This study describes the role of Tudor-SN in optimal stress tolerance throughout the life cycle of Arabidopsis. It finds evidence suggestive of new mechanisms regulating the metabolism of mRNAs entering the secretory pathway.
Abstract
Tudor-SN (TSN) copurifies with the RNA-induced silencing complex in animal cells where, among other functions, it is thought to act on mRNA stability via the degradation of specific dsRNA templates. In plants, TSN has been identified biochemically as a cytoskeleton-associated RNA binding activity. In eukaryotes, it has recently been identified as a conserved primary target of programmed cell death–associated proteolysis. We have investigated the physiological role of TSN by isolating null mutations for two homologous genes in Arabidopsis thaliana. The double mutant tsn1 tsn2 displays only mild growth phenotypes under nonstress conditions, but germination, growth, and survival are severely affected under high salinity stress. Either TSN1 or TSN2 alone can complement the double mutant, indicating their functional redundancy. TSN accumulates heterogeneously in the cytosol and relocates transiently to a diffuse pattern in response to salt stress. Unexpectedly, stress-regulated mRNAs encoding secreted proteins are significantly enriched among the transcripts that are underrepresented in tsn1 tsn2. Our data also reveal that TSN is important for RNA stability of its targets. These findings show that TSN is essential for stress tolerance in plants and implicate TSN in new, potentially conserved mechanisms acting on mRNAs entering the secretory pathway.
INTRODUCTION
The importance of different facets of RNA metabolism during development and adaptation to environmental challenges of eukaryotic organisms is now well established. Notably, significant advances have been made in understanding the mechanisms by which small RNAs arise within the cell and assemble within RNA-induced silencing complexes (RISCs) to buffer transcriptome activity (Siomi and Siomi, 2009). In plants, genetic studies have demonstrated the relevance of small RNA pathways and RISC activity in development and stress responses and have revealed the broad impact of mRNA decay and ribonucleoparticle architecture in adaptation to abiotic challenges. In particular, a number of RNA-interacting proteins have been identified as the molecular determinants of abscisic acid (ABA) response phenotypes ranging from seed germination to flowering time (Kuhn et al., 2008; Zhang et al., 2008). Others are the products of genes regulated by abiotic stress or by ABA itself (Lorkovic, 2009). However, the evolutionary conservation of several of these proteins hints that there remain to be discovered fundamental, conserved mechanisms allowing stress adaptation through regulation of mRNA maturation, export, storage, decay, translation, or localization.
While studying gene expression in root apices adapted to progressive drought (Vartanian et al., 1994; M.W. Bianchi, C. Roux, and B. Seijo, unpublished data), we isolated transcripts of plant homologs of the RNA binding protein Tudor-SN (TSN). TSN was first described as a transcriptional coactivator in cultured animal cells (Tong et al., 1995; Dash et al., 1996; Leverson et al., 1998; Yang et al., 2002). Subsequently, it was shown to promote spliceosome assembly in vitro (Pham et al., 2004) and to copurify with the ARGONAUTE protein within the RISC (Caudy et al., 2003). The latter association was suggested to be of functional significance in nematodes, where, in some of the reported studies, TSN was shown to be required for the proper expression of small RNA targets (Caudy et al., 2003; Grishok et al., 2005; Kim et al., 2005). However, the presence of TSN in the RISC may be functionally distinct from ARGONAUTE activity, as TSN was associated to a nuclease activity specific for double-stranded RNA molecules containing G:U or I:U pairs in animal cells (Scadden, 2005; Yang et al., 2006). Both the sequence and the domain architecture of TSN are well conserved in eukaryotes, with the notable exception of Saccharomyces cerevisiae, which lacks a TSN gene, as well as the small RNA machinery altogether.
TSN proteins are characterized by four staphylococcal nuclease-like domains (SN), followed by a Tudor domain interdigitated with a fifth, C-terminal SN domain (Figure 1A). Recent crystallography studies suggest that double-stranded nucleic acid substrates are captured by a tandem repeat of the SN domain, while an aromatic cage in the Tudor domain binds the dimethylarginines present in posttranslationally modified protein partners (Li et al., 2008; Friberg et al., 2009). Interestingly, Sundström et al. (2009) have recently reported that in animals and plants, TSN is targeted at multiple sites by caspases and metacaspases, respectively, during programmed cell death, underlining the likely importance of TSN function in eukaryotes. In plants, TSN was first identified as an RNA binding activity present in the cytoskeletal, endoplasmic reticulum (ER), and ribosomal fractions of extracts from rice (Oryza sativa) seeds and pea (Pisum sativum) seedlings (Sami-Subbu et al., 2001; Abe et al., 2003). More recent studies have suggested that TSN participates in the localization of prolamine mRNAs in rice endosperm (Wang et al., 2008), where these transcripts are transported to specific sites of translation on the ER. Finally, evidence indicates that Arabidopsis thaliana plants with attenuated TSN levels are impaired in pollen viability, as a consequence of ectopic cell death (Sundström et al., 2009).
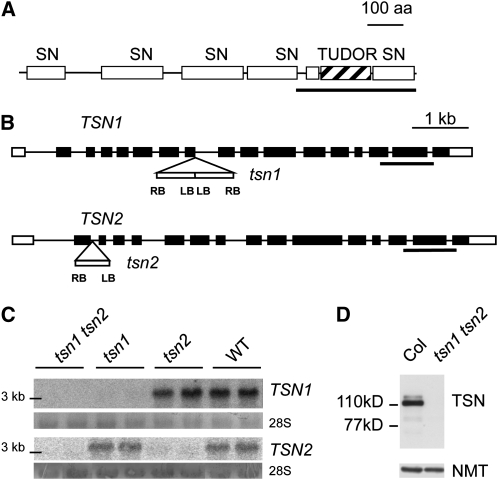
Figure 1.
Reverse Genetics of Arabidopsis TSN Genes.
(A) Domain architecture of TSN proteins. The schematic shows the SN domains and the tudor domain, which is interdigitated with the fifth SN domain. The fragment produced in Escherichia coli for the generation of TSN antibodies is underlined. aa, amino acids.
(B) Intron-exon structure of TSN1 and TSN2. The position and structure of T-DNA insertions in the tsn1 and tsn2 alleles are indicated (not to scale; details in Methods). White boxes, untranslated regions; black boxes, exons. LB and RB, left border and right border of the T-DNA.
(C) Blots of seedling RNA. Genotypes, products from the tsn2 × tsn1 cross are indicated at the top (two lines per genotype are shown). Probes employed, indicated at the right, correspond to regions underlined in (B). Note the absence of signal in tsn1 tsn2. The 28S rRNA band is shown below each hybridization as a control. WT, wild type.
(D) Protein blot of seedling extracts. Genotypes are indicated at the top, and antibodies employed are on the right. Note the absence of signal in tsn1 tsn2. Positions of two molecular weight markers are indicated on the left. Detection of N-myristoyltransferase (NMT) serves as a control.
Despite the important roles for this protein that have been proposed based on the biochemical work, phenotypes of TSN-null mutants in any eukaryote are not known to date. Large-scale mutagenesis programs in Schizosaccharomyces pombe (Decottignies et al., 2003), Caenorhabditis elegans (Grishok et al., 2005; Kim et al., 2005), and Drosophila melanogaster (Dorner et al., 2006) have classed TSN mutants as viable, without further description. In this study, we identify an Arabidopsis TSN-null mutant, characterize its physiological and molecular phenotypes, and study expression patterns and the subcellular localization of TSN. The data obtained show the importance of TSN for stress adaptation, allow an evaluation of TSN involvement in RISC activity, and implicate TSN in the stabilization of a subset of salt stress–responsive mRNAs entering the secretory pathway.
RESULTS
Identification and Complementation of a TSN-Null Mutant in Arabidopsis
In Arabidopsis, two highly similar genes, designated here as TSN1 (AT5G07350) and TSN2 (AT5G61780), encode 84% identical proteins of 990 and 984 amino acids, respectively (an alignment with plant and human TSNs is shown in Figure 1 online). T-DNA mutants for TSN1 and TSN2, in the Wassilewskija (Ws) and Columbia (Col-0) backgrounds, respectively, were isolated (Figure 1B) and crossed to generate a tsn1 tsn2 double mutant. The tsn1 tsn2 mutant was devoid of TSN mRNA and protein (Figures 1C and 1D). The double homozygous mutant was 3 to 5 times underrepresented in the progeny derived from self-fertilization of individuals carrying the homozygous mutation for either locus and heterozygous for the other mutation (see Supplemental Table 1 online). Nonetheless, the double homozygous mutant was viable and fertile. In this work, phenotypes of tsn1 tsn2 were compared with sibling lines complemented by either a TSN1 or a TSN2 transgene (TSN1EC and TSN2EC lines) segregated from the same population, so that the mutant and complemented control have similar genetic backgrounds (additional information is in Methods and Supplemental Figure 2 online).
