In arbuscular mycorrhizal (AM) symbiosis, development of arbuscules, the structures responsible for mineral nutrient transfer to the plant, requires host plant functions. This work identifies two half-ABC transporters that are widely conserved in the vascular plants. Their interaction and function in the peri-arbuscular membrane is required for arbuscule development and a functional AM symbiosis.
Abstract
In the symbiotic association of plants and arbuscular mycorrhizal (AM) fungi, the fungal symbiont resides in the root cortical cells where it delivers mineral nutrients to its plant host through branched hyphae called arbuscules. Here, we report a Medicago truncatula mutant, stunted arbuscule (str), in which arbuscule development is impaired and AM symbiosis fails. In contrast with legume symbiosis mutants reported previously, str shows a wild-type nodulation phenotype. STR was identified by positional cloning and encodes a half-size ATP binding cassette (ABC) transporter of a subfamily (ABCG) whose roles in plants are largely unknown. STR is a representative of a novel clade in the ABCG subfamily, and its orthologs are highly conserved throughout the vascular plants but absent from Arabidopsis thaliana. The STR clade is unusual in that it lacks the taxon-specific diversification that is typical of the ABCG gene family. This distinct phylogenetic profile enabled the identification of a second AM symbiosis-induced half-transporter, STR2. Silencing of STR2 by RNA interference results in a stunted arbuscule phenotype identical to that of str. STR and STR2 are coexpressed constitutively in the vascular tissue, and expression is induced in cortical cells containing arbuscules. STR heterodimerizes with STR2, and the resulting transporter is located in the peri-arbuscular membrane where its activity is required for arbuscule development and consequently a functional AM symbiosis.
INTRODUCTION
The majority of the vascular flowering plants, including most crop species of agronomic significance, are able to develop symbiotic associations with arbuscular mycorrhizal (AM) fungi. The symbiosis develops in the roots where the AM fungi deliver phosphate and nitrogen to the root cortex and in return obtain carbon from the plant (Smith and Read, 2008). Fossils indicate that early land plants formed associations with AM-like fungi, and it has been proposed that the symbiosis may have enabled plants to transition to terrestrial habitats (Remy et al., 1994). Surveys of extant plant species indicate that the ability to form AM symbioses has been maintained in the angiosperm lineage, and there is evidence of the symbiosis in the lycophytes and some bryophyte lineages (Wang and Qiu, 2006; Ligrone et al., 2007). In general, the symbiosis is beneficial for plant growth and has a significant impact on plant biodiversity and ecosystem productivity (van der Heijden et al., 1998) and is an integral component of sustainable agriculture.
To form AM symbiosis, the two symbionts undergo a series of coordinated, developmental transitions that enable the fungus to enter the root cortex and establish highly branched hyphae called arbuscules in the root cells (Bonfante-Fasolo, 1984; Parniske, 2008). The symbiosis is initiated with communication through diffusible signals. The fungus perceives the presence of a plant root through strigolactones in the root exudates, and these molecules elicit an increase in fungal metabolism and vigorous hyphal branching (Akiyama et al., 2005; Besserer et al., 2006). This is accompanied by the production of myc factor(s), as yet unknown fungal signal molecules that trigger calcium oscillations and priming of the root cells (Kosuta et al., 2003, 2008; Navazio et al., 2007). Contact between the hyphae and root cells is followed by hyphopodia formation on the root surface; meanwhile, the underlying epidermal cells undergo cytoskeletal alterations, including the formation of a prepenetration apparatus that enables the fungal hyphae to pass through the epidermal cell (Genre et al., 2005). Once in the cortex, the fungus grows in the intercellular spaces of the root and also within the root cells, where the fungal hyphae differentiate to form arbuscules, extensively branched, specialized hyphae that function in the delivery of mineral nutrients to the root cell (Maeda et al., 2006; Javot et al., 2007). Arbuscule development is accompanied by significant alterations to the cortical cell, some of which occur prior to fungal entry into the cell, and several lines of evidence suggests that a short-distance mobile signal initiates reprogramming of the cortical cell potentially preparing it for the entering hypha (Liu et al., 2003; Genre et al., 2008). During arbuscule development, a plant-derived membrane, the peri-arbuscular membrane develops around the branching hypha and separates the fungus from the plant cell cytoplasm (Bonfante-Fasolo, 1984; Pumplin and Harrison, 2009). Phosphate transport proteins essential for symbiotic Pi transfer to the plant cell reside in this membrane (Harrison et al., 2002; Parniske, 2008) Apart from the Pi transporters, relatively little is known about the peri-arbuscular membrane. It is physically connected to the plasma membrane of the cell, but its origins, lipid content, and other transport activities have not been described.
Development of the symbiosis is regulated at least in part by the plant, and the initial stages of the symbiosis are controlled by a symbiosis signaling pathway (Parniske, 2008). In legumes, this pathway is required also for symbiosis with nitrogen-fixing rhizobia, and as a consequence seven components of this common symbiosis signaling pathway have now been identified. These include a receptor kinase (Endre et al., 2002; Stracke et al., 2002), channels (Ané et al., 2004; Imaizumi-Anraku et al., 2005), a calcium calmodulin-dependent kinase (Ané et al., 2004; Basu et al., 2005; Tirichine et al., 2006), and components of the nuclear pore complex (Kanamori et al., 2006; Saito et al., 2007). Mutations in any of these components disrupt fungal entry into the root cortex, and the fungus is restricted to the epidermal cell layers. By contrast, the genes that control the cortical phase of AM symbiosis, including arbuscule formation and establishment of the symbiotic interface, are largely unknown. One common symbiosis signaling mutant, cyclops, shows a reduction in both nodule numbers and arbuscule formation, indicating that the common symbiosis signaling pathway is necessary also for the cortical phase of symbiosis (Messinese et al., 2007; Yano et al., 2008). Three genes whose expression impact arbuscule development or maintenance have been identified via reverse genetics. RNA interference (RNAi)-induced silencing of a gene of unknown function, Vapyrin, prevents arbuscule formation, while silencing of a subtilisin protease reduces arbuscule numbers (Pumplin et al., 2009; Takeda et al., 2009), and silencing of a phosphate transporter results in premature arbuscule degeneration (Maeda et al., 2006; Javot et al., 2007). Several mutants that show altered arbuscule phenotypes have been identified in forward genetic screens, but so far, the genes responsible are unknown (Paszkowski et al., 2006; Sekhara et al., 2007). Gene expression studies indicate considerable changes in the cortical cells throughout arbuscule development and point to the existence of a complex cell autonomous and cell nonautonomous signaling occurring in the vicinity of the growing hyphae (Liu et al., 2003, 2007; Küster et al., 2007; Gutjahr et al., 2008; Gomez et al., 2009). Lysophosphatidylcholine (LPC) may be one of these signals, as it was shown to elicit AM symbiosis-specific Pi transporter gene expression in potato (Solanum tuberosum) and tomato (Solanum lycopersicum; Drissner et al., 2007).
The common symbiosis signaling pathway was uncovered through genetic screens for mutants defective in nodulation with subsequent analyses of their mycorrhizal phenotypes (Parniske, 2008). To identify genes involved in arbuscule formation, we undertook a genetic screen for mutant impaired in arbuscule development. This screen led to the identification of a mutant, stunted arbuscule (str), in which arbuscule development is initiated but subsequently arrests and AM symbiosis fails. Positional cloning and characterization of STR, along with the identification of a second transporter, STR2, reveals an ancient and highly conserved pair of ABC transporters of the ABCG subfamily essential for AM symbiosis.
