Six genes encode acyl-CoA binding proteins (ACBPs) in Arabidopsis. This study suggests a role for Arabidopsis ACBP3 as a phospholipid binding protein in the regulation of leaf senescence by modulating membrane phospholipid metabolism and the stability of autophagy-related protein ATG8.
Abstract
In Arabidopsis thaliana, a family of six genes (ACBP1 to ACBP6) encodes acyl-CoA binding proteins (ACBPs). Investigations on ACBP3 reported here show its upregulation upon dark treatment and in senescing rosettes. Transgenic Arabidopsis overexpressing ACBP3 (ACBP3-OEs) displayed accelerated leaf senescence, whereas an acbp3 T-DNA insertional mutant and ACBP3 RNA interference transgenic Arabidopsis lines were delayed in dark-induced leaf senescence. Acyl-CoA and lipid profiling revealed that the overexpression of ACBP3 led to an increase in acyl-CoA and phosphatidylethanolamine (PE) levels, whereas ACBP3 downregulation reduced PE content. Moreover, significant losses in phosphatidylcholine (PC) and phosphatidylinositol, and gains in phosphatidic acid (PA), lysophospholipids, and oxylipin-containing galactolipids (arabidopsides) were evident in 3-week-old dark-treated and 6-week-old premature senescing ACBP3-OEs. Such accumulation of PA and arabidopsides (A, B, D, E, and G) resulting from lipid peroxidation in ACBP3-OEs likely promoted leaf senescence. The N-terminal signal sequence/transmembrane domain in ACBP3 was shown to be essential in ACBP3-green fluorescent protein targeting and in promoting senescence. Observations that recombinant ACBP3 binds PC, PE, and unsaturated acyl-CoAs in vitro and that ACBP3 overexpression enhances degradation of the autophagy (ATG)-related protein ATG8 and disrupts autophagosome formation suggest a role for ACBP3 as a phospholipid binding protein involved in the regulation of leaf senescence by modulating membrane phospholipid metabolism and ATG8 stability in Arabidopsis. Accelerated senescence in ACBP3-OEs is dependent on salicylic acid but not jasmonic acid signaling.
INTRODUCTION
In plants, senescence occurs at the ultimate stage in leaf development and precedes cell death (Lim et al., 2007). Yellowing, the most striking phenotypic change during leaf senescence, results from the loss in chlorophyll due to the disintegration of chloroplasts and disruption in photosynthesis. This is accompanied by other dramatic changes in cellular metabolism in senescing leaves, such as hydrolysis of membrane lipids, proteins, and nucleic acids and remobilization of the hydrolyzed nutrients to young leaves, developing seeds, and other storage tissues (Buchanan-Wollaston, 1997; Nam, 1997; Quirino et al., 2000; Lim et al., 2007). Other than being age dependent, leaf senescence can also be induced by abiotic and biotic factors including extended darkness, drought, extreme high or low temperature, nutrient deficiency, oxidative stress, and pathogen infections (Lim et al., 2007). Internal signals including abscisic acid, salicylic acid, ethylene, and jasmonates also regulate leaf senescence (Lim et al., 2007).
The identification of >800 senescence-associated genes (SAGs) and a complex molecular network of pathways during leaf senescence in Arabidopsis thaliana suggest that large-scale cellular changes occur (Lim et al., 2007). Early events in senescence include membrane deterioration and leakiness, hydrolysis of membrane lipids, and production of free fatty acids and sterols (Thompson et al., 2000; Hopkins et al., 2007). Participating lipid-degrading enzymes include phospholipase D (PLD), lipoxygenase, acyl hydrolase, and possibly other lipases (Fan et al., 1997; Hong et al., 2000; Thompson et al., 2000; He and Gan, 2002). Chloroplast lipids are degraded by galactolipase, acyl hydrolase, and other lipases (Woolhouse, 1984; Padham et al., 2007). Free fatty acids, free radicals, and lipid peroxides cause deterioration of the bilayer membrane, promoting leakiness and loss of selective permeability (Thompson et al., 1998).
The senescence-associated gene SAG101 is expressed at the onset of leaf senescence and encodes a putative membrane acyl hydrolase likely involved in the initial hydrolysis of membrane phospholipids (He and Gan, 2002). An mRNA encoding an Arabidopsis plastidic triacylglycerol lipase was upregulated in senescing leaves and flower petals (Padham et al., 2007). Suppression of phospholipase PLDα, acyl hydrolase SAG101, or a membrane-bound lipase associated with membrane lipid hydrolysis and metabolism inhibited senescence in transgenic Arabidopsis (Fan et al., 1997; Thompson et al., 2000; He and Gan, 2002). Specifically, antisense suppression of PLDα delayed abscisic acid- and ethylene-induced leaf senescence but did not alter age-dependent senescence, suggesting that PLDα is associated with phytohormone-regulated, rather than age-dependent, senescence (Fan et al., 1997). Transgenic Arabidopsis plants expressing antisense SAG101 were arrested in age-dependent leaf senescence, whereas those overexpressing SAG101 exhibited premature leaf senescence (He and Gan, 2002). Also, antisense suppression of a membrane lipid lipase in transgenic Arabidopsis retarded senescence and enhanced seed yield (Thompson et al., 2000). In addition, the onset of leaf senescence is affected by modifications in the expressions of enzymes of phospholipid biosynthesis. For example, the silencing of the gene encoding phosphoethanolamine N-methyltransferase, an enzyme in the biosynthesis of choline (a vital precursor of phosphatidylcholine), culminated in early senescence in transgenic Arabidopsis (Mou et al., 2002).
ACBPs are a family of proteins that facilitate the binding of long-chain acyl-CoA esters at the conserved acyl-CoA binding domain (Xiao and Chye, 2009). In Arabidopsis, the 10-kD ACBP6 and five larger proteins (ACBP1 to ACBP5) that each contain an acyl-CoA binding domain have been shown to bind different acyl-CoA esters with varying affinities, implying that they do not have redundant roles in vivo (Engeseth et al., 1996; Chye, 1998; Chye et al., 1999, 2000; Leung et al., 2004, 2006; Gao et al., 2009; Xiao et al., 2009). Furthermore, the presence of other structural domains, ankyrin repeats in ACBP1 and ACBP2 and kelch motifs in ACBP4 and ACBP5, enable them to interact with protein partners (Li and Chye, 2004; Li et al., 2008; Gao et al., 2009). Some of these ACBP mRNAs are induced by abiotic and biotic stresses (Chen et al., 2008; Li et al., 2008; Xiao et al., 2008a; Gao et al., 2009). Our recent work has revealed that cytosolic ACBP6 binds phosphatidylcholine (PC) in vitro and that ACBP6 overexpressors show increased expression of PLDδ and enhanced freezing tolerance, indicating the involvement of plant ACBPs in phospholipid metabolism (Chen et al., 2008). The increased tolerance to heavy metal stress of plants overexpressing membrane-associated ACBP1 and ACBP2 is likely related to their ability to bind acyl-CoAs and to facilitate repair of the phospholipid bilayer membrane (Xiao et al., 2008a; Gao et al., 2009).
Arabidopsis ACBP3 was demonstrated to be extracellularly targeted by transient expression of fluorescence-tagged ACBP3 C-terminal truncated fusions in tobacco (Nicotiana tabacum) BY-2 cells and onion (Allium cepa) epidermal cells (Leung et al., 2006), but its biological functions were undefined. In this study, we show that ACBP3 mRNA is upregulated by dark treatment and in senescing Arabidopsis rosettes. Overexpression of ACBP3 accelerated both dark-induced and age-dependent leaf senescence, whereas downregulation of ACBP3 delayed dark-induced senescence. These findings indicate an important role of Arabidopsis ACBP3 in leaf senescence.
RESULTS
The ACBP3 Transcript Is Developmentally Regulated in Rosettes and Is Induced by Dark Treatment
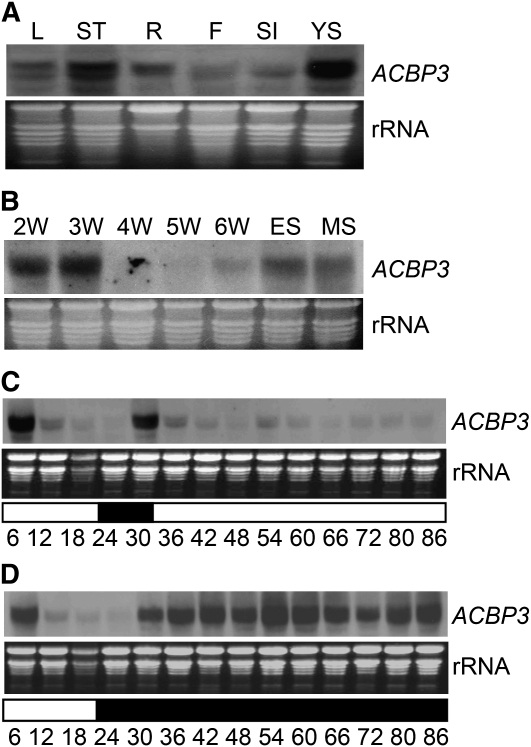
On RNA gel blot analysis, ACBP3 mRNA was expressed in 6-week-old rosettes, stalks, and roots and 1-week-old young seedlings, with relatively lower abundance in flowers and siliques 6 to 8 d after fertilization (Figure 1A). Its expression in 2- to 5-week-old rosettes peaked at 3 weeks and then decreased (Figure 1B). Subsequently, ACBP3 expression again increased from 6-week-old fully expanded rosettes through early and mid senescence (ES and MS; Figure 1B).
Figure 1.
RNA Gel Blot Analyses Showing ACBP3 Expression at Different Stages of Arabidopsis Rosette Development under LDs Followed by LL or DD Treatment.
(A) Total RNA isolated from 6-week-old leaves (L), stalks (ST), roots (R), and flowers (F), siliques (SI) 6 to 8 d after fertilization, and 1-week-old young seedlings (YS).
(B) Total RNA isolated from 2-week-old (2W), 3-week-old (3W), 4-week-old (4W), 5-week-old (5W), and 6-week-old (6W) rosettes as well as from rosettes in early-stage senescence (ES) and mid-stage senescence (MS). ES corresponds to plants aged 49 to 50 d after sowing with ∼25% yellow leaf area, and MS corresponds to 50 to 53 d after sowing with ∼50% yellow leaf area as previously defined (van der Graaff et al., 2006).
(C) and (D) Expression of ACBP3 under LD (first 30 h) followed by LL (C) or DD (D) treatment. Arabidopsis plants were germinated and grown for 4 weeks under 16 h light (06:00 to 21:00)/8 h dark (21:00 to 06:00) cycles and then shifted to LL or DD. Total RNA was extracted from rosettes harvested at 6-h intervals. Ethidium bromide–stained rRNA is shown below the blots to indicate the relative amount of total RNA loaded per lane. Bottom numbers indicate time of treatment. White and black bars indicate light and dark periods, respectively.
Some mRNAs encoding ACBPs are regulated by light; ACBP3 mRNA decreased in the light (06:00 to 21:00) and was elevated following a dark period (21:00 to 06:00) (Xiao et al., 2009). To investigate the effect of continuous darkness (DD) and continuous light (LL) on ACBP3 expression, LD-grown rosettes were harvested every 6 h under DD or LL. RNA gel blot analysis confirmed that the ACBP3 mRNA decreased in LL, dampening-off to a much reduced peak at 54 h (Figure 1C). By contrast, plants transferred to DD displayed increased expression (Figure 1D), suggesting that ACBP3 is downregulated in LL but upregulated in DD. Its induced expression during DD and senescence is consistent with microarray data on ACBP3 expression (see Supplemental Figure 1 online; van der Graaff et al., 2006). Taken together, these results support a role for ACBP3 in leaf senescence.
ACBP3 Overexpression Accelerates Starvation-Induced Leaf Senescence
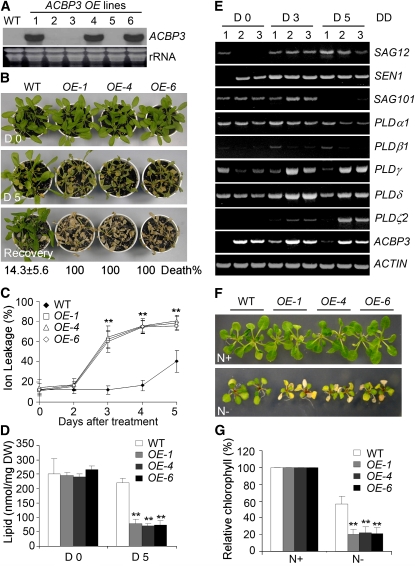
To address the effect of ACBP3 overexpression on dark-induced leaf senescence, ACBP3-overexpressor lines (ACBP3-OEs) were generated. Three independent T2 transgenic lines that accumulated ACBP3 mRNA were identified via RNA gel blot analysis (Figure 2A),and their resultant T3 stable transgenic plants were subsequently used. Under normal growth conditions (LD), 3-week-old independent ACBP3-OEs (OE-1, OE-4, and OE-6) were similar to the wild type (Figure 2B, top). When plants were transferred to DD, all ACBP3-OEs showed enhanced sensitivity, with leaf vitrification after 5 d (Figure 2B). Following a 3-d recovery under LD, differences in death rates between the wild type (14.3% ± 5.6%) and ACBP3-OEs (100%) were significant (P < 0.01; Figure 2B).
Figure 2.
Overexpression of ACBP3 Accelerates Starvation-Induced Leaf Senescence.
