This study demonstrates that an RNA-dependent RNA polymerase involved in trans-acting small interfering RNA production plays a role in establishing adaxial-abaxial polarity in rice floral organs. A model is presented for anther patterning in rice.
Abstract
Establishment of adaxial-abaxial polarity is essential for lateral organ development. The mechanisms underlying the polarity establishment in the stamen remain unclear, whereas those in the leaf are well understood. Here, we investigated a rod-like lemma (rol) mutant of rice (Oryza sativa), in which the development of the stamen and lemma is severely compromised. We found that the rod-like structure of the lemma and disturbed anther patterning resulted from defects in the regulation of adaxial-abaxial polarity. Gene isolation indicated that the rol phenotype was caused by a weak mutation in SHOOTLESS2 (SHL2), which encodes an RNA-dependent RNA polymerase and functions in trans-acting small interfering RNA (ta-siRNA) production. Thus, ta-siRNA likely plays an important role in regulating the adaxial-abaxial polarity of floral organs in rice. Furthermore, we found that the spatial expression patterns of marker genes for adaxial-abaxial polarity are rearranged during anther development in the wild type. After this rearrangement, a newly formed polarity is likely to be established in a new developmental unit, the theca primordium. This idea is supported by observations of abnormal stamen development in the shl2-rol mutant. By contrast, the stamen filament is likely formed by abaxialization. Thus, a unique regulatory mechanism may be involved in regulating adaxial-abaxial polarity in stamen development.
INTRODUCTION
Lateral organs, such as leaves and floral organs, develop from the flank of the meristem. Establishment of adaxial-abaxial polarity is essential for the proper development of the lateral organs in plants (Husbands et al., 2009). In Arabidopsis thaliana, HOMEODOMAIN-LEUCINE ZIPPERIII (HD-ZIPIII) genes, such as PHABULOSA (PHB), are required for specification of adaxial cell fate, whereas KANADI (KAN) and YABBY genes are involved in that of abaxial cell fate (Siegfried et al., 1999; Bowman, 2000; Kerstetter et al., 2001; McConnell et al., 2001). Small RNAs, such as miR166, act as the abaxial determinant by suppressing HD-ZIPIII function through posttranscriptional regulation (Emery et al., 2003; Kidner and Martienssen, 2004). AUXIN RESPONSE FACTOR3 (ARF3; also known as ETTIN [ETT]) and ARF4 are required for abaxial identity (Pekker et al., 2005; Garcia et al., 2006). In snapdragon (Antirrhinum majus), PHANTASTICA plays a crucial role in adaxial cell fate (Waites and Hudson, 1995; Waites et al., 1998).
Genes encoding proteins that function in the biosynthesis of trans-acting small interfering RNA (ta-siRNA) are likely to be also involved in the control of adaxial-abaxial polarity in lateral organs (reviewed in Chitwood et al., 2007; Husbands et al., 2009; Voinnet, 2009). ta-siRNAs are derived from TAS transcripts, which are first processed by microRNA-guided cleavage and transcribed by RNA-DEPENDENT RNA POLYMERASE6 (RDR6) together with the zinc-finger protein SUPRESSOR-OF-GENE-SILENCING3 (SGS3) (Yoshikawa et al., 2005). The resulting double-stranded RNAs are processed into 21-nucleotide ta-siRNAs by DICER-LIKE4 (DCL4). The ta-siRNAs are then loaded into an RNA-induced silencing complex containing ARGONAUTE (AGO) protein and function in the cleavage of target mRNA (Allen et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). The abaxial determinants ETT and ARF4 are targeted by a class of ta-siRNAs, termed ta-siR2141 and ta-siR2142 (so called tasiR-ARF), produced from TAS3 (Allen et al., 2005). The initial step of TAS3 RNA cleavage requires a unique AGO protein, AGO7 (Montgomery et al., 2008).
In grasses, such as rice (Oryza sativa) and maize (Zea mays), genes involved in the ta-siRNA pathway, in addition to genes homologous to Arabidopsis KAN and HD-ZIPIII (Juarez et al., 2004; Candela et al., 2008; Zhang et al., 2009), seem to have essential roles in the regulation of adaxial-abaxial polarity in leaf development (reviewed in Chitwood et al., 2007; Husbands et al., 2009). Mutants of rice shoot organization1 (sho1), sho2, and maize leaf bladeless1 (lbl1) form abnormal shoots, in which the adaxial identities of the leaves are partially compromised (Timmermans et al., 1998; Itoh et al., 2000). Rice SHO1 and SHO2 encode proteins closely related to Arabidopsis DCL4 and AGO7, respectively (Liu et al., 2007; Nagasaki et al., 2007). Maize LBL1 encodes a homolog of Arabidopsis SGS3 (Nogueira et al., 2007). One of the alleles of the SHO1 locus, dcl4-1, produces abnormal lemma, probably caused by the loss of adaxial identity (Liu et al., 2007). In these mutants, loss of adaxial identity is associated with a reduction in the levels of HD-ZIPIII expression and an increase in the accumulation of miR166. Mutations in the SHOOTLESS2 (SHL2) gene, which encodes a protein similar to Arabidopsis RDR6, cause failure of shoot apical meristem (SAM) initiation in the embryo (Allen et al., 2005; Nagasaki et al., 2007). Weak alleles of shl2 exhibit shoot and leaf phenotypes similar to those in sho1 and sho2 (Satoh et al., 2003). In shl2, downregulation of HD-ZIPIII and upregulation of miR166 have also been observed, as in other mutants of the ta-siRNA pathway (Nagasaki et al., 2007). Thus, all genes related to the ta-siRNA pathway are likely to be involved in the establishment of adaxial-abaxial polarity in rice and maize.
By contrast, mutants associated with the ta-siRNA pathway, such as dcl4, rdr6, and sgs3, show no obvious phenotype in leaf polarity in Arabidopsis, although the change from the juvenile to the adult phase is promoted in these mutants (Peragine et al., 2004; Xie et al., 2005; Yoshikawa et al., 2005; Hunter et al., 2006). When these mutants are combined with either asymmetric leaves1 (as1) or as2, polarity defects in the leaves become evident, suggesting that the ta-siRNA pathway is also associated with the regulation of adaxial-abaxial polarity in Arabidopsis (Li et al., 2005; Garcia et al., 2006; Xu et al., 2006). ta-siARFs produced from TAS3 target the transcripts from ETT and ARF4 (Allen et al., 2005). TAS3 is expressed in the adaxial domain of the leaf, and the expression of ETT and ARF4 is upregulated in rdr6, sgs3, and ago7 mutants (Garcia et al., 2006). Recently, it was shown that ta-si RNA moved intercellularly, suggesting that small RNAs function as a mobile morphogenetic signal (Chitwood et al., 2009).
