermore, Cd2+ was also observed to affect nitrogen distribution in various plant species. In Cd2+-treated tomato (Solanum lycopersicum) seedlings, the total amino acid content increased in phloem and roots, together with an increase of glutamate dehydrogenase, cytosolic glutamine synthetase, and asparagine synthetase expression, suggesting that nitrogen was recycled and translocated from shoots to roots as a Cd2+ protection and storage strategy that may preserve roots (Chaffei et al., 2004).
In Cd2+-treated pea (Pisum sativum) plants, a similar NO3− distribution pattern was observed, and the authors proposed a model in which inhibited transpiration led to less nitrate transport to shoots and, thus, to nitrate accumulation in roots, resulting in a smaller decrease in root nitrate content under Cd2+ stress (Hernandez et al., 1997). Our data suggest that, in addition to this model of general reduction of transpiration-mediated nutrient translocation from roots to shoots, the NRT1.8-regulated nitrate accumulation in roots may represent a major mechanism in the process of nitrate distribution under Cd2+ stress, because the wild-type Ws maintained a higher root nitrate proportion than the nrt1.8-1 mutant when plants were exposed to Cd2+ stress (Figure 8B). In addition, among all known nitrate assimilation pathway genes, only NRT1.8 is dramatically upregulated in response to Cd2+ stress (see Supplemental Table 1 online; Figure 6). Thus, these data suggest that the alteration of the root/shoot nitrate ratio by NRT1.8 may represent an active regulatory process, rather than a passive consequence of inhibited transpiration.
Possible Interaction of Nitrate Distribution and Cd2+ Tolerance
Our findings raise the question of how nitrate distribution could affect Cd2+ tolerance. One possible explanation could be a mechanism similar to that observed for organic nitrogen redistribution, in which amino acids are transported from shoots to roots to preserve roots as nutritional safeguard organs, thus allowing future recovery (Chaffei et al., 2004). However, nitrate represents only a small proportion of all the nitrogen sources in roots, the direct nutritional effect of which might be subtle compared with that of organic nitrogen. Thus, a more plausible model is that NO3− serves as a signal molecule that enhances NR activity at elevated concentrations (Hernandez et al., 1995; Stitt, 1999). When recycled to roots, the proportionally increased nitrate induces NR and NiR activity otherwise inhibited by Cd2+, permitting nitrate uptake and assimilation to some extent. In NRT1.8 overexpression lines, CHL1, nitrate reductase 1 (NIA1), nitrite reductase (NIR), and cytosolic glutamine synthase 1 (GSR1) were constitutively detected in roots and shoots under normal conditions (see Supplemental Figures 5C and 5D online). Interestingly, when exposed to Cd2+, lower reduction in CHL1, NIA1, NIR, and GSR1 mRNA levels was observed in roots of NRT1.8 overexpression lines compared with the wild type (see Supplemental Figures 5D and 5F online), as would be predicted by this model. The observation that Cd2+ sensitivity in nrt1.8-1 is nitrate concentration dependent (Figure 7; see Supplemental Figure 4 online) and that a wild-type copy of NRT1.8 rescues the phenotype further supports this hypothesis. Consistently, the nrt1.5 mutant appears to exhibit increased tolerance to Cd2+ compared with the wild-type control (see Supplemental Figure 7 online).