(A) RNA gel blot analysis on the wild type and ACBP3-OEs (OE-1 to OE-6), three of which show higher ACBP3 expression than the wild type. Total RNA (30 μg/lane) stained with ethidium bromide before blotting (bottom).
(B) Images of wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants before (D 0) and 5 d after DD treatment (D 5), followed by a 3-d recovery at LD (Recovery). The numbers ± sd (n = 20) below the images indicate the death rates following recovery.
(C) Electrolyte leakage of wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants after DD treatment for 0, 2, 3, 4, and 5 d. Asterisks indicate significant difference from the wild type (P < 0.01). Values are means ± sd (n = 3) of three independent experiments.
(D) Membrane lipid content of wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants before (D 0) and 5 d after DD treatment (D 5). Asterisks indicate significant difference from the wild type at day 5 (P < 0.01). Values are means ± sd (n = 3). DW, dry weight.
(E) RT-PCR analyses showing expression of SAG12, SEN1, SAG101, PLDα1, PLDβ1, PLDγ, PLDδ, PLDζ2, and ACBP3 in wild-type and ACBP3-OE (OE-1 and OE-4) plants after DD treatment for 0, 3, and 5 d. The ACTIN mRNA was probed as a control. 1, wild type; 2, OE-1; 3, OE-4.
(F) Image of wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants grown for 2 weeks on MS (N+) or nitrogen-lacking MS medium (N−).
(G) Relative chlorophyll contents of wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants grown on MS (N+) or nitrogen-lacking MS medium (N−). Total chlorophyll content was measured and normalized per gram fresh weight of sample, and relative chlorophyll content under nitrogen starvation “N−” was expressed as a percentage of the value for the same line grown under MS “N+” (set to 100%). Values are means of three independent experiments. Bars represent sd (n = 3).
Membrane electrolyte leakage and membrane lipid content, two well-established markers for assessment of membrane integrity in senescent leaves (Fan et al., 1997), were measured. A twofold increase in electrolyte leakage occurred only on day 5 in DD-treated wild-type leaves (Figure 2C). By contrast, electrolyte leakage in ACBP3-OEs was significantly greater (P < 0.01) than the wild type, 3, 4, and 5 d following DD (Figure 2C). The membrane lipid content in rosettes 5 d after DD treatment decreased significantly (P < 0.01) more in OE-1 (64% decrease compared with day 0), OE-4 (68% decrease), and OE-6 (67% decrease) than in the wild type (12% decrease) (Figure 2D).
The expression of genes encoding senescence-associated proteins SAG12, SEN1, and SAG101, three widely used leaf senescence marker genes in Arabidopsis (He and Gan, 2002; Xiao et al., 2004; Thompson et al., 2005), was determined. The expression of senescence-associated PLDα1 (Fan et al., 1997) and four other PLDs (PLDβ1, PLDγ, PLDδ, and PLDζ2), which are upregulated during age-dependent or dark-induced senescence (see Supplemental Figure 1 online; van der Graaff et al., 2006), was also tested. RT-PCR analyses using total RNA from 3-week-old wild type, OE-1, and OE-4 showed that SAG12, SEN1, and PLDβ1 mRNAs were induced in all lines, 3 and 5 d after DD treatment, while SAG101 mRNA was induced in 3 d (Figure 2E). Whereas PLDγ was downregulated in the wild type after DD, it was upregulated in ACBP3-OE lines (Figure 2E). PLDδ and PLDζ2 mRNAs were more highly expressed on day 3 than day 5 in the wild type but were upregulated on both days in ACBP3-OEs (Figure 2E). Measured as a control, the ACBP3 mRNA was dark induced on days 3 and 5 in the wild type and was constitutively expressed in the OE lines. Elevation of SAG12, SEN1, SAG101, PLDδ, PLDγ, and PLDζ2 mRNAs in ACBP3-OEs over the wild type likely caused accelerated dark-induced senescence in the ACBP3-OEs.
To examine the role of ACBP3 during nutrient starvation, 1-week-old LD-grown wild type and ACBP3-OEs on Murashige and Skoog (MS) medium were transferred to either MS or MS medium lacking nitrogen. After 2 weeks of nitrogen starvation, the cotyledons and the first true leaves of ACBP3-OEs were yellow, whereas those of the wild type remained green (N- in Figure 2F). Phenotypic differences were absent when grown on MS medium (N+ in Figure 2F). Under nitrogen starvation conditions, the relative chlorophyll contents of ACBP3-OEs were significantly lower than the wild type (Figure 2G). These results demonstrate that ACBP3 overexpression promoted both DD and nitrogen starvation-induced leaf senescence.
ACBP3-OEs Display Premature Leaf Senescence
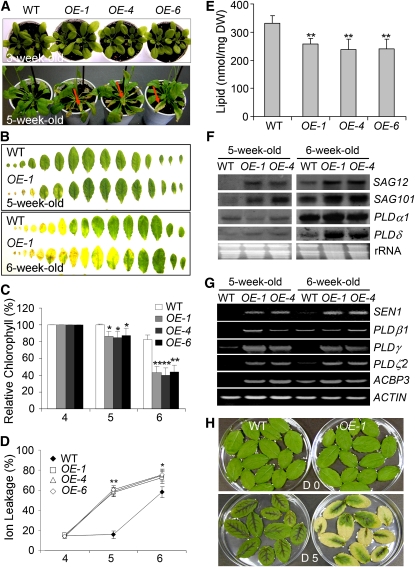
When we investigated if ACBP3-OEs were altered in age-dependent leaf senescence, differences were absent between soil-grown ACBP3-OEs and the wild type in the first 3 weeks (Figure 3A), but bottom rosette leaves in all 5-week-old ACBP3-OEs, but not the wild type, had yellowed, indicating the onset of leaf senescence (Figure 3A). Approximately 80% (P < 0.05) of rosette leaves in 6-week-old ACBP3-OEs were yellow in comparison to the wild type, of which the majority (∼60%) of leaves remained green (Figure 3B). Measurements of chlorophyll content of 4-, 5-, and 6-week-old rosettes from the wild type and ACBP3-OEs revealed a significant decline at weeks 5 and 6 in OEs in comparison to the wild type (Figure 3C). Premature leaf senescence in ACBP3-OE rosettes was accompanied by increased electrolyte leakage at weeks 5 and 6 (Figure 3D) and decreased membrane lipid content at week 6 (Figure 3E) in comparison to the wild type.
Figure 3.
Age-Dependent Senescence Phenotype in the ACBP3-Overexpressing Transgenic Plants.
(A) Comparison in phenotypes of LD-grown 3- and 5-week-old wild type and ACBP3-OEs (OE-1, OE-4, and OE-6). Arrows indicate the senescent rosettes of ACBP3-OEs.
(B) Comparison of all leaves from the rosettes of 5- and 6-week-old wild-type and ACBP3 OE-1 plants.
(C) Relative chlorophyll contents of 4-, 5-, and 6-week-old wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants. Total chlorophyll content was measured and normalized per gram fresh weight of sample. Chlorophyll content of 4-week-old plants was set at 100%, and values were expressed relative to their corresponding genotypes to obtain their relative contents. Asterisks indicate significant difference from the wild type at the same age (*P < 0.05 or **P < 0.01). Values are means ± sd (n = 3) of three independent experiments.
(D) Electrolyte leakage of 4-, 5-, and 6-week-old wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants. Asterisks indicate significant difference from the wild type at the same age (*P < 0.05 or **P < 0.01). Values are means ± sd (n = 3) of three independent experiments.
(E) Membrane lipid content of 6-week-old wild-type and ACBP3-OE (OE-1, OE-4, and OE-6) plants. Asterisks indicate significant difference from the wild type (P < 0.01). Values are means ± sd (n = 3). DW, dry weight.
(F) RNA gel blot analyses showing expression of SAG12, SAG101, PLDα1, and PLDδ in 5- and 6-week-old wild-type and ACBP3-OE (OE-1 and OE-4) plants. Total RNA (30 μg/lane) stained with ethidium bromide before blotting (bottom).
(G) RT-PCR analyses showing expression of SEN1, PLDβ1, PLDγ, PLDζ2, and ACBP3 in 5- and 6-week-old wild type and ACBP3-OE (OE-1 and OE-4). The ACTIN mRNA was used as a control (below).
(H) Phenotype of detached leaves from 4-week-old ACBP3 OE-1 in comparison to the wild type. Photos were taken at 0 and 5 d after detachment.
Analysis of total rosette RNA from 5- and 6-week-old OEs (OE-1 and OE-4) and the wild type showed expression of senescence marker genes SAG12, SAG101, and SEN1 in 5-week-old OE plants but not in the wild type (Figures 3F and 3G). Also, PLDδ, PLDβ1, PLDγ, and PLDζ2 mRNAs were upregulated in these OEs (Figures 3F and 3G). By contrast, PLDα1 expression did not vary between the wild type and OEs (Figure 3F). In 6-week-old plants, the expression of SAG12, SAG101, SEN1, PLDα1, PLDδ, and PLDζ2 was upregulated in comparison to 5-week-old plants, and SAG12, SAG101, PLDδ, SEN1, and PLDζ2 mRNA levels were higher in the ACBP3-OEs (Figures 3F and 3G). Although PLDγ expression was downregulated in 6-week-old plants, it was higher in the ACBP3-OEs (Figure 3G). When premature leaf senescence was assessed in detached leaves, leaves from 4-week-old OE-1 showed accelerated senescence marked by yellowing 5 d after detachment, while those of the wild type remained dark green (Figure 3H).
Characterization of an acbp3 T-DNA Insertional Mutant
A T-DNA insertional mutant of ACBP3 (designated acbp3) was identified to examine further the role of ACBP3 in leaf senescence. PCR analysis followed by DNA sequence analysis confirmed the T-DNA insertion in the mRNA splicing site of the second intron of ACBP3 (see Supplemental Figure 2A online). RNA gel blot analysis indicated expression of truncated ACBP3 mRNAs (see Supplemental Figure 2B online). RT-PCR followed by DNA sequencing analyses revealed a 1.0-kb truncated mRNA with the second exon deleted and a 4.5-kb mRNA inclusive of T-DNA sequences, generated from splicing at the right border of the inserted T-DNA (see Supplemental Figure 2C online). Subsequently, the acbp3 mutant was complemented, and ACBP3 expression in three independent putative complemented transformants (acbp3-C1, acbp3-C2, and acbp3-C3) was confirmed by RT-PCR analysis (see Supplemental Figure 2D online).
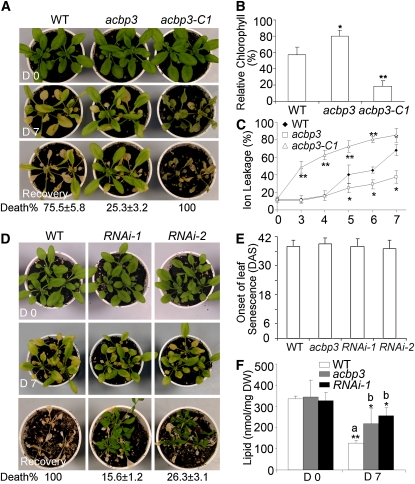
When LD-grown 3-week-old wild-type and acbp3 mutant plants were transferred to DD, wild-type leaves yellowed after 7 d, while most mutant leaves remained green (Figure 4A). After recovery under LD, ∼80% of the wild type died, while 75% (P < 0.05) of mutants survived (Figure 4A). The complementation lines were similarly treated, and a representative line (Figure 4A) shows reversal of the acbp3 delayed leaf senescence phenotype back to the wild-type phenotype. When the chlorophyll content and electrolyte leakage of the wild type, acbp3 mutant, and acbp3-C1 before and after DD treatment were measured, the relative chlorophyll content in the mutant was significantly higher than in either the wild type or acbp3-C1 (Figure 4B). Electrolyte leakage in the acbp3 mutant was significantly lower (P < 0.05) at 5 to 7 d after DD than in the wild type, while those of acbp3-C1 were significantly greater (P < 0.01) than the wild type at 3 to 6 d (Figure 4C).
Figure 4.
Phenotypes of the acbp3 Mutant and ACBP3 RNAi Transgenic Arabidopsis.
(A) Phenotype of 3-week-old wild-type, acbp3 mutant, and acbp3-C1 seedlings before (D 0) and after 7 d of DD treatment (D 7), followed by recovery for 3 d at LD (Recovery). The numbers ± sd (n = 20) below the images represent the death rates following recovery.
(B) Relative chlorophyll contents of 3-week-old wild type, acbp3 mutant, and acbp3-C1 at 7 d after DD treatment. The chlorophyll contents were expressed relative to the values for the same line obtained before DD treatment. Asterisks indicate significant difference from the wild type (*P < 0.05 or **P < 0.01). Values are means ± sd (n = 3) of three independent experiments.
(C) Electrolyte leakage of the wild type, acbp3 mutant, and acbp3-C1 after DD treatment for 0, 3, 4, 5, 6, and 7 d. Asterisks indicate significant difference from the wild type on the same day (*P < 0.05 or **P < 0.01). Values are means ± sd (n = 3) of three independent experiments.
(D) Phenotype of 3-week-old seedlings of wild-type and ACBP3 RNAi lines (RNAi-1 and RNAi-2) before (D 0) and after 7 d of DD treatment (D 7), followed by recovery for 10 d at LD (Recovery).
(E) Onset of age-dependent leaf senescence in LD-grown wild-type, acbp3 mutant, and ACBP3 RNAi lines (RNAi-1 and RNAi-2). Bars represent sd (n = 10). DAS, days after sowing.