RESULTS
A Medicago truncatula Mutant with a Defect in AM Symbiosis but Not in Nodulation
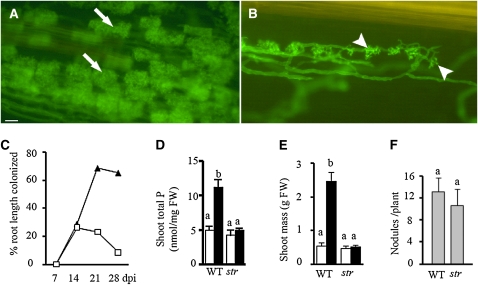
To identify AM symbiosis mutants, M2 seedlings from a M. truncatula ethyl methanesulfonate mutant population were inoculated with Glomus versiforme. Three weeks later, part of the root system was excised, stained, and examined microscopically for aberrant mycorrhizal associations. The screen yielded a mutant in which arbuscule development was impaired and the arbuscules were tiny and shrivelled relative to those in wild-type (A17) roots. The mutant was named str for stunted arbuscule. In str mycorrhizal roots, stunted arbuscules were present generally in one or two files of cortical cells, and the colonization levels in str roots were low (Figures 1A to 1C). str mycorrhizal plants did not show AM symbiosis-associated increases in shoot Pi content or shoot growth (Figures 1D and 1E) that occur in wild-type roots. To date, all of the AM symbiosis mutants reported in legumes are also impaired in their ability to form symbiosis with nitrogen-fixing bacteria and are referred to as common symbiosis mutants (Parniske, 2008). To assess the nodulation phenotype of str, plants were inoculated with Sinorhizobium meliloti and then examined at 21 d after inoculation for the presence of nodules. Both str and wild-type roots showed pink nodules, and there was no significant difference in nodule numbers on str and wild-type roots (Figure 1F; see Supplemental Figure 1A online). SYTO 13 staining of sectioned nodules indicated that the nodules were colonized by S. meliloti, and nodules from str roots did not differ in appearance from those of the wild type (see Supplemental Figures 1B to 1E online). In both the wild type and str, SYTO 13 staining revealed cells full of elongated rod-shaped bacteroids (see Supplemental Figures 1C and 1E online). This morphology is typical of the terminally differentiated bacteroids of M. truncatula/S. meliloti symbiosis (Haynes et al., 2004). Consistent with the presence of differentiated nitrogen-fixing bacteroids, wild-type and str plants grown for 21 d in the absence of nitrogen had green shoots and did not display symptoms of nitrogen deprivation, which further suggests that the symbioses with S. meliloti were functional (see Supplemental Figure 1A online). Thus, in contrast with the legume AM symbiosis mutants reported so far, STR is not a common symbiosis protein required for both nodulation and AM symbiosis, but instead appears to be required uniquely for AM symbiosis.
Figure 1.
The str Phenotype.
(A) and (B) Fluorescence microscopy image of wild-type (A) and str (B) roots colonized with G. versiforme. Roots were stained with WGA-alexafluor 488 to enable visualization of the fungus. Bar = 10 μm.
(C) Colonization levels. Closed triangles are wild-type roots, and open squares are str roots after inoculation with G. versiforme.
(D) and (E) Phosphate content and growth response. Mock-inoculated plants (white bars) and plants inoculated with G. versiforme at 42 d after inoculation (black bars). n = 12 plants for each line. Different letters indicate significant differences (P < 0.001). FW, fresh weight. Error bars represent sd.
(F) Nodulation. Nodule numbers in str and the wild type 21 d after inoculation with S. melliloti do not differ significantly (P > 0.05). n = 10 plants for each line. Error bars represent sd.
A closer investigation of AM symbiosis in str revealed that the initial phases of the symbiosis, namely, hyphopodia formation and fungal entry into the cortex, occur as in wild-type roots (see Supplemental Figure 1F online). Arbuscule development is initiated correctly, including the expression of genes typically associated with arbuscule formation (see Supplemental Figure 1G online), but in str roots, arbuscule development is notably retarded relative to wild-type roots. Arbuscules in str do not achieve full size or branch density, and 48 to 72 h after inoculation (HAI) they lose turgor, shrink, and growth ceases (Figures 2A to 2C). The tiny, shriveled arbuscules persist in the cortical cells. Vital staining and expression of an SCP promoter-green fluorescent protein (GFP) marker both indicate that the cortical cells remain alive. Therefore, the death of the arbuscule is not a consequence of loss of viability of the root cell (Figure 2D).
Figure 2.
Arbuscule Development in str and Wild-Type M. truncatula Roots.
(A) to (C) G. versiforme arbuscules. Left, center, and right panels show confocal laser scanning microscopy images, light-field images, and an overlay, respectively. The fluorescent green signal arises from WGA-Alexafluor 488 staining of the fungal cell wall. Arrowheads indicate the plant cell wall. Bars = 10 μm. a, arbuscules; ih, intercellular hyphae.
(A) Arbuscules in the wild type at 48 HAI.
(B) Arbuscules in str at 48 HAI.
(C) Two stunted arbuscules in str at 72 HAI (arrows).
(D) GFP fluorescence from a M. truncatula SCP1 promoter-GFP fusion in str cortical cells containing stunted arbuscules indicates that cortical cells remain alive. Panels show bright-field (left) and confocal laser scanning microscopy images (middle and right), respectively. The middle and right panels show optical sections through a cortical cell with a stunted arbuscule at different depths, revealing GFP in the cytosol and nucleus (n).
Positional Cloning of STR
A positional cloning approach was taken to identify STR. In a mapping population (F2) derived from a cross between str and M. truncatula genotype A20, str segregated as a single, recessive trait and showed linkage to markers on the south arm of chromosome 4 (see Supplemental Table 1 online). Fine mapping, involving the genotyping of 8712 individuals coupled with phenotypic analysis of recombinants, placed str to the south of marker 003C06 at the extreme end of chromosome 4 (Figure 3A; see Supplemental Figure 2 online). Despite the extensive mapping population, there were no recombination events that delimited a flanking marker to the south of str. The mapping data place str in a 230-kb region covered by three BACs or in a region of unknown length between the BAC contig and the telomere (see Supplemental Figure 2 online). The 230-kb region is predicted to contain 65 genes, 13 of which are associated with transposable elements. Analyses of candidate genes from this region indicated that transcript levels for one gene, predicted to encode an ATP binding cassette (ABC) transporter, were higher in A17/G. versiforme mycorrhizal roots relative to levels in the mock-inoculated controls. Furthermore, transcript levels for this ABC transporter were reduced in str relative to A17. Sequencing the ABC transporter gene from str revealed a C-to-T transition at nucleotide 1903 downstream of the first ATG codon. This base change results in the conversion of Gln-635 to a stop codon and creates a truncated protein predicted to lack the last three transmembrane domains of the protein (Figures 3B and 3C). No other mutations were detected in the coding sequence or in 2000 bp of upstream flanking DNA. Introduction of the wild-type ABC transporter gene into str roots complemented the str phenotype and restored arbuscule development and wild-type colonization. In str root systems transformed with the empty vector, arbuscules were stunted with an average length of 15 μm, while arbuscules in str root systems expressing a wild-type copy of STR showed an average length of 47 μm. The latter is indistinguishable from arbuscules in the wild type where the average arbuscule length was 45 μm (Figures 3D and 3E). The infection units in str roots expressing STR were significantly longer than in str transformed with the empty vector, indicating that the fungus continues to proliferate in the cortex (Figure 3F). Complementation of the str phenotype was observed in 42 independent transgenic root systems, and for a selection of these, the presence of the STR allele was confirmed by sequencing (see Supplemental Table 2 online). Together, the mapping, sequencing, and complementation data indicate that the ABC transporter is STR.
Figure 3.
Positional Cloning of STR.
(A) Genetic mapping indicates that STR is located south of QZ-10E12. cM, centimorgan.
(B) Diagram of STR showing four exons (black rectangles) and three introns (diagonal lines). White star indicates a C-to-T mutation at position 1903 in str.
(C) Conserved domains of STR. A, Q, C, B, D, and H represent Walker A, Q-loop, ABC signature, Walker B, D-loop, and H-loop domains, respectively (11).Transmembrane α-helices are denominated as 1 to 6. Asterisk indicates the translation stop in str.
(D) to (F) Complementation of str.
(D) str roots transformed with a wild-type copy of STR (left panel) and empty vector (right panel), respectively, showing arbuscules (a) and internal hyphae (ih) at 28 d after inoculation. Bar = 100 μm.