Floral organs are thought to be modified leaves. In contrast with flattened organs, such as the sepal and petal, the stamen is morphologically different from the leaf. The stamen consists of an anther in the distal part and a filament in the proximal part. In the anther, four microsporangia (pollen sacs) are positioned in two pairs. The pollen sacs of each pair are adjacent to each other and share a common dehiscence zone (the stomium). Each pair is a structural unit of the anther, called a theca, and combined with a connective. The filament is a thin and radially symmetrical structure (Goldberg et al., 1993; Hufford, 1996). Thus, the stamen that is differentiated from a primordium develops into two distinct parts: the bilaterally symmetrical anther and the radially symmetrical filament. The molecular genetic mechanism underlying the establishment of polarity in the leaf is well understood, as described above. This mechanism seems to be applicable to the sepal and the petal in the flower (McConnell and Barton, 1998; Sawa et al., 1999a; Pekker et al., 2005). In addition, studies indicate that a similar mechanism is involved in the development of inner nonflattened organs, such as the stamen, carpel, and ovule (Eshed et al., 1999, 2001; Kelley et al., 2009). For example, anthers are radialized in kan, phb-1d, and filamentous flower (fil) mutants (Chen et al., 1999; Sawa et al., 1999a; Eshed et al., 2001, 2004). However, the mechanism underlying the establishment of adaxial-abaxial polarity in the stamen has not yet been elucidated in detail.
In this study, we focused on a mutant, named rod-like lemma (rol), in which lateral organs such as the lemma and stamen fail to establish proper adaxial-abaxial polarity. Gene isolation revealed that the rol mutant is caused by a weak mutation in the SHL2 gene that encodes an Arabidopsis RDR6-like protein. The weak mutation in SHL2 (ROL) provides a unique opportunity to examine the regulation of polarity of lateral organs by genes in the ta-siRNA pathway during the reproductive phase in rice. Using this mutant, we found that stamen development is under the unique regulation of adaxial-abaxial polarity, which acts differently between the anther and the filament. The adaxial-abaxial polarity is rearranged at an early stage of anther development in the distal region. Initially, the polarity of the stamen primordium is the same as that in ordinary flattened lateral organs. After the rearrangement, however, the primordium divides into two units that have independent adaxial-abaxial polarity. The polarity units correspond to the two thecae, and the anther develops based on this polarity. In the proximal region, by contrast, the filament is formed by abaxialization.
RESULTS
The rol Mutation Is Associated with Loss of Adaxial-Abaxial Polarity in the Lemma
We isolated a recessive mutant, named rol, which showed pleiotropic phenotypes in both the inflorescence (panicle) and spikelet (Figure 1; see Supplemental Figure 1 online). In a rol panicle, the branching pattern was disturbed and the number of spikelets was decreased (see Supplemental Figure 1 online). In some cases, a branch terminated without developing spikelets by forming a dome-like structure at its end. In rol spikelets, morphological defects were observed in the lemma, palea, and stamen, and a lack of organs or reduction in their number was also observed.
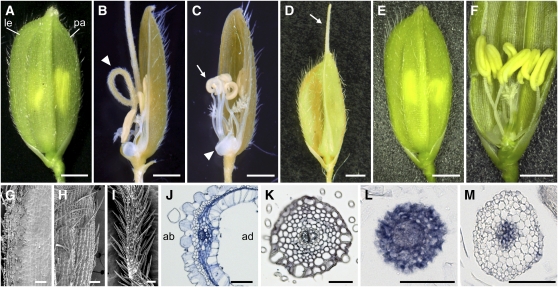
Figure 1.
Phenotypes of the rol Mutant.
(A) A wild-type spikelet. le, lemma; pa, palea.
(B) A rol spikelet with the rod-like lemma (arrowhead).
(C) A rol spikelet without a lemma. Arrowhead and arrow indicate a rudimentary organ (arrested lemma) and curled anthers, respectively.
(D) A rol spikelet with an elongated awn (arrow).
(E) and (F) rol spikelets rescued by introducing an 8.6-kb genomic fragment of the SHL2 gene. In the rescued transgenic plant, the development of the lemma and stamens is similar to that in the wild type.
(G) and (H) Adaxial (G) and abaxial (H) epidermal surface of the lemma in the wild type.
(I) Epidermal surface of the rod-like lemma in rol.
(J) A cross section of the lemma in the wild type. ab, abaxial side; ad, adaxial side.
(K) A cross section of the rod-like lemma in rol.
(L) and (M) In situ localization of ETT3 (L) and PHB3 (M) in the rod-like lemma of rol.
Bars = 1 mm in (A) to (F), 100 μm (G) to (I), and 50 μm (J) to (M).
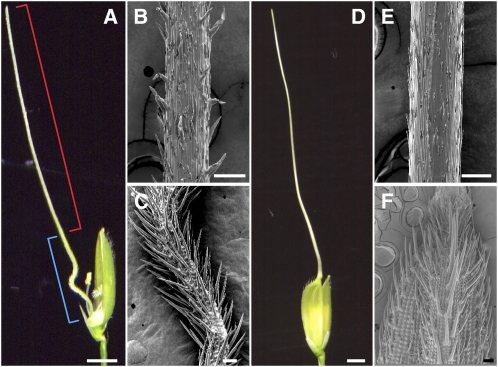
A prominent phenotype of rol spikelets was a needle-like structure at the lemma position (Figures 1B and 2A, Table 1). The needle-like structure seemed to be divided into two parts along the distal-proximal axis. In the distal part, the epidermal surface of the needle-like structure resembled those of the awns formed in Kasalath, an indica variety (the awn is reduced in most japonica) (Figure 2). By contrast, the proximal part showed morphological features resembling those of the abaxial surface, rather than the adaxial surface, of the lemma (Figures 1G to 1I, 2C, and 2F); that is, convex cells (tubercles) and many trichomes were formed in both the lemma and the proximal part of the needle-like structure. Cells similar to those at the adaxial surface in the wild-type lemma were not observed in this needle-like structure. A cross section of the proximal part of the needle-like structure showed radial symmetry. The epidermal cells were arranged in a circular shape, and their morphologies were similar to those of abaxial cells of the wild-type lemma (Figure 1K). These observations suggested that the proximal part of the needle-like structure corresponded to a lemma lacking adaxial identity (i.e., rod-like lemma). In addition to the needle-like structure, the lemmas of rol exhibited aborted development and an elongated awn (Figures 1C and 1D, Table 1).
