This work describes a high-throughput screen for mutants affected in their ability to release hydrogen cyanide using the model legume Lotus japonicus. Mutants revealed unexpected specificity in the enzymes of both synthesis and degradation of cyanogenic glucosides and closely related compounds. This may offer flexibility in the production of deterrents to herbivores by this species.
Abstract
Cyanogenesis, the release of hydrogen cyanide from damaged plant tissues, involves the enzymatic degradation of amino acid–derived cyanogenic glucosides (α-hydroxynitrile glucosides) by specific β-glucosidases. Release of cyanide functions as a defense mechanism against generalist herbivores. We developed a high-throughput screening method and used it to identify cyanogenesis deficient (cyd) mutants in the model legume Lotus japonicus. Mutants in both biosynthesis and catabolism of cyanogenic glucosides were isolated and classified following metabolic profiling of cyanogenic glucoside content. L. japonicus produces two cyanogenic glucosides: linamarin (derived from Val) and lotaustralin (derived from Ile). Their biosynthesis may involve the same set of enzymes for both amino acid precursors. However, in one class of mutants, accumulation of lotaustralin and linamarin was uncoupled. Catabolic mutants could be placed in two complementation groups, one of which, cyd2, encoded the β-glucosidase BGD2. Despite the identification of nine independent cyd2 alleles, no mutants involving the gene encoding a closely related β-glucosidase, BGD4, were identified. This indicated that BGD4 plays no role in cyanogenesis in L. japonicus in vivo. Biochemical analysis confirmed that BGD4 cannot hydrolyze linamarin or lotaustralin and in L. japonicus is specific for breakdown of related hydroxynitrile glucosides, such as rhodiocyanoside A. By contrast, BGD2 can hydrolyze both cyanogenic glucosides and rhodiocyanosides. Our genetic analysis demonstrated specificity in the catabolic pathways for hydroxynitrile glucosides and implied specificity in their biosynthetic pathways as well. In addition, it has provided important tools for elucidating and potentially modifying cyanogenesis pathways in plants.
INTRODUCTION
Cyanogenesis is the release of hydrogen cyanide (HCN) upon tissue damage. It functions as a defense mechanism against herbivores and is found in ferns, angiosperms, and gymnosperms (Hegnauer, 1977; Zagrobelny et al., 2008). Cyanogenesis usually results from the enzymatic degradation of cyanogenic glucosides, although the identity of cyanogenic glucosides varies between plants and depends on the amino acids used for their synthesis. Many crop species produce cyanogenic glucosides, including wheat (Triticum aestivum), maize (Zea mays), sorghum (Sorghum bicolor), and barley (Hordeum vulgare), and this trait may have contributed to their initial selection for domestication as it provides a degree of natural pest and herbivore resistance (Jones, 1998). However, cyanogenesis can be a significant social and economic problem and can limit the use of particular harvests for human or animal consumption. For example, cassava (Manihot esculenta) is a staple crop in many developing countries, especially in sub-Saharan Africa, but adequate food processing is required to remove its cyanogenic glucosides before human consumption. Many processed foods containing cassava have a final cyanide content well above the FAO/WHO's recommended safety limit of 10 ppm (Montagnac et al., 2009). This is particularly problematic during years with low rainfall when the cyanide content of cassava roots increases. Acute and chronic cyanide poisoning resulting in severe neurological diseases, including paralysis and visual impairment (Konzo disease), are problems in parts of tropical Africa, especially when dietary protein levels are low (Nhassico et al., 2008). Similarly, forage grasses and legumes are also often cyanogenic and require careful agricultural management to avoid toxicity to cattle (Wheeler and Mulcahy, 1989). Consequently, there has been considerable interest in attempting to breed low or acyanogenic cultivars of forage crops.
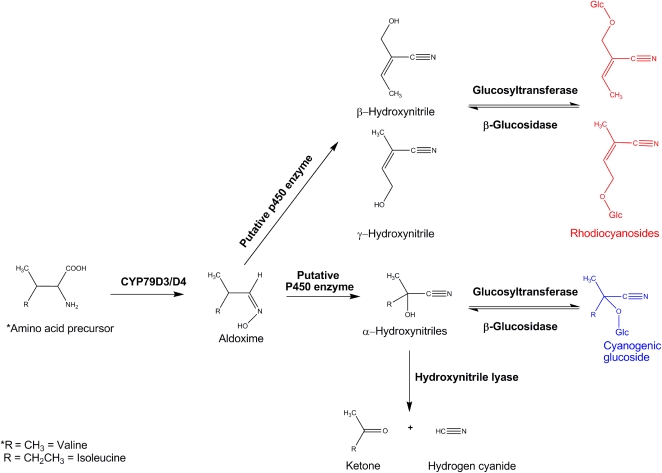
Current understanding of synthesis of cyanogenic glucosides (α-hydroxynitrile glucosides) is based largely on biochemical data from sorghum. The cyanogenic glucoside in sorghum, dhurrin, is derived from the aromatic amino acid Tyr. Synthesis of dhurrin requires two multifunctional cytochrome P450 enzymes, CYP79A1 and CYP71E1, and a UDPG-glucosyltransferase (UGT85B1) (Jones et al., 1999; Bak et al., 2006). Ectopic expression of the genes encoding these enzymes in Arabidopsis thaliana, which does not produce cyanogenic glucosides, resulted in dhurrin production (Tattersall et al., 2001). Members of the CYP79 family also catalyze the first step in cyanogenic glucoside production, the conversion of the amino acid precursor to an oxime intermediate, in other cyanogenic plants despite having different amino acids that serve as precursors for cyanogenic glucosides in these species (Figure 1). CYP79E1 and CYP79E2 are involved in the synthesis of triglochinin and taxiphyllin (both derived from Tyr) in the sea arrow grass (Triglochin maritime; Nielsen and Møller, 2000). CYP79D1 and CYP79D2 are active in the synthesis of linamarin and lotaustralin (derived from the aliphatic amino acids Val and Ile, respectively) in cassava (Andersen et al., 2000). In the noncyanogenic plant, Arabidopsis, members of the CYP79 family catalyze the conversion of amino acids to aldoximes as intermediates in the biosynthesis of glucosinolates, and glucosinolate biosynthesis is thought to have evolved from cyanogenic glucoside biosynthesis (Bak et al., 2006; Halkier and Gershenzon, 2006).
Figure 1.
Biosynthesis and Catabolism of Cyanogenic Glucosides (α-Hydroxynitrile Glucosides) and Rhodiocyanosides in L. japonicus.
Cyanogenic glucosides are α-hydroxynitrile glucosides and release HCN on hydrolysis. Rhodiocyanosides are β- and γ-hydroxynitrile glucosides and consequently are noncyanogenic. The first step in biosynthesis is catalyzed by the cytochrome P450s CY79D3 and CYP79D4 (which are active in different parts of the plant), but gene products catalyzing other steps of synthesis have not been identified.
Cyanogenic glucosides and the corresponding β-glucosidases that hydrolyze them are stored in separate cellular compartments (Morant et al., 2008a, 2008b). Tissue damage results in the mixing of these components. The resulting aglycone may dissociate spontaneously or may require the activity of α-hydroxynitrilases to release HCN (Figure 1). Specific cyanogenic β-glucosidases have been identified from several plant species. Sorghum contains two isozymes of the cyanogenic β-glucosidase dhurrinase: Dhr1 and Dhr2 (Hösel et al., 1987). Both enzymes show high specificity for dhurrin but differ in their expression patterns and reactivity toward synthetic substrates (Hösel et al., 1987; Cicek and Esen, 1998). A single cyanogenic β-glucosidase called linamarase is present in white clover (Trifolium repens), and segregation of alleles of the gene encoding linamarase (Li) underpins some of the natural variation in cyanogenesis in this species (Dunn and Hughes, 1983; Oxtoby et al., 1991; Olsen et al., 2007).
Like cassava, the model legume Lotus japonicus produces linamarin and lotaustralin cyanogenic glucosides derived from Val and Ile, respectively. It also produces related γ- and β-hydroxynitrile glucosides, called rhodiocyanosides, which do not release cyanide upon hydrolysis (Forslund et al., 2004). Of the cyanogenic glucosides, lotaustralin is predominant, occurring at ∼10-fold higher levels in leaves than linamarin. Rhodiocyanoside A is the predominant, noncyanogenic hydroxynitrile glucoside and accumulates to levels similar to those of lotaustralin. Several enzymes involved in the metabolism of cyanogenic hydroxynitrile glucosides have been identified in L. japonicus (Figure 1). Two genes, CYP79D3 and CYP79D4, encode cytochrome P450 enzymes that catalyze the conversion of both Val and Ile to their corresponding aldoximes, the first intermediates in the biosynthesis of linamarin and lotaustralin, respectively (Forslund et al., 2004).
For the breakdown of cyanogenic glucosides in L. japonicus, a hydroxynitrile glucoside-cleaving β-glucosidase activity that prefers rhodiocyanosides and lotaustralin to linamarin as substrate has been purified from leaves (Morant et al., 2008a). Two β-glucosidases, BGD2 and BGD4, which hydrolyzed cyanogenic glucosides and rhodiocyanoside A following heterologous expression in Arabidopsis, were identified (Morant et al., 2008a).