In optimal in vitro conditions, tsn1 tsn2 seedlings showed a moderate reduction (10 to 15%) in root growth when compared with lines complemented by either TSN1 or TSN2 (Figures 2A and 2B). This was accompanied by a quantitatively comparable cell elongation defect (see Supplemental Figure 3 online). A slight but statistically significant growth defect was also apparent in tsn1 and tsn2 single mutants, compared with their respective wild-type counterparts (Figures 2A and 2B). Protein blot analysis employing an antiserum recognizing TSN1 and TSN2 equally well (see Methods) showed that complemented lines express a protein product of the expected size, whereas the signal is completely absent in tsn1 tsn2 (Figure 2C).
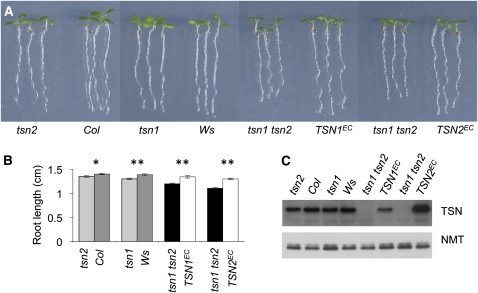
Figure 2.
Root Growth of tsn1, tsn2, and tsn1 tsn2 Mutants.
(A) Picture of representative 5-d-old seedlings. In this and following figures, tsn1 and tsn2 are compared with their respective wild-type ecotype, whereas the double mutant phenotype is reported employing two representative pairs of tsn1 tsn2 and nearly isogenic TSN1EC or TSN2EC control lines (the latter obtained by transformation of tsn1 tsn2 with TSN wild-type loci; see text).
(B) Root length of 5-d-old seedlings. Note that a slight but statistically significant root elongation defect of the single mutants is enhanced in tsn1 tsn2. Standard error bars are indicated (n = 43 to 70). *, **: Student's t test, P values < 0.05 or < 0.001, respectively.
(C) Detection of TSN by protein blot. An expected band of ∼110 kD is present in the TSNEC plants and completely absent in tsn1 tsn2 (TSN2EC lines systematically overproduced TSN compared with TSN1EC plants). Detection of N-myristoyltransferase (NMT) serves as a control.
Redundant Expression Patterns of the At-TSN1 and At-TSN2 Genes
To establish the expression domains of TSN, green fluorescent protein (GFP) translational fusions driven by the putative TSN promoters were introduced into wild-type Col-0 plants and into the double mutant. Very similar patterns of fluorescence were detected for TSN1 and TSN2 constructs (as well as with TSNpro:GFP transcriptional fusions; see Supplemental Figure 4 online) with accumulation maxima in the cap and elongation zone of the root apices (Figure 3A). TSN protein could also be detected by protein blot in leaves, flowers, siliques, and mature seeds (Figure 3B), which also correlates with the TSN1 and TSN2 transcript expression profiles available in microarray databases (Schmid et al., 2005; see Supplemental Figure 5 online).
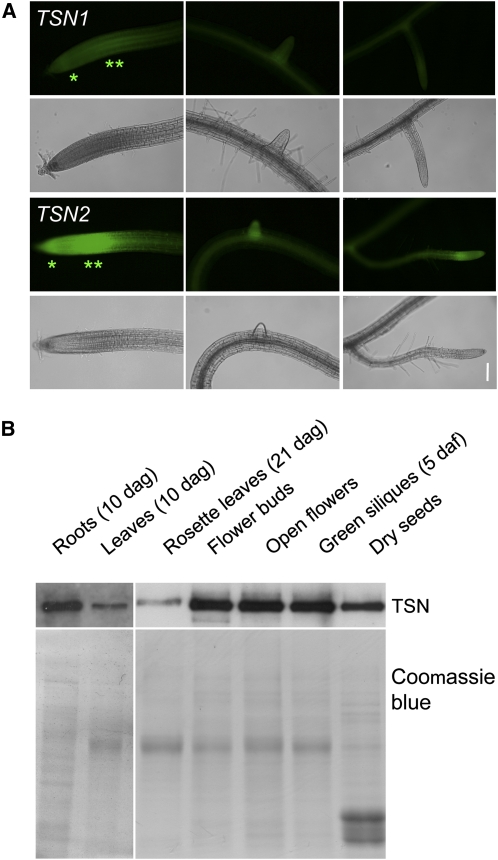
Figure 3.
TSN1 and TSN2 Expression Patterns.
(A) GFP patterns of TSN1pro:GFP:TSN1 (TSN1) and TSN2pro:GFP:TSN2 (TSN2) translational fusions in 10-d-old roots. Epifluorescence signals are shown above the corresponding visible light images. Expression maxima locate to the root cap (asterisk) and elongation zone (double asterisk) of the primary apex (left panels) and in lateral root tips, with the TSN2 construct producing a stronger signal.
(B) Detection of TSN by protein blots in different organs of Col-0 plants. Coomassie blue staining of a twin gel serves as a control. dag, days after germination; daf, days after flowering.
[See online article for color version of this figure.]
The Double Mutant tsn1 tsn2 Is Hypersensitive to Stress
The mild phenotypes initially observed in tsn1 tsn2 were surprising, since the striking evolutionary conservation of TSN can be explained only by persistent and important selective pressures. Standard laboratory conditions, however, clearly have little in common with natural environments and can mask essential functions. Indeed, severe phenotypes were revealed by submitting seedlings to long-term salinity stress on media supplemented with 100 mM NaCl to mimic prolonged drought. In these conditions, growth and survival rates of tsn1 and tsn2 mutants were not significantly different from those of the wild type. By contrast, tsn1 tsn2 was severely affected (Figure 4A). In addition, a large portion (50%) of tsn1 tsn2 seedlings died shortly after transfer to higher NaCl concentrations (150 mM; Figure 4B), while plants complemented by TSN1 showed a 87% survival rate. TSN2EC lines failed to complement this phenotype (see Supplemental Figure 6 online). Therefore, TSN function is required for stress adaptation in Arabidopsis seedlings, and the roles of TSN1 and TSN2 may not be completely redundant at high stress levels.
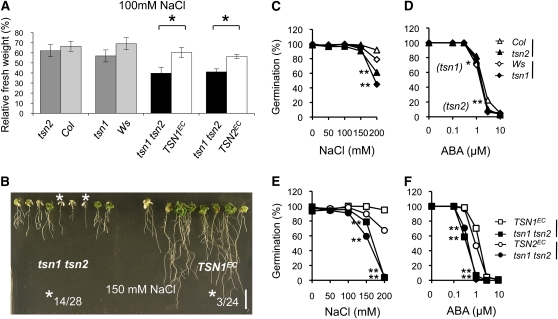
Figure 4.
Salt Hypersensitivity of tsn1 tsn2 and Germination Sensitivity of Single and Double Mutants to Salt and ABA.
(A) Fresh weight of plants grown for 17 d on growth medium (GM) supplemented with 100 mM NaCl, reported as percentage of fresh weight on GM. Seedlings were germinated on GM and grown for 4 d before transfer to salt-supplemented or control plates. Standard error bars are indicated (n = 25 to 30). *, Student's t test, P values < 0.05.
(B) Representative picture of seedlings grown for 17 d on 150 mM NaCl plates. Note that large rates of mortality occur in tsn1 tsn2 seedlings (white asterisk) soon after transfer to salt-containing media and that growth of surviving individuals is severely stunted. These phenotypes are complemented in controls transformed with the TSN1 locus (TSN1EC). The ratio of plants scored as dead to the total seedlings analyzed is indicated for each genotype. The ratios are significantly different: χ2 test of independence, P value < 0.001. Bar = 1 cm.
(C) to (F) Germination on increasing ABA or NaCl concentrations. Germination of the double mutant is clearly hypersensitive to both NaCl (E) and ABA (F). Note that both phenotypes are suppressed in the complemented lines and are already detectable in the single mutants ([C] and [D]). Lines at the sides of genotypes identify mutant/control pairs. *, **: P values < 0.05 or < 0.001, respectively, indicating significant difference from corresponding controls, χ2 test of independence.