Additionally, the Cd2+ allocation to shoots in nrt1.8-1 plants may partially contribute to the Cd2+ sensitivity in the nrt1.8-1 mutant (Figure 9). Cd2+ is highly toxic to plants, and most Cd2+ accumulates in roots to protect important organs (Gong et al., 2003). When NRT1.8 was disrupted, more Cd2+ accumulated in mutant shoots than in wild-type shoots (Figure 9A), which thus would lead to greater Cd2+ toxicity in mutant plants. How nitrate distribution would affect Cd2+ allocation remains an open question. A previous study showed that in Brassica juncea, Cd2+ transport in the xylem sap was coordinated with nitrogen ligands (Salt et al., 1995). Additionally, nitrogen-containing molecules including phytochelatins and GSH have been detected concomitantly with Cd2+ transport in the vasculature (Gong et al., 2003; Chen et al., 2006; Li et al., 2006; Mendoza-Cozatl et al., 2008). Together, these data indicate that nitrate or nitrate-derived peptide and oligopeptide-assisted Cd2+ distribution may occur, permitting nitrogen-coordinated Cd2+ recycling and reduced Cd2+ accumulation in shoots in the wild type. However, it is worth noting that this process is not likely to be regulated by NRT1.8 directly, or at least not by NRT1.8-mediated transport of GSH or phytochelatins, the most studied Cd2+ chelators, because we could not obtain positive data showing that NRT1.8 could transport these chemicals (see Supplemental Figure 6 online).
In summary, NRT1.8 encodes a nitrate transporter that mediates nitrate removal from the xylem sap, and functional characterization of the NRT1.8 gene has revealed an important correlation between nitrate allocation and Cd2+ tolerance. Our research further indicates that nitrate allocation may be of essential importance for plant responses to environmental signals.
METHODS
Plant Materials, Growth Conditions, and Cd2+ Sensitivity Analyses
Arabidopsis thaliana plants were grown in quarter-strength sterile hydroponic solution at 22°C with 16-h-light/8-h-dark cycles as described (Arteca and Arteca, 2000; Gong et al., 2003). At 3 to ∼4 weeks of age, plants were exposed to treatments as indicated. For Cd2+ sensitivity analyses, seedlings were grown for 7 d on quarter-strength Murashige and Skoog basal medium (Sigma-Aldrich) with 1 g/L MES, 0.8% (w/v) Suc, and 1.5% Bacto agar and then transferred to plates with basal medium or medium with either elevated nitrate levels or combinations of nitrate and 50 μM Cd2+ as indicated. Plants grew vertically for an additional 7 to 10 d, at which point root elongation was determined
RNA Gel Blotting and RT-PCR/Quantitative RT-PCR
Four-week-old hydroponically grown plants were exposed to different level of Cd2+ stress for specified time intervals and other stresses as stated. Alternatively, plants were subjected to 3-d nitrate starvation, transferred to solutions with different levels of nitrate as indicated, and incubated for 2 h. Total RNA was extracted from roots and shoots using TRIzol reagent (Invitrogen) following the manufacturer's instructions and then separated in a formaldehyde/agarose gel and RNA gel blotting, probe labeling with isotope P32, and hybridization were performed according to standard protocols suggested by the manufacturers (GE Healthcare). X-ray film autoradiography was performed according to the manufacturer's protocol (Kodak) with an overnight exposure time to ensure linear detection of the indicated hybridization signals. First-strand cDNA was synthesized from DNaseI-digested total RNA using M-MLV reverse transcriptase (Promega), and PCR was performed on a PerkinElmer GeneAmp 9700 with indicated cycles (pilot experiments were performed to determine the optimal cycles for efficient and linear amplification of indicated genes) using Ex-Taq DNA polymerase (TaKaRa); PCR products were separated on 1% agarose gel and stained with ethidium bromide. Quantitative RT-PCR was performed on a Corbett Research Rotor-Gene 3000 thermal cycler using SYBR Premix Ex-Taq (TaKaRa) according to manufacturers' protocols; the primers used were as follows: NRT1.8 (forward, 5′-GGCTTCAGATTCTTGGATAG-3′; reverse, 5′-AACCACAGAGTAGAGGATGG-3′), NRT1.5 (forward, 5′-TGTCATTGGACTTTCATCGC-3′; reverse, 5′-CCCACAACCTCTTGGTCTAATC-3′). Actin2 was used as an internal reference gene (forward, 5′-AGGTATCGCTGACCGTATGAG-3′; reverse, 5′-CATCTGCTGGAATGTGCTGA-3′).