(F) Membrane lipid content of the wild type, acbp3 mutant, and ACBP3 RNAi-1 line before (D 0) and after 7 d of DD treatment (D 5). “a” indicates significant difference of DD-treated wild type (D 7) from that of LD wild type (D 0; P < 0.01); “b” indicates significant difference of DD-treated acbp3 and RNAi-1 from DD-treated wild type (D 7; P < 0.05). Values are means ± sd (n = 3). DW, dry weight.
Downregulation of ACBP3 by RNA Interference Delays Dark-Induced Senescence
Detection of truncated mRNAs in the acbp3 T-DNA insertional mutant prompted us to use an RNA interference (RNAi) approach to generate ACBP3-RNAi transgenic lines. RNA gel blot analysis using total RNA from 3-week-old rosettes of wild-type and RNAi lines (RNAi-1 and RNAi-2) before and 24 h after DD treatment, as well as from 6-week-old plants, showed very low ACBP3 expression in both wild-type and RNAi lines (see Supplemental Figure 2E online), noticeable downregulation in RNAi lines 24 h after dark treatment (see Supplemental Figure 2F online), and marked downregulation in 6-week-old senescing rosettes (see Supplemental Figure 2G online). Since ACBP1 expression did not vary between wild-type and ACBP3-RNAi lines (see Supplemental Figures 2E to 2G online), downregulation in the RNAi lines was deemed ACBP3 specific. Similar to the acbp3 mutant, no obvious developmental abnormalities were observed in LD-grown RNAi seedlings (Figure 4D, top row). We also failed to find any significant phenotypic difference in age-dependent leaf senescence among wild-type, acbp3 mutant, and RNAi lines (Figure 4E). However, 7-d DD-treated RNAi-1 and RNAi-2 lines exhibited a delay in leaf senescence (Figure 4D) with death rates (after 10 d of recovery) significantly lower than that of the wild type (P < 0.05; Figure 4D), suggesting that RNAi of ACBP3 arrested dark-induced leaf senescence. Decreases in rosette membrane lipid content of the acbp3 mutant (36%) and ACBP3 RNAi-1 line (22%) 7 d after DD treatment were significantly (P < 0.01) less than that of the wild type (62%) (Figure 4F). Taken together, we conclude that ACBP3-KOs (acbp3 and RNAi) inhibited the effects of DD-induced leaf senescence.
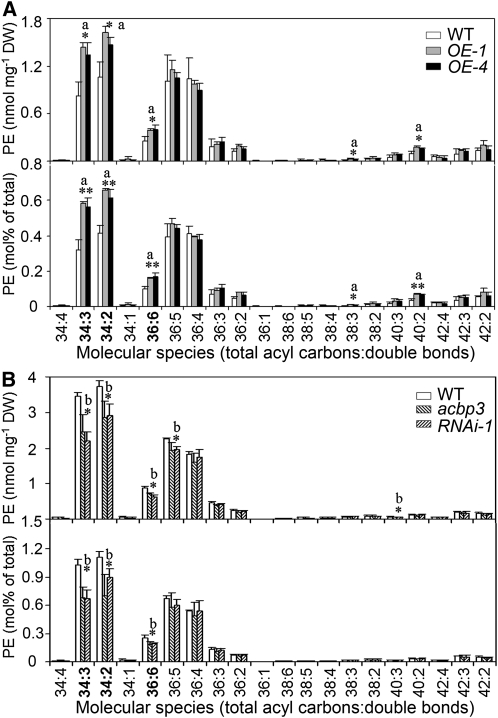
Altered Expression of ACBP3 Is Correlated with Changes in Phosphatidylethanolamine Composition
To correlate ACBP3 expression and membrane lipid composition, total lipids were extracted from rosettes of 3-week-old LD-grown wild-type, ACBP3-OE (OE-1 and OE-4), and ACBP3-KO (acbp3 and RNAi-1) plants, followed by analysis of individual lipid molecular species using electrospray ionization–tandem mass spectrometry. Here and throughout, individual acyl chains for Arabidopsis leaf membrane lipid molecular species are indicated based on determination by Devaiah et al. (2006). Results showed that the phosphatidylethanolamine (PE) content of LD-grown rosette leaves from wild-type, ACBP3-OE, and ACBP3-KO plants were in the order: ACBP3-OEs > wild type > ACBP3-KOs (Table 1). Moreover, particular PE molecular species were differentially affected by ACBP3 overexpression (OE-1 and OE-4) and suppression (acbp3 and RNAi-1) (Figure 5). In particular, PE molecular species 34:3 (16:0-18:3)-, 34:2 (16:0-18:2)-, and 36:6 (di18:3)-PE were higher in the ACBP3-OE plants and lower in the ACBP3-KO plants relative to the wild type (Figures 5A and 5B). Differences in PE composition were further confirmed by calculation of PE composition (mol % of membrane lipids analyzed) as well as PE/PC ratios (Table 1, Figure 5). By contrast, few differences were observed between rosettes of the wild type and ACBP3-OEs or the wild type and ACBP3-KOs in other lipid species, including digalactosyldiacylglycerol (DGDG), monogalactosyldiacylglycerol (MGDG), phosphatidylglycerol (PG), PC, phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidic acid (PA), lysoPC, lysoPE, and lysoPG (Table 1; see Supplemental Figures 3 and 4 online).
Table 1.
Total Amount of Lipid in Each Head Group Class in 3-Week-Old Leaves of the Wild Type, ACBP3-OEs (OE-1 and OE-4), and ACBP3-KOs (acbp3 and RNAi-1) Grown under LDs
| Lipid Class | Wild Type-1 | OE-1 | OE-4 | Wild Type-2 | acbp3 | RNAi-1 |
| DGDG | 38.6 ± 9.8 | 39.7 ± 1.1 | 36.9 ± 2.5 | 42.6 ± 1.3 | 46.7 ± 16.6 | 45.3 ± 9.8 |
| MGDG | 172.9 ± 24.0 | 168.6 ± 7.1 | 165.6 ± 7.3 | 224.6 ± 13.8 | 223.7 ± 32.5 | 212.3 ± 26.9 |
| PG | 5.3 ± 0.5 | 4.7 ± 0.5 | 5.6 ± 1.5 | 18.2 ± 1.0 | 25.8 ± 12.9 | 22.4 ± 2.5 |
| PC | 22.5 ± 6.5 | 20.0 ± 1.2 | 19.2 ± 1.0 | 26.3 ± 0.8 | 25.7 ± 1.3 | 25.7 ± 2.8 |
| PE | 5.0 ± 1.4 | 6.8 ± 0.3a* | 6.2 ± 0.5 | 13.8 ± 0.2 | 10.9 ± 1.8b* | 10.9 ± 0.8b** |
| PI | 7.2 ± 1.2 | 6.7 ± 0.3 | 6.0 ± 0.4 | 8.0 ± 0.3 | 8.2 ± 1.3 | 9.6 ± 2.6 |
| PS | 0.18 ± 0.09 | 0.17 ± 0.04 | 0.21 ± 0.03 | 1.1 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.1 |
| PA | 0.06 ± 0.03 | 0.13 ± 0.04 | 0.10 ± 0.02 | 0.43 ± 0.07 | 0.95 ± 0.76 | 0.78 ± 0.41 |
| LysoPG | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.03 ± 0.03 | 0.03 ± 0.01 | 0.04 ± 0.01 | 0.05 ± 0.02 |
| LysoPC | 0.05 ± 0.04 | 0.05 ± 0.00 | 0.04 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| LysoPE | 0.07 ± 0.03 | 0.10 ± 0.01 | 0.09 ± 0.00 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.02 |
| PE mol % | 2.14 ± 0.14 | 2.74 ± 0.04a** | 2.60 ± 0.23a* | 4.13 ± 0.13 | 3.06 ± 0.70b* | 3.36 ± 0.54b* |
| PE/PC | 0.24 ± 0.04 | 0.34 ± 0.01a* | 0.32 ± 0.02a* | 0.53 ± 0.02 | 0.42 ± 0.06b* | 0.42 ± 0.04b* |
Values are means ± sd (nmol/mg dry weight; n = 3). Significant differences in OEs or KOs from the wild type are in bold. The PE composition (mol %) normalized with total membrane lipids and the PE/PC ratios are shown in the last two rows.
Value higher when compared to wild type-1 plants in the same experiment (*P < 0.05; **P < 0.01).
Value lower when compared to wild type-2 plants in the same experiment (*P < 0.05; **P < 0.01).
Figure 5.
Changes in PE Composition in ACBP3-OEs (OE-1 and OE-4) and ACBP3-KOs (acbp3 and RNAi-1).
(A) Content of PE molecular species of 3-week-old wild type and ACBP3-OEs (OE-1 and OE-4) under LDs.
(B) PE content of 3-week-old wild type and ACBP3-KOs (acbp3 and RNAi-1) under LD. “a” indicates value of OEs higher than the wild type; “b” indicates value of KOs lower than the wild type (*P < 0.05 or **P < 0.01). Values are the means ± sd (n = 3). DW, dry weight.
Subsequently, lipid profiling was performed on lipids extracted from rosettes of 3-week-old DD-treated wild type, ACBP3-OEs, and ACBP3-KOs as well as 6-week-old wild type and ACBP3-OEs. Following DD treatment, all wild type, ACBP3-OEs, and ACBP3-KOs showed decreases or a tendency toward decrease in galactolipids DGDG and MGDG (Tables 1 and 2); this reflected changes in multiple molecular species (see Supplemental Figures 4 and 5 online). Overall, the decreases in DGDG and MGDG in ACBP3-OEs were greater than in the wild type, while such decreases in ACBP3-KOs were less than in the wild type. Lower levels of galactolipids compared with the wild type were also observed in 6-week-old rosettes of OEs (Table 1; see Supplemental Figure 5 online).
Table 2.
Total Amount of Lipid in Each Head Group Class in 3- or 6-Week-Old Leaves of the Wild Type, ACBP3-OEs, and ACBP3-KOs Grown under LDs or DDs
| Lipid Class | 3 Weeks Old/DD |
6 Weeks Old/LD |
3 Weeks Old/DD |
||||||
| Wild Type-1 | OE-1 | OE-4 | Wild Type-2 | OE-1 | OE-4 | Wild Type-3 | acbp3 | RNAi-1 | |
| DGDG | 20.2 ± 1.3 | 5.4 ± 2.9b** | 4.7 ± 0.5b** | 53.5 ± 2.9 | 39.6 ± 2.4b** | 35.6 ± 4.4b** | 9.6 ± 3.2 | 13.7 ± 6.1 | 18.1 ± 3.7a* |
| MGDG | 152.2 ± 9.5 | 49.1 ± 34.7b** | 37.7 ± 7.0b** | 221.6 ± 24.0 | 170.2 ± 20.9b* | 155.5 ± 31.4b* | 59.4 ± 9.3 | 144.4 ± 33.0a* | 166.7 ± 32.9a* |
| PG | 5.7 ± 1.2 | 3.7 ± 1.0 | 5.5 ± 0.4 | 18.0 ± 1.5 | 13.5 ± 1.4b** | 11.2 ± 1.4b** | 11.5 ± 1.2 | 13.5 ± 1.3a* | 19.1 ± 0.4a** |
| PC | 30.6 ± 1.5 | 13.5 ± 5.0b** | 14.3 ± 1.3b** | 16.5 ± 0.5 | 14.6 ± 2.7 | 15.1 ± 2.3 | 29.5 ± 6.6 | 30.7 ± 2.4 | 31.5 ± 2.8 |
| PE | 5.4 ± 0.2 | 3.1 ± 1.0b* | 3.2 ± 0.6b** | 10.1 ± 0.5 | 10.8 ± 0.5 | 10.7 ± 0.7 | 7.9 ± 1.8 | 9.7 ± 0.4 | 9.2 ± 1.5 |
| PI | 6.9 ± 0.3 | 2.5 ± 0.8b** | 2.3 ± 0.6b** | 10.6 ± 0.4 | 7.6 ± 0.6b** | 7.1 ± 1.1b** | 5.7 ± 0.7 | 6.2 ± 0.2 | 8.9 ± 1.1a* |
| PS | 0.33 ± 0.02 | 0.28 ± 0.17 | 0.31 ± 0.03 | 1.34 ± 0.18 | 1.11 ± 0.04b* | 1.01 ± 0.13b* | 1.2 ± 0.1 | 1.0 ± 0.2 | 1.1 ± 0.1 |
| PA | 0.06 ± 0.02 | 1.57 ± 0.55a** | 2.02 ± 0.20a** | 0.46 ± 0.07 | 1.04 ± 0.07a** | 1.25 ± 0.35a** | 1.44 ± 1.15 | 1.12 ± 1.57 | 1.52 ± 0.40 |
| LysoPG | 0.02 ± 0.02 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.04 ± 0.03 | 0.06 ± 0.01 | 0.05 ± 0.02 | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.02 |
| LysoPC | 0.08 ± 0.01 | 0.12 ± 0.01a* | 0.11 ± 0.01a** | 0.04 ± 0.01 | 0.07 ± 0.01a** | 0.07 ± 0.01a** | 0.15 ± 0.03 | 0.10 ± 0.04 | 0.14 ± 0.02 |
| LysoPE | 0.20 ± 0.01 | 0.12 ± 0.02 | 0.16 ± 0.03 | 0.08 ± 0.03 | 0.11 ± 0.01 | 0.13 ± 0.02 | 0.32 ± 0.05 | 0.37 ± 0.15 | 0.65 ± 0.11 |
| PE mol % | 2.45 ± 0.24 | 4.28 ± 0.94a* | 4.51 ± 0.59a** | 3.04 ± 0.23 | 4.18 ± 0.35a** | 4.48 ± 0.42a** | 6.30 ± 1.72 | 4.71 ± 1.38 | 3.65 ± 0.88 |
| PE/PC | 0.18 ± 0.01 | 0.24 ± 0.01a** | 0.22 ± 0.0a* | 0.61 ± 0.03 | 0.75 ± 0.11a* | 0.71 ± 0.08a* | 0.27 ± 0.03 | 0.32 ± 0.03 | 0.29 ± 0.04 |
Values are means ± sd (nmol/mg dry weight; n = 3). Significant differences in OEs or KOs from the wild type are in bold. The PE composition (mol %) normalized with total membrane lipids and the PE/PC ratios are shown in the last two rows.