To date, genetic analyses of cyanogenesis have focused mainly on natural variation in white clover, in which both cyanogenic and acyanogenic individuals may be found. It is thought that opposing selection pressures maintain this polymorphism; while cyanogenesis provides protection against generalist herbivores, it may reduce fitness in colder climates, under drought conditions, or as a result of resource allocation trade-offs under normal conditions or at times of environmental stress (Daday, 1965; Hayden and Parker, 2002; Olsen and Ungerer, 2008). Two genetic loci underlie the natural variation in cyanogenesis in white clover. The Ac locus determines the presence or absence of the cyanogenic glucosides, lotaustralin and linamarin, and encodes a cytochrome P450 of the CYP79D subfamily (Olsen et al., 2008). The Li locus encodes a β-glucosidase, linamarase, which hydrolyzes cyanogenic glucosides, resulting in the release of HCN. Acyanogenic clover accessions can lack functional genes at either or both loci (Olsen et al., 2007). Although studies of white clover have focused on the presence or absence of cyanogenesis, the existence of quantitative variation in cyanogenesis has also been reported (Hayden and Parker, 2002). European populations of Lotus corniculatus also show natural variation for cyanogenesis, with acyanogenic accessions lacking the cyanogenic glucosides, the β-glucosidase activity, or both (Armstrong et al., 1913; Jones, 1977).
We have undertaken a targeted genetic screen to identify cyanogenesis-deficient mutants in L. japonicus. A range of mutants affecting both the biosynthesis and the breakdown of cyanogenic glucosides was obtained, providing unique tools for the comprehensive genetic dissection of the cyanogenesis process. We describe the method for high-throughput screening for cyanogenesis-deficient mutants, a method that could be adapted easily for screening for acyanogenic mutants of crops. Our screen identified novel genetic loci involved in both synthesis and breakdown of cyanogenic glucosides. A detailed characterization of one group of breakdown mutants demonstrated that, in contrast with previous reports, the only β-glucosidase required for cyanogenesis in leaves of L. japonicus is BGD2. The closely related β-glucosidase, BGD4, is specific for hydrolysis of rhodiocyanosides.
RESULTS
Isolation of cyanogenesis deficient Mutants in L. japonicus
Despite the existing biochemical understanding of the synthesis and breakdown of cyanogenic glucosides, a genetic dissection in a single species is important for several reasons: to identify any additional steps in both the biosynthetic and breakdown pathways, to identify regulators of cyanogenesis, and to establish the relationship between production of cyanogenic and noncyanogenic hydroxynitrile glucosides. In addition, the development of a robust forward-screening method will establish the feasibility of screening for acyanogenic mutants of crops such as sorghum. Among the resources available for studying L. japonicus are many wild accessions collected from the various regions of Japan (Legume Base, Miyazaki University, Japan). We grew several individuals of each of 80 accessions and found all of them to contain cyanogenic glucosides, although metabolic profiling did identify accessions that lacked rhodiocyanosides, which are related noncyanogenic hydroxynitrile glucosides (Bjarnholt et al., 2008). Assays of cyanogenesis revealed all 80 accessions to be cyanogenic, although there was variation in the amounts of cyanide produced over a standardized period of time. Consequently, we decided to undertake a forward genetic screen for mutants impaired in cyanogenesis using the accession Miyakojima MG-20, a standard experimental line that has been used for determining the genome sequence of L. japonicus (Sato et al., 2008).
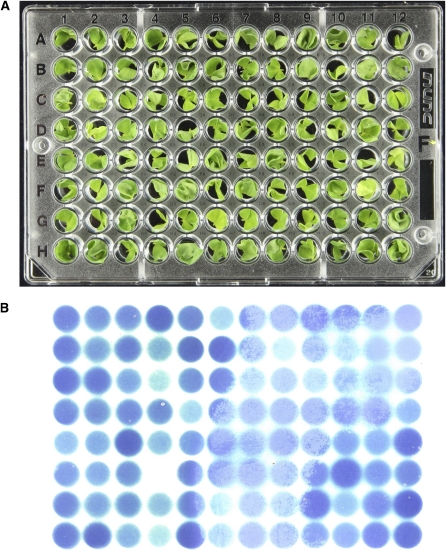
To enable processing of a large number of samples, we developed a microtiter plate–based screen for cyanide release using the cyanide-sensitive paper developed by Feigl and Anger (1966). Whatman 3MM paper is pretreated with a solution of tetra base (4,4’-methylenebis-(N,N-dimethylaniline)) and copper(II) ethylacetoacetate dissolved in chloroform. The dried paper turns blue following oxidation of the tetra base when it comes in contact with the HCN gas that is produced. This method allows easy identification of mutants in a high-throughput screen and has adequate sensitivity and reliability to identify both completely acyanogenic individuals and those with reduced cyanogenesis. A total of 7000 seeds from the MG-20 accession were mutagenized using 1% ethyl methanesulfonate (EMS). M2 seeds of 3652 M1 families were sown in 286-well trays, each row containing seeds from a single M1 family. Six weeks after germination, 12 M2 plants from each family were tested for cyanogenesis by detaching the apical trifoliate leaves and placing them in the wells of a 96-well microtiter plate (Figure 2A). The first unfolded trifoliate leaf is the most cyanogenic in L. japonicus and gave the best reproducibility in scoring individuals. Leaves were frozen at −80°C overnight and then thawed at room temperature. Cyanide production was assayed following this single freeze-thaw cycle by immediately placing a piece of Feigl-Anger paper between the lid and the microtiter plate. A clear blue color developed in under 3 h over each well containing a cyanogenic leaf (Figure 2B). The color intensity was indicative of the amount of HCN produced (see Supplemental Figure 1 online).
Figure 2.
High-Throughput Screening for cyd Mutants in L. japonicus Using Feigl-Anger Paper.
(A) Microtitre plate containing expanded apical leaves from individual Lotus plants.
(B) Feigl-Anger paper exposed to the plate shown in (A) for 2 h following a single freeze-thaw cycle. The blue spots of the image are shown as a mirror image so that they correspond directly with the leaves in the microtiter plate. Well F4 contains a leaf from a cyd1 mutant plant, resulting in the absence of a blue spot.
Of ∼44,000 M2 plants tested, 226 were scored initially as showing no or reduced cyanogenesis. These plants were subsequently reevaluated by assaying a second leaf. This resulted in the selection of 100 plants, representing 69 distinct M2 families, which were repotted to allow further growth and seed production. After ∼1 month, newly developed leaves of the repotted plants were tested twice more. Just one sibling was chosen from families from which multiple M2 individuals were selected, as they most likely represented the same mutational event. This resulted in the final selection of 44 independent mutant lines that consistently showed a cyanogenesis-deficient (cyd) phenotype. The level of cyanogenesis observed in the mutants on the basis of the Feigl-Anger paper varied from reduced color to no detectable color formation. Further biochemical and genetic analysis of these mutants allowed their classification into distinct phenotypic groups on the basis of their metabolite profiles.
Characterization and Classification of cyd Mutants by Metabolic Profiling
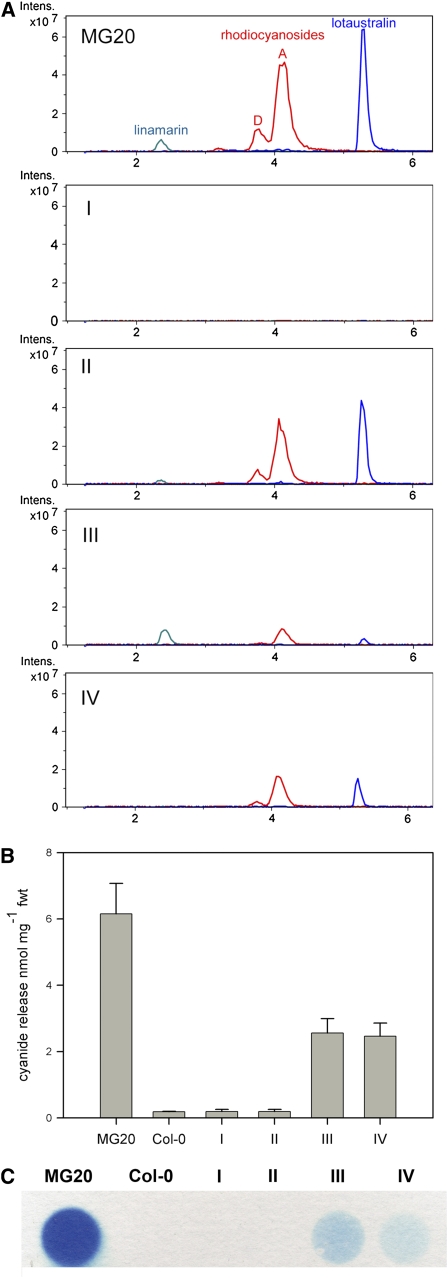
As the screening method selected for loss of cyanide production, mutants that affected either the production or the breakdown of cyanogenic glucosides were expected from the screen. These were distinguished by metabolic profiling; in the first category, the levels of cyanogenic glucosides should be reduced, while in the second category, cyanogenic glucoside content should be unaffected. Extracts were made from leaves of plants that were approximately 6 weeks old by boiling them in 85% methanol. The cyanogenic glucoside/rhodiocyanoside content of leaf extracts from each mutant was determined by liquid chromatography–mass spectrometry (LC-MS). Based on the metabolic profiles and the level of cyanogenesis, we were able to place the mutants into four general classes (Table 1). Representative LC-MS chromatograms for each mutant class are shown in comparison to the MG-20 parental line in Figure 3A.