Histochemical Analysis and Tissue Sectioning
A 1309-bp genomic fragment immediately upstream from the NRT1.8 start codon was amplified by PCR with forward primer (5′-TTAGATTGTTCGATTCAGATTTAGG-3′) and reverse primer (5′-AGGATCCCATAGATTCGATTTGGTGTAGAGAT-3′), which was then cloned into the vector pGEM-T Easy (Promega), digested with HindIII and BamHI, and subsequently cloned into binary expression vector pBI101-Hm2. The resulting construct pBHNRT was then transformed into Columbia-0 by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). GUS staining was performed for 16 h with 2 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide), 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 0.2% Triton X-100, and 50 mM NaPO4, pH 7.2 (68.4 parts of Na2HPO4 with 31.6 parts of NaH2PO4). The reaction was stopped by 70% ethanol. Tissue sectioning (8 μm) was prepared for dark-field microscopy as described (Johnson et al., 2002).
In Situ Hybridization
One-week-old seedlings grown on quarter-strength Murashige and Skoog medium were transferred to nitrogen-depleted medium and grown for 3 d. They then were subjected to nitrate induction as indicated for 2 h. Tissue sectioning, digoxigenin labeling of RNA probe, and in situ hybridization were performed as described (Long and Barton, 1998) with minor modifications. A gene-specific fragment containing the 390-bp (983 to 1372) coding region of NRT1.8 was amplified by PCR (forward, 5′-TGACTCAAGTCGAAGAAGTGAAA-3′; reverse, 5′-TTGAGATCGAAGTAGCACTTTCC-3′) and cloned into pGEM T Easy (Promega). Sense and antisense probes were in vitro synthesized using T7 and SP6 primers according to the manufacturer's instructions.
EGFP Fusion and Subcellular Localization
The stop codon of NRT1.8 was mutagenized by PCR amplification to introduce a KpnI site. This site was then used to make an in-frame NRT1.8:EGFP fusion construct. The final construct 35S/NRT1.8:EGFP-pGreenII and the empty vector 35S/EGFP-pGreenII were transiently expressed in onion epidermal cells using a particle gun–mediated system (PDS-1000/He; Bio-Rad). These constructs were also transiently expressed in Arabidopsis protoplasts via polyethylene glycol–mediated transformation (Yoo et al., 2007). The bombarded cells and transformed protoplasts were held in the dark at 22°C for 12 h followed by GFP imaging using confocal microscopy (Carl Zeiss; LSM 510 Meta) as described (Guo et al., 2003).
Functional Characterization of NRT1.8 in Xenopus laevis Oocytes
The NRT1.8 cDNA was subcloned as a BamHI-EcoRI fragment into the oocyte expression vector pOO2 (Ludewig et al., 2002), and cRNA was synthesized using the Ambion mMessage mMachine kit. Xenopus oocytes were isolated and maintained as described (Osawa et al., 2006) with the following modifications: oocytes were defolliculated for 30 to 90 min and were injected with 23 to 46 ng of NRT1.8 cRNA on the same day or the day following isolation; oocytes were incubated in a ND-96 Ringer solution with antibiotics (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, 2.5 mM sodium pyruvate, 10 μg/mL streptomycin sulfate, and 50 μg/mL gentamycin sulfate, pH 7.4). Oocytes were voltage clamped 1 to 2 d after injection essentially as described (Osawa et al., 2006). Voltage clamp recordings were initiated in a bath solution containing 230 mM mannitol, 0.15 mM CaCl2, and 10 mM MES/Tris, pH 5.5 (Huang et al., 1999). High- and low-affinity nitrate uptake assays were performed as described (Huang et al., 1999). Nitrate was added to the bath solution as HNO3 at the indicated concentrations.