(E) Length of G. versiforme arbuscules in str transformed with STR transgene (str/VSTR) (n = 50, 10 plants), empty vector (str/V) (n = 32, 5 plants), and in the wild type (WT) (n = 28, five plants). The wild type and str/VSTR do not differ significantly (P > 0.05). Cortical cell lengths do not differ between str (49.3 ± 1.5 μm, n = 24) (five plants) and wild-type plants (47.8 ± 1.3 μm, n = 28) (five plants).
(F) Lengths of infection units in str transformed with a wild-type STR transgene (str/VSTR) (n = 25, five plants) are significantly longer than those in str transformed with empty vector (str/V) (n = 35, five plants) at 28 d after inoculation. Different letters indicate a significant difference (P < 0.001). Error bars represent se.
STR Is Highly Conserved in the Angiosperms and Defines a Novel Clade in the ABCG Subfamily of ABC Transporters
ABC proteins are found in a wide range of organisms, and most of them function as transporters. They are modular proteins made up of combinations of two core elements, a transmembrane domain comprised of five to six transmembrane α-helices, and a nucleotide binding fold that mediates ATP hydrolysis to energize transport. The full-size ABC transporter molecules have two transmembrane domains and two nucleotide binding domains (Martinoia et al., 2002; Rea, 2007; Yazaki et al., 2009). In addition to the full ABC transporters, there are half-transporters that possess only one nucleotide binding domain and one transmembrane domain. These proteins form either homo- or heterodimers to create a complete transporter (Rees et al., 2009). In plants, the ABC proteins belong to a very large superfamily, which is further divided in subfamilies based on the number and organization of the membrane and nucelotide binding domains (Martinoia et al., 2002; Rea, 2007; Yazaki et al., 2009). STR shares sequence similarity with the half-transporter proteins of the ABCG subfamily. The ABCG subfamily is the largest of the ABC transporter subfamilies in plants with 29 half-transporter members in Arabidopsis thaliana (Rea, 2007; Verrier et al., 2008; Yazaki et al., 2009). Potential STR orthologs were identified in all vascular plant species for which a whole genome sequence is available but not in Arabidopsis, a nonmycorrhizal species (Figure 4A). Additionally, an ortholog was readily identified in the genome sequence of Selaginella moellendorffii but not in Physcomitrella patents, although orthologs of other ABCG transporters were present. The ability to form AM symbioses with arbuscules is widespread in the extant lycophytes, including S. moellendorffii, and has been reported in two bryophyte lineages, the liverworts and hornworts, but not in bryopsids (mosses) such as P. patens (Wang and Qiu, 2006; Ligrone et al., 2007).
Figure 4.
Phylogenetic Tree and Expression Patterns of Selected ABCG Transporters.
(A) Phylogenetic tree of ABCG family transporters of Arabidopsis and M. truncatula, with a focus on STR and STR2 and their orthologs. The shaded clusters include putative orthologs of STR and STR2, from plants with complete, or near complete, genome sequences. Gm, Glycine max; Sb, Sorghum bicolor; Os, Oryzae sativa; Pt, Populus trichocarpa; Vv, Vitis vinifera; Lj, Lotus japonicus; Sm, Selaginella moellendorffii. Branches with gray and black filled circles are ABCG family transporter proteins from Arabidopsis and M. truncatula, respectively. The naming system for the Arabidopsis transporters is as reported by Verrier et al. (2008). For M. truncatula, the number for each gene family member (e.g., 1, 2, 3, etc.) is determined by the order of identification. Mt ABCG10 and Mt ABCG13 are indicated with the numbers 10 and 13, respectively. MDR-1 is used as an outgroup. The bootstrap values for the branches are shown. Accession numbers are shown in Supplemental Table 4 online.
(B) Quantitative RT-PCR to monitor transcript levels of STR, STR2, PT4, ABGC10, and ABGC13 in M. truncatula roots. α-Tubulin is G. intraradices gene and provides an indication of the level of fungal RNA in the mycorrhizal root samples. PT4 is expressed only in mycorrhizal roots and serves as a marker of arbuscules. Roots were harvested at 7 d after contact with G. intraradices spores. The values represent transcript levels of each gene expressed relative to the transcript levels of M. truncatula EF-1α. Error bars represent sd (n = 3 independent biological replicates). WT, wild type; WT/Gi, wild type colonized with G. intraradices; str/Gi, str colonized with G. intraradices.
STR and its orthologs form a distinct clade in the ABCG transporter family (Figure 4A), and this clade is unusual in that there is no apparent taxon-specific expansion or diversification, which has occurred almost universally in remainder of the ABCG family (Crouzet et al., 2006; Sugiyama et al., 2006; Verrier et al., 2008) (Figure 4A). Instead, each plant species contains a single STR ortholog and those with duplicated genomes, such as soybean (Glycine max), contain two paralogs of STR. The presence of the S. moellendorffii ortholog in the STR clade supports an early origin for this protein (Kenrick and Crane, 1997), while the occurrence of orthologs in both monocotyledons and dicotyledons indicates that it was retained through vascular plant evolution.
Identification of STR2, a Second ABC Transporter with a Phylogenetic Profile That Mirrors That of STR
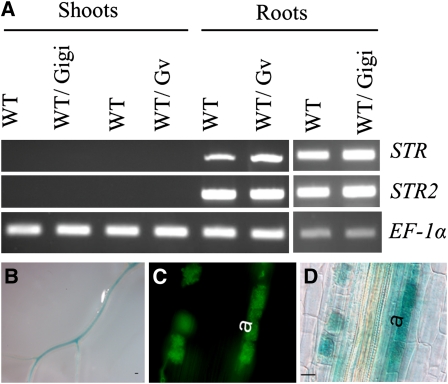
Further examination of the ABCG transporter subfamily in M. truncatula revealed that STR shares the highest identity with a second ABCG transporter, which we named STR2. STR2 and its orthologs form a sister clade to STR with a phylogenetic profile identical to that of STR and STR orthologs. Like STR, STR2 is expressed in roots and transcript levels increase significantly in mycorrhizal roots (Figure 4B). By contrast, two other ABCG transporters genes related to STR, ABCG13 and ABCG10, are expressed constitutively in roots, but transcript levels do not increase in mycorrhizal roots (Figure 4B). STR and STR2 transcripts were detected only in roots, and STR promoter-β-glucuronidase (GUS) and STR2 promoter-GUS fusions revealed that the two genes have identical expression patterns (Figures 5A to 5D). Both genes are expressed at low levels in the vascular tissue (Figure 5B), and following colonization with mycorrhizal fungi, their expression is induced specifically in cells with arbuscules (Figures 5C and 5D; see Supplemental Figure 3 online). Given that the half-transporters are predicted to function as dimers, STR2 is a possible partner of STR.
Figure 5.
Expression of STR and STR2.
(A) RT-PCR to detect STR and STR2 transcripts in RNA from roots and shoots of either mock-inoculated control wild-type plants (WT) or plants colonized by G. gigantea (WT/Gigi) or plants colonized by G. versiforme (WT/Gv). To survey the shoots for transcripts, PCR reactions included 35 cycles of amplification for STR and STR2, which resulted in near saturation for the root RNA samples and masks the AM symbiosis-induced expression. M. truncatula EF-1α was included as a control with 30 cycles of amplification.
(B) Whole roots showing GUS activity in the vascular tissue (V) in transgenic roots expressing STR promoter-UidA fusions. Roots were GUS stained for 24 h. Bar = 100 μm.
(C) and (D) Fluorescence microscopy images and corresponding bright-field images of roots colonized with G. versiforme reveal GUS expression in cortical cells with arbuscules (a). Roots were GUS stained for 6 h. Bar = 20 μm.
Knockdown of STR2 by RNAi Results in a Stunted Arbuscule Phenotype
To determine whether STR2 has a function in AM symbiosis, two independent RNAi constructs were generated and introduced into wild-type M. truncatula roots (see Supplemental Figure 4 online). Transgenic roots expressing either of two RNAi constructs designed to target STR2 showed a mycorrhizal phenotype identical to that of str. The fungus was able to penetrate the roots, but arbuscule development was impaired and tiny, stunted arbuscules were visible in the cortical cells (Table 1, Figures 6A and 6B). As observed in str, colonization levels were low (Table 1). RT-PCR analyses confirmed that roots expressing STR2 RNAi showed a reduction in STR2 transcript levels, while STR transcripts were not altered (Figures 6C to 6E). These data indicate that the RNAi constructs were specific for STR2 and that the stunted arbuscule phenotype can be attributed to a reduction in STR2 expression. Thus, STR2 is also essential for arbuscule development in AM symbiosis.