Value higher when compared to similarly treated wild-type plants in the same experiment (*P < 0.05; **P < 0.01).
Value lower when compared to similarly treated wild-type plants in the same experiment (*P < 0.05; **P < 0.01).
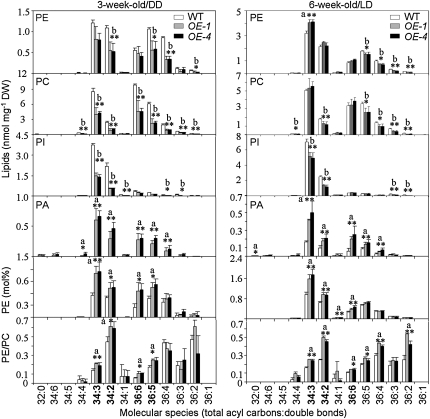
Among phospholipids, both DD-treated and 6-week-old ACBP3-OEs showed decreases in PE, PC, and PI and increases in PA (Tables 1 and 2, Figure 6), but these changes were not observed in DD-treated ACBP3-KOs (Table 2; see Supplemental Figure 4 online). In particular, DD-treated OEs exhibited significant loss in the phospholipid molecular species, PE (34:2, 36:2, 36:4, and 36:5), PC (34:2, 34:3, 34:4, 36:2, 36:3, 36:4, 36:5, and 36:6), and PI (34:1, 34:2, 34:3, 36:3, and 36:4), while the PA molecular species 34:2 (18:2-16:0)-, 34:3 (18:3-16:0)-, 36:4 (di18:2/18:3-18:1)-, 36:5 (18:2-18:3)-, and 36:6 (di18:3)-PA were higher in the OEs (Figure 6). Significant increases in particular PA species were accompanied by decreases in the corresponding PC species during DD treatment (Figure 6). Similarly, in 6-week-old OEs, higher levels of molecular species of 34:2- and 34:3-PA occurred along with lower levels of 34:2- and 34:3-PI, while higher levels of 36:4- and 36:5-PA were accompanied by lower levels of 36:4 and 36:5 in both PC and PE (Figure 6). These results suggest possible conversion of PC, PE, and PI species to PA in ACBP3-OEs. In addition, 34:3-PE was higher in 6-week-old OEs (Figure 6). Although the actual amount of PE species was lower in DD-treated and 6-week-old OEs than in the wild type, the mole percentage of PE was significantly higher than in the wild type; likewise, the PE/PC ratio was increased in OEs compared with the wild type in DD treatment and in 6-week-old OEs (Table 2, Figure 6).
Figure 6.
Phospholipid Composition of 3-Week-Old DD-Treated and 6-Week-Old LD-Grown Wild Type and ACBP3-OEs (OE-1 and OE-4).
PE, PC, PI, and PA contents of 3-week-old plants under DD (left column) and 6-week-old plants under LD (right column). The mol % of PE and the PE/PC ratios of each molecular species is shown in the bottom two graphs. “a” indicates value of OEs higher than the wild type; “b” indicates value of OEs lower than the wild type (*P < 0.05 or **P < 0.01). Values are the means ± sd (n = 3 or 4). DW, dry weight.
Taken together, the lipid profiling data indicate that ACBP3 overexpression is correlated with relative increases in PE and PA, at the expense of other phospholipids, particularly PC and PI; ACBP3 suppression is associated with a relative decrease in PE levels.
Changes in Oxidized Membrane Lipid Species and Acyl-CoA Esters in ACBP3-OEs
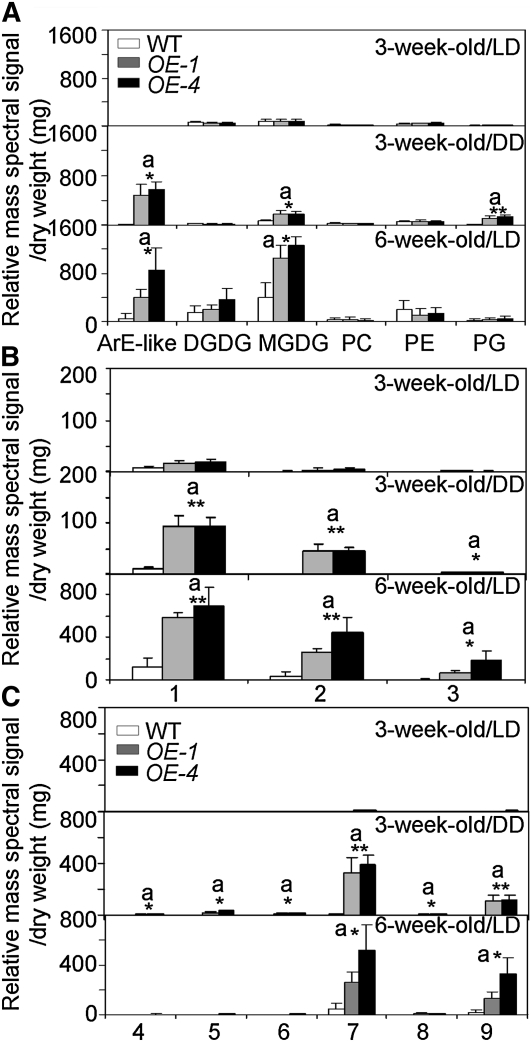
Galactolipids (MGDG and DGDG) that contain 12-oxophytodienoic acid (OPDA) and dinor-OPDA (dnOPDA) have been identified in Arabidopsis (Stelmach et al., 2001; Hisamatsu et al., 2003, 2005; Andersson et al., 2006; Buseman et al., 2006; Kourtchenko et al., 2007). Those species containing two or more OPDA or dnOPDA chains have been termed arabidopsides (Hisamatsu et al., 2003, 2005; Andersson et al., 2006; see Supplemental Table 1 online). To test whether the accelerated leaf senescence phenotype in ACBP3-OE rosettes is related to arabidopsides, we analyzed oxidized membrane lipids including galactolipids (MGDG and DGDG) and phospholipids (PC, PE, and PG) in DD-treated and 6-week-old ACBP3-OE rosettes in comparison to the wild type. Electrospray ionization–tandem mass spectrometry was employed to perform precursor scanning for detection of membrane lipids containing oxidized acyl species (Maeda et al., 2008). As shown in Figure 7A, in Arabidopsis rosettes, oxidized MGDG and arabidopside E-like species (i.e., acylated MGDGs containing at least one OPDA species) were significantly higher in 3-week-old DD-treated and 6-week-old LD-grown OE rosettes than in the wild type of the same age and treatment (P < 0.05). Levels of arabidopsides A, B, and D (compounds 1, 2, and 3; see Supplemental Table 1 online) were more than 9-, 43-, and 20-fold higher, respectively, in 3-week-old DD-treated ACBP3-OEs than in the wild type (Figure 7B). All acylated MGDG species (compounds 4 to 9; see Supplemental Table 1 online) were higher in leaves of ACBP3-OEs than in those of the wild type after DD treatment. In 6-week-old ACBP3-OEs, arabidopsides A, B, and D were 5-, 7-, and 19-fold higher than in the wild type (Figure 7B). Among the six acylated MGDGs, arabidopsides E and arabidopside G (compounds 8 and 10; see Supplemental Table 1 online) were significantly higher in rosettes of OE lines than in those of the wild type (Figure 7C). In addition, oxidized PG species, identified as OPDA/16:1, OPDA/16:0, 18:3-O/18:3, and 18:3-O/16:3, displayed significant increases in 3-week-old DD-treated (P < 0.05), but not in 6-week-old OE lines (see Supplemental Tables 2 to 4 online).
Figure 7.
Oxylipin-Containing Polar Lipid Species in 3-Week-Old DD-Treated and 6-Week-Old Wild Type and ACBP3-OEs.
(A) Total amount of arabidopside E-type (ArE-like) acylated MGDG and oxidized DGDG, MGDG, PC, PE, and PG complex lipids in 3-week-old LD-, 3-week-old DD-, and 6-week-old LD-treated wild type and ACBP3-OEs (OE-1 and OE-4).
(B) Content of complex lipids arabidopside A (1), arabidopside B (2), and arabidopside D (3) in 3-week-old LD-, 3-week-old DD-, and 6-week-old LD-treated wild type and ACBP3-OEs (OE-1 and OE-4).
(C) Content of arabidopside E-type (acylated MGDG) complex lipids (5 to 10) in 3-week-old LD-, 3-week-old DD-, and 6-week-old LD-treated wild type and ACBP3-OEs (OE-1 and OE-4). “a” indicates value of OEs higher than the wild type (*P < 0.05 or **P < 0.01). Values are the means ± sd (n = 3 or 4).
Furthermore, analyses of acyl-CoA content of rosettes from both acbp3 and RNAi-1 leaves did not reveal any differences in acyl-CoA profiles in comparison to the wild type (see Supplemental Figure 6 online). However, saturated 16:0- and 18:0-CoA species, as well as polyunsaturated 18:3-CoA, significantly increased in the two ACBP3-OEs, while other species such as 14:0-, 18:1-, 18:2-, and 20:0-CoA remained unchanged (see Supplemental Figure 6 online).
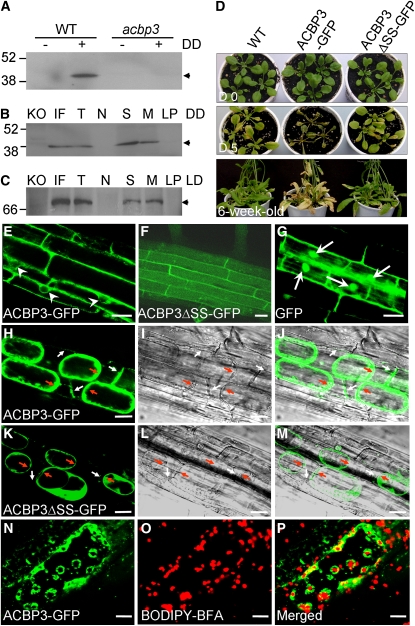
Analyses of Transgenic Arabidopsis Expressing ACBP3-GFP and ACBP3ΔSS-GFP
The subcellular localization of ACBP3 was reexamined by protein blot analysis using ACBP3-specific antibodies and stable transgenic lines expressing full-length ACBP3-GFP (for green fluorescent protein) because the previously reported (Leung et al., 2006) transiently expressed ACBP3-DsRed consisted of only the first 337 of 362 amino acids in ACBP3 and lacked the acyl-CoA binding domain. ACBP3-specific antibodies were observed to cross-react with total protein from DD-treated wild type, but not with that from either LD-grown wild type or the acbp3 mutant (Figure 8A). To demonstrate targeting of the ACBP3 native protein within the apoplast, the intercellular fluids (IFs), which contain extracellular-localized proteins in plant leaf cells, were isolated from DD-treated wild-type Arabidopsis and transgenic lines expressing the full-length ACBP3-GFP fusion. Protein blot analyses indicated that the 39.3-kD ACBP3 native protein and 66-kD ACBP3-GFP fusion protein were detected in the IF preparations using anti-ACBP3 (Figure 8B) and anti-GFP antibodies (Figure 8C), respectively. However, ACBP3 and ACBP3-GFP were present in both soluble and membrane fractions (Figures 8B and 8C).
Figure 8.
Analyses of Transgenic Arabidopsis Expressing ACBP3-GFP and ACBP3ΔSS-GFP.
(A) Protein blot analyses using ACBP3-specific antibodies on total proteins from 3-week-old wild-type and acbp3 mutant plants grown under LD (−) or DD (+) for 3 d showing the cross-reacting 39.3-kD ACBP3 band.
(B) and (C) Analyses using ACBP3- (B) or GFP-specific (C) antibodies on subcellular fractions from 3-week-old DD-treated wild-type (B) or LD-grown transgenic plants expressing ACBP3-GFP (C). Total proteins from acbp3 (KO) grown under similar conditions were used as controls. T, total proteins; N, nuclei; S, soluble proteins; M, membranes; LP, large particles including mitochondria, chloroplasts, and peroxisomes. Arrowheads indicate the cross-reacting 39.3-kD ACBP3 and 66-kD ACBP3-GFP bands.
(D) Comparison in phenotypes of 3-week-old -ype and transgenic Arabidopsis expressing ACBP3-GFP and ACBP3ΔSS-GFP before (Day 0) and after 5 d (Day 5) of DD treatment and of 6-week-old plants grown in LD.
(E) to (G) Confocal microscopy of primary root cells in transgenic Arabidopsis expressing ACBP3-GFP (E), ACBP3ΔSS-GFP (F), and GFP control (G). Image in (E) shows localization of ACBP3-GFP in the peripheries of the cell and nuclei (arrowheads), that in (F) shows ACBP3ΔSS-GFP localization in the cytosol, and that in (G) shows GFP localization in nuclei (arrows) and cytosol.