Table 1.
Classification of the L. japonicus Mutants Obtained in a Screen for Cyanogenesis Deficiency
| Mutant Class | Phenotype | Number of Families | Locus |
| Class I | Absence of CG and Rho; acyanogenic | 1 | cyd1 |
| Class II | Normal levels of CG and Rho; catabolic mutants | 10 | cyd2, cyd3 |
| Class III | Uncoupled biosynthesis, reduction in Ile-derived compounds | 7 | |
| Class IV | Reduction in CG and Rho | 26 |
This classification is based on both the level of cyanogenesis determined by Feigl-Anger paper and the LC-MS profiles. CG, cyanogenic glucosides; Rho, rhodiocyanosides.
Figure 3.
Phenotypic Classification of cyd Mutants.
(A) Relative intensities of representative extracted ion chromatograms following LC-MS of MG-20 and selected mutant lines representative of the different phenotypic classes (I to IV). Extracted ion peaks are for sodium adducts with the following molecular masses: linamarin, 270; lotaustralin, 284; and rhodiocyanosides, 282.
(B) Colorimetric determination of HCN production in single leaves of MG-20 and the representative mutant lines over 30 min. A leaf from Arabidopsis (Col-0) was included as a noncyanogenic control. Error bars indicate se of the mean of three biological replicates.
(C) Feigl-Anger paper exposure of single leaves from MG-20 and the representative mutant lines belonging to the different phenotypic classes. The second well contains an Arabidopsis leaf as a negative control.
Among the acyanogenic mutant lines, one mutant (assigned to phenotypic class I on the basis of its metabolite profile) contained neither the cyanogenic α-hydroxynitrile glucosides linamarin and lotaustralin nor the γ- and β-hydroxynitrile glucosides, rhodiocyanosides A and D (Table 1, class I; Figure 3A). This suggested that this mutant is defective in cyanogenic glucoside biosynthesis, possibly of the aldoxime precursor. Class II consisted of mutants that contained near-normal levels of cyanogenic glucosides and rhodiocyanosides, suggesting that their cyanogenesis-deficient phenotype was due to a defect in catabolism and a failure to release HCN (Table 1, class II; Figure 3A). This second class contained a total of 10 mutant lines, five of which were consistently scored as acyanogenic in the paper test. Members of phenotypic class III showed severely reduced levels of the Ile-derived lotaustralin but wild-type levels of the Val-derived linamarin (Table 1, class III; Figure 3A). Concomitant with the reduction in lotaustralin in class III mutants, there was also a reduction in the Ile-derived rhodiocyanosides A and D. Since this class of mutants showed differential effects on the Val- and Ile-derived cyanogenic glucosides, it was termed “uncoupled.” All seven mutant lines in this class showed a low but detectable level of cyanogenesis (Figure 3B). A fourth class of mutants showed reduced levels of cyanogenesis and contained reduced levels of all hydroxynitrile glucosides (Table 1, class IV; Figures 3A to 3C). This group is less well defined, and it is possible that weak alleles of the other classes are present within this group.
Leaves from mutants from these four phenotypic classes were reassessed for cyanogenesis by a colorimetric method that used a barbituric acid-pyridine reagent (Lambert et al., 1975). Leaves of the noncyanogenic plant Arabidopsis (Columbia-0) were used as negative control. This colorimetric assay involved homogenization of tissue in a buffer and freeze-thaw cycles, prior to adding the reagent (Halkier and Møller, 1989). This provided an independent quantitative analysis of cyanide production in the selected phenotypic classes compared with the MG-20 parental line (Figure 3B). There was a good correlation between the amount of staining observed using the Feigl-Anger paper and the quantitative data from the spectrophotometric assay (Figure 3C; see Supplemental Figure 1 online).
Complementation tests and genetic mapping using molecular markers confirmed that the single mutant assigned to class I was not allelic to mutants in class II. The single class I mutant was called cyd1 and segregated as a monogenic recessive.
The Catabolic Class of Mutants (Class II) Is Composed of Two Complementation Groups
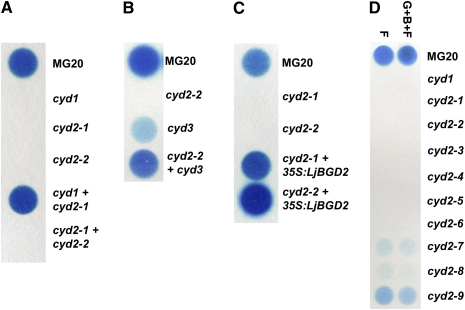
To characterize further the 10 mutant lines in class II, they were assigned to complementation groups. Biochemical complementation assays were performed by placing leaf tissue from two mutants in the same well of a microtiter plate and grinding in MES buffer, thereby mixing the homogenized tissue of the two mutants. The ability of the mix to release HCN was assayed using Feigl-Anger paper. The effectiveness of this assay is demonstrated in Figure 4A. Combining the cyd1 mutant, which completely lacks cyanogenic glucosides, with a class II mutant that contains wild-type cyanogenic glucoside levels restored cyanogenesis. When mutants in the same putative complementation group were mixed (for example, cyd2-1 and cyd2-2), no restoration of cyanogenesis was observed, as shown in Figure 4A. Testing combinations of the class II mutants for biochemical restoration of cyanogenesis suggested that they fell into just two groups. The largest biochemical complementation group was composed of nine putative mutations of a locus we named cyd2. One additional catabolic mutant was able to complement cyd2-2 biochemically and was named cyd3 (Figure 4B).
Figure 4.
Mutant Complementation Tests Using Biochemical Assays and Transgenic Plants.
(A) Biochemical complementation test of cyd1 and two cyd2 alleles.
(B) Biochemical complementation test of cyd2-2 with cyd3.
(C) Complementation of the cyd2-1 and cyd2-2 mutants transformed with a 35S:LjBGD2 construct.
(D) The level of cyanogenesis assayed in cyd1 and nine cyd2 alleles after tissue disruption using either a single freeze-thaw cycle only (F) or grinding in buffer followed by a freeze-thaw cycle (G+B+F).
Confirmation that cyd3 belongs to a complementation group distinct from cyd2 was obtained by genetic complementation tests. Cyanogenesis was fully restored in the F1 of reciprocal crosses between cyd2-4 and cyd3 and a cross between cyd2-5 and cyd3. F2 progeny from these crosses were tested for cyanogenesis using Feigl-Anger paper and segregated 1:1 for the acyanogenic compared with the wild-type phenotype (12 acyanogenic plants:14 cyanogenic plants in the F2 generation), demonstrating the involvement of two unlinked genetic loci. Crosses between cyd2 alleles showed no restoration of cyanogenesis in the F1 progeny.
Cyd2 Encodes BGD2
We focused our attention on identifying the gene lying at the cyd2 locus. The chromosomal location of the cyd2 locus was established by creating an F2 mapping population from a cross between cyd2-4 and L. japonicus cv Gifu B-129 (Handberg and Stougaard, 1992). F2 plants were assayed for cyanogenesis and showed the segregation of a single recessive locus. We used genetic markers polymorphic between MG-20 and Gifu (Legume Base, Miyazaki University, Japan) to show close linkage between the acyanogenic phenotype determined by cyd2-4 and marker TM0707 on chromosome 3 at 37.6 centimorgans (0 recombinants in 48 chromosomes). Morant et al. (2008a) identified two L. japonicus genes encoding enzymes with hydroxynitrile glucoside-cleaving activity by protein purification and amino acid sequencing. These genes, encoding β-glucosidases BGD2 and BGD4, were potential candidate genes for cyd2. While BGD4 is located on chromosome 5, BGD2 is located on chromosome 3 at ∼33.2 centimorgans on BAC TM0568. As this position corresponded well to the map location determined for cyd2, further evidence that BGD2 lies at the cyd2 locus was obtained by functional complementation of the cyd2-1 and cyd2-2 mutants following stable transformation of the BGD2 cDNA from MG-20 driven by the cauliflower mosaic virus (CaMV) 35S promoter. All kanamycin-resistant transformants showed restoration of cyanogenesis (Figure 4C).
Assessment of the Degree of Cyanogenesis in the cyd2 Alleles
The level of cyanogenesis in the first expanded apical leaf of each allele of cyd2 was determined using Feigl-Anger paper. Two methods of sample processing were used. The first method followed our procedure from the screen, in which leaf tissue was disrupted by a single freeze-thaw cycle in the absence of buffer (F in Figure 4D).The second method involved grinding in MES buffer followed by a freeze-thaw cycle (G+B+F in Figure 4D). Two leaves of each line were halved and divided between the two treatments, assuring that any variation observed between the treatments was not attributed to differences in the amount or quality of the plant material. Cyanogenesis was determined following exposure to Feigl-Anger paper for 2 h. Both processing methods produced similar results with slightly more HCN detected following a freeze-thaw cycle. In a series of repeated experiments, leaves of the mutants cyd2-1 to cyd2-5 were scored consistently as acyanogenic (Figure 4D). The cyd2-6 to cyd2-8 mutants showed weak or occasionally no detectable cyanogenesis, whereas the cyd2-9 mutant reproducibly showed a reduced level of cyanogenesis. We determined that the detection limit of the Feigl-Anger paper was 3 nmol HCN by measuring HCN release following acidification of KCN standards in 1 M NaOH (see Supplemental Figure 1A online). The more sensitive colorimetric assay also showed no cyanogenesis in the cyd2-1 to cyd2-5 mutants. Cyanogenesis for the cyd2-6 mutant was estimated at 7.3 nmol HCN leaf−1 h−1 (11.4 μg HCN g−1 fresh weight [FW]), close to the detection limit of the Feigl-Anger paper. The colorimetric assay estimated cyanogenesis in cyd2-9 as 36.8 nmol HCN leaf−1 h−1 (57 μg HCN g−1 FW) and in the MG-20 parental line as 231.4 nmol HCN leaf−1 h−1 (379 μg HCN g−1 FW). Consequently, cyd2-6, cyd2-7, cyd2-8, and cyd2-9 were assessed as weak alleles, whereas cyd2-1 through cyd2-5 represent complete blocks in HCN release from cyanogenic glucosides.