Table 1.
Arbuscule and Infection Unit Lengths in Transgenic Roots Expressing RNAi Constructs That Target STR2
| Vector Control (V) | STR2 RNAi-1 | STR2 RNAi-2 | |
| Arbuscule length (μm)a | 47.0 ± 1.2 (n = 55) | 20.9 ± 0.8 (n = 45) | 18.5 ± 1.1 (n = 36) |
| Infection unit length (μm)a | 2876.5 ± 383.7 (n = 19) | 907.3 ± 180.0 (n = 13) | 991.2 ± 108.4 (n = 15) |
Roots were analyzed 28 d after inoculation.
Figure 6.
Mycorrhizal Phenotype of STR2 RNAi Roots.
(A) Fluorescence microscopy image of a root expressing a STR2 RNAi construct colonized with G. versiforme. Roots were stained with WGA-Alexafluor 488. Bar = 10 μm.
(B) Confocal laser scanning microscopy image showing an arbuscule in transgenic roots expressing a STR2 RNAi construct. Internal hyphae (ih) and a stunted arbuscule (a) with shriveled branches (arrow) are representative of arbuscules in the STR2 RNAi roots. Bar = 10 μm.
(C) to (E) Real-time quantitative RT-PCR analysis of STR2 RNAi roots. Data shown are averages obtained from three to four independent transformants per line. White bars represent wild-type roots transformed with pHellsgate8-GUS as a control (V), and red bars represent wild-type roots transformed with STR2 RNAi-1 transgene (R1) and STR2 RNAi-2 transgene (R2), respectively. Different letters indicate samples that differ significantly from each other (P < 0.001). STR transcript levels (E) in STR2 RNAi roots are not affected in noncolonized roots (P > 0.05). In graphs (C) to (E), transcripts are expressed as a ratio relative to EF-1α transcripts. Error bars represent sd.
The STR2 RNAi constructs were introduced into str also. However, the phenotype of STR2 RNAi roots in the str background did not differ from that of str, and loss of both genes gave no apparent additive effect.
STR and STR2 Proteins Interact and Are Located in the Peri-Arbuscular Membrane
The half-ABC transporters form either homo- or heterodimers to create a full transporter molecule capable of transport (Martinoia et al., 2002; Rea, 2007; Rees et al., 2009). To determine if STR and STR2 dimerize and, simultaneously, to identify the membrane in which they are located, we used bimolecular fluorescence complementation (BiFC) (Hu et al., 2002). The N- and C-terminal halves of yellow fluorescent protein (YFP) were fused to the N-terminal ends of STR (YFPN-STR) and STR2 (YFPC-STR2), respectively. Each transgene was expressed from its native promoter. Additionally, we examined the ability of STR to form homodimers by creating STR constructs with the N- and C-terminal halves of YFP. In each case, the various combinations of constructs were introduced into wild-type (A17) and str roots on a single T-DNA. In str roots, the STR BiFC transgenes complemented the str mutation and restored wild-type mycorrhizal colonization, which indicates that the YFP-tagged STR proteins are functional (Figure 7A). Following colonization by G. versiforme, str roots and A17 roots expressing both the YFPN-STR and YFPC-STR2 transgenes showed YFP fluorescence on the plant peri-arbuscular membrane that surrounds the arbuscule. By contrast, roots expressing only STR split YFP fusions, YFPN-STR and YFPC-STR, or a single fusion control (YFPC-STR) showed no signal. These data indicate that STR forms heterodimers with STR2 but does not homodimerize. In roots expressing both STR and STR2 (YFPN-STR and YFPC-STR2) transgenes, YFP signal was detected on young arbuscules with few dichotomous branches as well as densely branched arbuscules. The signal was strong in the region of the membrane around the arbuscule branches and weak or absent from the trunk region. The peri-arbuscular membrane is continuous with the plasma membrane of the cell, but the STR/STR2 YFP signal was not detected on the plasma membrane around the periphery of the cortical cell (Figures 7B to 7G). This asymmetric distribution is similar to that of the M. truncatula symbiotic Pi transporter PT4 (Harrison et al., 2002; Pumplin and Harrison, 2009). YFP fluorescence from YFPN-STR/YFPC-STR2 was not observed in other locations within the cell except in cells with degenerate arbuscules, where some YFP signal was apparent in the vacuole. Additionally, we were unable to detect a signal in the cells of the vascular tissue in either mock-inoculated or mycorrhizal roots. Analyses of STR and STR2 promoter-GUS fusions indicated that expression levels in the vascular tissue were lower than the colonized cortical cells, and it is possible that YFPN-STR/YFPC-STR2 proteins are present at levels below detection.
Figure 7.
BiFC to Show STR and STR2 Interaction and Location in the Peri-Arbuscular Membrane.
(A) Confocal laser scanning microscopy image showing complementation of str with the STR and STR2 BiFC constructs. Two cortical cells containing fully developed arbuscules (a) are highlighted. Roots were stained with WGA-Alexa Fluor 488 and shown in red pseudocolor. Bar = 100 μm.
(B) to (G) Confocal microscopy images showing YFP signal on peri-arbuscular membrane. Images are representative of arbuscules observed in roots of eight str and nine wild-type plants transformed with the STR/STR2 BiFC constructs. Distinct branches of the arbuscule are visible (arrowhead). There is no YFP signal in the plasma membrane around the periphery of the cell. The interaction and location of STR/STR2 YFP was observed in 15 transgenic plants during symbiosis with G. versiforme and G. intraradices (assessed at multiple times between 21 and 35 d after inoculation). Bars = 10 μm.
(B) and (E) Confocal laser scanning microscopy images.
(C) and (F) Corresponding differential interference contrast images.
(D) and (G) Overlay images show cortical cells containing arbuscules in roots expressing STR and STR2 BiFC constructs.
The finding that STR and STR2 interact is consistent with the identical AM symbiosis phenotypes of str mutant and STR2 RNAi roots and also with the lack of any apparent additive effect when STR2 was silenced in the str background. Considering the mutant phenotypes and location of the STR/STR2 in the peri-arbuscular membrane, we propose that the transporter functions to export an essential signal or nutrient to the interface with the fungus (see Supplemental Figure 6 online).
DISCUSSION
The AM symbiosis occurs broadly in the angiosperms and in the Arum-type AM symbioses that occur in most herbaceous plants, the arbuscules mediate mineral nutrient transfer to the plant and are critical for the symbiosis. Arbuscule development is a complex process that involves the terminal differentiation of the fungal hypha and concomitantly a transient cellular reorganization and transcriptional reprogramming of the root cortical cells. To identify components of this cellular program, we screened directly for mutants impaired in arbuscule formation and identified str. In str, arbuscule growth arrests at an early stage. The partially branched hypha shrivels and tiny, stunted arbuscules persist in the cortical cells. Microscopy analyses indicate that the arbuscule phenotype differs from that of the Pi transporter mutant, mtpt4, where arbuscule development occurs normally, but arbuscule degeneration is triggered prematurely (Javot et al., 2007). The contrasting phenotypes of the str and mtpt4 mutants provide insights into arbuscule biology, and the absence of arbuscule degeneration in str supports the hypothesis that degeneration is a programmed event that occurs after arbuscule maturity rather than a default pathway triggered simply by a failure in arbuscule development.
Positional cloning revealed that STR encodes a novel ABC transporter. ABC transporters are found in a wide range of organisms from all kingdoms of life. They transport a diverse array of molecules, including ions, nutrients, secondary metabolites, and lipids across various cellular membranes. In plants, the ABC transporter gene family is greatly expanded relative to other organisms, and the Arabidopsis and rice (Oryza sativa) genomes are predicted to encode over 120 ABC proteins which are further classified into several subfamilies (Higgins, 1992; Schulz and Kolukisaglu, 2006; Rea, 2007).