Figure 2.
Comparison of the Needle-Like Structure in the rol Mutant and the Awn in Kasalath (indica).
(A) A spikelet with a needle-like structure in the rol mutant. The red bracket indicates the distal part, which has an awn identity. The blue bracket indicates the proximal part, which has lemma identity.
(B) and (C) Scanning electron microscopy images of the distal (B) and proximal (C) part of the needle-like structure in the rol mutant.
(D) to (F) A spikelet with a long awn in Kasalath.
(E) and (F) Scanning electron microscopy images of the awn (E) and the lemma (F) of Kasalath.
Bars = 2 mm (A) and (D) and 100 μm (B), (C), (E), and (F).
Table 1.
Percentage of Abnormal Lemmas in the rol Mutant
| Rod-Like | Aborted | Normal Shape with the Awn | Normal |
| 66.2 | 28.4 | 5.4 | 0 |
n = 500.
Next, we examined the spatial expression patterns of marker genes for adaxial-abaxial polarity. We used ETT3 (Os01g0753500), which is an ortholog of Arabidopsis ETT and is known as Os ETT3 (Sato et al., 2001; Nagasaki et al., 2007), and PHB3 (Os03g0640800), which is closely related to Arabidopsis PHB and was temporarily named OSHB3 (Zhong and Ye, 2004). In the wild-type lemma, ETT3 and PHB3 were expressed in some parts of the abaxial and adaxial sides, respectively, and in vascular bundles (see Supplemental Figure 2 online). In the rol mutant, however, ETT3 was expressed in all tissues of the rod-like lemma (Figure 1L). On the other hand, PHB3 expression was not detected in the epidermal and subepidermal tissues but was restricted to the vascular tissue (Figure 1M). Taken altogether, these analyses of the rod-like lemma suggest that the rol mutant has a defect in adaxial-abaxial polarity, and the lemma is abaxialized in this mutant.
rol Mutation Disturbs Anther Patterning
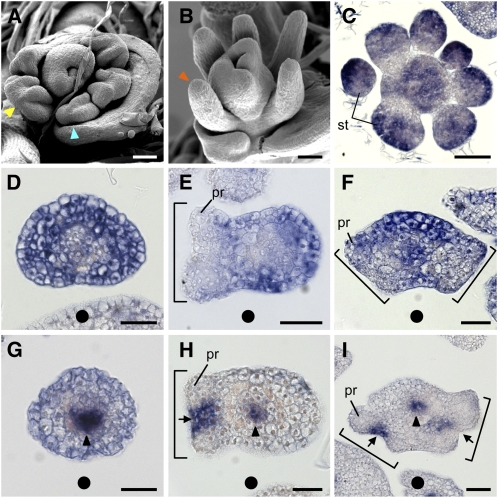
In the wild type, the anther has a bilaterally symmetrical structure and consists of two thecae, each of which contains two pollen sacs (Figures 3A, 3E, and 4A). The thecae are joined by the connective.
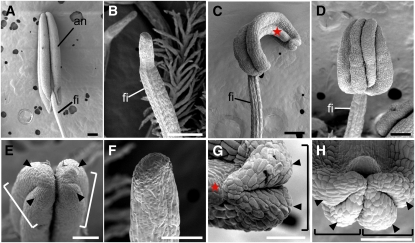
Figure 3.
Scanning Electron Microscopy Images of the Stamen in the Wild Type and rol.
(A) and (E) Wild type.
(B) and (F) Pin-like stamen.
(C) and (G) OT-type stamen with only one theca.
(D) and (H) ATT-type stamen showing four pollen sacs localized adaxially.
(E) to (H) Close-up views of (A) to (D), respectively.
Arrowheads and stars indicate the pollen sac and the connective, respectively. Brackets indicate a theca. an, anther; fi, filament. Bars = 200 μm in (A) to (D) and 100 μm in (E) to (H).
[See online article for color version of this figure.]
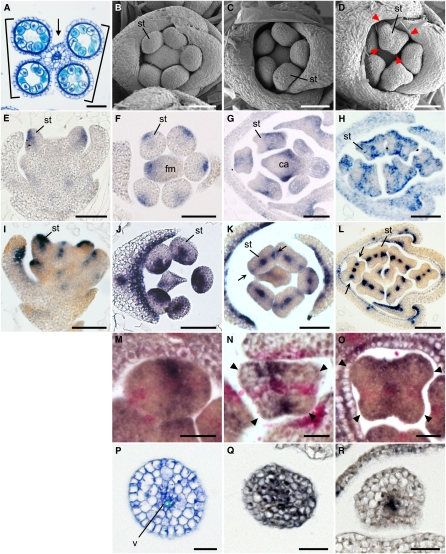
Figure 4.
Spatial Expression Pattern of ETT1 and PHB3 during Stamen Development in the Wild Type.
(A) Cross-section of an anther. Arrow and bracket indicate the connective and the thecae, respectively.
(B) to (D) Spikelets at the early developmental stages in the wild type. Arrowheads indicate protrusions in the stamen primordia.
(E) to (H) Spatial expression patterns of ETT1 in a longitudinal section (E) and cross sections ([F] to [H]) of the spikelet.
(I) to (L) Spatial expression patterns of PHB3 in a longitudinal section (I) and cross sections ([J] to [L]) of the spikelet. Arrows indicate the PHB3 expression domain in the lateral region of the stamen primordium.
(M) to (O) Two-color in situ hybridization in the anther. Purple, ETT1; pink, PHB3. Arrowheads indicate protrusions in the stamen primordia ([N] and [O]).