Characterization of the Mutations in the cyd2 Alleles
The BGD2 gene was sequenced in the nine cyd2 mutant alleles. Point mutations were observed in the BGD2 sequences for all cyd2 alleles, further verifying that BGD2 is the gene affected in the cyd2 mutants (Figure 5A). As expected for EMS-induced mutations, which result from the alkylation of guanine, all the point mutations identified in the BGD2 gene were G/C-to-A/T transitions. In cyd2-2, the guanine at the donor splice site of intron 11 was replaced by adenine. Quantitative RT-PCR failed to resolve any transcript for BGD2 in leaves (see Supplemental Figure 2 online). This suggested that the cyd2-2 mutation resulted in mis-splicing of the BGD2 transcript, causing it to be eliminated in leaves (probably through a process of nonsense-mediated decay). This allele could therefore be considered an RNA null and its acyanogenic phenotype indicative of complete loss of hydroxynitrile glucoside-cleaving β-glucosidase activity.
Figure 5.
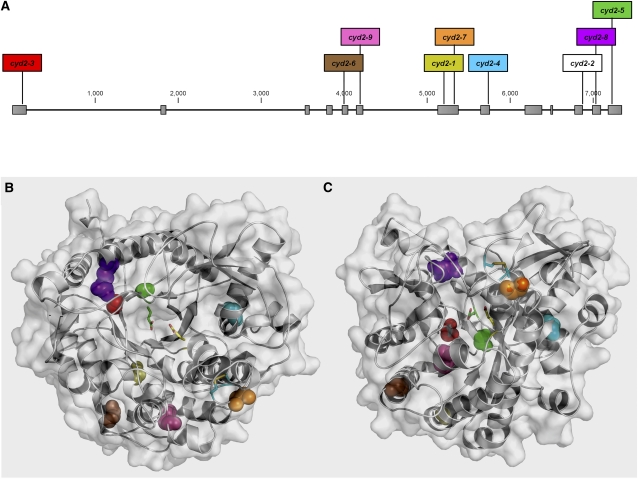
Location of Mutations in BGD2 Reducing or Eliminating Breakdown of Cyanogenic Glucosides in Leaves of L. japonicus.
(A) Schematic of BGD2 showing intron/exon structure (to scale) and the location of the cyd2 mutations. The allele names are colored to correspond to the altered residues shown in (B) and (C). cyd2-2 is shown in white because this mutation affects the splice site between exon 11 and intron 11.
(B) and (C) Space filling structural models of BGD2 showing the location of residues altered in cyd2 mutant alleles. Each BGD2 model is represented as a white ribbon and coated with a transparent surface to depict surface topology. The catalytic nucleophile E420 (green sticks) and the proton donor E208 (yellow sticks) are shown to depict the position of the active site, and the disulphide bridge between C227 and C335 (cyan sticks) is shown to reveal its proximity to G238. The residues of deleterious mutations are shown, color-coded as in (A). Top view is in (B) and side view in (C).
The other eight mutations in the different cyd2 alleles were substitutions of amino acids highly conserved in plant β-glucosidase enzymes; this conservation is graphically represented by sequence logos in Supplemental Figure 3 online. The sequences of a large number of plant β-glucosidases have been collected for multiple sequence alignment and graphically represented using WebLogo (www.p450.kvl.dk/BGD.shtml; Crooks et al., 2004; Morant et al., 2008a). Sequence logos indicate the degree of sequence conservation, and the frequency of a particular amino acid residue is represented by the height of its symbol (Schneider and Stephens, 1990). The observed amino acid substitutions in BGD2 were as follows: cyd2-1 caused an amino acid substitution at position 197, changing a Gly to a Glu (G197E), cyd2-3 caused a G51R amino acid substitution, cyd2-4 G305E, cyd2-5 G465E, cyd2-6 G138E, cyd2-7 G238D, cyd2-8 R442K, and cyd2-9 P166S (Figure 5A). Because four of these alleles were acyanogenic and phenotypically equivalent to cyd2-2 (Figure 4D), we conclude that these mutations eliminated the hydroxynitrile glucoside-cleaving β-glucosidase activity of BGD2 in vivo.
To understand why these mutations affected BGD2 enzyme activity, structural models of BGD2 were generated using two different modeling algorithms and the crystal structure of the cyanogenic β-glucosidase, linamarase (Tr CBG), from white clover, which has a TIM-barrel fold consisting of eight alternating β-strands and α-helices (β/α)8 (Barrett et al., 1995). The high degree of similarity between the template and BGD2 (70% identity) together with the convergence of the models (see Supplemental Figures 4A and 4B online) gave high credibility to the modeling, in particular regarding the positions of the various residues. The model of BGD2 from the Swiss-Model algorithm (see Methods) was used for further analysis. The model quality was further supported by an excellent overlay with the Tr CBG template, which showed the model and template were essentially identical with a root mean square deviation of 0.084 Å for 3144 atoms (0.19 Å for 1912 Cα atoms) included in the alignment. The only marked deviation in the conformation of the main chain was in the solvent accessible loop region A359-H364 in the BGD2 model, which is able to adopt a different conformation (see Supplemental Figures 4A and 4B online).
In general, the amino acids of BGD2 mutated in the cyd2 alleles were solvent inaccessible and not in direct proximity to the active site (Figures 5B and 5C), suggesting that the loss of activity of the mutated BGD2 versions results from structural changes affecting the catalytically competent conformation. In BGD2, both G51 and G465 are located within the hydrophobic core of the protein at a distance of ∼11 Å from the catalytic nucleophile. G51 is positioned on the first β-strand of the TIM-barrel fold, while G465 is on the adjacent last β-strand. The amino acid substitutions in cyd2-3 (G51R) and cyd2-5 (G465E) introduce large charged side chains in this tightly packed hydrophobic environment. The Gly to Glu substitutions in cyd2-1 (G197E) and cyd2-6 (G138E) cannot be accommodated in the available space and would cause major steric clashes most likely to elicit conformational destabilization. G305 is situated in the center of an α-helix, and its lack of a side chain allows the introduction of a sharp kink at this position (Figure 5C). Thus, the G305E substitution in cyd2-4 will distort this conformation and also result in repulsion with the nearby negative charges of D301 and D309 on the same α-helix. G238 is situated on a surface-accessible loop at a distance of 7 Å to the disulphide bridge between C227 and C235 (Figures 5B and 5C). The G238D substitution in cyd2-7 may result not only in the loss of the disulphide bridge but also in disruption of the salt bridge between R266 and E243, destabilizing this region close to the entry position of the active site. R442 mediates an ionic network interaction with E425 and D439, and although the R442K substitution in cyd2-8 is conservative, it is highly likely to result in the loss of this ionic network with significant destabilization as a consequence.
The cyd2-9 mutant was the most cyanogenic of the nine cyd2 alleles (Figure 4D). P166 is tightly packed in a hydrophobic pocket and makes van der Waals interactions with W121 and P122 on a different part of the structure as well as with P168 and L169 on the α-helix following P166. The cis conformation of Pro side chains generally results in significant rigidification of the polypeptide chain. While the P166S substitution in cyd2-9 can be accommodated within the available space, it is predicted to result in increased flexibility. Consistent with this idea is that cyd2-9 is the mutant that retains most BGD2 activity.
BGD4 Cannot Hydrolyze Cyanogenic Glucosides in L. japonicus
The recovery of nine alleles of cyd2/BGD2 deficient in cyanogenesis, but none affecting the gene encoding the closely related protein BGD4, suggested that BGD4 cannot hydrolyze cyanogenic glucosides in L. japonicus as suggested by Morant et al. (2008a). The cyd3 mutant line showed no nucleotide changes in BGD4, so Cyd3 represents a novel genetic locus in the catabolism of cyanogenic glucosides.