STR and its partner, STR2, which we identified by its relatedness and identical phylogenetic profile, are half-transporters of the ABCG subfamily. The ABCG subfamily is the largest but most poorly characterized ABC transporter subfamily in plants. There are 29 half-transporter members in Arabidopsis and 30 in rice (Higgins, 1992; Schulz and Kolukisaglu, 2006; Rea, 2007). ABC transporters from other subfamilies, such as the PDR subfamily, have roles in defense against plant pathogens, but transporters in the ABCG subfamily have not been implicated previously in plant–microbe interactions (Jasinski et al., 2001; van den Brule et al., 2002; Stein et al., 2006). In Arabidopsis, two ABCG transporters are involved in long-chain fatty acid transport and loss of function impairs wax deposition in the cuticle (Pighin et al., 2004; Bird et al., 2007). A third transporter is implicated in the transport of xenobiotics (Mentewab and Stewart, 2005). Currently, the roles of the other 26 half-transporters of the ABCG subfamily are undetermined. By contrast, humans have only five ABCG half-transporters, and they are relatively well characterized because altered function is associated with a variety of diseases including atherosclerosis, sitosterolemia, and Tangier's disease (Schmitz et al., 2001; Kusuhara and Sugiyama, 2007). The substrates of the mammalian ABCG transporters include cholesterol, sterols, and other lipid molecules, but also structurally unrelated anticancer drugs (Allikmets et al., 1998; Schmitz et al., 2001).
STR and STR2, and their respective putative orthologs, form two sister clades in the ABCG family, and the clades are unusual in that they lack the taxon-specific expansion that is widespread in this family. We analyzed plant species for which a whole genome sequence is available, and each diploid species has a single representative in the STR and STR2 clades, while those with duplicated genomes contain two representatives. The only exception is Populus trichocarpa, which has two copies of STR, but only one copy of STR2. It was proposed that P. trichocarpa went through whole-genome duplication and only a small portion of the duplicated genes survived in the genome (Tuskan et al., 2006). It may be that the second copy of STR2 was lost in this process. Apart from P. trichocarpa, STR and STR2 orthologs have identical phylogenetic profiles (see Supplemental Figure 5 online), and orthologs of both genes are absent in Arabidopsis, a species that does not form AM symbiosis. This suggests that AM symbiosis may have provided the driving force for the maintenance of these proteins through evolution. The similarity of the STR and STR2 phylogenetic trees is a hallmark of proteins that have undergone coevolution (Pazos and Valencia, 2008). The experimental data indicating that the proteins interact and function together further supports this, and STR and STR2 provide interesting opportunities for molecular evolution studies.
STR and STR2 are expressed only in the roots and show identical spatial expression patterns. Both promoters are active at a low level in the vascular tissue and highly active in cortical cells containing arbuscules. Similar to PT4, expression in the cortex is restricted to cells containing arbuscules, suggesting that it is triggered by a cell autonomous signal. During the positional cloning of STR, several thousand str plants were grown under a range of environmental conditions; however, a phenotype other than the mycorrhizal phenotype was not observed. It is possible the nonmycorrhizal function is redundant or that these environmental conditions were not suitable to reveal a phenotype.
The ABCG transporters dimerize to create a full-size transporter. Given the characteristics of STR and STR2, it seemed possible that they would function together. BiFC analysis with constructs expressed from the native promoters revealed that STR did not form homodimers but is able to heterodimerize with STR2. This finding is consistent with the str mutant and STR2 RNAi phenotypes, and together the phenotype and interaction data indicate that STR and STR2 function as a heterodimer. Loss of either partner impairs heterodimer formation. Similar observations have been made for the human ABCG5 and ABCG8 proteins, which interact to form a heterodimer and loss of either half-transporter results in sitosterolemia (Graf et al., 2002; Wittenburg and Carey, 2002). The STR/STR2 heterodimer resides in the peri-arbuscular membrane at the symbiotic interface between the cortical cell and the arbuscule. Based on the transport activity of most eukaryotic ABC transporters, it seems likely that STR/STR2 is an export pump, and the presence of STR/STR2 only in the peri-arbuscular membrane suggests that it functions specifically to export a substrate molecule from the cortical cell to the peri-arbuscular apoplastic space (see Supplemental Figure 6 online). This apoplastic space, which is delimited by the arbuscule membrane and arbuscule cell wall and the peri-arbuscular membrane is ∼100 nm wide and contains matrix material similar in composition to that of a primary plant cell wall (Bonfante-Fasolo and Perotto, 1992). It has an acidic pH, and it was shown recently that a subtilisin protease is secreted there (Smith et al., 2001; Takeda et al., 2009). Molecules secreted to this apoplastic space would be readily accessible to the AM fungus. Given the phenotype of the str mutant and the suggestion that transport is directed toward the AM fungal symbiont, it seems possible that STR/STR2 exports a signal molecule or possibly a nutrient essential for arbuscule growth. Currently, we know relatively little about the AM fungi, and it is difficult to predict what molecules might be essential for arbuscule development. The primary sequences of the ABC transporters provide few clues as to the substrate that they transport (Martinoia et al., 2002; Rea, 2007), but by extrapolation from the activities of the mammalian ABCG transporters and the three characterized ABCG proteins from Arabidopsis, the STR/STR2 substrate molecule might be a lipid but could also be a secondary metabolite (Allikmets et al., 1998; Schmitz et al., 2001; Pighin et al., 2004; Mentewab and Stewart, 2005; Bird et al., 2007). It was shown recently that LPC serves as a signal to induce symbiotic Pi transporter gene expression (Drissner et al., 2007). However, LPC seems an unlikely substrate for STR/STR2, as the M. truncatula symbiotic Pi transporter PT4 is expressed appropriately in str. A secondary metabolite is possible, and although the range of plant secondary metabolites is huge, the conservation of STR and STR2 across the angiosperms suggests that the substrate should be conserved; therefore, it is unlikely to be a secondary metabolite that is unique to a particular plant family. One possible class of substrates are strigolactones, terpenoid plant hormones that control plant development and serve as signals in the presymbiotic phase of AM symbiosis (Akiyama et al., 2005; Besserer et al., 2006; Gomez-Roldan et al., 2008; Umehara et al., 2008). Strigolactones present in root exudates activate metabolism of germinating AM fungi spores and induce branching of AM fungal hyphae prior to contact with the root (Besserer et al., 2006, 2008; Akiyama and Hayashi, 2008). Currently, it is not known what induces the dichotomous branching of the intracellular hyphae to create the arbuscule. Although a role for strigolactones in arbuscule development has not been demonstrated, the slow growth and reduced branching phenotype of arbuscules in str leads us to speculate that strigolactones might be involved. A localized supply of strigolactone could trigger the same elevated metabolism and extreme branching that is seen in the external hyphae. Strigolactones are potent plant hormones and have strong effects on both monocots and dicots at extremely low concentrations (Gomez-Roldan et al., 2008; Umehara et al., 2008). If strigolactones are involved in stimulating arbuscule branching, it would be necessary for the plant to ensure that their export to the AM fungus was tightly regulated to avoid unintended effects on plant development. The spatially restricted location of the STR/STR2 transporter in the peri-arbuscular membrane, particularly around the arbuscule branches, would ensure transport to the arbuscule apoplastic space and not to the general apoplast at the interface with adjacent cortical cells. While currently speculative, these ideas about possible substrates provide avenues for future research.
In summary, STR and STR2 encode novel half-ABC transporters and define two new sister clades within the ABCG subfamily of the ABC transporter gene family. The two proteins interact and the heterodimer resides in the peri-arbuscular membrane, where its activity is essential for arbuscule development in AM symbiosis. Unlike the symbiotic phosphate transporters, M. truncatula PT4 and rice PT11 (Harrison et al., 2002; Paszkowski et al., 2002), that import phosphate across the peri-arbuscular membrane into the cortical cell, we predict that the STR/STR2 transporter exports its substrate from the cortical cell to the peri-arbuscular apoplastic space. The data provide new insights into plant proteins required for AM symbiosis and pave the way for the identification of signals or nutrient molecules essential for AM symbiosis.