To explore further the implication of TSN in salt stress tolerance, we also followed seed germination of each of the single and double TSN mutants. Germination of all mutants was significantly salt hypersensitive, with tsn1 tsn2 showing the strongest phenotype, failing almost completely to germinate at 200 mM NaCl. Both TSN1EC and TSN2EC lines were efficiently complemented (Figures 4C and 4E). Next, we sought to establish whether these phenotypes reflected a deregulation in the response to ABA, the major negative regulator of germination and an important mediator of abiotic stress tolerance. Interestingly, germination of tsn1 tsn2 was indeed clearly ABA hypersensitive, being almost completely inhibited at 1 μM, a concentration having little or no effect on the single mutants and the complemented lines (Figures 4D and 4F). TSN protein levels were largely unchanged by short-term (up to 48 h) NaCl or ABA treatments of wild-type, tsn1, or tsn2 seedlings (see Supplemental Figure 7 online), indicating the absence of a direct regulation of protein abundance. We noticed, however, that a TSN1 Ser residue shown to be phosphorylated in a proteomic study (de la Fuente van Bentem et al., 2006) is conserved among plant homologs (S974; see Supplemental Figure 1 online), suggesting possible posttranslational modulations of TSN activity.
These results show that TSN is required for germination as well as for growth and survival during high salinity stress. We also observed that germination and growth of tsn1 tsn2 are still stress hypersensitive, albeit to a lesser extent and without increased mortality, when NaCl is replaced by mannitol (see Supplemental Figure 8 online). Therefore, this suggests that TSN function is required for adequate responses to both the salt toxicity and the osmotic components of high salinity stress.
Reduced Fitness of Soil-Grown tsn1 tsn2 Plants
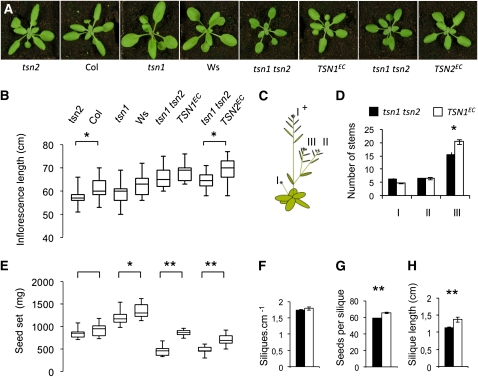
In agreement with the sensitivity of the tsn1 tsn2 seedlings to abiotic stress, double mutant plants cultivated in the greenhouse often produced smaller rosettes and inflorescences, shorter and poorly filled siliques, and, consistently, fewer seeds in comparison to control plants (see Supplemental Figure 9 online). In the more controlled environment of a growth chamber, tsn1 tsn2 siliques were completely filled, and tsn1 tsn2 plants displayed normal growth traits except for moderate reductions in silique length and tertiary branching (Figure 5). Seed set, however, was still significantly and reproducibly affected, with the double mutant producing from 54 to 70% of the seed mass of control lines. Furthermore, a deficiency in seed set was already present in tsn1 (Figure 5E).
Figure 5.
Reduced Fitness of Soil-Grown tsn1 tsn2 Plants.
(A) Rosette morphology (no significant differences were measured) of plants in the growth chamber, under optimal conditions.
(B) Main inflorescence length (boxes contain the two central quartiles separated by the median; whiskers extend to the minimum and maximum values observed).
(C) Illustration depicting the positions of (I) primary, (II) secondary, and (III) tertiary stems considered for branching analysis.
(D) to (H) Fitness traits (n = 12 plants): (D) branching; (E) seed production per plant (seed set); (F) silique density on the main inflorescence (I+); (G) seeds per silique (n = 24); and (H) silique length (n = 24). Standard error bars are shown. In these conditions, siliques were completely filled. *, **: Significant difference: t test P values < 0.05 and < 0.001, respectively.
[See online article for color version of this figure.]
TSN Is Heterogeneously Distributed in the Cytoplasm and Transiently Relocates to a Diffuse Pattern in Response to Salt Stress
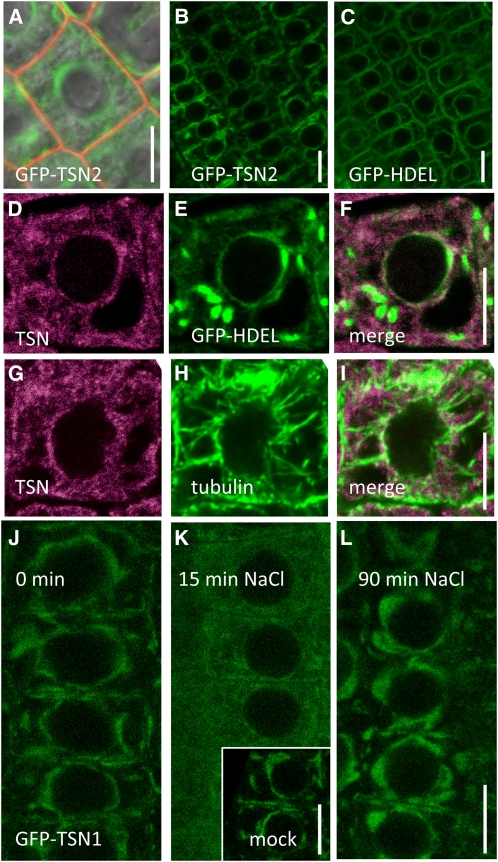
The above physiological analysis of tsn1 tsn2 revealed that TSN is required for the biologically and agronomically important traits of stress tolerance and plant fitness. As a first step toward the determination of the molecular processes affected by TSN, we examined its subcellular localization. In all cells observed, TSN1pro:GFP:TSN1 and TSN2pro:GFP:TSN2 lines displayed the same expression pattern. The GFP signal was clearly excluded from the nucleus and accumulated heterogeneously in the cytosol, but in patches around the nucleus and in the cell periphery, as particularly evident in root epidermal cells in the division zone (Figure 6A). These translational fusions suppressed the root elongation and germination defects of tsn1 tsn2 (see Supplemental Figure 10 online), indicating correct localization and functionality.
Figure 6.
TSN Subcellular Localization in Root Epidermal Cells.
All micrographs represent root epidermal cells at the basal end of the division zone. Bars = 5 μm.
(A) Signal of TSN2pro:GFP:TSN2 translational fusion (Col background), merged with the image of propidium iodide–stained cell walls (red signal) and with the visible light image. Note that TSN is excluded from the nucleus and dispersed heterogeneously throughout the cytosol, with a fraction accumulating in a perinuclear fashion. GFP patterns are indistinguishable in the tsn1 tsn2 background and/or with TSN1 fusions (see below).
(B) and (C) Comparison of ER-associated GFP and GFP-TSN signals. Note that ER-confined GFP:HDEL and the GFP-TSN are both found in the perinuclear region.
(D) to (F) Immunolocalization of TSN in Col roots expressing a GFP marker of the ER lumen (GFP-HDEL). TSN signal (D), GFP-HDEL signal (E), and merge (F). Partial colocalization is particularly apparent around the nucleus. Fusiform GFP structures are ER bodies (Matsushima et al., 2003).
(G) to (I) TSN and tubulin signals covisualized by immunolocalization in Col roots. The two patterns are clearly distinct, although compatible with partial colocalization.
(J) to (L) Effect of high salinity on TSN subcellular localization at 0 min (J), 15 min (K), and 90 min (L) after transfer to media supplemented with 150 mM NaCl. Note that, at 15 min, the GFP signal is homogeneously dispersed throughout the cytosol. At 90 min, the localization of TSN returns to that of untreated plants.
The observed perinuclear pattern is characteristic of an association with the nuclear envelope and the ER (Figure 6C; see, for example, Hong et al., 1999; Ridge et al., 1999; Dharmasiri et al., 2006). Therefore, TSN was also immunolocalized in plants expressing a GFP-HDEL fusion, which labels the ER lumen and ER bodies, present in epidermal root cells in our growth conditions (Haseloff et al., 1997; Matsushima et al., 2003). Partial colocalization of the TSN and GFP-HDEL signals was consistently observed in the perinuclear region (Figures 6D to 6F). Therefore, in specific areas of the cell, a fraction of the TSN is in close proximity with ER subcompartments. Since TSN was reported to interact with the cytoskeleton (Sami-Subbu et al., 2001; Abe et al., 2003; Chuong et al., 2004; Wang et al., 2008), a similar colocalization was performed using anti-α-tubulin antibodies (Figures 6G to 6I). Tubulin and TSN patterns are obviously distinct, but do show regions of overlap where the two proteins might interact. We next examined whether TSN subcellular localization could be dynamically regulated in response to salt stress, since TSN is functionally important for high salinity tolerance (see above). Indeed, both TSN1 and TSN2 GFP fusions were found to be dramatically and transiently redistributed within minutes upon imposition of NaCl stress. Within 15 min, the GFP signal shifted from a patchy, mostly perinuclear pattern to a signal homogenously diffused throughout the cytosol and then returned to its initial state within ER regions at later time points (Figures 6J to 6L).