(H) to (M) Plasmolysis of primary root cells in ACBP3-GFP (H) and ACBP3ΔSS-GFP (K). Image in (H) shows association of ACBP3-GFP with the apoplast (white arrows) and membranes (red arrows) and that in (K) shows localization of ACBP3ΔSS-GFP in the cytosol. (I) and (L) show transmitted light images of cells shown in (H) and (K), respectively. (J) and (M) show the merged images of (H) and (I), and (K) and (L), respectively. The red arrows indicate the plasma membrane, and the white arrows indicate the cell wall.
(N) to (P) Confocal images showing the intracellular colocalization (P) of ACBP3-GFP (N) and the BODIPY-BFA–labeled ER/Golgi complex (O) in 3-d-old hypocotyl cells of transgenic plants expressing ACBP3-GFP. Bars = 20 μm in (E) to (P).
To investigate the significance of the ACBP3 signal sequence (amino acids 1 to 26)/transmembrane domain (amino acids 7 to 26) in targeting, stable transgenic Arabidopsis lines expressing ACBP3ΔSS-GFP, which lacks the signal sequence, were generated. These plants were phenotypically similar to the wild type when grown under LDs (Figure 8D, top row). DD (5-d)-treated 3-week-old ACBP3-GFP plants showed symptoms of accelerated leaf senescence similar to that of ACBP3-OEs, while the ACBP3ΔSS-GFP lines behaved like the wild type (Figure 8D, middle row). Almost all rosettes of 6-week-old ACBP3-GFP lines had yellowed, whereas wild-type and ACBP3ΔSS-GFP rosettes remained green (Figure 8D, bottom row), suggesting that proper targeting is essential for ACBP3-accelerated senescence.
Confocal microscopy showed that fluorescence from ACBP3-GFP was localized at the cell periphery in the primary root cells (Figure 8E). By contrast, the GFP-transformed control was expressed in both nuclei and cytosol (Figure 8G). The signal sequence/transmembrane domain is deemed essential in ACBP3 targeting because ACBP3ΔSS-GFP–transformed plants showed green fluorescence in the cytosol (Figure 8F). To confirm ACBP3-GFP localization further, the primary root cells of ACBP3-GFP and ACBP3ΔSS-GFP plants were subjected to plasmolysis. GFP fluorescence was detected in the apoplast and membrane in ACBP3-GFP root cells (Figures 8H to 8J) but not in ACBP3ΔSS-GFP cells (Figures 8K to 8M). To verify the intracellular fluorescence of ACBP3-GFP (Figure 8E), hypocotyl cells of 3-d-old ACBP3-GFP transgenic seedlings were stained with the Golgi-labeling dye BODIPY-BFA, which labels the endoplasmic reticulum (ER)/Golgi complex (BFA compartments) in living cells (Deng et al., 1995). Intracellular fluorescent ACBP3-GFP predominantly colocalized and accumulated in the periphery of ER/Golgi complex (Figures 8N to 8P). These results suggest that the ACBP3 signal sequence/transmembrane domain functions in targeting and that ACBP3 localizes to the extracellular space, intracellular membranes, and the ER/Golgi complex.
Recombinant ACBP3 Binds PC, PE, and Polyunsaturated Acyl-CoA Esters in Vitro
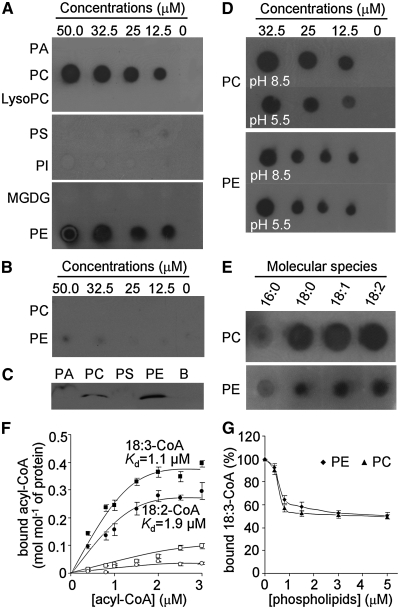
We further performed in vitro filter binding assays to test the interactions between ACBP3 and various phospholipids and galactolipids. Recombinant proteins consisting of a His-tag fused to the ACBP3 acyl-CoA binding domain (amino acids 216 to 362; rACBP3) or to a His-tagged deletion mutant lacking the acyl-CoA binding domain (amino acids 30 to 243; rACBP3ΔACB) were expressed and purified. Filter binding assays indicated that rACBP3 binds PC and PE (Figure 9A), and removal of the acyl-CoA binding domain in rACBP3ΔACB disrupted binding (Figure 9B). By using an independent liposome binding assay, rACBP3 was also shown to bind PC and PE liposomes but did not bind liposomes consisting of PA and PS (Figure 9C). Since the buffers used for both filter binding and liposome binding assays were at neutral pH (7.5) and ACBP3 was localized to both the apoplast and the ER/Golgi, the effect of different pH values on ACBP3/phospholipid binding was examined. ACBP3 was observed to bind to PC and PE at either acidic (5.5) or alkaline (8.5) pH (Figure 9D). When binding of molecular species of PC or PE was subsequently tested, rACBP3 was observed to bind all species of PC (di 16:0-, 18:0-, 18:1-, and 18:2-PC) and PE (di 16:0-, 18:0-, 18:1-, and 18:2-PE) tested (Figure 9E).
Figure 9.
His-Tagged ACBP3 Recombinant Protein Interacts with PC and PE in Vitro.
(A) and (B) Binding of full-length rACBP3 (A) and deletion mutant rACBP3ΔACB (B) to lipids on filters. Various concentrations (0, 12.5, 25, 32.5, and 50 μM) of lipids (PA, PC, lysoPC, PS, PI, MGDG, and PE) were spotted onto nitrocellulose and incubated with 1 μg/mL of purified rACBP3 (A) or rACBP3ΔACB (B).
(C) rACBP3/liposome binding assay. Liposomes (100 μg) consisting of PA, PC, PS, or PE were incubated with 5 μg/mL of purified rACBP3 protein. B, blank control.
(D) The effects of different pH values on rACBP3/lipid binding. Various concentrations (0, 12.5, 25.0, and 32.5 μM) of PC and PE were spotted onto nitrocellulose and incubated with 1 μg/mL of purified rACBP3 protein in either pH 8.5 or 5.5.
(E) Effect of PC and PE acyl species on rACBP3/lipid binding. Fifty micromolar lipids (di16:0-, di18:0-, di18:1-, and di18:2-PC/PE) spotted onto nitrocellulose were incubated with 1 μg/mL of purified rACBP3 protein. The rACBP3/lipid binding reactions in (A) to (E) were detected by immunoblotting with horseradish peroxidase (HRP)–conjugated anti-penta-His antibodies.
(F) Lipidex assays of rACBP3 and rACBP3ΔACB incubated with [14C]18:2-CoA and [14C]18:3-CoA. Squares indicate binding of rACBP3 (closed) and rACBP3ΔACB (open) to [14C]18:3-CoA, and circles indicate binding of rACBP3 (closed) and rACBP3ΔACB (open) to [14C]18:2-CoA. Data are average of three independent experiments. Bars represent sd (n = 3).
(G) Displacement of [14C]18:3-CoA by PC and PE liposomes in Lipidex competition assays. Different concentrations of PC and PE liposome (0, 0.4, 0.8, 1.5, 3.0, and 5.0 μM) were incubated with 0.8 μM [14C]18:3-CoA and 0.2 μM rACBP3. Assays were performed in triplicates, with blanks, at each concentration of liposome. The bound acyl-CoAs in the presence of PC and PE liposome were expressed relative to the value obtained from reaction containing 0 μM liposome (100%). Data are average of three independent experiments. Bars represent sd (n = 3).
Lipidex assays have shown that rACBP3 binds polyunsaturated [14C]20:4-CoA and shows low affinities to either saturated or monounsaturated acyl-CoAs (Leung et al., 2006). Using similar assays (Figure 9F), rACBP3 also binds other two polyunsaturated acyl-CoAs, [14C]18:2-CoA and [14C]18:3-CoA, with the dissociation constants (Kd) of 1.9 and 1.1 μM, respectively, and removal of the acyl-CoA binding domain (rACBP3ΔACB) abrogated binding. Lipidex competition assays were used to determine if di18:2-PC and di18:2-PE liposomes could compete with [14C]18:3-CoA in binding rACBP3. The binding of [14C]18:3-CoA to rACBP3 decreased in the presence of both di18:2-PC and di18:2-PE liposomes (Figure 9G), implying that binding was displaced by the di18:2-PC and di18:2-PE liposomes. These results suggest that ACBP3 specifically binds PC, PE, and polyunsaturated acyl-CoA esters via the acyl-CoA binding domain.
ACBP3 Overexpression Disrupts Autophagosome Formation and Enhances Degradation of ATG8
Conjugation of the ubiquitin-like protein ATG8 and PE produces an ATG8-PE complex that is essential for autophagosome formation (Kirisako et al., 1999; Yoshimoto et al., 2004; Fujioka et al., 2008; Chung et al., 2009). To investigate whether the senescence phenotypes of ACBP3-OEs and ACBP3-KOs (acbp3 and RNAi-1) are related to changes in PE, transgenic Arabidopsis lines expressing the autophagy marker GFP-ATG8e fusion (Contento et al., 2005) were generated (designated GFP-ATG8e) and crossed to ACBP3-OE (OE-1) and ACBP3-RNAi (RNAi-1) plants to produce OE-1/GFP-ATG8e and RNAi-1/GFP-ATG8e. When seedlings were grown on nitrogen-lacking MS medium for 14 d, the primary root lengths of OE-1 were significantly shorter (P < 0.05) than the wild type, while those of GFP-ATG8e and RNAi-1 were longer (P < 0.05; see Supplemental Figure 7A online). In comparison, the root growth of OE-1/GFP-ATG8e was similar to OE-1, and RNAi-1/GFP-ATG8e to GFP-ATG8e and RNAi-1 (see Supplemental Figure 7A online). DD-treated and 5-week-old OE-1/GFP-ATG8e plants also displayed an accelerated leaf senescence phenotype like OE-1 (see Supplemental Figures 7B and 7C online).
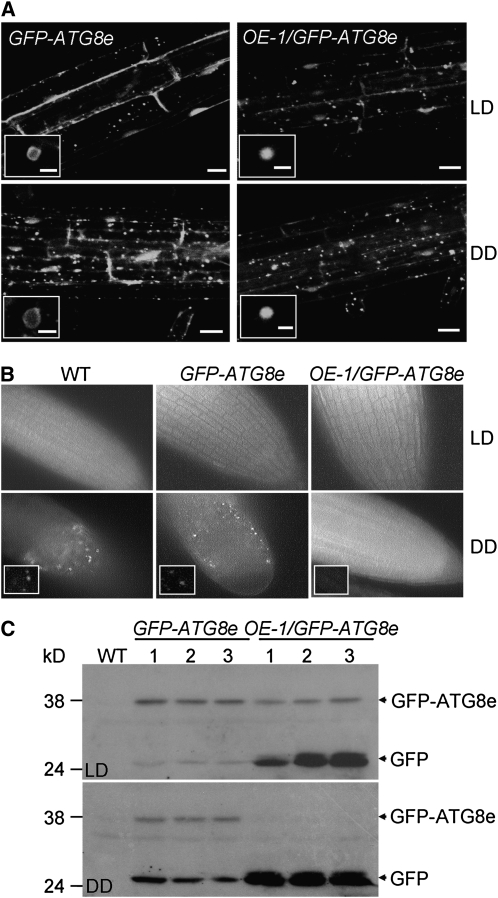
When roots of 1-week-old seedlings of GFP-ATG8e and OE-1/GFP-ATG8e grown on MS medium were transferred to MS under LD or DD for 3 d and then observed by confocal microscopy, fluorescence of GFP-ATG8e was associated with the plasma membrane as well as in many ring-shaped and punctuate structures representing the autophagosomes under LD (Yoshimoto et al., 2004). These structures were more abundant in the root cells upon DD treatment (Figure 10A). Fewer of these autophagosome structures were detected in root cells of OE-1/GFP-ATG8e under either LD or DD, although some small dotted structures were evident (Figure 10A). These results were further confirmed by analysis of similarly treated wild-type, GFP-ATG8e, and OE-1/GFP-ATG8e seedlings following by staining with monodansylcadaverine (MDC), a fluorescent dye labeling autophagosomes in Arabidopsis cells (Contento et al., 2005). Numerous MDC-stained autophagosomes accumulated in root cells of wild-type and GFP-ATG8e seedlings after 3 d of DD treatment, and autophagosome formation was disrupted in OE-1/GFP-ATG8e root cells (Figure 10B).
Figure 10.
ACBP3-OE Disrupts Autophagosome Formation and Enhances GFP-ATG8e Degradation.
(A) GFP fluorescence in GFP-ATG8e and OE-1/GFP-ATG8e root epidermal cells. One-week-old MS-grown GFP-ATG8e and OE-1/GFP-ATG8e seedlings were grown under LDs or DDs for 3 d. Bars = 25 μm. Insets show ring and dot structures at high magnification (bars = 1 μm).
(B) MDC staining of root tips. One-week-old wild-type, GFP-ATG8e, and OE-1/GFP-ATG8e seedlings were grown under LDs or DDs for 3 d, followed by staining with MDC. Insets show MDC-stained autophagosomes at high magnification.
(C) Processing of GFP-ATG8e proteins in wild-type, GFP-ATG8e, and OE-1/GFP-ATG8e plants under LDs and upon 5 d of DD treatment using anti-GFP antibodies. The C-terminal processed GFP-ATG8e and its degraded form leaving only the GFP are indicated on the right.