METHODS
Plant and AM Fungal Growth Procedures
Medicago truncatula, cultivar Jemalong (A17), and str mutant plants were grown as described previously (Liu et al., 2007). The maintenance of Glomus versiforme, Glomus intraradices, and Gigaspora gigantea was as described previously (Liu et al., 2007). Spore preparation, surface sterilization, and inoculation procedures were as described previously (Bécard and Fortin, 1988; Harrison and Dixon, 1993; St-Arnaud et al., 1996; Liu et al., 2004, 2007). Roots were stained in 0.2 μg/mL WGA-Alexafluor 488 (Molecular Probes), and colonization levels (Figure 1C) were assessed by the gridline intersect method (McGonigle et al., 1990). For analyses shown in Figure 2, a synchronized inoculation system was used (Lopez-Meyer and Harrison, 2006). Plants were harvested at 24, 48, and 72 HAI (three plants per line at each time point). For gene expression analyses shown in Figure 4B, seedlings were planted into cones with spores placed in a sand layer positioned 6 to 7 cm below the point of planting. M. truncatula roots reach the spores 7 d after planting during which time spores have germinated. Roots are harvested at 7 d after contact with AM fungal spores and 14 d after planting. At this early harvest time (7 d after contact), the colonization levels in A17 and str are comparable (also shown in Figure1C), and str has some actively developing arbuscules. Mycorrhiza-associated growth responses and phosphate benefit experiments were as described previously (Javot et al., 2007). Plants were harvested at 42 d after inoculation, and phosphate content was assessed by colorimetric assay (Ames, 1966). str was backcrossed to the wild type (A17) twice, and phenotypic analyses were performed on the backcrossed (BC1 or BC2) generations. Most experiments were performed with G. versiforme and G. intraradices. There were no obvious differences in the str phenotype observed in these two different symbioses. Large quantities of G. intraradices spores are easier to acquire.
Nodulation Assays
Plants were inoculated with Sinorhizobium meliloti strain 2011 as described (Liu et al., 2003). Ten plants of each line (M3 and BC2 F3 generations) were harvested at 21 d after inoculation, and nodules were counted and examined microscopically. Nodules were fixed in 50 mM PIPES, pH 6.8, and 1% glutaraldehyde for 3 h and rinsed overnight in 50 mM PIPES. Nodules were hand-sectioned down the longitudinal axis with a double-edge razor blade and stained for 10 min with 5 μM SYTO 13 in 50 mM PIPES, pH 8.5. Nodules were viewed by confocal microscopy as described (Haynes et al., 2004). SYTO 13 was excited at 488 nm, and fluorescence emitted between 505 and 530 nm was collected.
Cell Viability Assays
Cell viability was assessed by succinate dehydrogenase staining as reported previously (Javot et al., 2007). Also, the M. truncatula SCP1-GFP transgene (Liu et al., 2003) was introduced into the str background, and GFP fluorescence was used to monitor cell viability.
Large-Scale DNA Extractions and Marker Analysis
DNA was extracted using a modified CTAB DNA miniprep protocol from an Arabidopsis laboratory manual (Weigel and Glazebrook, 2002). Primers for simple sequence repeat markers and cleaved-amplified polymorphic sequence markers and the conditions for their use are shown in Supplemental Table 3 online.
RNA Extraction, RT-PCR Analysis, and Real-Time Quantitative PCR Analysis
Total RNA was extracted using TRI reagent (Invitrogen). The primers and conditions for the RT-PCR and real-time PCR analysis are described in Supplemental Table 3 online. Quantitative RT- PCR analysis was performed essentially as described by Liu et al. (2007) with the following modifications. Primers were designed using an online real-time PCR primer design tool (http://www.idtdna.com/Scitools/Applications/RealTimePCR/). PCR reactions were performed in a total volume of 10 μL, which contained 5 μL 2× SYBR Green PCR master mix, 150 nmoles of each primer, and 20 ng of cDNA template. The PCR program was as follows: 95°C for 5 min, 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. Dissociation curve analysis was performed with the thermal cycle of 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s, to confirm specificity. The data were analyzed with Applied Biosystems SDS 2.2 software. The M. truncatula elongation factor 1-α (EF-1α) gene was included to enable normalization of transcript levels of all genes assayed.
Synthesis of Radioisotope Labeled Probes and BAC Library Hybridization
Primers for the generation of a BAC end probe corresponding to BAC mth2-7D10 and the conditions for probe generation are described in Supplemental Table 3 online. One set of MT_ABb (mth2) library filters were hybridized as described in the online protocol from Clemson University Genomics Institute (https://www.genome.clemson.edu/protocols.shtml).
Complementation Analysis
A 6169-bp HindIII fragment was digested from mth2-53O24 BAC DNA. The fragment, which contains the STR gene and 2237 bp of 5′ flanking DNA and 802 bp 3′ flanking DNA was cloned into binary vector pCambia 2301. The construct was transformed into str mutant roots, and composite plants with transgenic roots systems were inoculated with G. versiforme and harvested 4 weeks after inoculation. Transgenic roots also express GUS as a consequence of a 35S-GUS gene carried on the T-DNA. Half of the selected roots were stained for GUS activity and with WGA-Alexafluor 488 as described (Harrison et al., 2002). The other halves of selected roots were used for DNA and RNA isolation, followed by PCR amplification of STR and sequencing to confirm transformation.
Generation of Composite Plants with Transgenic Roots by Agrobacterium rhizogenes–Mediated Transformation
Binary vectors were transformed into A17 roots by A. rhizogenes–mediated transformation as described by Boisson-Dernier et al. (2001) with modifications described by Liu et al. (2003).
Arbuscule Size and Infection Unit Length Measurement
Image J software (http://rsb.info.nih.gov/ij/) was used to measure cell length, arbuscule length, and length of infection units. Infection units were selected at random, and arbuscules located in the middle of infection units were measured. For RNAi roots, an average of five arbuscules per infection unit from 10 randomly selected infection units were measured for arbuscule size. Sixteen infection units were measured for the infection unit length.
Statistical Analysis
Data were analyzed by analysis of variance, followed by Turkey's HSD test, to test differences between plant genotypes and treatment. All data presented are means ± sd, except for Figure 3 and Table 1, where the se is shown.
Phylogenetic Analysis
ABCG protein amino acid sequences were aligned using ClustalW program from workbench (http://sdsc.workbench.edu). The phylogenetic tree was constructed using Phylip (Felsenstein, 1989) neighbor-joining method with bootstrap values from 1000 neighbor-joining bootstrap replicates. The tree was visualized using the program TreeView (Page, 1996).
STR (MtABCG1) and STR2 (MtABCG2) sequences have been deposited in the National Center for Biotechnology Information under the accession numbers FJ659114 to FJ659117.
STR2 RNAi Constructs
Two independent STR2 RNAi constructs were generated. The regions corresponding to 1068 to 1361 and 2218 to 2499 nucleotides (relative to the ATG start codon) of STR2 were amplified and introduced into pHellsgate 8 (Helliwell et al., 2002) to create the STR2 RNAi-1 (R1) and STR2 RNAi-2 (R2) constructs, respectively. Primers used for generating RNAi constructs are shown in Supplemental Table 3 online. Composite plants with transgenic roots were generated and inoculated with G. versiforme spores. Roots were harvested at 2 and 4 weeks after inoculation, stained, and the mycorrhizal phenotype assessed microscopically. RNA was extracted from roots 2 weeks after inoculation for real-time quantitative RT-PCR analysis.
STR and STR2 Promoter-UidA Constructs
A fragment containing 2010 bp of the STR 5′ proximal region was generated by PCR. Restriction sites XbaI and HindIII were included at the ends of the fragment. This was used to replace the 35S promoter upstream of the UidA gene in the pCAMBIA2301 vector (CAMBIA). Likewise, an XbaI-HindIII fragment containing 2000 bp of the STR2 5′ proximal region was used to create the STR2 promoter-GUS construct. The primers are listed in Supplemental Table 3 online. Binary vectors were transformed into A17 roots by A. rhizogenes–mediated transformation (Boisson-Dernier et al., 2001). Composite plants with transgenic roots were inoculated with G. versiforme spores and harvested at 4 weeks after inoculation. Roots were stained for GUS activity as described (Harrison et al., 2002).