Isolation and Complementation of T-DNA Insertion Line
The nrt1.8-1 mutant was isolated from the ALPHA T-DNA insertion population generated by the Arabidopsis Knockout Facility at the University of Wisconsin (Krysan et al., 1999). Primers used for PCR screening were NRT1.8 forward primer (5′-AGTTAGACAGTTTGAGGTTTGCACTCAAG-3′), reverse primer (5′-GTAGAATCTCTCCAAGTGTCCTTTGTTGA-3′) and T-DNA left border primer JL202 (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′). RT-PCR was performed to verify knockout of the NRT1.8 transcript using forward primer (5′-CTTTATCTACTGGTGCGTTATTGCT-3′) and reverse primer (5′-ATAGAGATTCAACTCTTCATGGTGC-3′). To make a complementation construct, a 2055-bp fragment containing the NRT1.8 genomic sequence (−2 to 2053) was PCR amplified (forward, 5′-GGATCCATGGATCAAAAAGTTAGACAGTT-3′; reverse, 5′-ATTTAGCTCCTGACTCAGACTTC-3′), sequenced, and subcloned into pBI101-Hm2 that was predigested with BamHI and SacI. The resulting construct was further digested with HindIII and BamHI, into which the 1309-bp NRT1.8 promoter fragment was inserted. The final construct containing both the NRT1.8 promoter and coding regions was then used to transform the nrt1.8-1 mutant as described above. Transgenic lines showing hygromycin resistance were selected and evaluated for Cd2+ sensitivity. The nrt1.5 T-DNA insertion lines were ordered from the ABRC, and homozygous lines were screened and characterized by PCR as described (Krysan et al., 1999).
Determination of Nitrate and Cd2+ Levels
The nrt1.8-1 mutant and its wild-type control Ws and complementation line NRT1.8/nrt1.8 were grown in hydroponic solution for 4 weeks as described above and then transferred to hydroponic solution containing 10 mM nitrate or treatments as indicated for nrt1.5 mutants and its wild-type Columbia-0. After 72 h incubation or as indicated, all rosette leaves were removed, the inflorescence stems were cut using a sharp razor, and xylem sap was collected for 6 h as described (Sunarpi et al., 2005). Alternatively, 3-week-old plants were exposed to 20 μM Cd2+ when indicated and grown another 3 d before tissue harvest. To collect enough xylem sap for determination of Cd2+ concentrations, plants were instead exposed to a much less toxic Cd2+ treatment as indicated. Nitrate was extracted and determined by HPLC (Agilent 1200 series) using a PARTISIL 10 strong anion exchanger column (Whatman) as described (Chiu et al., 2004). Cd2+ was extracted and quantified using inductively coupled plasma–mass spectrometry as described (Gong et al., 2003) with minor modification.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CHL1 (At1g12110), NRT1.2 (At1g69850), NRT1.3 (At3g21670), NRT1.4 (At2g26690), NRT1.5 (At1g32450), NRT1.6 (At1g27080), NRT1.7 (At1g69870), NRT1.8 (At4g21680), AT1G18880, AT1G22540, AT1G22550, AT1G22570, AT1G27040, AT1G33440, AT1G52190, AT1G59740, AT1G62200, AT1G68570, AT1G69860, AT1G72130, AT1G72140, AT2G02020, AT2G02040 (PTR2B), AT2G37900, AT2G38100, AT2G40460, AT3G01350, AT3G16180, AT3G25260, AT3G25280, AT3G43790, AT3G45650, AT3G45660, AT3G45680, AT3G45690, AT3G45700, AT3G45710, AT3G45720, AT3G47960, AT3G54140 (PTR1), AT5G01180, AT5G11570, AT5G13400, AT5G19640, AT5G28470, AT5G46040, AT5G46050 (PTR3), AT5G62680, AT5G62730, AT3G54450, AT1G72120, AT1G72125, AT3G53960, NIA1 (At1g77760), NIR (At2g15620), GSR1 (At5g37600), and Actin2 (At3g18780). All microarray data have been deposited in the National Center for Biotechnology Information/Gene Expression Omnibus database under the accession number GSE22114.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Isolation and Characterization of the T-DNA Insertion Line nrt1.8-1 and Its Complementation Lines.
Supplemental Figure 2. NRT1.8 Expression Was Upregulated in Response to External NO3− Concentration.
Supplemental Figure 3. NRT1.8 Is a Low-Affinity Nitrate Transporter.
Supplemental Figure 4. Cadmium Sensitivity in nrt1.8-1 Was Increased with Higher Nitrate Level.