(P) Cross section of a filament.
(Q) and (R) Expression of ETT1 (Q) and PHB3 (R) in the filament.
ca, carpel; fm, floral meristem; st, stamen; v, vascular tissue. Bars = 50 μm in (A) to (L) and 20 μm in (M) to (R).
In the rol mutant, a reduction in the number of pollen sacs and abnormal positioning of the thecae were observed. The phenotypes of rol stamens were classified into three types in addition to the wild-type stamen (Table 2). The first type of rol stamen, the most severe phenotype, produced no pollen sacs, and the distal end of this stamen had a pin-like structure (pin-like type; Figures 3B and 3F). The second type of rol stamen had only two pollen sacs, which often curled (Figures 3C and 3G). These two pollen sacs seemed to correspond to a theca. A putative connective tissue was observed at the opposite side of the theca. Therefore, it is likely that this type of stamen consists of one theca and a connective (one-theca [OT]-type). The third type of rol stamen had an abnormal arrangement of pollen sacs, although four pollen sacs were produced (Figures 3D and 3H). In this type, all pollen sacs (two thecae) were located on the adaxial side of the stamen (adaxially localized two-thecae [ATT]-type). We did not observe a stamen with one or three pollen sacs in rol spikelets (Table 2). Therefore, these results suggest that the theca is a developmental unit of the anther.
Table 2.
Percentage of Abnormal and Wild-Type Stamens in the rol Mutant
| Anther Type | Pin | – | OT | – | ATT | Wild Type |
| No. of pollen sacs | 0 | 1 | 2 | 3 | 4 | 4 |
| Percentage (n = 2238) | 2.5 | 0 | 20.7 | 0 | 34.1 | 42.8 |
Next, we compared stamen primordia at an early developmental stage. In the wild type, stamen primordia were found to be round in the early stages of development and subsequently became rectangular. The four corners of the rectangular stamen protruded and later developed into four pollen sacs (Figures 4B to 4D). In a rol spikelet, stamen primordia had no, two, or four protrusions, the number of which corresponded to that of pollen sacs in mature anthers (Figures 5A and 5B). When four protrusions were observed, they were localized on the adaxial side. Taken together, anther patterning was disturbed in rol and was likely to be associated with defects in adaxial-abaxial polarity (see Discussion). The polarity along the proximal-distal axis seemed to be normal in the rol stamen because the anther and filament formed on the apical and basal parts of the stamen, respectively.
Figure 5.
Scanning Electron Microscopy Images of Stamen Primordia and Spatial Expression Pattern of ETT1 and PHB3 during Stamen Development in rol.
(A) and (B) Scanning electron microscopy images of stamen development. Red, blue, and yellow arrowheads indicate pin-like, OT-type, and ATT-type stamen, respectively.
(C) An early stage of stamen development.
(D) and (G) Pin-like stamen primordia.
(E) and (H) Primordia of OT-type stamen.
(F) and (I) Primordia of ATT-type stamen. Brackets indicate a theca ([E], [F], [H], and [I]) and arrows indicate the region where the stomium differentiates later ([H] and [I]). Black arrowheads indicate the procambium ([G], [H], and [I]). Solid circles indicate the direction of the center of the flower.
pr, protrusion that differentiates later into the pollen sac; st, stamen. Bars = 50 μm in (A) to (C) and 20 μm in (D) to (I).
Rearrangement of Adaxial-Abaxial Polarity during Stamen Development
We then examined the expression patterns of marker genes for adaxial-abaxial polarity during stamen development. In the wild type, expression of ETT1 (Os05g0563400), which is a major ETT gene expressed in the stamen, was restricted to the abaxial domains of the stamen primordia at an early stage before the four corners of the primordia were visible (Figures 4E and 4F). After formation of the four corners, ETT1 transcripts were detected in both the adaxial and abaxial domains of the initial axis (Figure 4G), although this expression appeared to be patchy at a later stage (Figure 4H). The ETT1-expressing domains corresponded to the regions between the two thecae and the abaxial side of the pollen sac (see Discussion) when the anther matured.
PHB3 transcripts were detected in the adaxial domain and in the procambium of the stamen primordia in the early round stage (Figures 4I and 4J). After the stage at which stamen primordia became rectangular, PHB3 transcripts disappeared from the adaxial domain and were detected in the two lateral domains of the stamen primordia (Figures 4K and 4L, arrows). The PHB3 expression domain corresponded to the region between the pollen sacs within a theca in the mature anther. PHB3 transcripts were also detected in the procambium in the stamen primordia (Figures 4J to 4L). Thus, these results indicated that the spatial expression patterns of two marker genes, ETT1 and PHB3, were markedly changed and rearranged during anther patterning in the stamen primordia.
To confirm this change, we analyzed the spatial expression pattern of the two marker genes by two-color in situ hybridization, which made it possible to detect expression of the two genes simultaneously at a chosen developmental stage. The results revealed that the rearrangement of the expression patterns of the two genes was completed before the appearance of the four corners in the stamen primordium (Figures 4M to 4O). PHB3 transcripts (red) were detected in the two lateral domains and in the procambium. At the same stage, ETT1 transcripts (purple) were detected in both the adaxial and the abaxial domains of the stamen primordium. The results also indicated that the four corners became obvious at the regions between the expression domains of ETT1 and PHB3 (Figures 4M to 4O, arrowheads).
Taken together, these results indicate that rearrangement of the expression patterns of the two marker genes takes place before morphological changes of the stamen primordia, from the round shape to the rectangular shape. Therefore, it seems that the cell fates along the adaxial-abaxial axis are markedly changed during anther patterning. In addition, formation of the protrusions in the anther primordia seems to be closely associated with the boundaries between the expression domains of the two marker genes.
In the presumptive region where the filament would differentiate, ETT1 was expressed in the epidermal and subepidermal cell layers and in the procambium (Figure 4Q), whereas PHB3 expression was restricted to the procambium (Figure 4R). This result suggests that the filament may develop without adaxial identity.
Spatial Expression Patterns of ETT1 and PHB3 in the rol Stamen
In the rol mutant, the expression domain ETT1 was expanded widely in the stamen primordia during the early round stage (Figure 5C) compared with the wild type (Figure 4F). This result suggests that rol stamen primordia have a defect in adaxial-adaxial polarity from an early developmental stage.