STR and STR2 BiFC Constructs
The BiFC constructs were driven by the native STR and STR2 promoters and were introduced into roots on a single T-DNA. To make pSAT1-ProSTR:cEYFP-STRcDNA, two tandem 35S promoters in pSAT1-cEYFP-C1-B (GenBank accession number DQ168996 and ABRC stock number CD3-1070) were replaced by a 2010-bp PCR fragment amplified from the promoter region of STR, through AgeI and NcoI restriction sites. The STR full-length cDNA fragment was then digested with BglII and SalI and inserted into the MCS of this construct. The same strategy was used to make pSAT1-ProSTR2:nEYFP-STR2cDNA and pSAT1-ProSTR:nEYFP-STRcDNA from pSAT1-nEYFP-C1 (GenBank accession number DQ168995 and ABRC stock number CD3-1074), except that STR2 cDNA fragment was cut and inserted through BglII and EcoRI sites. To create pCambia-ProSTR:cEYFP-STRcDNA, the region containing STR promoter, cEYFP, and STR cDNA was released from pSAT1-ProSTR:cEYFP-STRcDNA through AgeI and NotI restrictions, blunt ended, and ligated into pCambia2301 that had been linearized by SmaI, blunt ended, and dephosphorylated. Finally, the region containing STR2 promoter, nEYFP, and STR2 cDNA was released from pSAT1-ProSTR2:nEYFP-STR2cDNA, blunt ended, and inserted into the T-DNA region of pCambia-ProSTR:cEYFP-STRcDNA. The region containing STR promoter, nEYFP, and STR cDNA from pSAT1-ProSTR:nEYFP-STRcDNA was also released and ligated into pCambia-ProSTR:cEYFP-STRcDNA to make STR/STR BiFC construct. pCambia-ProSTR:cEYFP-STRcDNA served as a single construct negative control. Eight str plants transformed with the STR/STR2 BiFC constructs showed complementation of the mycorrhizal phenotype. Six of the eight plants were examined by confocal microscopy, and YFP fluorescence, located on peri-arbuscular membrane, was observed. Nine wild-type plants transformed with the STR/STR2 BiFC constructs showed YFP fluorescence located on peri-arbuscular membrane. The emission profile of the fluorescence signals showed a near complete overlap with the EYFP emission profile from the Leica database (data not shown), confirming that the signal was YFP and arose from the interaction of STR and STR2. By contrast, complementation was also observed from eight str plants transformed with STR/STR BiFC construct; however, no YFP fluorescence was observed.
Confocal Laser Scanning Microscopy
Roots were imaged using a Leica SP5 confocal laser scanning microscope attached to a Leica DM6000 microscope. AlexaFluor 488 was excited at 488 nm, and fluorescence emitted between 505 to 582 nm was collected. YFP was excited with a green argon laser (514 nm), and emitted fluorescence between 526 and 566 nm was collected. Optical sections were acquired at 0.3- to 0.75-μm intervals, and Z series representing complete arbuscules were collected. Images shown in Figure 2 are 1.5-μm slices assembled from optical sections from the center of the arbuscule. Differential interference contrast images shown are a single plane through the arbuscule. Images were analyzed and assembled using Leica LAS software (Leica Microsystems).
In Figure 7A, the WGA-Alex fluor 488 signal is shown in red pseudocolor. In Figures 7B to 7G, images shown are single optical sections.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: STR (FJ659114), STR2 (FJ659116), PT4 (AY116211), M. truncatula ABCG10 (AC122164), M. truncatula ABCG13 (AC186136), and M. truncatula EF-1α (EX532628).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Symbiotic Phenotypes of str and A17.
Supplemental Figure 2. Diagram to Illustrate Mapping of STR on Chromosome 4.
Supplemental Figure 3. Histochemical Staining for GUS Activity in M. truncatula Roots Expressing a STR2 Promoter-UidA Fusion Construct.
Supplemental Figure 4. Alignment of STR and STR2 Nucleotide Sequences in the Regions Targeted for RNA Interference.
Supplemental Figure 5. Phylogenetic Trees of STR and STR2 Orthologs.
Supplemental Figure 6. Diagram Illustrating the Location of STR/STR2 on the Peri-Arbuscular Membrane in a Cortical Cell Containing an Arbuscule.
Supplemental Table 1. str Is Linked to Four Markers on Chromosome 4.
Supplemental Table 2. Sequence Confirmation That a Wild-Type STR Transgene Is Expressed in the Complemented str Roots.
Supplemental Table 3. Primer Sequences and PCR Conditions.
Supplemental Table 4. Accession Numbers of Genes Shown in Figure 4A.
Supplemental Data Set 1. Text File of Alignment Used to Generate Figure 4A.
Supplemental Data Set 2. Text File of Alignment Used to Generate STR Ortholog Tree in Supplemental Figure 5.
Supplemental Data Set 3. Text File of Alignment Used to Generate STR2 Ortholog Tree in Supplemental Figure 5.
Supplementary Material
Acknowledgments
We thank Aynur Cakmak for technical support, R.V. Penmetsa and D.R. Cook (UC Davis) for an initial determination of the nodulation phenotype of str, and Gabrielle Endre (University of Szeged) for initial mapping discussions. Funding for this project was provided by National Research Initiative Competitive Grants 2005-35319-15315 and 2008-35301-19039 from the USDA National Institute of Food and Agriculture.
References
- Akiyama K., Hayashi H. (2008). Plastid-derived strigolactones show the way to roots for symbionts and parasites. New Phytol. 178: 695–698 [DOI] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K.-I., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Allikmets R., Schriml L.M., Hutchinson A., Romano-Spica V., Dean M. (1998). A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 58: 5337–5339 [PubMed] [Google Scholar]
- Ames B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8: 115–118 [Google Scholar]
- Ané J.-M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Basu N.K., Kovarova M., Garza A., Kubota S., Saha T., Mitra P.S., Banerjee R., Rivera J., Owens I.S. (2005). Phosphorylation of a UDP-glucuronosyltransferase regulates substrate specificity. Proc. Natl. Acad. Sci. USA 102: 6285–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G., Fortin J.A. (1988). Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108: 211–218 [DOI] [PubMed] [Google Scholar]
- Besserer A., Becard G., Jauneau A., Roux C., Sejalon-Delmas N. (2008). GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148: 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Puech-Pagès V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., Portais J.C., Roux C., Bécard G., Séjalon-Delmas N. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D., Beisson F., Brigham A., Shin J., Greer S., Jetter R., Kunst L., Wu X.W., Yephremov A., Samuels L. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Becard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Bonfante-Fasolo P. (1984). Anatomy and morphology of VA mycorrhizae. Mycorrhizae VA, Powell C.L., Bagyaraj D.J., (Boca Raton, FL: CRC Press; ), pp. 5–33 [Google Scholar]
- Bonfante-Fasolo P., Perotto S. (1992). Plant and endomycorrhizal fungi: The cellular and molecular basis of their interaction. Molecular Signals in Plant-Microbe Communications, Verma D.P.S., (Boca Raton, FL: CRC Press; ), pp. 445–470 [Google Scholar]
- Crouzet J., Trombik T., Fraysse A.S., Boutry M. (2006). Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 580: 1123–1130 [DOI] [PubMed] [Google Scholar]
- Drissner D., Kunze G., Callewaert N., Gehrig P., Tamasloukht M., Boller T., Felix G., Amrhein N., Bucher M. (2007). Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318: 265–268 [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevel Z., Mihacea S., Kaló P., Kiss G. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP - Phylogeny inference package. Cladistics 5: 164–166 [Google Scholar]
- Genre A., Chabaud M., Faccio A., Barker D.G., Bonfante P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungus within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Timmers T., Bonfante P., Barker D.G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez S.K., Javot H., Deewatthanawong P., Torres-Jerez I., Tang Y., Blancaflor E.B., Udvardi M.K., Harrison M.J. (2009). Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Graf G.A., Li W.P., Gerard R.D., Gelissen I., White A., Cohen J.C., Hobbs H.H. (2002). Coexpression of ATP-binding cassette proteins ABCG5 and ABCG8 permits their transport to the apical surface. J. Clin. Invest. 110: 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Banba M., Croset V., An K., Miyao A., An G., Hirochika H., Imaizumi-Anraku H., Paszkowski U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.J., Dewbre G.R., Liu J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.J., Dixon R.A. (1993). Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol. Plant Microbe Interact. 6: 643–654 [Google Scholar]