TSN Is Dispensable for RISC Activity
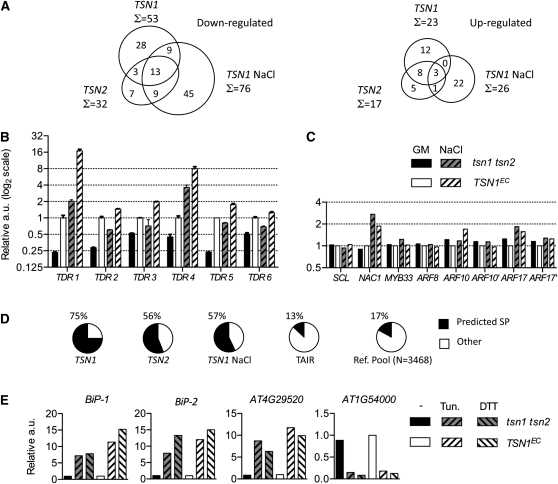
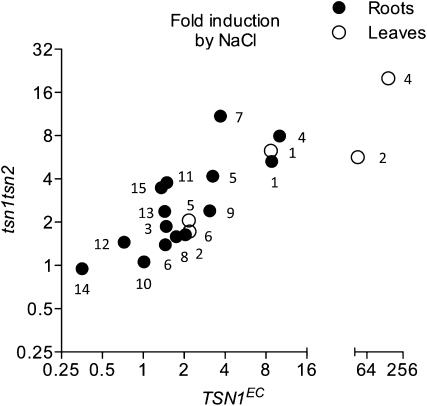
The elucidation of the molecular role of TSN proteins through identification of physiological targets or partners has been the focus of remarkable research efforts. Notably, several biochemical studies have suggested a direct interaction of TSN with RNA (Sami-Subbu et al., 2001; Scadden, 2005; Li et al., 2008; Paukku et al., 2008; Friberg et al., 2009) and, in animals, a role for TSN in the RISC (Caudy et al., 2003; Grishok et al., 2005). Therefore, a transcriptome analysis was undertaken to identify mRNAs whose abundance is regulated via TSN. Three sets of genome-wide profiling experiments were performed to compare the transcriptome of tsn1 tsn2 roots to that of plants complemented by either TSN1 or TSN2 in standard growth conditions or complemented by TSN1 under salt stress. For the latter experiment, we chose to employ seedlings 48 h after transfer to solid media supplemented with 100 mM NaCl. In these conditions, tsn1 tsn2 plants have not yet developed the general growth defects evident in the longer term experiments (see Supplemental Figure 11 online). Out of the ∼12,500 transcripts scored as expressed, 76 transcripts were underrepresented and 26 overrepresented (statistical significance threshold of Bonferroni's P < 0.05) in the mutant with respect to TSN1EC under salt stress (Figure 7A). Fewer deregulations were revealed on standard media or in the comparison with roots expressing TSN2, for the latter case possibly as a consequence of a less efficient complementation in the TSN2EC line. However, the intersection between the populations of TSN1 and TSN2-dependent transcripts indicate that the functional redundancy observed for the two genes in physiological tests persists at the molecular level. Complete lists of transcripts classed as differentially expressed by statistical analysis of transcriptome data (P < 0.05) are presented in Supplemental Data Sets 1A to 1C online.
Figure 7.
Root Transcriptome of tsn1 tsn2.
(A) Venn diagrams representing downregulated and upregulated transcripts in tsn1 tsn2 identified by whole genome comparisons (P < 0.05). tsn1 tsn2 was compared with TSN1EC or TSN2EC on standard media (TSN1 and TSN2) and with TSN1EC on media supplemented with 100 mM NaCl (TSN1 NaCl).
(B) Expression of TSN-dependent transcripts (TDR) in roots of 7-d-old seedlings grown in standard conditions (GM) or transferred at 5 d to media supplemented with 100 mM NaCl (NaCl). Values are reported in actin units on a log2 scale and are relative to TSN1EC levels on GM. Key is as in (C). Error bars are standard error of three qPCR replicates. One of three comparable biological replicates is shown.
(C) Characterized microRNA targets are not significantly affected in tsn1 tsn2. Samples are as in (B). qPCR primers surrounded the cleavage sites. Additional primers placed downstream of the cleavage site were employed for ARF10 and ARF17 (ARF10’ and ARF17’) to detect also the 3′ cleavage products. One of three comparable biological replicates is shown.
(D) Percentage of downregulated transcripts encoding proteins with a predicted signal peptide (SP) in the different microarray experiments, labeled as in (A). The frequencies of SP transcripts in a reference pool of TAIR transcripts within the same abundance and length ranges (Ref. Pool), or in all Arabidopsis mRNAs (TAIR), are included for comparison (see Methods).
(E) Markers of the UPR respond normally to chemical induction of UPR in roots of tsn1 tsn2 and TSN1EC. Levels are relative to the complemented control on standard media. One of two comparable biological repeats is shown. −, standard media; Tun., tunicamycin; a.u., actin units.
Known targets of small RNAs were little affected by the tsn1 tsn2 mutation, with only three transcripts appearing as underrepresented in tsn1 tsn2 out of the 70 to 90 targets scored as expressed in the different tsn1 tsn2 versus TSNEC comparisons (see Supplemental Data Set 1D online). Major perturbations of RISC cleavage activity in the tsn1 tsn2 mutant are therefore absent, a conclusion also confirmed by quantitative RT-PCR (qRT-PCR) on known ARGONAUTE1-dependent microRNA targets expressed in roots, employing primers flanking the characterized cleavage sites (Figure 7C). RNA gel blot analysis of the 3′ product generated by mir160-targeted cleavage of the ARF10 mRNA confirmed this observation (see Supplemental Figure 12 online). This is consistent with the absence, in the tsn1 tsn2 mutant, of the dramatic developmental abnormalities described in mutants affected in the AGO1 protein (Morel et al., 2002) or in other important steps of small RNA pathways, such as in the dcl1 mutants (Schauer et al., 2002) or in overexpressors of viral silencing inhibitors (Dunoyer et al., 2004). Furthermore, careful scrutiny of leaf morphology did not reveal the heterochronicity hallmarks characteristic of mutants of the trans-acting small interfering RNA pathways (such as zip-1/ago7, rdr6-11, and sgs3-11; Hunter et al., 2003; Gasciolli et al., 2005). Thus, TSN is, at least in plants, dispensable for global RISC activity. Further analysis of the transcriptome data, as shown below, proved instrumental in defining the molecular role of TSN in vivo.
A Distinct Set of mRNAs Entering the Secretory Pathway Is Affected in the tsn1 tsn2 Mutant
Transcripts underrepresented in tsn1 tsn2 in at least one microarray experiment with Bonferroni's P values < 10−12 are presented in Table 1. These mRNAs are ranked according to their fold reduction in the double mutant under NaCl stress, as established by qRT-PCR analysis on tsn1 tsn2 and TSN1EC seedlings. Downregulation in tsn1 tsn2 could be reliably reproduced by qRT-PCR (Figure 7B) for 15 of the 18 transcripts tested. For simplicity, we termed these transcripts TDR1 to 15, for TSN-Dependent RNA. Based on the patterns of changes in expression, two main groups, with some transcripts intermediate between them, can be distinguished: transcripts strongly downregulated under NaCl stress and comparably affected on standard media (TDR1 to 6), and mRNAs affected only in standard conditions (TDR11 to 15). The latter reach normal levels or are possibly slightly overexpressed in tsn1 tsn2 in response to high salinity. Ranking according to the impact of the tsn1 tsn2 mutation under stress appears therefore to be a good criterion to select TDRs more directly regulated by TSN.
Table 1.