Arabidopsis ATG8 proteins are processed by removal of their C-terminal portion to facilitate conjugation of the processed proteins with PE (Yoshimoto et al., 2004) followed by a second processing event that may result in degradation of ATG8 (Sláviková et al., 2005). To understand further the possible role of ACBP3 in regulating the ATG8 protein, posttranslational analyses of GFP-ATG8e fusion proteins in GFP-ATG8e as well as OE-1/GFP-ATG8e lines were conducted. Protein blot analysis using anti-GFP antibodies showed that under LD conditions, two bands were detected in GFP-ATG8e. The upper band corresponds to the expected C-terminal processed form of GFP-ATG8e and the lower band, to the GFP domain alone, which likely represents a degraded form of GFP-ATG8e. DD treatment enhanced conversion to the degraded form (Figure 10C). By contrast, the C-terminal processed bands of GFP-ATG8e in OE-1/GFP-ATG8e plants were much weaker than those of GFP-ATG8e under LD and were undetectable under DD, consistent with progressive degradation of GFP-ATG8e (Figure 10C).
The Early Senescence Phenotype in ACBP3-OEs Is Salicylic Acid Dependent
To investigate whether the early senescence phenotype in the ACBP3-OEs is salicylic acid (SA) or jasmonic acid (JA) dependent, we generated ACBP3-OE/coi1-2 and ACBP3-OE/npr1-5 double combinations by crossing the OE-1 plant with mutants known to inactivate SA and JA signaling pathways, respectively, npr1-5 (Shah et al., 1999) and coi1-2 (Xiao et al., 2004). Age-dependent early senescence in OE-1 was completely suppressed following crosses with the npr1-5 mutant, but not with the coi1-2 mutant, in both whole plants and detached leaves of the OE-1/npr1-5 line (Figures 11A and 11B). When plants were subjected to DD treatment, all OE-1/coi1-2 plants died (similar to OE-1), while OE-1/npr1-5 survived better than OE-1 and OE-1/coi1-2 (Figure 11C), suggesting that early senescence in ACBP3-OEs is dependent on the SA, but not the JA, signaling pathway.
Figure 11.
The Early Senescence Phenotype in ACBP3-OEs Is Suppressed by Inactivation of the SA Signaling Pathway.
(A) Phenotype of 5-week-old wild-type, OE-1, OE-1/coi1-2, coi1-2, OE-1/npr1-5, and npr1-5 plants under LDs. Arrows indicate the senescent leaves of OE-1 and OE-1/coi1-2.
(B) Phenotype of detached leaves from 4-week-old wild type, OE-1, OE-1/coi1-2, coi1-2, OE-1/npr1-5, and npr1-5. Photos were taken at 0 and 5 d after detachment.
(C) Images of 3-week-old wild-type, OE-1, OE-1/coi1-2, coi1-2, OE-1/npr1-5, and npr1-5 plants before (LD) and after 5 d of DD treatment.
DISCUSSION
We demonstrated here that the expression of Arabidopsis ACBP3 is affected by light/dark cycling and is developmentally regulated in rosette leaves. Upregulation of ACBP3 in extended darkness and in senescing leaves implies that ACBP3 is associated with starvation-induced and age-dependent leaf senescence. However, unlike SAG101 (He and Gan, 2002), ACBP3 expression is not senescence specific because ACBP3 is also strongly expressed in 2- and 3-week-old seedlings. High ACBP3 expression in early development as well as in senescence may be related to a greater need for lipid transport during these periods in the life cycle of the plant. Given that ACBP3 binds phospholipids and acyl-CoA esters, the membrane and apoplast localization of ACBP3-GFP further suggests that ACBP3 might participate in the biogenesis of membrane phospholipids and/or in cell wall development.
Proposed Role for ACBP3 in Leaf Senescence
Based on the abilities of rACBP3 to bind phospholipids PC, PE, and unsaturated acyl-CoAs, we propose that Arabidopsis ACBP3 regulates membrane lipid metabolism in leaves. A correlation between ACBP3 expression levels and PE composition, with increases seen in ACBP3-OEs and corresponding decreases seen in the ACBP3-KOs, suggests that ACBP3 indeed binds PE and regulates PE homeostasis in vivo. Changes in PE composition are reflected in the accelerated and delayed senescence phenotypes in the ACBP3-OEs and ACBP3-KOs, respectively. Although decreases in galactolipids and phospholipids were also observed in DD-treated and 6-week-old OEs, they may have arisen as a result of accelerated senescence. Further acyl-CoA profiling revealed that ACBP3 overexpression increased acyl-CoA pools, especially 16:0-, 18:0-, and 18:3-acyl-CoA content (see Supplemental Figure 6 online). Yurchenko et al. (2009) reported that Brassica napus ACBP overexpression in Arabidopsis seeds increased polyunsaturated (18:2 and 18:3) fatty acid composition and recombinant Bn-ACBP is essential for lysophosphatidylcholine acyltransferase activity by regulating exchange of the acyl group between the acyl-CoA pool and PC. Likewise, ACBP3 may play an important role in the exchange of acyl-CoAs (especially unsaturated acyl-CoAs) with PE in Arabidopsis.
Moreover, the phenotypes of accelerated starvation-induced and age-dependent senescence in the ACBP3-OEs resembled those of Arabidopsis atg2, atg5, atg7, atg10, and atg18a mutants, defective in ATG proteins essential for autophagosome formation during autophagy (Doelling et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Yoshimoto et al., 2009). Consistent with the atg2 and atg5 mutants (Yoshimoto et al., 2009), the early senescence phenotype of ACBP3-OEs is SA dependent and does not require an intact JA signaling pathway. Interestingly, yeast autophagic processes require the conjugation of two ubiquitin-like small proteins, Apg8 and Apg12, to their corresponding targets, phospholipid PE and Apg5, respectively, via an ATP-dependent reaction cascade involving the activating enzyme E1 (ATG7) and conjugating enzyme E2 (ATG3 and ATG10) (Mizushima et al., 1998; Ichimura et al., 2000). The yeast ATG8-PE complex likely binds the autophagic membrane by the lipid moiety and expands the membrane during autophagosome formation (Kirisako et al., 1999). These conjugation systems in yeast (ATG8-PE and ATG12-ATG5) are conserved in plants, including Arabidopsis (Yoshimoto et al., 2004; Fujioka et al., 2008), rice (Oryza sativa), and maize (Zea mays; Chung et al., 2009), and they participate in autophagic nutrient recycling throughout the life cycle of the plant, especially during senescence or under nutrient-limiting growth conditions. In addition, the ATG8-PE complex is reversibly deconjugated by the addition of PLD, which inhibits the formation of the ATG8-PE complex (Fujioka et al., 2008; Chung et al., 2009). We suggest that altered expression of ACBP3 may affect the availability of PE and/or the conjugation of Arabidopsis ATG8 proteins to PE.
In starvation-treated or senescing leaves, increased expression of ATGs including ATG8 promotes the autophagy pathway for nutrient recycling; this event may be subject to ACBP3 regulation via its competitive binding of PE. Meanwhile, the expression of PLDs, acyl hydrolases, and/or other yet unidentified membrane lipid lipases activates the degradation of membrane lipids, including both galactolipids and phospholipids. From lipid profiling, we observed that despite a general decline in most membrane lipid species in DD-treated wild type, polyunsaturated species containing 18:3 (such as 36:6, 34:3 in PC, and 36:6 in PI) significantly increased during DD treatment. The increased concentrations in the membrane of unsaturated membrane lipids would maintain membrane stability and integrity essential for an effective nutrient recycling during senescence, at least during the early stages (Lim et al., 2007). Thus, in wild-type Arabidopsis, these processes may be highly regulated and rate limited to maintain regular growth, proper nutrient recycling, and timely onset of senescence. However, in the ACBP3-OE lines, as a consequence of the constitutive binding of PE by ACBP3 in the DD-treated or premature senescing leaves, a shortage in free PE would impair ATG8 lipidation and indirectly disrupt autophagosome formation, resembling the atg5, atg7, atg10, and atg18a mutants in accelerating senescence (Doelling et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008).
How disruption of the Arabidopsis ATG system would accelerate starvation-induced and age-dependent senescence remains unclear. Alternative pathways may be activated upon loss of the ATG system, producing rapid degradation of cytosolic and chloroplast components and compromising the viability of the atg5, atg7, and atg10 mutants (Doelling et al., 2002; Thompson et al., 2005; Phillips et al., 2008). Since phospholipids, with the exception of PE and PA, generally decline in starvation-treated and premature senescing ACBP3-OE lines, the upregulation of senescence-related lipid-degrading enzymes, such as PLDs, lipoxygenase, acyl hydrolase, and other lipases (Woolhouse, 1984; Fan et al., 1997; Hong et al., 2000; Thompson et al., 2000; He and Gan, 2002; Padham et al., 2007), may present an alternative route in lieu of ATG. The accumulation of membrane degradation products such as PA and arabidopsides is consistent with their possible roles in accelerating leaf senescence in ACBP3-OEs. In comparison, reduced binding of PE during DD in the ACBP3-KOs results in proper autophagosome formation and recycling that subsequently improves their survival rate in extended darkness. In agreement with this, a similar improved survival phenotype under extended darkness has also been observed in the Arabidopsis ATG8f-overexpression line, which likely enhances the autophagy process during darkness (Sláviková et al., 2008). By contrast, changes in nitrogen-induced and age-dependent leaf senescence were not evident in ACBP3-KOs, suggesting that alternative ACBP3-independent mechanisms exist.
Potential Roles of PA and Arabidopsides in Promotion of Leaf Senescence
Our results support a relationship between PA, leaf senescence, and cell death. PA increases in response to stress and is a lipid mediator in numerous stress-related plant signaling pathways, such as freezing tolerance, pathogen infection, wounding, and oxidative stress (Munnik, 2001; Wang, 2004). Our findings indicate that, in addition to a decrease in membrane phospholipids (mainly PC and PI) as a consequence of senescence-mediated membrane deterioration, PA was 24- to 34-fold higher in ACBP3-OEs than the wild type. PA, but not other phospholipids, is known to induce leaf cell death and yellowing and elevates the levels of reactive oxygen species in both attached and detached Arabidopsis leaves (Park et al., 2004). Treatment with 16:0-18:2-PA (100 μM) has been demonstrated to enhance Philodendron cordatum leaf senescence by promoting chlorophyll breakdown (Hong et al., 2009). Both age-dependent and environmentally triggered leaf senescence culminates in programmed cell death. PA accumulation could have caused accelerated leaf senescence programmed cell death in the ACBP3-OEs.
The early-senescing leaves of ACBP3-OEs accumulated oxylipin-containing complex lipids, including arabidopsides A, B, D, E, and G. These oxidized lipids, resulting from lipid peroxidation of polyunsaturated fatty acids (Mosblech et al., 2009), may subsequently promote senescence; a previous report revealed that one of these complex lipids, arabidopside A, is known to induce leaf senescence in oat (Avena sativa; Hisamatsu et al., 2006). Arabidopsides also accumulate upon wounding treatment and during the hypersensitive response primarily by enzymatic reactions via LOX pathways (Andersson et al., 2006; Buseman et al., 2006; Kourtchenko et al., 2007). Lipoxygenase 2 (LOX2) is essential for arabidopside biosynthesis in response to wounding (Glauser et al., 2009). Here, ABCP3 is implicated in the accumulation of arabidopsides A, B, D, E, and G and several related acylated MGDGs, but their levels in ACBP3-KOs were not much different from the wild type in either LD-grown or DD-treated rosettes (see Supplemental Figure 8 online), suggesting that ACBP3 may not be directly essential for the in planta biosynthesis of arabidopsides. The proposed functions of arabidopsides in defense, in wounding (Andersson et al., 2006; Buseman et al., 2006; Kourtchenko et al., 2007), and in leaf senescence are all related to cell death induction; however, it is unclear whether arabidopsides induce senescence directly or through the production of jasmonates, which can also induce leaf senescence (He et al., 2002). Also, no difference was observed between ACBP3-OEs or ACBP3-KOs and wild-type plants in primary root length and lateral root development before and after Pi starvation (see Supplemental Figure 9 online), suggesting that ACBP3 may not be directly involved in switching extraplastidic membrane composition from phospholipids to galactolipids.
Proper Targeting Is Essential in Mediating ACBP3-Associated Leaf Senescence
Reports on the senescence-associated secretory phenotype in humans suggest that many secreted proteins/factors, such as insulin-like growth factor binding protein (IGFBP7), phospholipase A2 (PLA2G2A), and chemokines, are important regulators of senescence (Acosta et al., 2008; Kuilman et al., 2008; Wajapeyee et al., 2008; Augert et al., 2009). The overexpression of chemokine receptors (CXCR2 and IL6R) or the PLA2 receptor (PLA2R) promoted senescence, whereas their corresponding knockdown expression disrupted senescence (Acosta et al., 2008; Kuilman et al., 2008; Augert et al., 2009). The expression of secreted PLA2G2A was elevated over 20-fold in senescent cells, and its overexpression affected senescence in a PLA2R-dependent manner (Augert et al., 2009). Similar to these reported senescence-associated proteins, ACBP3 is also secreted. Our results indicate that ACBP3 is extracellularly targeted via the ER/Golgi pathway and its N-terminal signal sequence is essential for targeting. Although recombinant ACBP3 binds PC and PE in both acidic and alkaline buffers, only transgenic Arabidopsis expressing ACBP3-GFP (but not ACBP3ΔSS-GFP) exhibited premature senescence, suggesting that the proper targeting of ACBP3 is essential in mediating leaf senescence. Possibly, ACBP3 represents a potential component promoting leaf senescence via extracellular lipid metabolism in plants. Putative secretory phospholipases A2 (Wang, 2004), yet to be characterized, may participate in this extracellular pathway together with ACBP3. Furthermore, the Caenorhabditis elegans ACBP protein, MAA-1, which may partially resemble ACBP3 in its subcellular association with the Golgi apparatus and the endocytic pathway, regulates acyl-CoA–dependent endosomal vesicle formation and trafficking (Larsen et al., 2006). Taken together, we suggest that a similar function may be conserved in ACBPs across species since the absence of proper targeting in the ACBP3ΔSS-GFP transgenic plants impeded ACBP3 function in Arabidopsis.