Three types of expression patterns of marker genes were observed in association with the stamen primordia morphologies. In the rol stamen primordia with a pin-like structure, ETT1 was expressed in the entire region of the epidermal and subepidermal cell layers (Figure 5D). On the other hand, PHB3 transcripts were not detected in the outer layers of the stamen primordia and were restricted to the procambium (Figure 5G). In the rol stamen primordia with two protrusions, probably corresponding to an OT-type stamen, ETT1 transcripts were detected in the abaxial side of the protrusion and a putative connective region, which expanded in this type of stamen, and were excluded from the protrusions themselves (Figure 5E; see Discussion). PHB3 transcripts were detected in the domain between the two protrusions and in the procambium (Figure 5H). In the rol stamen primordia with four protrusions, probably corresponding to an ATT-type stamen, ETT1 transcripts were widely distributed in the abaxial domain of the primordium and in the domain between the two thecae (Figure 5F). PHB3 transcripts were detected in the domain between the two protrusions within each theca and in the procambium (Figure 5I).
Despite the aberrant morphologies of the rol stamen in terms of protrusions, the position of the expression domains of two markers is likely to be common to both the wild type and rol: ETT1 was expressed between thecae, whereas PHB3 was expressed between the pollen sacs in a theca. It should be noted that the protrusion was formed in the region between the expression domains of ETT1 and PHB3 even in the aberrant rol stamen primordia.
The rol Mutant Has a Defect in the Gene Encoding RNA-Dependent RNA Polymerase, Which Is Required for the ta-siRNA Pathway
We tried to isolate the gene responsible for the rol mutant by positional cloning to elucidate its molecular function. The putative ROL locus was mapped to a region between two molecular markers, RM7075 and RM5638, on chromosome 1. Because this region encompasses the centromere, it would be difficult to isolate this gene by a standard positional cloning method. We therefore focused on the fact that the rol plants developed filamentous structures in leaves at a low frequency, a phenotype similar to the leaf phenotype observed in shoot organization (sho) mutants (Itoh et al., 2000, 2008). The SHO genes are involved in the regulation of adaxial-abaxial polarity and encode proteins involved in the ta-siRNA pathway (Itoh et al., 2000, 2008; Nagasaki et al., 2007). A database search revealed that a putative gene encoding an RNA-dependent RNA polymerase (RdRP) involved in the ta-siRNA pathway was located in the region to which the putative ROL locus roughly mapped.
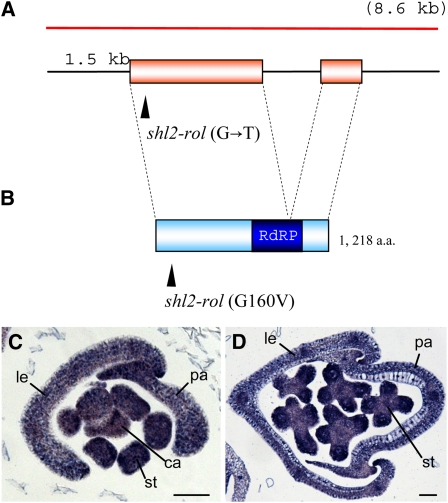
Next, we identified a nucleotide change that caused an amino acid substitution in the gene for this RdRP in the rol mutant (Figures 6A and 6B). Introduction of an 8.6-kb genomic fragment encompassing the RdRP gene rescued the mutant phenotype of the rol spikelet (Figures 1E and 1F). Therefore, we concluded that the rol phenotype was caused by a mutation in the gene encoding RdRP. This gene is closely related to RDR6 in Arabidopsis and has been reported as the SHL2 gene in rice (Nagasaki et al., 2007). Hereafter, we refer to the mutant (allele) as shl2-rol (an allele of the SHL2 locus) instead of rol.
Figure 6.
Schematic Representation of the SHL2 (ROL) Gene and Its Encoded Protein, and Spatial Expression Pattern of SHL2.
(A) The SHL2 gene. Boxes indicate coding regions. Arrowhead indicates the mutation site in the shl2-rol mutant. The red bar shows the 8.6-kb genomic fragment used for the complementation test. This fragment contains a promoter of ∼1.5 kb.
(B) Schematic representation of the SHL2 protein. Arrowhead indicates the amino acid substitution in the shl2-rol mutant. The blue box indicates the conserved RdRP domain.
(C) and (D) Spatial expression pattern of SHL2 in the spikelet. Bars = 50 μm. ca, carpel; le, lemma; pa, palea; st, stamen.
Loss-of-function mutation of SHL2 causes failure in the formation of the SAM in the embryo. Twelve shl2 alleles have been reported to date (Nagasaki et al., 2007). Most alleles have a nonsense mutation, frameshift mutation, or amino acid substitution in the conserved RdRP domain. By contrast, shl2-rol has an amino acid substitution in the N-terminal region far from the RdRP domain (Figure 6B), suggesting that the effect of this mutation is weak on RdRP activity. The shl2 mutants so far reported are embryonic or seedling lethal. Because of the weak mutation, shl2-rol may grow to the reproductive phase and show the unique spikelet phenotypes described above.
We examined the spatial expression pattern of SHL2 in the spikelet. SHL2 was expressed in all organs, including the stamen and lemma, from the early to the late stages (Figures 6C and 6D). In addition, the SHL2 signal was detected ubiquitously without any specific localization within the organs. This expression pattern suggests that SHL2 is not a primary factor that localizes the ETT transcripts in the patterns described above.
DISCUSSION
The SHL2 Gene Is Involved in the Regulation of Adaxial-Abaxial Polarity in the Floral Organs in Addition to in the Leaves
In this study, we revealed that the regulation of polarity along the adaxial-abaxial axis is disturbed during the development of lateral organs, such as the stamen and the lemma in the shl2-rol mutant. Gene isolation indicated that the shl2-rol mutant is caused by a mutation in the SHL2 gene, which encodes an RDR6-like protein that acts in the ta-siRNA pathway in Arabidopsis. Nagasaki et al. (2007) demonstrated that genes involved in the ta-siRNA pathway are essential for SAM formation in the rice embryo, and one of the shootless mutants (shl2) is caused by a defect in an ortholog of Arabidopsis RDR6. Accordingly, the gene that causes the rol phenotype is a novel allele of SHL2, in which the amino acid substitution (G160V) probably causes a weak defect in RdRP activity. This weak mutation in SHL2 seems to provide a unique opportunity to examine the regulation of polarity of lateral organs in the reproductive phase in rice because most known alleles of shl2 are embryonic lethal.