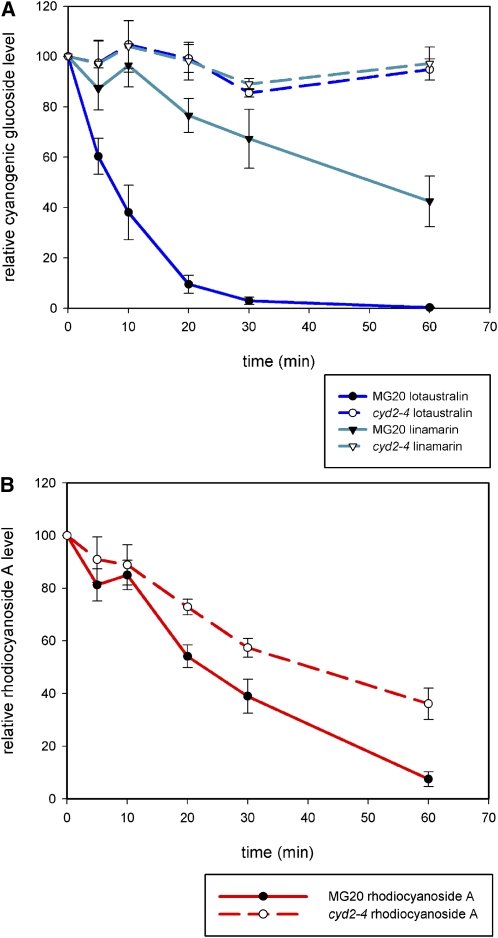
To confirm that BGD4 cannot hydrolyze linamarin and lotaustralin, we compared the breakdown of all hydroxynitrile glucosides in tissue homogenates of the leaves of the wild type, MG-20, and the cyd2-4 mutant line. A time course measuring the levels of hydroxynitrile glucosides in homogenized leaf tissue showed not only that both linamarin (added exogenously to enable accurate measurements) and lotaustralin were rapidly degraded in wild-type tissue, but also that their levels remained unchanged in homogenates of leaf tissue from the cyd2-4 mutant. Rhodiocyanoside A was degraded in the wild-type tissue and also in the cyd2-4 tissue homogenate, albeit at reduced rates (Figures 6A and 6B). In the absence of a standard for rhodiocyanoside D, it was not possible to determine the specificities of these activities for this minor component in leaves. These data suggested that cyd2 encodes the only β-glucosidase (BGD2) capable of cyanogenic glucoside breakdown in leaves of L. japonicus, whereas the breakdown of rhodiocyanoside A is, at least in part, conferred by a different β-glucosidase, specific for rhodiocyanosides.
Figure 6.
Hydroxynitrile Glucoside Hydrolyzing Activity in Leaves of the Wild Type (MG-20) and cyd2-4.
(A) Time course of catabolism of cyanogenic glucosides. Assays of leaf homogenates were spiked with purified linamarin to a final concentration of 200 μM to provide adequate levels to measure. Levels of hydroxynitrile glucosides were assessed by LC-MS and normalized to a spike of 4-methylthiobutyl glucosinolate (4-MTB). Data points were calculated as percentages of the initial levels of compounds detected. Error bars indicate se of the mean of three biological replicates.
(B) Time course of catabolism of rhodiocyanoside A in leaf homogenates. Levels of rhodiocyanoside A were measured and data points calculated as described in (A). Error bars indicate se of the mean of three biological replicates.
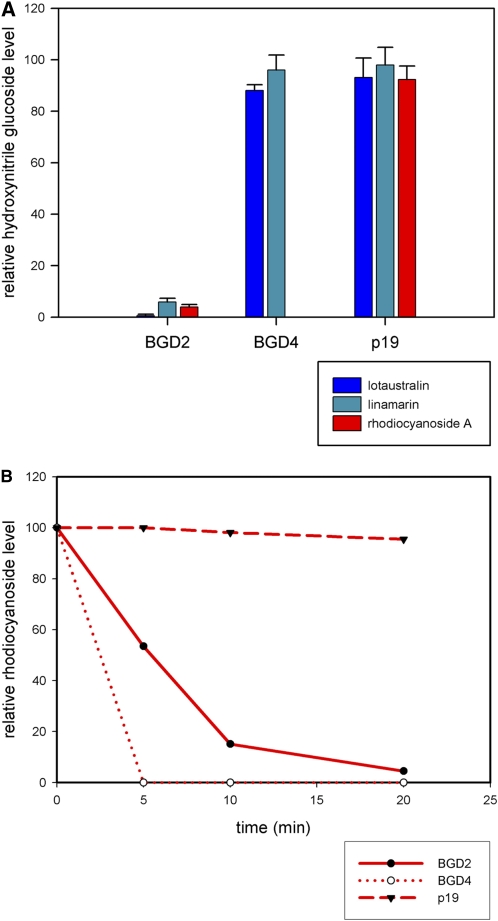
To investigate the likely substrate specificity of BGD2 and BGD4 further, cDNAs encoding the two β-glucosidases were expressed transiently in Nicotiana benthamiana by incorporating them into a vector driven by the CaMV 35S promoter. Since N. benthamiana makes no cyanogenic glucosides or related compounds such as glucosinolates, leaves of this species should provide a clean background for assaying the activity of the β-glucosidases. This was confirmed by assays of catabolic activity on uninoculated leaves or of leaves inoculated with the suppressor of gene silencing, p19 (Voinnet et al., 2003), using a range of possible glucoside substrates. Extracts of leaves inoculated with Agrobacterium tumifaciens carrying the vector for expression of BGD2 showed hydrolysis of cyanogenic glucosides (both linamarin and lotaustralin) and rhodiocyanoside A (Figure 7A). By contrast, extracts of leaves inoculated with Agrobacterium carrying the vector for expression of BGD4 showed no hydrolysis of linamarin or lotaustralin. The same extracts could hydrolyze rhodiocyanoside A efficiently, confirming the analysis of the hydrolytic activity in the leaves of the wild type and cyd2-4 mutants. In fact, the data from the MG-20 and cyd2-4 extracts suggested that BGD4 was more active in hydrolyzing rhodiocyanoside A than was BGD2 (Figures 6 and 7). This conclusion was supported by time-course analysis of BGD2 and BGD4 activities (produced transiently in N. benthamiana) on rhodiocyanoside A (Figure 7B). These data confirmed that BGD2 specifically hydrolyzes cyanogenic glucosides and can also hydrolyze rhodiocyanoside A, whereas BGD4 is specific for breakdown of rhodiocyanoside A. These substrate specificities of the enzymes involved in the breakdown pathways for hydroxynitrile glucosides in leaves of L. japonicus were revealed as a consequence of our genetic dissection of cyanogenesis.
Figure 7.
Activity of BGD2 and BGD4 Expressed Transiently in N. benthamiana on a Range of Different Potential Substrates.
(A) Relative activity of BGD2 and BGD4 compared with the control (empty vector plus p19 suppressor of gene silencing alone) shown by the relative amounts of different hydroxynitrile glucosides remaining after a 30-min incubation. Leaf disc extracts were spiked with hydroxynitrile glucosides to 200 μM and levels of compounds measured by LC-MS and normalized to a spike of 4MTB. Data are presented as percentages of the initial levels of compounds detected. Error bars indicate se of the mean of three biological replicates.
(B) Time courses of activity of BGD2, BGD4, and p19 control on rhodiocyanoside A, showing that BGD4 hydrolyses rhodiocyanoside A more rapidly than BGD2. Rhodiocyanoside A levels were measured and data points calculated as described in (A).
DISCUSSION
Cyanogenesis is a defense mechanism widely distributed in the plant kingdom and present in many major crop species. Our knowledge of cyanogenesis is limited to the biochemical identification and characterization of some key biosynthetic enzymes and to population studies in some cyanogenic species. Large variations in cyanogenic potential are observed in many plant species determined by genetic, environmental, and physiological factors that are only partially understood (Jones, 1998).
Breeding programs and transgenic approaches in several crops have been aimed at reducing or eliminating cyanogenesis, especially in tissues used for feed or food (Jørgensen et al., 2005). Initial biochemical approaches to investigate cyanogenesis have focused on sorghum and cassava due to the social and economic importance of these crops and the high levels of endogenous cyanogenesis in some cultivars (Hösel et al., 1987; Cicek and Esen, 1998; Jones et al., 1999; Jørgensen, et al., 2005; Bak, et al., 2006). However, these species are not suitable models for genetic approaches that aim to dissect the molecular mechanisms and regulatory pathways that control cyanogenic potential in plants. The legume L. japonicus was originally developed as a genetic model to study symbiotic nitrogen fixation (Handberg and Stougaard, 1992), and a range of molecular genetic tools is presently available for this species. Recent biochemical studies of cyanogenesis in L. japonicus have established this legume as a suitable system for the genetic approach presented here (Forslund et al., 2004). As it is closely related to the forage legume L. corniculatus, it is also a suitable model for plant–insect interaction studies, in particular with regards to specialized herbivores (Zagrobelny et al., 2007), and for testing methods for generating acyanogenic mutants for agricultural applications.
To enable the high-throughput analysis of the large number of plants required for a genetic screen, we developed a microtiter plate assay based on HCN detection by Feigl-Anger paper (Feigl and Anger, 1966). A single freeze-thaw cycle of leaf tissue without any tissue processing was sufficient and highly effective in detecting cyanogenesis. Cyanide detection with the Feigl-Anger paper correlated well with a colorimetric method of cyanide detection. Our results demonstrate the feasibility of isolating low and acyanogenic mutants by this method, which constitutes a relatively simple, high-throughput screening procedure that could be adapted for screening cyanogenic crops for acyanogenic mutants.