METHODS
Plant Materials and Growth Conditions
The coi1-2 (Xiao et al., 2004) mutant was obtained from D.X. Xie (Tsinghua University, Beijing, China) and the npr1-5 (Shah et al., 1999) mutant from The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org). The acbp3 T-DNA mutant was identified from the SALK collection (SALK_012290; http://signal.salk.edu) obtained from TAIR. All Arabidopsis thaliana wild-type (ecotype Columbia-0), acbp3, acbp3-complemented lines, ACBP3 RNAi lines, and ACBP3-OEs were grown under LD cycles (16 h light, 23°C/8 h dark, 21°C). The methods for identification of acbp3 mutant and generation of acbp3-complemented and ACBP3 RNAi lines are described in the Supplemental Methods online. For DD and LL treatments, 3-week-old LD-grown Arabidopsis was transferred to a growth chamber with constant darkness or light. Samples were collected and photographed at the indicated times. To calculate death rates (percentages) after DD treatment, 12 plants per genotype were DD treated for the indicated duration followed by LD recovery (3 d for ACBP3-OEs and acbp3 or 10 d for ACBP3-RNAi lines), and the average of two independent experiments was recorded. For nitrogen starvation treatment, 1-week-old seedlings LD grown on MS medium were transferred to either MS or nitrogen-deficient MS medium and grown under LD for 2 weeks.
RNA Gel Blot Analysis
Total RNA extraction and RNA gel blot analysis were performed as previously described (Xiao et al., 2008a). The gene-specific primers (for ACBP3, ML737/ML738; for SAG12, ML878/ML879; for PLDα1, ML921/ML922; for PLDδ, ML923/ML924; for SAG101, ML1024/ML1025) used to generate cDNA probes are summarized in Supplemental Table 5 online.
Generation of ACBP3-OE, ACBP3-GFP, ACBP3ΔSS-GFP, and GFP-ATG8e Transgenic Arabidopsis
The 1.1-kb BamHI-SalI full-length ACBP3 cDNA from pAT180 (Leung et al., 2006) was cloned into the BamHI-SalI site of binary vector pSa13 (Xiao et al., 2008a) to generate pAT343 in which ACBP3 is expressed from the cauliflower mosaic virus 35S promoter. Plasmid ACBP3-GFP (pAT314) was constructed by cloning a 1.1-kb RT-PCR ACBP3 cDNA fragment made with primers ML752 and ML738 (see Supplemental Table 5 online) into vector pGEM-T Easy to produce plasmid pAT312. The primers introduced two BamHI sites and removed the stop codon to facilitate generation of the GFP fusion. The BamHI-digested fragment from pAT312 was subsequently inserted into pBI-eGFP vector (Xiao et al., 2008b) that was BamHI digested and calf intestine alkaline phosphatase (Roche) treated. Plasmid ACBP3ΔSS-GFP (pAT313) was generated by cloning a 1.0-kb ACBP3 fragment lacking the signal sequence, which was PCR derived using ML753 and ML738 (see Supplemental Table 5 online) from pGEM-T Easy derivative pAT311. In primer ML753, the ATG sequence (bold in Supplemental Table 2 online) that encodes amino acid Met was introduced as a start codon. The 1.0-kb fragment from pAT311 was cloned into the BamHI site of pBI-eGFP, and orientations of the inserts were determined by enzyme digestion. To generate the GFP-ATG8e fusion, ATG8e cDNA was amplified by RT-PCR using primer pairs ML1088 and ML1089 (see Supplemental Table 5 online) and cloned into pGEM-T Easy to obtain pAT450. The XhoI-SacI fragment from pAT450 was subsequently inserted into similar sites on pBI-eGFP to produce pAT541 (GFP-ATG8e). In-frame translational fusions were confirmed by DNA sequence analysis.
The plant transformation vectors (pAT343, pAT313, pAT314, pAT541, and pBI-eGFP) were mobilized to Agrobacterium tumefaciens strain LBA4404 and introduced into wild-type Arabidopsis using the floral dip method (Clough and Bent, 1998). Putative transformants expressing pAT343, pAT313, pAT314, or pAT541 were selected on MS medium containing kanamycin (50 μg mL−1) and verified by PCR using a cauliflower mosaic virus 35S promoter-specific forward primer 35SB and gene-specific reverse primer ML738 (or ML1089 for pAT541), followed by RNA gel blot analysis using an ACBP3 full-length cDNA probe and/or protein blot analyses using anti-GFP antibodies (Invitrogen). Subsequently, the T2 homozygous lines were tested on kanamycin-containing MS medium, and the resistant plants were further analyzed.
To generate the ACBP3-OE/GFP-ATG8e, ACBP3-OE/coi1-2, and ACBP3-OE/npr1-5 double combinations, the OE-1 line was crossed to GFP-ATG8e, coi1-2 (Xiao et al., 2004), and npr1-5 (Shah et al., 1999) plants. ACBP3-OE/GFP-ATG8e plants were obtained by screening F2 seeds on MS medium containing kanamycin (50 μg mL−1) followed by PCR using 35SB paired with either ML738 or ML1089. The ACBP3-OE/coi1-2 line was generated by screening F2 seeds on MS medium containing kanamycin (50 μg mL−1) and MeJA (20 μM). The ACBP3-OE/npr1-5 line was obtained by selection of F2 seeds on MS medium containing kanamycin (50 μg mL−1) followed by PCR analyses to distinguish between the wild-type NPR1 and npr1-5 mutant allele according to Shah et al. (1999).
Measurement of Electrolyte Leakage and Chlorophyll Content
Electrolyte leakage measurements were performed following Welti et al. (2002). Briefly, conductivity was measured on plant samples in deionized water following gentle agitation at 23°C for 1 h. Subsequently, the solution was heated in a 100°C water bath for 10 min and cooled to 23°C to determine total ionic strength. Chlorophyll was extracted from Arabidopsis leaves by immersion in 1 mL of N,N-dimethylformamide for 48 h in the dark at 4°C. Absorbance was recorded at 664 and 647 nm, and total chlorophyll concentration was calculated (Xiao et al., 2004). The total chlorophyll content was measured and normalized per gram fresh weight of sample.
Analyses of Lipids and Oxidized Lipids
Total lipid extraction was performed according to Welti et al. (2002), and lipids were dissolved in chloroform for analysis. The profiles of membrane lipids were measured by an automated electrospray ionization–tandem mass spectrometry method as previously described (Devaiah et al., 2006) or by a slightly modified procedure described here (see Supplemental Methods online).
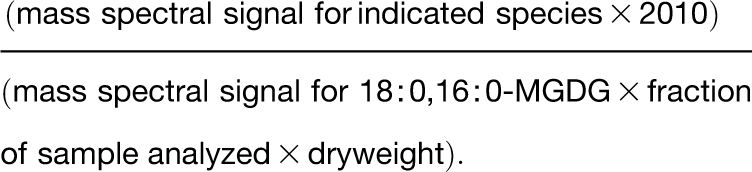
Oxidized lipid analysis was performed similarly to that previously described (Maeda et al., 2008) with the addition of scanning to higher mass range to detect acylated MGDG species as [M + OAc]− ions and use of the internal standard, 18:0,16:0-MGDG. Mass spectral signals were normalized to the signal for 2010 pmol 18:0,16:0-MGDG, an unnaturally occurring lipid species that was added to the portion of the sample being analyzed; relative mass spectral signal/dry weight was calculated by the following formula:
 |
Scans for precursors of negative acyl anions (m/z 291 for 18:4-O and 18:3-2O, m/z 293 for 18:3-O and 18:2-2O, m/z 295 for 18:2-O, and m/z 283 for internal standard fatty acyl chain 18:0) were used to detect species of each class. The nomenclature (e.g., 18:4-O) is acyl carbons:double bond equivalents:number of oxygens beyond the ester oxygens. 18:4-O has been identified as OPDA (Buseman et al., 2006). The analyzed molecular species of oxidized PC, PE, PG, nonacylated MGDG, and nonacylated DGDG were those analyzed previously (Maeda et al., 2008).
RT-PCR Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) from Arabidopsis rosettes. First-strand synthesis was performed using the Superscript first-strand synthesis system (Invitrogen). Gene-specific primers used for RT-PCR are shown in Supplemental Table 5 online. The numbers of cycles used in amplification with each primer pair were within the linear range (see Supplemental Table 5 online). Ethidium bromide staining was used to detect PCR products. Identical results were obtained from two independent biological replicates, one of which is shown.
Extraction of IFs, Subcellular Fractionation, and Protein Blotting Analyses
Extraction of IFs and subcellular fractionation of whole-plant protein from 3-week-old Arabidopsis were performed as previously described (Xia et al., 2004; Xiao et al., 2008b). IFs, total proteins, and different subcellular fractions were electrophoretically transferred to Hybond-C membrane (Amersham) using the Trans-Blot cell (Bio-Rad), and anti-GFP (Invitrogen; 1:3000) or anti-ACBP3 (1:10,000) antibodies were used in protein blot analyses. To generate ACBP3-specific antibodies, a synthetic peptide (DARVLESQRNFQVV corresponding to amino acids 59 to 72 of ACBP3) was used in rabbit immunization.
For GFP-ATG8e processing analysis, total proteins were extracted from 3-week-old wild-type, GFP-ATG8e, and OE-1/GFP-ATG8e rosettes at the indicated time points, and anti-GFP antibodies (Invitrogen; 1:3000) were used in protein blot analysis.
Microscopy Analyses
A Zeiss LSM 510 inverted confocal laser scanning microscope equipped with helium/neon lasers and multitracking was used for analysis of ACBP3-GFP localization following Li et al. (2008), and images were processed using LSM 510 software (Zeiss). Plasmolysis was performed on the primary root cells by incubating the samples in 10% (w/v) mannitol solution for 20 min. For BODIPY-BFA staining analyses, hypocotyls of 3-d-old transgenic seedlings expressing ACBP3-GFP were stained with 1 μM BODIPY 558/568-BFA (Invitrogen) for 20 min at room temperature and subsequently observed using the confocal laser scanning microscope.
MDC staining was performed as previously described (Contento et al., 2005). One-week-old seedlings were immersed in 0.05 mM MDC (Sigma-Aldrich) in PBS for 10 min followed by washing twice in PBS buffer and subsequently observed using fluorescence microscopy with a 4′,6-diamino-phenylindole–specific filter.
Lipidex and Lipid Binding Assays
The plasmid for rACBP3ΔACB expression was constructed by PCR amplification of a 0.7-kb ACBP3 fragment from pAT314 using primer pairs ML753 and ML407 (see Supplemental Table 5 online), followed by cloning into pGEM-T Easy vector to generate plasmid pAT474. Plasmid pAT475 was obtained by insertion of a 0.7-kb BamHI-EcoRI fragment from pAT474 into the same sites of the pRSET A vector (Invitrogen). Expression and purification of rACBP3 from pAT223 (Leung et al., 2006) and rACBP3ΔACB from pAT475 were performed according to Leung et al. (2006).
Lipidex, lipid binding, and Lipidex competition assays were performed as described previously (Leung et al., 2006; Chen et al., 2008, 2010). Purified rACBP3 and rACBP3ΔACB (at 0.2 μM final concentration) was mixed with acyl-CoA, at final concentrations ranging from 0.2 to 3.0 μM, and incubated for 30 min at 37°C. Subsequently, 400 μL of ice-cold 50% slurry of Lipidex-1000 in binding buffer were added. After centrifugation, a 200-μL aliquot of the supernatant analyzed for radioactivity counts using an LS 6500 liquid scintillation counter (Beckman). In Lipidex competition assays, PC or PE liposome was included in the binding buffer including both rACBP3 and radiolabeled acyl-CoA. In lipid binding assays, lipids at various concentrations were spotted onto nitrocellulose followed by incubation at room temperature for 1 h in the dark. The lipid-bound filter was blocked with TBS containing 1% nonfat milk for 1 h and then incubated with 1 μg/mL of purified rACBP3 or rACBP3ΔACB protein in blocking buffer for 2 h, and gently washed three times with TTBS (TBS plus 0.1% Tween 20), each for 10 min. After 1-h incubation with HRP-conjugated anti-His antibodies (Qiagen; 1:2000), the filter was again washed three times with TTBS, and then the reactions were detected using the ECL Western blotting detection kit (Amersham) following the manufacturer's protocol. LysoPC, PC, PA, di18:0-PC, and di18:2-PC were purchased from Sigma-Aldrich; PE, PI, PS, di16:0-PC, di18:1-PC, and di16:0-PE from Echelon Biosciences; and di18:0-PE, di18:1-PE, di18:2-PE, [14C]18:2-CoA, and [14C]18:3-CoA from Avanti Polar Lipids.