Supplemental Figure 5. Overexpression of NRT1.8 Alters the Expression of Key Genes in the Nitrate Assimilation Pathway.
Supplemental Figure 6. Analysis of Substrate Selectivity for NRT1.8.
Supplemental Figure 7. nrt1.5 Mutants Show Increased Tolerance to Cadmium Stress.
Supplemental Table 1. Transcriptomic Analysis of Nitrate Assimilation Pathway under Cd2+ Stress.
Supplemental Table 2. Accession Numbers of the Sequences Corresponding to Figure 1A.
Supplemental Data Set 1. Text File Corresponding to Figure 1A.
Supplementary Material
Acknowledgments
We thank Minjung Kim and Daniel Schachtman (Donald Danforth Plant Science Center) for providing NRT1.8 cloned into pOO2 and NRT1.8 cRNA, Xiao-Shu Gao (Core Facility, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences) for helping with confocal microscopy, and the University of California, San Diego, GeneChip core and CapitalBio Co. for help with microarray experiments. This research was supported by the National Basic Research Program of China (2009CB119000), the National Science Foundation of China (30770179), the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams and Chinese Academy of Sciences Innovation program (KSCX2-YW-N-056), and in part by the Shanghai Rising-Star Program (06QA14085) grants to J.-M.G. Initial NRT1.8 experiments of J.-M.G. were supported by National Institute of Environmental Health Sciences (P42 ESI0337), Department of Energy (DOE-DE-FG02-03ER15449), and National Science Foundation (MCB-0918220) grants to J.I.S.
References
- Andrews M. (1986). The partitioning of nitrate assimilation between root and shoot of higher plants. Plant Cell Environ. 9: 511–519 [Google Scholar]
- Arteca R.N., Arteca J.M. (2000). A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol. Plant. 108: 188–193 [Google Scholar]
- Beevers L., Hageman R. (1980). Nitrate and nitrite reduction. The Biochemistry of Plants 5: 116–168 [Google Scholar]
- Cerezo M., Tillard P., Filleur S., Munos S., Daniel-Vedele F., Gojon A. (2001). Major alterations of the regulation of root NO3− uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffei C., Pageau K., Suzuki A., Gouia H., Ghorbel M., Masclaux-Daubresse C. (2004). Cadmium toxicity induced changes in nitrogen management in Lycopersicon esculentum leading to a metabolic safeguard through an amino acid storage strategy. Plant Cell Physiol. 45: 1681–1693 [DOI] [PubMed] [Google Scholar]
- Chen A., Komives E., Schroeder J. (2006). An improved grafting technique for mature Arabidopsis plants demonstrates long-distance shoot-to-root transport of phytochelatins in Arabidopsis. Plant Physiol. 141: 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C., Lin C., Hsia A., Su R., Lin H., Tsay Y. (2004). Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Choi J.H., Jung H.Y., Kim H.S., Cho H.G. (2000). PhyloDraw: A phylogenetic tree drawing system. Bioinformatics 16: 1056–1058 [DOI] [PubMed] [Google Scholar]
- Chopin F., Orsel M., Dorbe M.-F., Chardon F., Truong H.-N., Miller A.J., Krapp A., Daniel-Vedele F. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S., Bent A. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford N. (1995). Nitrate: nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N., Glass A. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3: 389–395 [Google Scholar]
- Davenport R., Muñoz-Mayor A., Jha D., Essah P., Rus A., Tester M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30: 497–507 [DOI] [PubMed] [Google Scholar]
- De Angeli A., Monachello D., Ephritikhine G., Frachisse J.M., Thomine S., Gambale F., Barbier-Brygoo H. (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442: 939–942 [DOI] [PubMed] [Google Scholar]
- Fan S.-C., Lin C.-S., Hsu P.-K., Lin S.-H., Tsay Y.-F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B.G. (2000). Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Glass A.D., Shaff J., Kochian L. (1992). Studies of the uptake of nitrate in barley: IV. Electrophysiology. Plant Physiol. 99: 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Lee D., Schroeder J. (2003). Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10118–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Waner D., Horie T., Li S., Horie R., Abid K.B., Schroeder J. (2004). Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101: 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Young J., Crawford N. (2003). The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L., Garate A., Carpena-Ruiz R. (1997). Effects of cadmium on the uptake, distribution and assimilation of nitrate in Pisum sativum. Plant Soil 189: 97–106 [Google Scholar]