Transcripts Downregulated in tsn1 tsn2 Roots
| Expression Ratio |
|||||||||
| Microarray |
qRT-PCR |
||||||||
| TDRa | AGIb | TSN1c | TSN2d | NaCle | GMf | NaClf | TAIR Annotationg | SPh | CRPi |
| 1 | AT2G43510 | 0.3 | 0.1 | 0.1 | Defensin-like (DEFL), protease inhibitor | + | 700 | ||
| 2 | AT3G53980 | 0.2 | 0.2 | 0.2 | 0.5 | 0.3 | Protease inhibitor/lipid transfer protein | + | 4730 |
| 3 | AT2G16005 | 0.4 | 0.2 | 0.4 | 0.4 | MD-2 lipid recognition domain protein | + | − | |
| 4 | AT2G37870 | 0.3 | 0.6 | 0.4 | Protease inhibitor/lipid transfer protein | + | 4750 | ||
| 5 | AT3G04720 | 0.4 | 0.3 | 0.5 | Pathogenesis-related 4 PR4 | + | 5800 | ||
| 6 | AT1G17860 | 0.4 | 0.5 | 0.5 | Trypsin and protease inhibitor family | + | 6020 | ||
| 7 | AT5G42600 | 0.4 | 0.3 | 0.6 | Marneral synthase MRN1 | − | − | ||
| 8 | AT2G18370 | 0.4 | 0.6 | 0.6 | Protease inhibitor/lipid transfer protein | + | 4060 | ||
| 9 | AT2G33790 | 0.2 | 0.9 | 0.6 | Pollen Ole e 1 allergen protein | + | 3510 | ||
| 10 | AT1G12090 | 0.4 | 0.7 | 0.6 | Extensin-like protein ELP | + | 4820 | ||
| – | AT1G45015 | 0.4 | 1.2 | 0.7 | MD-2-lipid recognition domain protein | + | − | ||
| 11 | AT3G58550 | 0.3 | 0.3 | 0.4 | 1.0 | Protease inhibitor/lipid transfer protein | + | 4660 | |
| 12 | AT1G73330 | 0.4 | 0.3 | 0.5 | 1.0 | Protease inhibitor ATDR4 | + | 6080 | |
| 13 | AT5G05960 | 0.4 | 0.7 | 1.0 | Protease inhibitor/lipid transfer protein | + | 4730 | ||
| 14 | AT2G19970 | 0.3 | 0.4 | 1.1 | Pathogenesis-related protein, putative | + | − | ||
| 15 | AT3G08770 | 0.4 | 0.5 | 1.3 | Lipid transfer protein LTP6 | + | 3860 | ||
| – | AT3G22840 | 0.4 | 0.3 | 0.8 | 1.4 | Early light-inducible protein ELIP1 | − | − | |
| – | AT4G27080j | 0.3 | 0.4 | 0.2 | 1.0 | 1.0 | Protein disulfide isomerase-like | − | − |
| – | AT1G48760 | 0.4 | ndk | ndk | δ-Adaptin | − | − | ||
Only transcripts associated with probability scores <10−12 in at least one genome-wide tsn1 tsn2/TSNEC comparison are shown, with the corresponding high scoring ratios. Transcripts are ranked according to their downregulation in tsn1 tsn2 under NaCl stress as determined by qRT-PCR (see Supplemental Data Sets 1A to 1C online for complete listings).
TSN-dependent RNA number, assigned to the 15 transcripts whose deregulation was confirmed in at least two independent biological repeats by qRT-PCR.
Arabidopsis Genomic Initiative number.
tsn1 tsn2/TSN1EC expression ratio in microarray experiments using plants grown in standard conditions.
tsn1 tsn2/TSN2EC expression ratio in microarray experiments using plants grown in standard conditions.
tsn1 tsn2/TSN1EC expression ratio in microarray experiments using plants grown under high salinity stress.
tsn1 tsn2/TSN1EC expression ratios measured by qRT-PCR in at least two biological repetitions involving plants grown in standard conditions (GM) or under high salinity stress (NaCl).
Database annotation of the protein product.
Presence (+) or absence (−) of a predicted signal peptide.
When applicable, Cys-rich peptide family class (Silverstein, 2007).
AT4G27080 never showed differential expression in qRT-PCR assays (possible spot identity error on the array).
Not determined.
In contrast with the robust downregulation of underrepresented transcripts (Table 1, Figure 7B), deregulation of RNAs overrepresented in tsn1 tsn2 in the microarray experiments (nine transcripts with Bonferroni's P < 10−12) was observed by qRT-PCR in only ∼50% of the cases, across independent biological repetitions and different media batches (see Supplemental Data Sets 1A to 1C online). This suggests that overrepresented transcripts are more indirect and environment-dependent markers of increased stress in tsn1 tsn2. Two of the more consistently upregulated transcripts (AT4G13770 and AT5G23020) encode enzymes of the glucosinolate biosynthetic pathway, suggesting that production of these stress-related metabolites could be upregulated in the mutant. Since inspection of all of the proteins predicted from transcripts overrepresented in tsn1 tsn2 did not yield additional common functional traits, these mRNAs were not investigated further in this study. By contrast, a striking feature was readily apparent among downregulated transcripts, even when enlarging the analysis to include all underrepresented mRNAs (P < 0.05). Downregulated transcripts were strongly and significantly enriched (20- to 57-fold; see Supplemental Data Sets 1A to 1C online) in mRNAs encoding Cys-rich peptides (Silverstein 2007) and, more generally, in transcripts predicted to be translated on the ER (i.e., encoding proteins with a putative signal peptide) (Figure 7D).
Secretory pathways are known to be downregulated, at both transcriptional and posttranscriptional levels, during the unfolded protein response (UPR), a cellular reaction to the stress-induced accumulation of misfolded proteins in the ER lumen (Ron and Walter, 2007; Vitale and Boston, 2008). Indeed, levels of all of five TDRs tested (TDR1, 2, 5, 11, and 14) were found to be reduced by chemical induction of the UPR (see Supplemental Figure 13 online). Therefore, we asked whether downregulation of mRNAs encoding specific groups of secreted proteins in the tsn1 tsn2 mutant could be ascribed to a global perturbation of the UPR. We monitored the expression of known molecular markers of this response (Martinez and Chrispeels, 2003) in tsn1 tsn2. On standard media, transcripts levels of genes encoding ER luminal binding proteins (BiP-1 and BiP-2) and of an additional UPR-responsive marker (AT4G29520) were at the same low levels in the double mutant and in the complemented control. When the UPR was activated by the presence of tunicamycin or DTT, all three mRNAs were similarly induced in both genotypes. In addition, the opposite and expected behavior was seen with the UPR-repressed marker AT4G54000 (Figure 7E). These data rule out a constitutive activation of the unfolded protein response in tsn1 tsn2.
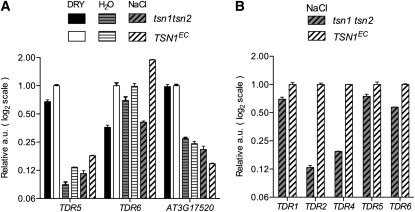
Together, these results show that TSN is required for optimal expression of a specific set of mRNAs encoding putatively secreted proteins in Arabidopsis roots. Next, we investigated whether levels of TDRs were also dependent on TSN in aerial parts and seeds, since tsn1 tsn2 plants showed NaCl hypersensitivity at the whole seedling level and at germination. Among the 15 transcripts listed in Table 1, TDR5 and 6 are reported to be significantly expressed in mature seeds and during germination, whereas TDR1, 2, and 4 to 6 are expressed in leaves (Schmid et al., 2005). Strikingly, TDR6 levels in tsn1 tsn2 were 3- to 5-fold reduced in mature seeds and in seeds imbibed in 200 mM NaCl (Figure 8A), while levels of TDR2 and 4 showed 5- to 10-fold reductions in the aerial parts of mutant seedlings under salt stress (Figure 8B). The observation that transcripts identified as TSN dependent in roots can be affected also in other tissues of tsn1 tsn2 plants suggests that the observed decrease in TDR levels is closely connected to the action of TSN. If this is the case, TDRs should share characteristics able to explain the salt sensitivity of the tsn1 tsn2 mutant and their connection with the TSN protein. The first point was addressed by probing the response of TDRs to NaCl stress.
Figure 8.
Impact of tsn1 tsn2 on TDRs Expressed in Seeds and Aerial Parts.
(A) Transcript levels for two seed expressed TDRs were quantified in mature seeds (DRY), after 24 h of imbibition in water or after 24 h of imbibition in 200 mM NaCl (NaCl). Values, plotted on a log2 scale, are relative to levels in mature seed of the control. Note the 3- to 4-fold reduction of TDR6 levels in mature seeds and in NaCl imbibed seeds of the double mutant. Late embryogenesis abundant mRNA AT3G17520 was included as a control. Seeds were pooled from 12 plants per genotype and showed >90% germination in water.