- Haynes J.G., Czymmek K.J., Carlson C.A., Veereshlingam H., Dickstein R., Sherrier D.J. (2004). Rapid analysis of legume root nodule development using confocal microscopy. New Phytol. 163: 661–668 [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Wesley S.V., Wielopolska A.J., Waterhouse P.M. (2002). High-throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Higgins C.F. (1992). ABC transporters - From microorganisms to man. Annu. Rev. Cell Biol. 8: 67–113 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZip and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Jasinski M., Stukkens Y., Degand H., Purnelle B., Marchand-Brynaert J., Boutry M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13: 1095–1107 [PMC free article] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P., Crane P.R. (1997). The origin and early evolution of plants on land. Nature 349: 33–39 [Google Scholar]
- Kosuta S., Chabaud M., Lougnon G., Gough C., Dénarié J., Barker D.G., Bécard G. (2003). A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E.D. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küster H., Vieweg M.F., Manthey K., Baier M.C., Hohnjec N., Perlick A.M. (2007). Identification and expression regulation of symbiotically activated legume genes. Phytochemistry 68: 8–18 [DOI] [PubMed] [Google Scholar]
- Kusuhara H., Sugiyama Y. (2007). ATP-binding cassette, subfamily G (ABCG family). Pflugers Arch. 453: 735–744 [DOI] [PubMed] [Google Scholar]
- Ligrone R., Carafa A., Lumini E., Bianciotto V., Bonfante P., Duckett J.G. (2007). Glomeromycotean associations in liverworts: A molecular cellular and taxonomic analysis. Am. J. Bot. 94: 1756–1777 [DOI] [PubMed] [Google Scholar]
- Liu J., Blaylock L., Endre G., Cho J., Town C.D., VandenBosch K., Harrison M.J. (2003). Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of the arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Blaylock L., Harrison M.J. (2004). cDNA arrays as tools to identify mycorrhiza-regulated genes: identification of mycorrhiza-induced genes that encode or generate signaling molecules implicated in the control of root growth. Can. J. Bot. 82: 1177–1185 [Google Scholar]
- Liu J., Maldonado-Mendoza I., Lopez-Meyer M., Cheung F., Town C.D., Harrison M.J. (2007). The arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 50: 529–544 [DOI] [PubMed] [Google Scholar]
- Lopez-Meyer M., Harrison M.J. (2006). An experimental system to synchronize the early events of development of the arbuscular mycorrhizal symbiosis. Molecular Plant-Microbe Interactions, Vol. 5, Federico Sánchez C.Q., López-Lara I.M., Geiger O., (St. Paul, MN: International Society for Molecular Plant-Microbe Interactions; ), pp. 546–551 [Google Scholar]
- Maeda D., Ashida K., Iguchi K., Chechetka S., Hijikata A., Okusako Y., Deguchi Y., Izui K., Hata S. (2006). Knockdown of an arbuscular mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol. 47: 807–817 [DOI] [PubMed] [Google Scholar]
- Martinoia E., Klein M., Geisler M., Bovet L., Forestier C., Kolukisaoglu U., Muller-Rober B., Schulz B. (2002). Multifunctionality of plant ABC transporters - More than just detoxifiers. Planta 214: 345–355 [DOI] [PubMed] [Google Scholar]
- McGonigle T.P., Miller M.H., Evans D.G., Fairchild G.L., Swan J.A. (1990). A new method that gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Mentewab A., Stewart C.N. (2005). Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat. Biotechnol. 23: 1177–1180 [DOI] [PubMed] [Google Scholar]
- Messinese E., Mun J.H., Yeun L.H., Jayaraman D., Rouge P., Barre A., Lougnon G., Schornack S., Bono J.J., Cook D.R., Ane J.M. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Navazio L., Moscatiello R., Genre A., Novero M., Baldan B., Bonfante P., Mariani P. (2007). A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol. 144: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Paszkowski U., Jakovleva L., Boller T. (2006). Maize mutants affected at distinct stages of the arbuscular mycorrhizal symbiosis. Plant J. 47: 165–173 [DOI] [PubMed] [Google Scholar]
- Paszkowski U., Kroken S., Roux C., Briggs S.P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F., Valencia A. (2008). Protein co-evolution, co-adaptation and interactions. EMBO J. 27: 2648–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin J.A., Zheng H.Q., Balakshin L.J., Goodman I.P., Western T.L., Jetter R., Kunst L., Samuels A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Pumplin N., Harrison M.J. (2009). Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 151: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Mondo S.J., Topp S., Starker C.G., Gantt J.S., Harrison M.J. (2009). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61: 482–494 [DOI] [PubMed] [Google Scholar]
- Rea P.A. (2007). Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Rees D.C., Johnson E., Lewinson O. (2009). ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 10: 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy W., Taylor T.N., Hass H., Kerp H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G., Langmann T., Heimerl S. (2001). Role of ABCG1 and other ABCG family members in lipid metabolism. J. Lipid Res. 42: 1513–1520 [PubMed] [Google Scholar]
- Schulz B., Kolukisaglu H.U. (2006). Genomics of plant ABC transporters: The alphabet of photosynthetic life forms or just holes in membranes? FEBS Lett. 580: 1010–1016 [DOI] [PubMed] [Google Scholar]
- Sekhara R., Schorderet M., Feller U., Reinhardt D. (2007). A petunia mutant affected in intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi. Plant J. 51: 739–750 [DOI] [PubMed] [Google Scholar]
- Smith S.E., Dickson S., Smith F.A. (2001). Nutrient transfer in arbuscular mycorrhizas: How are fungal and plant processes integrated? Aust. J. Plant Physiol. 28: 683–694 [Google Scholar]
- Smith S.E., Read D.J. (2008). Mycorrhizal Symbiosis. (San Diego, CA: Academic Press; ). [Google Scholar]
- St-Arnaud M., Hamel C., Vimard B., Caron M., Fortin J.A. (1996). Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol. Res. 100: 328–332 [Google Scholar]
- Stein M., Dittgen J., Sanchez-Rodriguez C., Hou B.H., Molina A., Schulze-Lefert P., Lipka V., Somerville S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szcyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Sugiyama A., Shitan N., Sato S., Nakamura Y., Tabata S., Yazaki K. (2006). Genome-wide analysis of ATP-binding cassette (ABC) proteins in a model legume plant, Lotus japonicus: Comparison with Arabidopsis ABC protein family. DNA Res. 13: 205–228 [DOI] [PubMed] [Google Scholar]
- Takeda N., Sato S., Asamizu E., Tabata S., Parniske M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58: 766–777 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- van den Brule S., Muller A., Fleming A.J., Smart C.C. (2002). The ABC transporter SpTUR2 confers resistance to the antifungal diterpene sclareol. Plant J. 30: 649–662 [DOI] [PubMed] [Google Scholar]
- van der Heijden M.G.A., Klironomos J.N., Ursic M., Moutoglis P., Streitwolf-Engel R., Boller T., Wiemken A., Sanders I.R. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72 [Google Scholar]
- Verrier P.J., et al. (2008). Plant ABC proteins - A unified nomenclature and updated inventory. Trends Plant Sci. 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Wang B., Qiu Y.L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16: 299–363 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Wittenburg H., Carey M.C. (2002). Biliary cholesterol secretion by the twinned sterol half-transporters ABCG5 and ABCG8. J. Clin. Invest. 110: 605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K., Shitan N., Sugiyama A., Takanashi K. (2009). Cell and molecular biology of ATP-binding cassette proteins in plants. Int. Rev. Cell Mol. Biol. 276: 263–299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.