Further analysis of the selected mutant lines by LC-MS analysis of cyanogenic glucoside and rhodiocyanoside content allowed for their classification. A large number of mutants showed reduced cyanogenic glucoside content, and one of the mutants (class I: cyd1) completely lacked cyanogenic glucosides. These mutations may affect enzymes catalyzing various synthetic steps or their regulation by genetic, physiological, or environmental factors. The isolation of one mutant line completely lacking cyanogenic glucosides in leaves demonstrated the feasibility of this screening strategy for improvement of cyanogenic crops used for food or feed. The class III or “uncoupled” mutants showed a specific reduction in Ile-derived compounds but contained normal levels of the Val-derived cyanogenic glucoside, linamarin. These mutants question the view that a single metabolon (Olsen et al., 2008) or the same set of enzymes (Collinge and Hughes, 1984) are responsible for the synthesis of both forms of cyanogenic glucoside. Biosynthetic steps specific to the Ile-derived compounds may be defective in these mutants, or these mutations may affect genes involved specifically in the synthesis of Ile, although such mutants would probably have pleiotropic phenotypes, which were not observed in the uncoupled class of mutants. Because these mutants show reductions in both lotaustralin and the noncyanogenic rhodiocyanosides, they confirm the close relationship that must exist between the biosynthetic pathways for these compounds (Bjarnholt et al., 2008). Mutations selectively impacting the production of the noncyanogenic rhodiocyanosides but not the cyanogenic glucosides would not have been detected in this screen for cyanogenesis-deficient mutants. Interestingly, LC-MS screens of accessions of Lotus revealed natural variation that did uncouple cyanogenic glucoside and rhodiocyanoside production. The MG-74 accession lacked rhodiocyanosides A and D but contained both lotaustralin and linamarin (Bjarnholt et al., 2008). The loss of rhodiocyanoside production in MG-74 could be assigned to a single recessive trait, suggesting that, despite the likely sharing of some biosynthetic steps, at least one gene product separates the production of rhodiocyanosides from lotaustralin in Lotus. As linamarin is a relatively minor cyanogenic compound in L. japonicus, it is unlikely that our screen would have identified mutants affected specifically in linamarin levels while containing normal levels of lotaustralin.
Ten mutants contained wild-type levels of all hydroxynitrile glucosides but were scored as low or acyanogenic in the high-throughput screen. These mutants were likely to be defective in the catabolism of cyanogenic glucosides and fell into two complementation groups: cyd2 and cyd3. The cyd2 mutants were analyzed in detail. Two hydroxynitrile glucoside-cleaving β-glucosidases, BGD2 and BGD4, had been identified in Lotus by biochemical methods (Morant et al., 2008a). The genes encoding these enzymes were strong candidates for the cyd2 and cyd3 loci, and cyd2 mutants mapped closely to the gene encoding BGD2 on chromosome 3. All nine cyd2 alleles showed sequence changes in BGD2 consistent with EMS mutagenesis. Six of the eight mutations caused by amino acid changes in BGD2 involved substitution of conserved Gly residues by charged amino acids. Several factors may contribute to the strong bias for Gly substitutions. Gly residues represent the largest fraction of the amino acids that are highly conserved in β-glucosidase enzymes, and Gly codons are very suitable targets for EMS mutagenesis in part because each Gly codon has at least two guanine bases. The observed Gly substitutions all involve changing this small amino acid to a charged amino acid such as Glu and are likely to affect enzyme structure, stability, and consequently function (Perry et al., 2009). The modeling of the observed amino acid changes on the BGD2 protein structure also suggested that these changes would affect folding and stability of the enzyme, rather than directly affecting the active sites.
Somewhat surprisingly, the screen did not identify mutants in BGD4. Our data demonstrated that mutations in BGD2 are sufficient to eliminate cyanogenesis in leaves of Lotus and that BGD4 is unable to compensate for loss of BGD2 activity. As we identified nine independent mutants in the BGD2 gene, it is unlikely that mutants in BGD4 affecting cyanogenesis would have been missed. Given that BGD4 is expressed in leaves at the same time as BGD2 (Morant et al., 2008a), our genetic data suggested that BGD4 is not biochemically equivalent to BGD2 despite the close similarity in the primary amino acid sequences of the two proteins. Analysis of hydrolytic activity of wild-type and cyd2 mutant lines showed that the BGD2 gene encodes the only β-glucosidase with activity on cyanogenic glucosides in leaves of L. japonicus, although this enzyme also has some activity on rhodiocyanosides. Biochemical analysis of BGD4 showed that this β-glucosidase was unable to hydrolyze linamarin or lotaustralin but, by contrast, was able to degrade rhodiocyanoside A efficiently. Subsequent to our undertaking this research, the Arabidopsis lines expressing BGD2 and BGD4, used by Morant et al. (2008a), were reexamined, and both were found to be expressing BGD2. Consequently, the specificities reported for BGD4 by Morant et al. (2008a) are incorrect, as confirmed by our genetic analysis and by protein expression in N. benthamiana.
The existence of two very closely related β-glucosidases with only one involved in cyanogenesis has also been suggested in white clover (Olsen et al., 2007). The Li gene in clover encodes linamarase, the β-glucosidase responsible for the breakdown of cyanogenic glucosides. Most, or all, of the Li gene is absent from the genomes of acyanogenic clover accessions unable to breakdown cyanogenic glucosides. A closely related β-glucosidase gene, encoding a protein with ∼92% amino acid identity to linamarase, was consistently present in both cyanogenic and acyanogenic accessions. The biological role of this Li paralog is presently unknown, although its expression could not be detected in leaves (Olsen et al., 2007).
Our genetic dissection of cyanogenesis in L. japonicus has revealed that despite sharing some enzymic steps, both the synthesis of the different hydroxynitrile glucosides and their breakdown can be separated genetically. The class III mutants affect the accumulation of lotaustralin but not linamarin, showing that it is possible to uncouple the accumulation of these two cyanogenic glucosides, despite the suggestion that the same set of enzymes or a single metabolon (Collinge and Hughes, 1984; Olsen et al., 2008) are responsible for the synthesis of both forms. The natural variants that accumulate cyanogenic glucosides but no rhodiocyanosides (for example, MG-74; Bjarnholt et al., 2008) demonstrate that cyanogenic glucoside synthesis can be uncoupled from rhodiocyanoside synthesis. The specificity of the activities of BGD2 and BDG4 in the catabolism of hydroxynitrile glucosides shows that breakdown of rhodiocyanosides can be uncoupled from hydrolysis of cyanogenic glucosides. The possibility of uncoupling both synthesis and breakdown of the different types of hydroxynitrile glucosides offers flexibility in the accumulation and breakdown of these metabolites. The potential for metabolic flexibility means that different hydroxynitrile glucosides could serve different biological roles in plants, including acting as deterrents to herbivores in different cells of the plant or in response to different environmental signals or stresses.
METHODS
Plant Material
For the mutant screen, Lotus japonicus cv MG-20 (Legume Base, Miyazaki University, Japan) M2 seeds were sown in 286-well horticultural trays as 13 × 22 rows containing Levington F1 compost in a greenhouse during late summer without supplementary lighting (minimum day/night temperature, 22/18°C). Each 13-well row contained seeds from a single M2 family. Seeds were scarified and oversown and then thinned or transplanted to give a single plant per well. The terminal three leaflets of a single leaf from each M2 plant were harvested into 96-well plates that could be closed with a lid. Putative mutants were grown further in 7-cm pots of compost for further testing and seed collection under supplementary sodium lighting to provide an 18-h daylength.
For all other experiments, L. japonicus cv MG-20 and the cyd mutants derived from the screen were germinated from seed on filter paper and grown in soil in a greenhouse at 22°C. Nicotiana benthamiana plants were germinated from seed on filter paper and grown in soil in a greenhouse at 22°C.
Genetic Screen for Mutants in Cyanogenesis
Feigl-Anger paper for the detection of HCN was prepared by separately dissolving 5 g of copper ethylacetoacetate (Alfa Aesar) and 5 g of tetra base 4,4’-methylenebis (N,N-dimethylaniline) (Sigma-Aldrich) in 0.5 liters each and combining both solutions. Whatman 3MM paper cut to 8 × 12-cm size was wetted with the solution and dried. The paper was stored at 4°C until use. The first expanded apical leaf of each M2 plant was collected in 96-well microtiter plates that could be closed with a lid. The tissue was disrupted by a single freeze-thaw cycle and HCN production detected by overlaying the plate with a sheet of Feigl-Anger paper. Lids were weighed down because a tight fit between plate, paper, and lid was necessary for the development of discrete spots above each well. A permanent record of the detection paper was made by scanning the paper immediately after exposure because the blue color fades with time.
Complementation of cyd2 Mutants with 35S:LjBGD2
An expression construct of the BGD2 cDNA under the control of the CaMV 35S promoter/terminator elements was constructed by Gateway recombination into pJAM1502 (Luo et al., 2007). The coding region of BGD2 was amplified by PCR from L. japonicus MG-20 cDNA. The primers used contained attB1 and attB2 gateway cloning sites and were forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCACTCAACACGTTCTTG-3′, and reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAATATCTTTTAAGAAAGTTT-3′. The construct was transferred into Agrobacterium tumefaciens (AGL1) by electroporation. Transformation of L. japonicus was based on hypocotyl explants as described by Lombari et al. (2005).
Sequencing of BGD2 and BGD4 in cyd Mutants
Sections of the BGD2 gene were amplified from genomic DNA in five separate PCR reactions to provide complete coverage of the 13 exons and confirm the identity of the mutations in the cyd2 plant lines. Fragments were cloned into pGEM-T Easy (Promega) and the sequence verified in several clones of each mutant. Similarly, sections of both BGD2 and BGD4 genes were amplified and determined to be wild-type sequence in the cyd3 line.