(B) Impact, under NaCl stress, of the tsn1 tsn2 mutation on TDRs known to be present in aerial parts. Note the strong downregulation of TDR2 and TDR4 in aerial parts. A full representation of this experiment can be found in Supplemental Figure 14B online. Error bars are standard errors of three qPCR measurements on two biological repeats. a.u., actin units.
Stress Regulation of TSN-Dependent mRNAs
The expression of TDRs was examined under the same conditions in which the NaCl sensitivity phenotype was observed in tsn1 tsn2 seedlings. Most TDRs (11/15) were found to be upregulated by high salinity treatment, in the roots and aerial parts of both tsn1 tsn2 and control plants (Figure 9). Importantly, the transcriptional response to salt stress is globally normal in tsn1 tsn2, a notion confirmed by the correct regulation in tsn1 tsn2 of RD29A and RD29B, two well-characterized stress and ABA-responsive markers (see Supplemental Figure 14A online). Therefore, the results presented so far suggest that TSN is required for specific types of mRNAs entering the secretory pathways to attain, or maintain (for TDRs not upregulated by NaCl), sufficient levels for adaptive responses. Interestingly, aerial parts of tsn1 tsn2 seedlings show reduced TDR levels only when under stress (see Supplemental Figure 14B online), which is consistent with this idea.
Figure 9.
Salt Regulation of TSN-Dependent Transcripts in Roots and Aerial Parts.
Induction factors of TDR 1-15 in TSN1EC and tsn1 tsn2 roots (closed circles) and aerial parts (open circles), 48 h after transfer of 5-d-old seedlings to solid media supplemented with 100 mM NaCl. Values, plotted on log2 scales and numbered according to Table 1, are the average of qRT-PCR data from two biological repetitions.
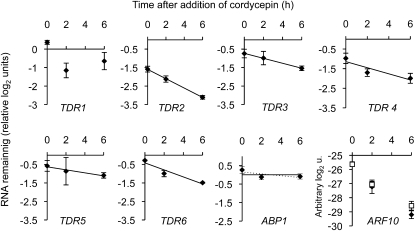
TSN-Dependent RNAs Are Destabilized in tsn1 tsn2
Since TSN displays RNA binding and nuclease activities and is excluded from the nucleus, we asked next whether TSN acts at the posttranscriptional level. RNA decay kinetics of TDR1 to 6 were measured in root tissue, under NaCl stress (Figure 10). In the liquid culture conditions required for application of the transcription inhibitor cordycepin, the mRNA levels of these TDRs, with the exception of TDR1, were still affected by the tsn1 tsn2 mutation and decayed in good agreement with a first order reaction. Their degradation rates were specifically and reproducibly increased in the mutant. This was in contrast with mRNA levels of ABP1, which also is a secreted protein. Therefore, the observed downregulation of TDRs in the absence of TSN can be explained, at least in part, by a selective destabilization of the transcript.
Figure 10.
TSN-Dependent Transcripts Are Destabilized in tsn1 tsn2 Roots.
mRNA remaining in roots of tsn1 tsn2 after 0, 2, and 6 h incubation in the presence of cordycepin. Values are relative to levels in TSN1EC and expressed in log2 units, except when noted otherwise. TDR2 to 6 are degraded more rapidly in tsn1 tsn2, in contrast with ABP1 (encoding a secreted protein) included as a control. Data were normalized to the geometric average of ACTIN and IAA2 levels. ARF10 mRNA levels are reported in arbitrary log2 units (u.; negatives of qPCR crossing points), without any normalization. ARF10 levels decrease steeply and similarly in tsn1 tsn2 (filled diamonds) and TSN1EC (open squares), indicating the efficiency of transcriptional inhibition by cordycepin. All experiments were performed in liquid media supplemented with 100 mM NaCl, as described in Methods. TDR1 behaves differentially in liquid conditions (no downregulation at 0 h). Error bars are standard errors of three qPCR measurements on two pooled biological repeats.
DISCUSSION
This study allowed us to define an important role for TSN at the whole plant level. Absence of TSN protein drastically affected germination, seedling growth, survival, and fitness in unfavorable environmental conditions, revealing that optimal stress tolerance throughout the life cycle is the trait responsible for the evolutionary conservation of this RNA binding protein, at least in plants. The two Arabidopsis TSN genes appeared largely functionally redundant, with TSN1 showing moderate predominance. This was best seen in the comparison of single mutants to their corresponding wild-type ecotype in germination assays (Figure 4C) and indicated by the complementation efficiency of the double mutant by expressing either the TSN1 or TSN2 wild-type transgene (Figures 4B and 4E).
Several RNA-interacting proteins have already been identified as being involved in ABA-regulated germination by forward genetic screens for hypersensitive mutants (Kim et al., 2007; Kuhn et al., 2007, 2008). Therefore, the ABA hypersensitivity of tsn1 tsn2 germination and the observed strong requirement for TSN for germination under salt stress (Figures 4E and 4F) may reflect an involvement of TSN in ABA-regulated reprogramming of mRNA metabolism during seed maturation and germination (Rajjou et al., 2004; Holdsworth et al., 2008), as suggested by the dramatic deregulation of TDR6 in seeds. In addition, the requirement for TSN for the survival of seedlings at the onset of salt stress (Figure 4B) suggests its implication in analogous processes critical for adaptation to these conditions (Ma et al., 2006; Jiang et al., 2007). Indeed, the induction of TSN-dependent transcripts by high salinity (Figure 9) suggests that the NaCl hypersensitivity of tsn1 tsn2 is caused, at least to some extent, by insufficient levels of salt-regulated mRNAs important for tolerance. Such transcripts are well represented among Cys-rich proteins, the main targets of TSN action in Arabidopsis roots; several members of this family of secreted peptides are involved in abiotic and biotic stress responses (Silverstein, 2007). Little is known about the precise function of the identified TSN-dependent genes. However, it is tempting to connect the presence of numerous protease inhibitors (Table 1) among the proteins encoded by TDRs to the antiapoptotic role recently proposed for TSN in eukaryotes (Sundström et al., 2009).
Strikingly, most of the transcripts identified as downregulated in tsn1 tsn2 roots are predicted to encode proteins translated on the ER (Figure 7D). Several converging lines of evidence suggest a link between TSN activity and levels of mRNAs entering the secretory pathway. First, presence of a putative signal peptide is the most prominent common trait of TSN-dependent transcripts (see Supplemental Data Sets 1A to 1C online), in which we did not detect other globally shared motifs in the primary sequence nor in the promoter regions of corresponding genes. Secondly, previous functional connections between TSN and secretion activity, while few, have been reported in animal systems: lowering of TSN levels by antisense RNA reduced lipoprotein secretion in rat hepatocytes (Palacios et al., 2006), and TSN overexpression increased mRNA levels and translation of the signal peptide containing human angiotensin II type 1 receptor. The latter effect was independent from Hs-Argonaute2 and required the interaction of TSN with the 3′ untranslated region (Paukku et al., 2008). This observation suggests that additional factors, besides the presence of a signal peptide in the predicted protein product, determine whether a transcript will be subject to TSN-mediated regulation. The identification of a relatively small number of TSN-dependent mRNAs in Arabidopsis roots, mostly encoding Cys-rich peptides secreted to the cell wall, is consistent with this idea.
The third line of evidence linking TSN with levels of ER-translated mRNAs is provided by Wang et al. (2008), who have shown that rice endosperm tissue underexpressing TSN contains lower levels of storage protein transcripts, which are also translated on the ER. These authors proposed that TSN plays an important role in the transport and localization of storage protein mRNAs in rice. These transcripts are selectively transported to specific ER regions, where their translated products either accumulate locally and form protein bodies (prolamine) or enter a vesicular pathway to the storage vacuole (glutelin). A fraction of the TSN-containing cytosolic granules observed in rice albumen cells was cotransported with GFP-tagged prolamine mRNA. Interestingly, levels of both glutelin and prolamine transcripts were reduced upon partial downregulation of TSN in albumen tissue by RNA interference (Wang et al., 2008). Finally, the subcellular localization of TSN in plants is also compatible with an interaction with ER-associated transcripts: we observed a cytosolic signal dynamically associated with ER territories (Figure 6), and Wang et al. (2008) showed that a fraction of Os-TSN is present on the outer surface of ER storage protein bodies in rice albumen. In mammalian cells, ER association has been reported in specific cell types (Keenan et al., 2000; Low et al., 2006), and TSN is particularly abundant in tissues with high secretory activity (Broadhurst and Wheeler, 2001; Broadhurst et al., 2005).