- Hernandez L., Ramon A., Carpenaruiz R., Garate A. (1995). Evaluation of nitrate nutrition indexes in maize leaves - Metabolic nitrate, total nitrate content and nitrate reductase activity. J. Plant Nutr. 18: 869–887 [Google Scholar]
- Horie T., Horie R., Chan W.-Y., Leung H.-Y., Schroeder J.I. (2006). Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 47: 622–633 [DOI] [PubMed] [Google Scholar]
- Huang N., Liu K., Lo H., Tsay Y. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kolevski B., Smyth D. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P., Young J., Sussman M. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang Y., Okamoto M., Crawford N., Siddiqi M., Glass A. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dankher O., Carreira L., Smith A., Meagher R. (2006). The shoot-specific expression of {gamma}-glutamylcysteine synthetase directs the long-distance transport of thiol-peptides to roots conferring tolerance to mercury and arsenic. Plant Physiol. 141: 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Kuo H., Canivenc G., Lin C., Lepetit M., Hsu P., Tillard P., Lin H., Wang Y., Tsai C., Gojon A., Tsay Y. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Huang C., Tsay Y. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Barton M. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Ludewig U., von Wiren N., Frommer W. (2002). Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 277: 13548–13555 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Higher Plants, 2nd ed (London: Academic Press; ), pp. 231–255 [Google Scholar]
- Marschner H., Kirkby E.A.B.C., Engels C. (1997). Importance of cycling and recycling of mineral nutrients within plants for growth and development. Bot. Acta 110: 265–273 [Google Scholar]
- Maser P., et al. (2002). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Mendoza-Cozatl D., Butko E., Springer F., Torpey J., Komives E., Kehr J., Schroeder J. (2008). Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 54: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller I.S., Gilliham M., Jha D., Mayo G.M., Roy S.J., Coates J.C., Haseloff J., Tester M. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M., Filleur S., Fraisier V., Daniel-Vedele F. (2002a). Nitrate transport in plants: Which gene and which control? J. Exp. Bot. 53: 825–833 [DOI] [PubMed] [Google Scholar]
- Orsel M., Krapp A., Daniel-Vedele F. (2002b). Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa H., Stacey G., Gassmann W. (2006). ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem. J. 393: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I., Skurray R. (1994). The POT family of transport proteins. Trends Biochem. Sci. 19: 404. [DOI] [PubMed] [Google Scholar]
- Ren Z., Gao J., Li L.G., Cai X., Huang W., Chao D., Zhu M., Wang Z., Luan S., Lin H.X. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Salt D., Prince R., Pickering I., Raskin I. (1995). Mechanisms of cadmium mobility and accumulation in Indian mustard. Plant Physiol. 109: 1427–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanita di Toppi L., Gabbrielli R. (1999). Response to cadmium in higher plants. Environ. Exp. Bot. 41: 105–130 [Google Scholar]
- Smirnoff N., Stewart G. (1985). Nitrate assimilation and translocation by higher plants: Comparative physiology and ecological consequences. Physiol. Plant. 64: 133–140 [Google Scholar]
- Stitt M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Sunarpi, et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y.F., Chiu C.C., Tsai C.B., Ho C.H., Hsu P.K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay Y.-F., Schroeder J.I., Feldmann K.A., Crawford N.M. (1993). The herbicide sensitivity gene CHL1 of arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Wang R., Liu D., Crawford N. (1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Okamoto M., Xing X., Crawford N.M. (2003). Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Tischner R., Gutierrez R.A., Hoffman M., Xing X., Chen M., Coruzzi G., Crawford N.M. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S., Cho Y., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.