Analyses of phenotypes and expression patterns of marker genes indicated that the rod-like lemma is likely to be completely abaxialized in the shl2-rol mutant. As discussed below, pleiotropic phenotypes observed in the shl2-rol stamen may be a consequence of the defects in adaxial-abaxial polarity. Polarity defects in the palea were not severe compared with those in the lemma in the shl2-rol mutant. It is possible that this phenotypic difference may be partially due to differences in the levels and expression patterns of four ETT genes between the lemma and palea.
Although most shl2 mutants are embryonic lethal because of a failure in SAM formation, the weakest allele shl2-8 produces leaves that are defective in adaxial identity (Satoh et al., 1999; Satoh et al., 2003). Therefore, SHL2 is likely to be involved in the regulation of adaxial-abaxial polarity in the lateral organs in the vegetative and reproductive phases in rice. This conclusion is consistent with the observations that genes involved in the ta-siRNA pathway, such as SHO1 and SHO2 in rice and lbl1 in maize, are responsible for the regulation of adaxial-abaxial polarity in the leaf (Itoh et al., 2000, 2008; Liu et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007). Furthermore, a dcl4 mutant corresponding to sho1 causes formation of a rod-like lemma as the shl2-rol does (Liu et al., 2007). Thus, the ta-siRNA pathway seems to be associated with the regulation of polarity of lateral organs throughout the life cycle in rice.
This finding is in contrast with the observation that mutations in genes in the ta-siRNA pathway cause no obvious phenotype of polarity defects in Arabidopsis (Peragine et al., 2004; Adenot et al., 2006). In rice shl2, sho1, and sho2/dcl4 and maize lbl1, the levels of tasiR-ARF and cleavage products of ETT are reduced, in association with upregulation of ETT or spatial expansion of ETT expression domains (Liu et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007). It is conceivable that the expansion of ETT expression domains observed in the shl2-rol mutant may result from partial failure of the mechanism that produces ta-siRNA.
Rearrangement of the Expression Patterns of ARF and PHB during Stamen Development
During anther development, a marked rearrangement of the spatial expression patterns of ETT1 and PHB3 was observed. ETT1 and PHB3 were initially expressed in the abaxial and adaxial domains of the stamen primordia, respectively. Subsequently, however, the expression patterns of both genes changed dramatically: the genes were expressed in positions that crossed at right angles. On the basis of these observations, we propose a model that explains anther patterning in relation to the regulation of adaxial-abaxial polarity (Figure 7).
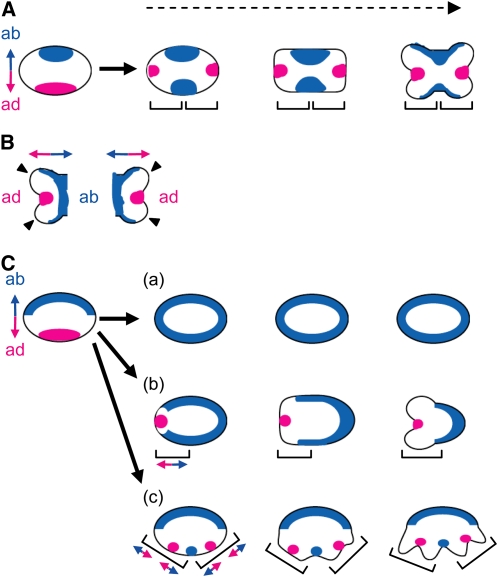
Figure 7.
Model of Anther Patterning in Rice.
(A) Model of anther patterning in the wild type.
(B) Adaxial-abaxial polarity in a theca. Arrowheads indicate the protrusions at the region between the domains of the adaxial and abaxial identities.
(C) Model of anther patterning in rol. (a) Pin-like stamen, (b) OT-type stamen, and (c) ATT-type stamen. The domains with the adaxial and abaxial identities represented by the expression domains of ETT1 and PHB3 are shown in red and blue, respectively. Each bracket represents a unit of the adaxial-abaxial polarity. Double-headed arrows represent the axis of adaxial-abaxial polarity. ab, abaxial side; ad, adaxial side. Dashed arrow indicates developmental progression.
In this model, adaxial-abaxial polarity is rearranged at an early stage of stamen development. Initially, adaxial-abaxial polarity is established in the stamen primordia as it is in other primordia of ordinary lateral organs, such as the leaf. Thus, adaxial identity is established in the domain adjacent to the meristem, and abaxial identity is established in the domain opposite to it. After the adaxial-abaxial rearrangement, a new polarity is established in a new unit, the theca primordium. This notion is consistent with the idea that the theca is a developmental unit, as shown by phenotypic analysis of the shl2-rol stamen. In the region between the domains bearing the adaxial and abaxial identities, protrusions are formed that go on to form the pollen sacs. After the adaxial-abaxial rearrangement, the anther seems to develop according to the new polarity, as discussed below. The ETT1 expression domain would correspond to the abaxial domain of the theca and the connective that is formed between thecae, whereas the PHB3 expression domain would correspond to the adaxial domain where the stomium differentiates.
The polarity change appears to be very rapid. Despite many efforts, we could not detect marker expression demonstrating the transition stage of polarity rearrangement. In addition, the complementary expression pattern of ETT1 and PHB3 was maintained during anther development, except for their expression in vascular tissue, where both genes were expressed, suggesting that cell fate along the adaxial-abaxial axis is strictly regulated. In proper adaxial-abaxial patterning during leaf development, positional information is required. It is assumed that crosstalk may exist between the miR166-mediated regulation of HD-ZIPIII and the tasiR-ARF–mediated regulation of ETT, and small RNA seems to be involved in this crosstalk (Chitwood et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007). In addition, small RNAs are thought to act as a mobile signal between the adaxial and abaxial domains (Chitwood et al., 2009). Therefore, it is possible that such intercellular communication via small RNAs may be implicated in the rapid and exact rearrangement of adaxial-abaxial polarity in anther patterning.