Which molecular function underlies the dependency on TSN for specific sets of mRNAs remains an open question. Although the possibility of an implication in RNA silencing mechanisms has been evoked for TSN by several studies in animals, none of the observed molecular and physiological phenotypes of the tsn1 tsn2 mutant in this study are reminiscent of a known effect of loss of RISC-directed cleavage activity. Interestingly, Scadden (2008) has recently shown that TSN can act, in transfected animal cells, as a global regulator of gene expression, through selective degradation of double-stranded RNAs enriched in G:U pairs. These TSN substrates were capable of inducing the assembly of translationally inactive cytosolic ribonucleoparticles, associated with mRNA instability (Scadden, 2008). It will be important to assess whether such posttranscriptional regulatory mechanisms are conserved in plants and whether they could target specific sets of mRNAs in physiological conditions. If conserved, such processes could contribute to the dramatic reprogramming of gene expression taking place during adaptation of plants and germinating seeds to stress, two phenomena affected in the tsn1 tsn2 mutant. Interestingly, the TSN-dependent modulation of mRNA levels described by Scadden (2008) was moderate (2- to 3-fold) and in the same range as those measured in this study, as well as in a study employing reporter gene expression in animal cells (Paukku et al., 2008). This could suggest that analysis of total cellular RNA may average out stronger effects focused, perhaps within specialized RISC complexes, on a subpopulation of transcripts. Interestingly, we observed a rapid and transient displacement of TSN in response to salt stress, consistent with a role in reprogramming mRNA metabolism for stress tolerance. The identification within TSN of the molecular determinants of this dynamic localization may help determine not only its functional significance, but also the specific subcellular compartment and ribonucleoparticle complexes associated with TSN. In parallel, the identification of sequences required for destabilization of TDRs in tsn1 tsn2 will be instrumental in exploring the RNA–protein interactions tethering, perhaps through multiple steps, TSN-dependent RNAs to TSN.
In conclusion, our data provide a physiological function at the whole-plant level for TSN and define TSN genes as important determinants of yield and adaptive processes in plants. Furthermore, they also point to new mechanisms regulating the metabolism of mRNAs entering the secretory pathway. The tools provided by this work should prove instrumental in further exploration of TSN function at the molecular and cellular levels.
METHODS
Plant Material
A tsn1 RNA-null mutant was isolated by PCR from the Wisconsin University collection of T-DNA insertions lines (Sussman et al., 2000). After backcross to wild-type Ws, a line (line A3.4.11.2.2) showing single T-DNA insertion inheritance was crossed to a single insertion, RNA-null tsn2 line, obtained from the Salk collection of T-DNA mutants (line Salk_052200; Alonso et al., 2003). Three tsn1 tsn2 double mutants were identified by genotyping 180 F2 seedlings by PCR, and additional tsn1 tsn2 lines were similarly isolated in the F3. Columbia lines expressing free-GFP and HDEL-GFP under control of the 35S promoter were described by Lee et al. (2007) and Haseloff et al. (1997), respectively.
Constructs
Complementation
To complement the tsn1 tsn2 double mutant, TSN1 and TSN2 genomic fragments (TAIR 9.0 coordinates: chromosome 5, regions 2319549-2326809 and 24837448-24844381, respectively) were amplified from Col-0 by PCR (using Ex Taq enzyme; http://www.takara-bio.us). Oligonucleotides employed are listed in Supplemental Data Set 1E online (Oligos 1 to 4). TSN1 and TSN2 amplification products were cloned by KpnI-XbaI and KpnI-BamHI digestion, respectively, of the pFP100 binary vector (Bensmihen et al., 2004) to generate pFPTSN1 and pFPTSN2. These constructs include 1933 and 1790 nucleotides upstream of the translation initiation sites, and 551 and 185 nucleotides downstream of the respective polyadenylation sites, as defined in the TAIR 9.0 release. Constructs were verified by sequencing.
GFP Fusions
To construct the translational fusions driven by their own promoter TSN1pro:GFP:TSN1 and TSN2pro:GFP:TSN2, full-length cDNAs (Accession Numbers pda06063 and pda12297) were obtained from RIKEN (Seki et al., 2002). TSN open reading frames were amplified by PCR (Oligos 5 to 8; see Supplemental Data Set 1E online) and inserted by Gateway BP reaction in pDONR201 (Invitrogen). Subsequent LR reactions with the pSBRIGHT binary vector (Bensmihen et al., 2005) allowed the generation of GFP:TSN translational fusions. Finally, TSN promoter regions, amplified from pFPTSN1 and pFPTSN2, using oligonucleotides with 5′ extensions unmasking HindIII sites after BsaI digestion (Oligos 9 to 12; see Supplemental Data Set 1E online), were inserted into the upstream HindIII site of the pSBRIGHT GFP:TSN binary vectors.
Plant Transformation
Arabidopsis thaliana plants were transformed via Agrobacterium tumefaciens (AGL1 strain) by the floral dip method (Steven and Clough, 1998). Seeds of primary pFPTSN1 and pFPTSN2 (see above) transformants were screened for expression of At2S3:GFP (the visual marker carried by the pSBRIGHT T-DNA; Bensmihen et al., 2004) by epifluorescence microscopy. TSN1pro:GFP:TSN1 and TSN2pro:GFP:TSN2 transformants were similarly isolated by screening for GFP expression at the seedling stage, when in the tsn1 tsn2 background, or for Basta resistance when in the Col-0 background.
Mutant Complementation and Phenotyping Strategy
As schematized in Supplemental Figure 2 online, tsn1 tsn2 plants (F3 generation of the tsn2 × tsn1 cross) were transformed with pFPTSN1 or pFPTSN2, and single-copy T2(F5) families were isolated. Within each subsequent T3(F6) family, tsn1 tsn2 plants homozygous for an ectopic copy of the TSN1 or TSN2 locus (termed TSN1EC and TSN2EC) were isolated, as were untransformed tsn1 tsn2 siblings. In this way, independent pairs of tsn1 tsn2 and TSNEC lines were obtained and bulk propagated since. We refer to them as “nearly isogenic pairs” to emphasize the fact that within each pair of lines, the genetic backgrounds of the tsn1 tsn2 mutant and the TSNEC complemented control are much more similar (98% homozygosity) than among wild-type and tsn1 tsn2 descendants of the initial tsn2 (Col-0) × tsn1 (Ws) cross.
Phenotypes first observed in tsn1 tsn2 F3 lines by comparison with cosegregating wild-type plants were subsequently confirmed using at least two nearly isogenic pairs each for complementation by TSN1 or TSN2. Except when otherwise noted, results are shown for two representative pairs of lines allowing comparison of the double mutant to plants complemented by TSN1 (pair 3.2: tsn1 tsn2, line A13.3.11; TSN1EC, line A13.3.2) or TSN2 (pair 9.4: tsn1 tsn2, line A10.9.5; TSN2EC, line A10.9.4). Phenotyping was also performed on single tsn1 and tsn2 mutants compared with their respective Ws and Col-0 wild-type ecotypes.
Plant Growth and Physiological Tests
Seeds were harvested from mutant and control plants systematically cocultivated in growth chambers (22°C, 16 h light/8 h dark photoperiod, 400 to 500 μE·m−2·s−1, 70% relative humidity) and were used <6 months after harvest. Seeds were surface sterilized as described (Bechtold and Pelletier, 1998) and plated onto growth medium (GM) prepared as follows: half-strength Murashige and Skoog medium (Sigma-Aldrich M-5524; Murashige and Skoog, 1962), adjusted to pH 5.7 with 0.5 g/L MES and KOH, was supplemented with 150 mM sucrose and 0.5 or 1% w/v Vitro Agar (Kalys) for horizontal or vertical culture, respectively. After 48 h of stratification at 4°C in the dark, plates, shelled with gas-permeable medical tape (Urgo Medical), were transferred to a culture room at 22°C under continuous light (300 μE·m−2·s−1). For growth and seed set evaluation, plants were grown in a growth chamber at 22°C with a 16-h-light/8-h-dark photoperiod (300 μE·m−2·s−1) and 65% relative humidity. Plants were grown in the greenhouse, in the same conditions, for genetic crosses, selection of transformants, stock amplification, and the measurements shown in Supplemental Figure 9 online.
Root Elongation Assays
Four-day-old seedlings grown vertically on GM were transferred onto test media, and the positions of root apices were recorded onto the back of the plate with a pen. At the end of the experiment, plates were scanned and root length was measured using ImageJ software (NIH). Two plates containing both mutant and control seedlings were employed per experimental point, and 30 to 40 roots per genotype were measured. Experiments were repeated at least four times.