Liposomes were prepared according to Sano et al. (1998). After washing twice with ice-cold TBS buffer (50 mM Tris-HCl, pH 7.0, and 0.1 M NaCl), the precipitated liposomes (100 μg) consisting of PA, PC, PI, or PE were incubated with 5 μg/mL of purified rACBP3 protein at 30°C for 30 min. The liposome/protein mixtures were washed three times and analyzed by SDS-PAGE to detect the rACBP3 protein by immunoblotting with HRP-conjugated anti-His antibodies (Qiagen; 1:2000).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank databases under the following accession numbers: ACBP1(At5g53470; NM_124726), ACBP3 (At4g24230; NM_118556), SAG12 (At5g45890; NM_123957), SAG101 (At5g14930; NM_121497), SEN1 (At4g35770; NM_119743), PLDα1 (At3g15730; NM_112443), PLDβ1 (At2g42010; NM_129765), PLDγ (At4g11850; NM_117255), PLDδ (At4g35790; NM_119745), PLDζ2 (At3g05630; NM_111436), and ATG8e (At2G45170; NM_180100).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Levels of ACBP3, PLDα1, PLDβ1, PLDδ, PLDγ, and PLDζ2 during Age-Dependent and Dark-Induced Senescence.
Supplemental Figure 2. Characterization of the acbp3 T-DNA Mutant and ACBP3-RNAi Transgenic Arabidopsis.
Supplemental Figure 3. Lipid Profiles of 3-Week-Old Wild Type and ACBP3-OEs (OE-1 and OE-4) Grown under LDs.
Supplemental Figure 4. Lipid Profiles of 3-Week-Old Wild Type and ACBP3-KOs (acbp3 and RNAi-1) Grown under LD Followed by DD Treatment for 7 d.
Supplemental Figure 5. Contents of Galactolipids and Lysophopholipids in Wild Type and ACBP3-OEs (OE-1 and OE-4).
Supplemental Figure 6. Acyl-CoA Content in Rosettes of 3-Week-Old Wild Type, ACBP3-KOs (acbp3 and RNAi-1), and ACBP3-OEs (OE-1 and OE-4).
Supplemental Figure 7. Phenotypes of OE-1/GFP-ATG8e and RNAi-1/GFP-ATG8e.
Supplemental Figure 8. Oxylipin-Containing Polar Lipid Contents in 3-Week-Old LD-Grown and DD-Treated ACBP3-KOs (acbp3 and RNAi-1).
Supplemental Figure 9. Root Growth of Wild Type, ACBP3-OEs, and ACBP3-KOs in Phosphorous-Deficient Medium.
Supplemental Table 1. OPDA-Containing Galactolipids.
Supplemental Table 2. Amount of Oxylipin-Containing Polar Lipid Species in 3-Week-Old Leaves of the Wild Type and ACBP3-OEs Grown under LDs.
Supplemental Table 3. Amount of Oxylipin-Containing Polar Lipid Species in 3-Week-Old Leaves of Wild-Type and ACBP3-OEs Plants Grown under LDs Followed by 5-d DD Treatment.
Supplemental Table 4. Amount of Oxylipin-Containing Polar Lipid Species in 6-Week-Old Leaves of the Wild Type and ACBP3-OEs Grown under LDs.
Supplemental Table 5. Nucleotide Sequences of Primers Used in This Study.
Supplemental Methods.
Supplemental References.
Acknowledgments
We thank M. Roth (Kansas Lipidomics Research Center) for lipid profiling, the ABRC for provision of npr1-5 and acbp3 mutant seeds, D.X. Xie (Tsinghua University) for the coi1-2 mutant, and S.F. Chen (University of Hong Kong) for provision of HPLC. This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region, China (Project HKU7047/07M), and the University of Hong Kong (Project 10208034, postdoctoral fellowship to S.X. and studentships to W.G., Q.-F.C., S.-X.Z., S.-W.C., and J.M.). Lipid profiling was performed at the Kansas Lipidomics Research Center, where method development and instrument acquisition were supported by the National Science Foundation (EPS 0236913, MCB 0455318, and DBI 0521587), the Kansas Technology Enterprise Corporation, the Kansas IDeA Network of Biomedical Research Excellence (INBRE) of the National Institutes of Health (P20RR16475), and Kansas State University.
References
- Acosta J.C., et al. (2008). Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133: 1006–1018 [DOI] [PubMed] [Google Scholar]
- Andersson M.X., Hamberg M., Kourtchenko O., Brunnström Å., McPhail K.L., Gerwick W.H., Göbel C., Feussner I., Ellerstrom M. (2006). Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: Formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopside E. J. Biol. Chem. 281: 31528–31537 [DOI] [PubMed] [Google Scholar]
- Augert A., Payré C., de Launoit Y., Gil J., Lambeau G., Bernard D. (2009). The M-type receptor PLA2R regulates senescence through the p53 pathway. EMBO Rep. 10: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. (1997). The molecular biology of leaf senescence. J. Exp. Bot. 48: 181–199 [Google Scholar]
- Buseman C.M., Tamura P., Sparks A.A., Baughman E.J., Maatta S., Zhao J., Roth M.R., Esch S.W., Shah J., Williams T.D., Welti R. (2006). Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 142: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.F., Xiao S., Chye M.L. (2008). Overexpression of the Arabidopsis 10-kilodalton acyl-Coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol. 148: 304–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.F., Xiao S., Qi W., Mishra G., Ma J., Wang M., Chye M.L. (2010). The Arabidopsis acbp1acbp2 double mutant lacking the acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol. 186: 843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye M.L. (1998). Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Mol. Biol. 38: 827–838 [DOI] [PubMed] [Google Scholar]
- Chye M.L., Huang B.Q., Zee S.Y. (1999). Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J. 18: 205–214 [DOI] [PubMed] [Google Scholar]
- Chye M.L., Li H.Y., Yung M.H. (2000). Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol. Biol. 44: 711–721 [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Deng Y., Bennink J.R., Kang H.C., Haugland R.P., Yewdell J.W. (1995). Fluorescent conjugates of brefeldin A selectively stain the endoplasmic reticulum and Golgi complex of living cells. J. Histochem. Cytochem. 43: 907–915 [DOI] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Engeseth N.J., Pacovsky R.S., Newman T., Ohlrogge J.B. (1996). Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch. Biochem. Biophys. 331: 55–62 [DOI] [PubMed] [Google Scholar]
- Fan L., Zheng S., Wang X. (1997). Antisense suppression of phospholipase Da retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Noda N.N., Fujii K., Yoshimoto K., Ohsumi Y., Inagaki F. (2008). In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283: 1921–1928 [DOI] [PubMed] [Google Scholar]
- Glauser G., Dubugnon L., Mousavi S.A., Rudaz S., Wolfender J.L., Farmer E.E. (2009). Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J. Biol. Chem. 284: 34506–34513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Xiao S., Li H.Y., Tsao S.W., Chye M.L. (2009). Arabidopsis acyl-CoA-binding protein ACBP2 interacts with a heavy-metal-binding protein ATFP6. New Phytol. 181: 89–102 [DOI] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D.F., Gan S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Gan S. (2002). A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu Y., Goto N., Hasegawa K., Shigemori H. (2003). Arabidopsides A and B, two new oxylipins from Arabidopsis thaliana. Tetrahedron Lett. 44: 5553–5556 [Google Scholar]
- Hisamatsu Y., Goto N., Hasegawa K., Shigemori H. (2006). Senescence-promoting effect of arabidopside A. Z. Naturforsch C 61: 363–366 [DOI] [PubMed] [Google Scholar]
- Hisamatsu Y., Goto N., Sekiguchi M., Hasegawa K., Shigemori H. (2005). Oxylipins arabidopsides C and D from Arabidopsis thaliana. J. Nat. Prod. 68: 600–603 [DOI] [PubMed] [Google Scholar]
- Hong J.H., Chung G.H., Cowan A.K. (2009). Delayed leaf senescence by exogenous lyso-phosphatidylethanolamine: Towards a mechanism of action. Plant Physiol. Biochem. 47: 526–534 [DOI] [PubMed] [Google Scholar]
- Hong Y., Wang T.W., Hudak K.A., Schade F., Froese C.D., Thompson J.E. (2000). An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proc. Natl. Acad. Sci. USA 97: 8717–8722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M., Taylor C., Liu Z., Ma F., McNamara L., Wang T.W., Thompson J.E. (2007). Regulation and execution of molecular disassembly and catabolism during senescence. New Phytol. 175: 201–214 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtchenko O., Andersson M.X., Hamberg M., Brunnström A., Göbel C., McPhail K.L., Gerwick W.H., Feussner I., Ellerström M. (2007). Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: Jasmonate signaling dependence. Plant Physiol. 145: 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Vredeveld L.C., Douma S., van Doorn R., Desmet C.J., Aarden L.A., Mooi W.J., Peeper D.S. (2008). Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Larsen M.K., Tuck S., Faergeman N.J., Knudsen J. (2006). MAA-1, a novel acyl-CoA-binding protein involved in endosomal vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell 17: 4318–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K.C., Li H.Y., Mishra G., Chye M.L. (2004). ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol. Biol. 55: 297–309 [DOI] [PubMed] [Google Scholar]
- Leung K.C., Li H.Y., Xiao S., Tse M.H., Chye M.L. (2006). Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta 223: 871–881 [DOI] [PubMed] [Google Scholar]
- Li H.Y., Chye M.L. (2004). Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 54: 233–243 [DOI] [PubMed] [Google Scholar]
- Li H.Y., Xiao S., Chye M.L. (2008). Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 59: 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Maeda H., Sage T.L., Isaac G., Welti R., Dellapenna D. (2008). Tocopherols modulate extraplastidic polyunsaturated fatty acid metabolism in Arabidopsis at low temperature. Plant Cell 20: 452–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. (1998). A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Mosblech A., Feussner I., Heilmann I. (2009). Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47: 511–517 [DOI] [PubMed] [Google Scholar]
- Mou Z., Wang X., Fu Z., Dai Y., Han C., Ouyang J., Bao F., Hu Y., Li J. (2002). Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 14: 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T. (2001). Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 6: 227–233 [DOI] [PubMed] [Google Scholar]
- Nam H.G. (1997). The molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 8: 200–207 [DOI] [PubMed] [Google Scholar]
- Padham A.K., Hopkins M.T., Wang T.W., McNamara L.M., Lo M., Richardson L.G., Smith M.D., Taylor C.A., Thompson J.E. (2007). Characterization of a plastid triacylglycerol lipase from Arabidopsis. Plant Physiol. 143: 1372–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Gu Y., Lee Y., Yang Z., Lee Y. (2004). Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 134: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B.F., Noh Y.S., Himelblu E., Amasino R.M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 7: 278–282 [DOI] [PubMed] [Google Scholar]
- Sano H., Kuroki Y., Honma T., Ogasawara Y., Sohma H., Voelker D.R., Akino T. (1998). Analysis of chimeric proteins identifies the regions in the carbohydrate recognition domains of rat lung collectins that are essential for interactions with phospholipids, glycolipids, and alveolar type II cells. J. Biol. Chem. 273: 4783–4789 [DOI] [PubMed] [Google Scholar]
- Shah J., Kachroo P., Klessig D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Sláviková S., Ufaz S., Avin-Wittenberg T., Levanony H., Galili G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59: 4029–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach B.A., Muller A., Hennig P., Gebhardt S., Schubert-Zsilavecz M., Weiler E.W. (2001). A novel class of oxylipins, sn1-O-(12-oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl diglyceride, from Arabidopsis thaliana. J. Biol. Chem. 276: 12832–12838 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Taylor C., Wang T.W. (2000). Altered membrane lipase expression delays leaf senescence. Biochem. Soc. Trans. 28: 775–777 [PubMed] [Google Scholar]
- Thompson J.E., Froese C.D., Madey E., Smith M.D., Hong Y. (1998). Lipid metabolism during plant senescence. Prog. Lipid Res. 37: 119–141 [DOI] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U.I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N., Serra R.W., Zhu X., Mahalingam M., Green M.R. (2008). Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2004). Lipid signaling. Curr. Opin. Plant Biol. 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H.E., Rajashekar C.B., Williams T.D., Wang X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Woolhouse H.W. (1984). The biochemistry and regulation of senescence in chloroplasts. Can. J. Bot. 62: 2934–2942 [Google Scholar]
- Xia Y., Suzuki H., Borevitz J., Blount J., Guo Z., Patel K., Dixon R.A., Lamb C. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23: 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Chen Q.F., Chye M.L. (2009). Light-regulated Arabidopsis ACBP4 and ACBP5 encode cytosolic acyl-CoA-binding proteins that bind phosphatidylcholine and oleoyl-CoA ester. Plant Physiol. Biochem. 47: 926–933 [DOI] [PubMed] [Google Scholar]
- Xiao S., Chye M.L. (2009). An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol. Biochem. 47: 479–484 [DOI] [PubMed] [Google Scholar]
- Xiao S., Dai L.Y., Liu F.Q., Wang Z.L., Peng W., Xie D.X. (2004). COS1: An Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant senescence and defense. Plant Cell 16: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Gao W., Chen Q.F., Ramalingam S., Chye M.L. (2008a). Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J. 54: 141–151 [DOI] [PubMed] [Google Scholar]
- Xiao S., Li H.Y., Zhang J.P., Chan S.W., Chye M.L. (2008b). Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Mol. Biol. 68: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko O.P., Nykiforuk C.L., Moloney M.M., Ståhl U., Banaś A., Stymne S., Weselake R.J. (2009). A 10-kDa acyl-CoA-binding-protein (ACBP) from Brassica napus enhances acyl exchange between acyl-CoA and phosphatidylcholine. Plant Biotechnol. J. 7: 602–610 [DOI] [PubMed] [Google Scholar]