Reconstitution of Adaxial-Abaxial Polarity during Stamen Development
Close relationships between lateral lamina growth and adaxial-abaxial polarity in leaf development have been widely accepted (Waites and Hudson, 1995; McConnell and Barton, 1998; McHale and Marcotrigiano, 1998; Timmermans et al., 1998). Failure to establish adaxial-abaxial polarity inhibits lateral lamina growth and produces radialized leaves. Likewise, the rod-like lemma may be formed by a failure in the outgrowth of lateral domains in shl2-rol. It seems likely that a similar mechanism may underlie anther development. Stamen primordia emerge initially as spherical structures. After rearrangement of adaxial-abaxial polarity, four protrusions emerge and then differentiate into pollen sacs. The regions where the initial protrusions form lie between the two domains that express the adaxial or abaxial gene markers. Thus, initiation of the protrusions and subsequent pollen sac differentiation in anther development are likely to be analogous to the formation of the outgrowth and lamina expansion in leaf development.
The pleiotropic phenotypes observed in the shl2-rol stamen would result from failure of the proper establishment of adaxial-abaxial polarity (Figures 3 and 7C). If there was a serious defect in the establishment of polarity, then no protrusions would be formed in the stamen primordium, and pin-like stamens would develop. In the case of an intermediate defect, only half of the stamen primordia would establish polarity to form one theca, and an OT-type stamen would be produced. In the case of a weak defect, two thecae might develop according to the two polarity units, although the abaxial domain, which corresponds to the region between the thecae, would be expanded (ATT-type stamen). Thus, the phenotype of the stamen and the spatial expression patterns of the marker genes are in agreement with each other in the shl2-rol mutant.
The expression pattern of the marker genes suggested that the filament lacks adaxial identity in the wild-type stamen. It is likely that the anther and the filament develop as independent polarity units. The filament would not be affected by the shl2-rol mutation, probably because the main effect of this mutation is loss of adaxial identity.
Once adaxial-abaxial polarity is established, this polarity is maintained in flattened lateral organs, such as leaves and petals, throughout their development. In the stamen, however, regulation of adaxial-abaxial polarity is likely to differ in the distal and proximal parts. In anther development in the distal part of the stamen, rearrangement of the polarity occurs after the initial establishment of adaxial-abaxial polarity, and the polarity unit seems to change from the stamen primordium to the theca. The anther may develop in accordance with the new polarity. By contrast, polarity may be lost in the proximal part of the stamen; thus, the filament that lacks adaxial identity seems to develop as a filamentous structure.
In Arabidopsis, ETT is initially expressed in the abaxial domain of the stamen primordia in a pattern highly similar to ETT3 in rice (Sessions et al., 1997). Later, ETT expression is detected in four domains, including the region between the thecae (interthecal furrows). PHB is initially expressed on the adaxial side of the stamen primordium and is later expressed in the domain between the pollen sacs within the theca (Dinneny et al., 2006). These expression patterns partially resemble those of ETT1 and PHB3 in rice and suggest that the expression domain of ETT and PHB is rearranged during stamen development in Arabidopsis, as in rice. In addition, kan, phb-d, and fil form radialized pin-like stamens, suggesting that failure to establish adaxial-abaxial polarity is closely related to loss of the anther (Chen et al., 1999; Sawa et al., 1999a; Eshed et al., 2001, 2004). Therefore, the model of stamen development that we propose here might be generally applicable to stamen development in angiosperms. ETT and FIL are initially expressed in the same pattern in the abaxial domains of the stamen primordia, but their expression domains differ at later stages of development: ETT is expressed between the thecae, whereas FIL is expressed in the connective (Sessions et al., 1997; Chen et al., 1999; Sawa et al., 1999b; Siegfried et al., 1999). Thus, the role of these abaxial determinants might be differentiated at the later stages of stamen development in Arabidopsis.
METHODS
Plant Materials
A novel mutant (named rol in this study) was found in M2 plants that had been mutagenized during tissue culture from Nipponbare. Nipponbare was used as a wild-type strain for comparing phenotypes and for in situ expression analysis. Kasalath, a cultivar of Oryza sativa spp indica, was used for observation of the awn.
Scanning Electron Microscopy
Young panicles and flowers were fixed in 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.2, at 4°C overnight. They were then dehydrated in a graded ethanol series, and 100% ethanol was replaced with 3-methylbutyl acetate. Samples were dried at their critical point, sputter-coated with platinum, and observed with a scanning electron microscope (model JSM-820S; JEOL) at an accelerating voltage of 5 kV.
In Situ Hybridization
Rice ETT1 (Os05g0563400) and ETT3 (Os01g0753500) have been previously described by Sato et al. (2001) and Nagasaki et al. (2007), respectively. Rice PHB3 (Os12g0612700) corresponds to OSHB3 (Zhong and Ye, 2004). To make probes for rice ETT1, ETT3, PHB3, and SHL2, partial cDNA fragments were amplified with the primers listed in Supplemental Table 1 online and cloned into the pCRII vector (Invitrogen). Using these plasmids as templates, a region containing the partial cDNA sequences and both T7 and SP6 promoter sequences was amplified with M13 forward and reverse primers. After removal of the primers, the resulting PCR products (100 to 200 ng) were used for RNA transcription. Synthesis of DIG-labeled RNA probes, in situ hybridization, and immunological detection were performed by the methods described by Suzaki et al. (2004).
For two-color in situ hybridization, an RNA probe for PHB3 was labeled using a fluorescein isothiocyanate (FITC) RNA labeling kit (Roche). Double hybridization with this FITC-labeled PHB3 probe and a DIG-labeled ETT1 probe was performed using the method described by Kouchi et al. (1995). The sections were hybridized at 55°C with a mixture of the two probes. After hybridization, the FITC-labeled probe was detected by incubation with an antifluorescein antibody conjugated to alkaline phosphatase (Roche) in combination with Fast Red TRInaphtol AS-MX (red) (Sigma-Aldrich) at 37°C for 4 to 6 h. To inactivate the alkaline phosphatase, the slides were incubated twice in 2× SSC at 68°C for 1.5 h each. The second detection was performed using anti-DIG antibody conjugated with alkaline phosphatase (Roche) in combination with NBT/BCIP solution (purple) (Roche). The slides were incubated at 37°C for 12 h. The slides were then mounted with glycerin, and signals were observed under a light microscope (BX-50; Olympus).