Homology Modeling of Mutations Affecting BGD2
Homology models were generated using both the first approach mode on the Swiss Model server (http://swissmodel.expasy.org/; Arnold et al., 2006) and the HHpred server (homology detection and structure prediction by HMM-HMM comparison, http://toolkit.tuebingen.mpg.de/hhpred#) (Sali et al., 1995) using the crystal structure of the glycoside hydrolase family 1 β-glucosidase (Tr CBG; linamarase) from white clover ((Trifolium repens); Barrett et al., 1995) as a template. Molecular rendering and structural alignments and measurements were performed using Pymol v0.99 software (http://www.pymol.org/) and Swiss Pdb viewer 3.7 (http://www.expasy.org/spdbv/) (Guex and Peitsch, 1997).
Quantitative Real-Time PCR
Total RNA was prepared from 100 mg of plant tissue using an RNeasy plant mini kit (Qiagen) with on-column DNaseI digestion following the suppliers protocol. For cDNA synthesis, 1 to 2 μg of total RNA were reverse transcribed using 50 μM oligo (dT)20 and SuperScript III reverse transcriptase (Invitrogen) following the supplier's protocol. Transcript levels of BGD2 were measured by real-time PCR using a SYBR green-based method on a Rotor-Gene 6000 (version 1.7) instrument (Corbett Research). The PCR reactions (20 μL) contained 400 nM of each primer, 5 μL of cDNA (diluted 1:10 in water), and 1× DyNAmo Flash SYBR Green mix (Finnzymes). Thermal cycling conditions were 95°C for 7 min followed by 35 cycles of 95°C for 15 s, 55°C for 30 s, and 65°C for 30 s followed by a final extension at 65°C for 5 min and a melt cycle from 60 to 95°C. Primers for Lj Actin were forward, 5′-ATCATACCTTCTATAACGAGCTTC-3′, and reverse, 5′-GTGGCTGACACCATCACCAGAATC-3′. Primers for BGD2 were forward, 5′-ATGGTATATGGATCCACTGACGT-3′, and reverse, 5′-TATGGGATTCCCGTTACGCTCAC-3′. The primer sets gave single PCR products that were verified by melt curve analysis at the end of each PCR run. Relative transcript levels of cDNA templates were calculated by comparing threshold values to standard curves of a purified PCR fragment of the gene of interest and of the reference gene (Takos et al., 2006). Relative expression ± se was calculated using three technical replicates for each mutant line.
Colorimetric Determination of Cyanogenic Potential
The cyanogenic potential of Lotus plants was quantified based on the colorimetric method described by Lambert et al. (1975) as modified by Halkier and Møller (1989). Single apical leaves were weighed and then macerated in 200 μL of 20 mM MES, pH 6.0, and subjected to three freeze-thaw cycles using liquid nitrogen. The samples were incubated for 30 min at 30°C and then frozen in liquid nitrogen. While samples thawed at room temperature, 40 μL of 6 M NaOH was added. A 60-μL aliquot from each sample was transferred to a 96-well microtiter plate. Sequentially the following reagents were added: 12.5 μL of glacial acetic acid, 50 μL of N-chlorosuccinimide (1 mg mL−1) and succinimide (2.5 mg mL−1), and 50 μL of pyridine (30% v/v) in 467 mM barbituric acid. After a 5-min incubation at room temperature, wells were scanned between 450 and 700 nm with a peak reading made at 584 nm. Released cyanide was calculated against standards of KCN made in 1 M NaOH. Activity ± se was calculated using three biological replicates.
Chemicals
Rhodiocyanoside A and lotaustralin were purified by preparative HPLC (Bjarnholt et al., 2008). Linamarin was purchased from AG Scientific. The 4-methylthiobutyl glucosinolate (4MTB) was a gift from Barbara Halkier.
LC-MS Analysis
For hydroxynitrile glucoside extraction, the first expanded trifoliate leaf from the top was boiled with 300 μL 85% methanol (60 to 70°C) in a water bath for 5 min and cooled on ice. The extract was filtered through 45-μm Ultrafree-MC Durapore PVDF filters (Millipore) and stored in glass containers at −20°C prior to analysis.
Analytical LC-MS was performed using an Agilent 1100 Series LC (Agilent Technologies) coupled to a Bruker HCT-Ultra ion trap mass spectrometer (Bruker Daltonics). A Zorbax SB-C18 column (Agilent; 1.8 μM, 2.1 × 50 mm) was used at a flow rate of 0.2 mL min−1, and the oven temperature was maintained at 35°C. The mobile phases were as follows: A, water with 0.1% (v/v) HCOOH and 50 μM NaCl; and B, acetonitrile with 0.1% (v/v) HCOOH. The gradient program was as follows: 0 to 0.5 min, isocratic 2% B; 0.5 to 7.5 min, linear gradient 2 to 40% B; 7.5 to 8.5 min, linear gradient 40% to 90% B; 8.5 to 11.5 isocratic 90% B; 11.60 to 17 min, isocratic 2% B. The flow rate was increased to 0.3 mL min−1 in the interval 11.2 to 13.5 min. The mass spectrometer was run in positive electrospray mode.
Biochemical Complementation Assay
Putative mutant plants were classified into biochemical groups by macerating together single apical leaves from two different mutant plants in 200 μL of 20 mM MES, pH 6.5, in a 96-well microtiter plate. Cyanogenesis was detected by exposure of the plate to Feigl-Anger paper for 1 to 2 h. Biochemical complementation was assessed by comparison to cyanogenesis from single leaves from each putative mutant tested.
Assays of Hydrolysis of Hydroxynitrile Glucosides in Wild-Type (MG-20) and cyd2-4 Leaves
Apical leaves were ground in 300 μL of 20 mM MES pH 6.5. During the incubations, 25 μL samples were taken and hydrolysis was terminated by addition of 140 μL of methanol at different time points. Samples were analyzed for hydroxynitrile glucoside levels by LC-MS. Incubations were run at 30°C for a total of 1 h. Homogenates were spiked with exogenous linamarin (200 μM) to ensure adequate levels to record hydrolytic activity of this minor constituent of the hydroxynitrile glucoside profile. Incubations were spiked to 200 μM with 4MTB, which is resistant to hydrolysis in Lotus extracts, and was used to correct data for volume losses during the sample preparation for LC-MS.
Transient Expression of BGD2 and BGD4 in Leaves of N. benthamiana Plants
An expression construct for the BGD4 cDNA under the control of the CaMV 35S promoter/terminator in pJAM1502 (Luo et al., 2007) was constructed as described for the BGD2 cDNA expression vector. The primers used for BGD4 amplification from cDNA contained attB1 and attB2 Gateway cloning sites and were forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCACTCAACACGTTCTTA-3′, and reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAATATCTTTTGAGAAAGTTC-3′. Overnight cultures of A. tumefaciens (AGL1) containing expression constructs for BGD2 and BGD4 and the gene silencing inhibitor protein 19 (Voinnet et al., 2003) were grown. The cells were resuspended in 50 mL 10 mM MgCl2, 10 mM MES, and 100 μM acetosyringone and incubated for 4 h.
Leaves of 3-week old N. benthamiana were infiltrated with A. tumefaciens (AGL1) containing the expression constructs described above. Leaves were always coinfiltrated with the expression construct containing the p19 gene to prevent silencing of the transgenes. After 4 d, the leaf material was collected and used for quantification of the activities of BGD2 and BGD4. Leaf discs were cut from infiltrated leaves, ground in 20 mM MES, pH 6.5, and spiked with exogenous hydroxynitrile glucosides (200 μM). Hydroxynitrile glucosides remaining after specified incubation times were measured by LC-MS and compared with initial levels measured as described for the hydrolysis experiments in Lotus leaves. The tobacco extracts were spiked with 4MTB at 200 μM, which was resistant to hydrolysis, and used to normalize data for volume losses during the sample preparation for LC-MS.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Lj BGD4, ACD65509; Lj BGD2, ACD65510; and Tr CBG, CAA40057. Lj Actin primers were based on an EST sequence, BP036880, from L. corniculatus flower buds.
Author Contributions
A.T., D.L., D.S., and F.R. performed the molecular and biochemical analyses of the cyd mutant lines, L.M. undertook the cyd2 transgenic complementation assays, and A.T., F.R., D.L., and C.E.O. undertook metabolite analysis of hydroxynitrile glucosides. M.A.H. modeled the Lj BGD2 protein on Tr CBG and interpreted the likely effects of the various cyd2 mutations on protein organization. M.S.M. synthesized some of the hydroxynitrile glucosides used in this work. T.L.W. undertook the mutagenesis and coordinated the screening of the M2 generation, which was performed by T.L.W., F.R., A.T., L.M., and C.M. The original project was conceived and planned by F.R., C.M., and T.L.W. F.R., A.T., and C.M. wrote the manuscript with considerable help from all the other authors.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Sensitivity of HCN Detection between the Feigl-Anger Paper and the Colorimetric Assay.
Supplemental Figure 2. qRT-PCR Analysis of Transcript Levels of BGD2 in Leaves of cyd2 Mutants.