Isolation of the Gene Responsible for the rol Mutation
A putative rol locus was mapped using an F2 population derived from the rol mutant and Kasalath. The locus was mapped to a region between the cleaved-amplified polymorphic sequence markers RM7075 and RM5638 on chromosome1 using 28 rol homozygotes. SHL2, which encodes RdRP (Nagasaki et al., 2007), which is similar to Arabidopsis RDR6 and is involved in the ta-siRNA pathway, was identified in this region by a database search. The genomic sequence of the SHL2 locus in the rol mutant was determined by direct sequencing after PCR amplification. Sequence analysis revealed that a mutation causing an amino acid substitution occurred in this gene, suggesting that the rol mutation is caused by a mutation in SHL2.
For complementation analysis, an 8.6-kb fragment encompassing the RdRP gene was amplified using Nipponbare genomic DNA as a template and cloned into a pENTR 2B vector (Invitrogen). For transformation of rice, a pBI-Hm12-GW plasmid containing the Gateway rfC cassette (Invitrogen) (Yoshida et al., 2009) was used. By an LR recombination reaction, the 8.6-kb fragment was transferred into pBI-Hm12-GW. The recombinant plasmid was introduced into Agrobacterium tumefaciens strain EHA101 and transformed into the rol mutant by the method described by Hiei et al. (1994).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SHL2 (AB353923), ETT1 (AB071290), ETT3 (AK072330), and PHB3 (AK102183).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of the rol Mutant.
Supplemental Figure 2. Spatial Expression Patterns of ETT3 and PHB3 in Wild-Type Spikelet.
Supplemental Table 1. Primers Used to Make in Situ Probes.
Acknowledgments
We thank M. Tasaka for valuable comments, Y. Iwasaki for help with our work, and M. Harada and K. Ohsawa for technical assistance. This research was supported in part by Grants-in-Aid for Scientific Research from a Ministry of Education, Culture, Sports, Science and Technology (MEXT) (20380005 and 21027005 to H.-Y.H.), the Global Center of Excellence (COE) Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from MEXT (to T.T.), and a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (to T.T.).
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Bowman J.L. (2000). The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3: 17–22 [DOI] [PubMed] [Google Scholar]
- Candela H., Johnston R., Gerhold A., Foster T., Hake S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 20: 2073–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Atkinson A., Otsuga D., Christensen T., Reynolds L., Drews G.N. (1999). The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126: 2715–2726 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Guo M., Nogueira F.T., Timmermans M.C.P. (2007). Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development 134: 813–823 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T.S., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C.P. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Weigel D., Yanofsky M.F. (2006). NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Bowman J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 9: 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Goldberg R.B., Beals T.P., Sanders P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hufford L. (1996). Origin and early evolution of angiosperm stamens. The Anther: Form, Function, and Phylogeny, D'Arcy W.G., Keating R.C., (Cambridge, UK: Cambridge University Press; ), pp. 58–91 [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A.Y., Chitwood D.H., Plavskin Y., Timmermans M.C.P. (2009). Signals and prepatterns: new insights into organ polarity in plants. Genes Dev. 23: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J.I., Sato Y., Nagato Y. (2008). The SHOOT ORGANIZATION2 gene coordinates leaf domain development along the central-marginal axis in rice. Plant Cell Physiol. 49: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Itoh J.I., Kitano H., Matsuoka M., Nagato Y. (2000). SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12: 2161–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C.P. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kelley D.R., Skinner D.J., Gasser C.S. (2009). Roles of polarity determinants in ovule development. Plant J. 57: 1054–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kouchi H., Sekine M., Hata S. (1995). Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell-cycle. Plant Cell 7: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xu L., Wang H., Yuan Z., Cao X.F., Yang Z.N., Zhang D.B., Xu Y.Q., Huang H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 17: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., et al. (2007). Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McHale N.A., Marcotrigiano M. (1998). LAM1 is required for dorsoventrality and lateral growth of the leaf blade in Nicotiana. Development 125: 4235–4243 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Nagasaki H., Itoh J., Hayashi K., Hibara K., Satoh-Nagasawa N., Nosaka M., Mukouhata M., Ashikari M., Kitano H., Matsuoka M., Nagato Y., Sato Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T.S., Madi S., Chitwood D.H., Juarez M.T., Timmermans M.C.P. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Nishimura A., Ito M., Ashikari M., Hirano H.-Y., Matsuoka M. (2001). Auxin response factor family in rice. Genes Genet. Syst. 76: 373–380 [DOI] [PubMed] [Google Scholar]
- Satoh N., Hong S.K., Nishimura A., Matsuoka M., Kitano H., Nagato Y. (1999). Initiation of shoot apical meristem in rice: Characterization of four SHOOTLESS genes. Development 126: 3629–3636 [DOI] [PubMed] [Google Scholar]
- Satoh N., Itoh J., Nagato Y. (2003). The SHOOTLESS2 and SHOOTLESS1 genes are involved in both initiation and maintenance of the shoot apical meristem through regulating the number of indeterminate cells. Genetics 164: 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Ito T., Shimura Y., Okada K. (1999a). FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11: 69–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Kanaya E., Morita E.H., Okada K. (1999b). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Suzaki T., Sato M., Ashikari M., Miyoshi M., Nagato Y., Hirano H.-Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Timmermans M.C.P., Schultes N.P., Jankovsky J.P., Nelson T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125: 2813–2823 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Waites R., Selvadurai H.R.N., Oliver I.R., Hudson A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789 [DOI] [PubMed] [Google Scholar]
- Xie Z.X., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Yang L., Pi L.M., Liu Q.L., Ling Q.H., Wang H., Poethig R.S., Huang H. (2006). Genetic interaction between the AS1-AS2 and RDR6–SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 47: 853–863 [DOI] [PubMed] [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H.-Y. (2009). The homeotic gene LONG STERILE LEMMA (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 106: 20103–20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.H., Xu Q., Zhu X.D., Qian Q., Xue H.W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.Q., Ye Z.H. (2004). amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45: 369–385 [DOI] [PubMed] [Google Scholar]