Supplemental Figure 3. Position and Identity of the Mutations in the BGD2 Gene for the Eight cyd2 Alleles Affecting the Amino Acid Sequence of the Protein.
Supplemental Figure 4. Superimposition of the Structure of Tr CBG from White Clover and Lj BGD2 from L. japonicus.
Acknowledgments
We would thank Jillian Perry, Steve Mackay, Tracey Welham, Iain McRobbie, and Serena Broughton for help with the mutagenesis and screening and Steen Malmmose, Morten Stephensen, Danny Shamoon, and John Page for Lotus plant maintenance. We thank Naomi Geshi and Casper Søgaard for growing N. benthamiana plants. This work was supported financially by the Danmarks Grundforskningsfond (Danish National Research Foundation) under the “Niels Bohr Visiting Professorship” program awarded to C.M. and Birger Lindberg Møller. D.L. was supported by a PhD studentship funded jointly by the Niels Bohr program and the Faculty of Life Science, Copenhagen University. T.L.W. and C.M. acknowledge financial support from the core strategic grant from the Biotechnology and Biological Sciences Research Council to the John Innes Centre.
References
- Andersen M.D., Busk P.K., Svendsen I., Møller B.L. (2000). Cytochrome P-450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. J. Biol. Chem. 275: 1966–1975 [DOI] [PubMed] [Google Scholar]
- Armstrong H.E., Frankland Armstrong E., Horton E. (1913). Herbage Studies. II.- Variation in Lotus corniculatus and Trifolium repens (cyanophoric plants). Proc. R. Soc. Lond. B. Biol. Sci. 86: 262–269 [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bak B., et al. (2006). Cyanogenic glycosides: A case study for evolution and application of cytochromes P450. Phytochem. Rev. 5: 309–329 [Google Scholar]
- Barrett T., Suresh C.G., Tolley S.P., Dodson E.J., Hughes M.A. (1995). The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 3: 951–960 [DOI] [PubMed] [Google Scholar]
- Bjarnholt N., Rook F., Motawia M.S., Cornett C., Jørgensen C., Olsen C.E., Jaroszewski J.W., Bak S., Møller B.L. (2008). Diversification of an ancient theme: Hydroxynitrile glucosides. Phytochemistry 69: 1507–1516 [DOI] [PubMed] [Google Scholar]
- Cicek M., Esen A. (1998). Structure and expression of a dhurrinase (β-glucosidase) from sorghum. Plant Physiol. 116: 1469–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge D.B., Hughes M.A. (1984). Evidence that linamarin and lotaustralin, the two cyanogenic glucosides of Trifolium repens L., are synthesized by a single set of microsomal enzymes controlled by the Ac/ac locus. Plant Sci. Lett. 34: 119–125 [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daday H. (1965). Gene frequencies in wild populations of Trifolium repens L. IV. Mechanism of natural selection. Heredity 20: 355–365 [Google Scholar]
- Dunn M.A., Hughes M.A. (1983). Biochemical-characterization of the Li locus in white clover(Trifolium repens L). Heredity 50: 111–30 [Google Scholar]
- Feigl F., Anger V. (1966). Replacement of benzidine by copper ethylacetoacetate and tetra base as spot-test reagent for hydrogen cyanide and cyanogen. Analyst (Lond.) 91: 282–284 [DOI] [PubMed] [Google Scholar]
- Forslund K., Morant M., Jørgensen B., Olsen C.E., Asamizu E., Sato S., Tabata S., Bak S. (2004). Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 135: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Halkier B.A., Møller B.L. (1989). Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L) Moench and partial-purification of the enzyme-system involved. Plant Physiol. 90: 1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg K., Stougaard J. (1992). Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 2: 487–496 [Google Scholar]
- Hayden K.J., Parker I.M. (2002). Plasticity in cyanogenesis of Trifolium repens L.: Inducibility, fitness costs and variable expression. Evol. Ecol. Res. 4: 155–168 [Google Scholar]
- Hegnauer R. (1977). Cyanogenic compounds as systematic markers in Tracheophyta. Plant Syst. Evol. (suppl. 1): 191–209 [Google Scholar]
- Hösel W., Tober I., Eklund S.H., Conn E.E. (1987). Characterization of β-glucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench seedlings. Arch. Biochem. Biophys. 252: 152–162 [DOI] [PubMed] [Google Scholar]
- Jones D.A. (1977). On the polymorphism of cyanogenesis in Lotus corniculatus L. VII. The distribution of the cyanogenic form in Western Europe. Heredity 39: 27–44 [Google Scholar]
- Jones D.A. (1998). Why are so many food plants cyanogenic? Phytochemistry 47: 155–162 [DOI] [PubMed] [Google Scholar]
- Jones P.R., Møller B.L., Høj P.B. (1999). The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor. J. Biol. Chem. 274: 35483–35491 [DOI] [PubMed] [Google Scholar]
- Jørgensen K., Bak S., Busk P.K., Sørensen C., Olsen C.E., Puonti-Kaerlas J., Møller B.L. (2005). Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol. 139: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.L., Ramasamy J., Paukstelis J.V. (1975). Stable reagents for the colorimetric determination of cyanide by modified König reactions. Anal. Chem. 47: 916–918 [Google Scholar]
- Lombari P., Ercolano E., El Alaoui H., Chiurazzi M. (2005). Agrobacterium-mediated in vitro transformation. Lotus japonicus Handbook, A. Marquez J., (Dordrecht, The Netherlands: Springer; ), pp. 251–259 [Google Scholar]
- Luo J., et al. (2007). Convergent evolution in the BAHD family of acyl transferases identification and characterisation of anthocyanin acyl transferases from Arabidopsis thaliana Plant J. 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Montagnac J.A., Davis C.R., Tanumihardjo S.A. (2009). Processing techniques to reduce toxicity and antinutrients of cassava for use as a staple food. Compr. Rev. Food Sci. Food. Saf. 8: 17–27 [DOI] [PubMed] [Google Scholar]
- Morant A.V., Bjarnholt N., Kragh M.E., Kjærgaard C.H., Jørgensen K., Paquette S.M., Piotrowski M., Imberty A., Olsen C.E., Møller C.E., Møller B.L., Bak S. (2008a). The β-glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus. Plant Physiol. 147: 1072–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant A.V., Jørgensen K., Jørgensen C., Paquette S.M., Sánchez-Pérez R., Møller B.L., Bak S. (2008b). β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Nhassico D., Muquingue H., Cliff J., Cumbana A., Bradbury J.H. (2008). Rising African cassava production, diseases due to high cyanide intake and control measures. J. Sci. Food Agric. 88: 2043–2049 [Google Scholar]
- Nielsen J.S., Møller B.L. (2000). Cloning and expression of cytochrome P450 enzymes catalyzing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of cyanogenic glucosides in Triglochin maritime. Plant Physiol. 122: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K.M., Hsu S.-C., Small L.L. (2008). Evidence on the molecular basis of the Ac/ac adaptive cyanogenesis polymorphism in white clover (Trifolium repens L.). Genetics 179: 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K.M., Sutherland B.L., Small L.L. (2007). Molecular evolution of the Li/li chemical defense polymorphism in white clover (Trifolium repens L.). Mol. Ecol. 16: 4180–4193 [DOI] [PubMed] [Google Scholar]
- Olsen K.M., Ungerer M.C. (2008). Freezing tolerance and cyanogenesis in white clover (Trifolium repens L. Fabaceae). Int. J. Plant Sci. 169: 1141–1147 [Google Scholar]
- Oxtoby E., Dunn M.A., Pancoro A., Hughes M.A. (1991). Nucleotide and derived amino acid sequence of the cyanogenic β-glucosidase (linamarase) from white clover (Trifolium repens L.). Plant Mol. Biol. 17: 209–219 [DOI] [PubMed] [Google Scholar]
- Perry J., Welham T., Brachmann A., Binder A., Charpentier M., Groth M., Haage K., Markmann K., Wang T.L., Parniske M. (2009). TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective EMS alleles towards glycine replacements. Plant Physiol. 151: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995). Evaluation of comparative protein modelling by MODELLER. Proteins 23: 318–326 [DOI] [PubMed] [Google Scholar]
- Sato S., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990). Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A.M., Ubi E.B., Robinson S.P., Walker A.R. (2006). Condensed tannin biosynthesis genes are regulated separately from other flavonoid biosynthesis genes in apple fruit skin. Plant Sci. 170: 487–499 [Google Scholar]
- Tattersall D.B., Bak S., Jones P.R., Olsen C.E., Nielsen J.K., Hansen M.L., Høj P.B., Møller B.L. (2001). Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293: 1826–1828 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wheeler J.L., Mulcahy C. (1989). Consequences for animal production of cyanogenesis in sorghum forage and hay – A review. Trop. Grassl. 23: 193–202 [Google Scholar]
- Zagrobelny M., Bak S., Ekstrøm C.T., Olsen C.E., Møller B.L. (2007). The cyanogenic glucoside composition of Zygaena filipendulae (Lepidoptera: Zygaenidae) as effected by feeding on wild-type and transgenic Lotus populations with variable cyanogenic glucoside profiles. Insect Biochem. Mol. Biol. 37: 10–18 [DOI] [PubMed] [Google Scholar]
- Zagrobelny M., Bak S., Møller B.L. (2008). Cyanogenesis in plants and arthropods. Phytochemistry 69: 1457–1468 [DOI] [PubMed] [Google Scholar]