Cell Length Measurements
Root tips were photographed under bright-field microscopy, and the lengths of cells with an emerging root hair (Le et al., 2001) were measured with ImageJ. A total of 120 cells from 12 independent roots were measured per genotype.
Chemical Treatments
Seedlings were grown vertically for 6 d on sterile synthetic monofilament membranes (50-μm mesh; Buisine) laid on GM plates and transferred with the membrane onto the plate's lid. Appropriate solutions were gently applied to the seedlings, plates were closed upside-down, and roots harvested at the end of the incubation period. Tunicamycin and DTT were added at 5 g/mL and 10 mM in liquid GM, respectively, as already described (Martinez and Chrispeels, 2003). Cordycepin (100 μg/mL) was added in an incubation buffer already described (Johnson et al., 2000), supplemented with 100 mM NaCl. Environmental perturbations were minimized by starting longer treatments first, so that all time points could be collected simultaneously.
Salt and Mannitol Stress Treatments of Seedlings
To observe the effect of salinity or osmotic stress on growth and survival, 4-d-old seedlings germinated vertically on GM media were transferred to media supplemented with increasing concentrations of NaCl or mannitol and grown as described above. Nongrowing seedlings were scored as dead when aerial parts had become completely white. Growth was assessed through root elongation measurements in the first 3 d after transfer or by measurement of the fresh weight 17 d after transfer. For gene expression studies, seedlings were grown on nylon mesh (50 μm opening; Buisine) laid on vertical GM-agar plates to facilitate manipulation and quick harvest. At day 5, seedlings were transferred to plates supplemented with 100 mM NaCl or to control plates, and harvested 48 h later.
Germination Assays
Seeds were stratified on plates for 2 d at 4°C in the dark. Seventy-two hours after transfer to the growth chamber, seedlings with two open cotyledons were scored as germinated. To observe the effect of salinity or osmotic stress, increasing concentrations of NaCl or mannitol were included in the media prior to autoclaving. ABA stock solution was prepared in DMSO, and all plates prepared for ABA dilution series contained identical amounts of the solvent. Three plates, each containing both mutant and control lines (∼100 seeds/genotype), were scored per experimental point.
Growth, Branching, and Seed Set
In vitro–germinated 6-d-old seedlings were transferred to horticultural soil and placed randomly in the growth chamber to avoid position effects. The diameter of the rosette was measured on photographs taken of 3-week-old plants. Plants were grown until completion of flowering on the main inflorescence, the time at which watering was reduced and maintained for two additional weeks. Twelve plants per genotype were harvested for measurements when dry.
Antibodies and Protein Blotting
In order to express in Escherichia coli the TSN1 K709-R991 and TSN2 K705-G983 C-terminal domains, corresponding fragments of TSN1 and TSN2 coding sequences were amplified (Oligos 13 to 16 in Supplemental Data Set 1E online) and cloned in the NdeI site of pET15b (Novagen). Proteins were expressed in the Rosetta(DE3)pLysS BL21 derivative (Novagen) according to the manufacturer's instructions. After purification by nickel-agarose affinity chromatography followed by SDS-PAGE, equivalent quantities of the TSN1 and TSN2 peptides were coinjected into rabbits (Paris Anticorps).
For protein blotting, total proteins were extracted in 50 mM Tris HCl, pH 7.5, 1 mM EDTA, 0.1% Triton X-100, 10% glycerol, 1 mM DTT, and 0.2 mM AEBSF. After centrifugation at 4°C for 5 min at 13,000g, supernatants were used for protein quantification by the Bradford assay (Bradford, 1976). Two micrograms of total proteins were resolved by SDS-PAGE on 8% SDS-PAGE and electroblotted onto nitrocellulose Hybond C Extra (GE Life Sciences). When necessary, twin gels were run and stained with Coomassie Blue R 350, using standard procedures, to visualize total protein. Membranes were treated with the anti-TSN serum at 1:4000 dilution or anti-N-myristoyltransferase 1 IgGs (Pierre et al., 2007) as a control using standard methods. Peroxidase activity coupled to the goat anti-rabbit secondary antibody (Calbiochem; 1:20,000 dilution) was revealed with an enhanced chemiluminescence kit (GE Life Sciences or Millipore) following the manufacturer's instructions.
RNA
qRT-PCR
Total RNA was isolated from roots of in vitro–grown seedlings with the RNeasy plant mini hit (Qiagen) and subjected to DNase treatment (RNase-Free DNase set; Qiagen). RNA quality and quantity were determined by electrophoresis and spectrophotometry. For cDNA synthesis, 2 μg of total RNA was reverse transcribed in 20-μL reactions with the Superscript II enzyme and oligo(dT) following the manufacturer's instructions (Invitrogen). The reaction product (0.5 μL) served as template in a SYBR Green qPCR master mix (Roche) run on a LightCycler 2.0 (Roche), with primers listed in Supplemental Data Set 1E online. Quantifications were calculated as recommended by the manufacturer, using the second derivative maximum method. PCR conditions were 95°C for 10 min followed by cycling at 95°C for 5 s, 62°C for 5 s, and 72°C for 15 s. Fusion curves were characterized by 0.1°C·s−1 ramping from 60°C up to 95°C. Except when noted otherwise, data were normalized with respect to ACTIN2/8 (Charrier et al., 2002), and a minimum of three biological repeats was analyzed in duplicate or triplicate PCR reactions. For the RNA decay experiment in presence of cordycepin, data were first normalized to the geometric average of ACTIN2/8 and IAA2 and subsequently to values of the complemented control.
RNA Gel Blotting
Standard procedures were followed. After electrophoresis of 10 μg of total RNA in 1% agarose gels, RNA was transferred to a Hybond XL membrane (GE Life Sciences) and hybridized (Church and Gilbert, 1984) with α-dCTP32–labeled gene-specific probes generated by PCR from the TSN cDNA clones (Oligos 17 to 20 in Supplemental Data Set 1E online) or from an ARF10 cDNA clone (Oligos 31 and 32 in Supplemental Data Set 1E online). After washing at high stringency (2× 30’ at 2× SSC 0.1% SDS followed by 2× 15’ at 0.5× SSC 0.1% SDS, at 65°C), membranes were exposed to a phosphor screen, and signals visualized on a phosphor imager (STORM; GE Life Sciences). To serve as a loading control, rRNA signals were visualized by ethidium bromide staining of the agarose gels prior to blotting or by methylene blue staining of membranes, using standard methods.
Microarray Analysis
RNA was extracted from roots of 5-d-old seedlings or from 7-d-old seedlings 48 h after transfer to media supplemented with 100 mM NaCl. Twelve hybridizations were performed on the URGV Arabidopsis 25K CATMA_2.3 chip (Sclep et al., 2007), corresponding to two biological replicates, with dye swaps, of tsn1 tsn2 versus TSN1EC comparisons on standard and NaCl-supplemented media and of a tsn1 tsn2 versus TSN2EC comparison on standard media. Procedures employed for probe preparation, hybridization, scanning of the arrays, and image analysis employing GENEPIX PRO (Axon Instruments) have been previously described (Jammes et al., 2005). Normalization and statistical analysis were performed with the R package Anapuce (http://cran.r-project.org/web/packages/anapuce/index.html), as described in detail by Gagnot et al. (2008). For each tsn1 tsn2/TSNEC comparison, a paired t test was performed on the log ratios from four arrays (two dye swaps of two independent biological repeats), assuming that the variance of the log ratios was the same for all transcripts. P values were adjusted by the Bonferroni method, which controls the family-wise error rate. Transcripts with a signal ≥27 were considered as expressed and as differentially represented when associated with a Bonferroni P ≤ 0.05. Data have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (Edgar et al., 2002) and in the Complete Arabidopsis Transcriptome Database at URGV (CATdb; Gagnot et al., 2008).
Immunolocalization and Microscopy
Immunolocalization of TSN and α-tubulin in Arabidopsis roots was performed as described (Sauer et al., 2006). Washing steps, cell wall digestion, membrane permeabilization, and primary and secondary antibody treatments were performed with an automated device (InSituPro VS; Intavis). Rabbit anti-TSN serum was used at 1:4000 dilution, and mouse anti-α-tubulin monoclonal antibody (Neomarkers) was used at a final concentration of 0.5 μg/μL. Goat anti-rabbit secondary antibody coupled with Alexa488 (Invitrogen) and anti-mouse secondary antibody coupled with Alexa568 (Invitrogen) were used with a final concentration of 2 μg/mL.
Slides were observed and images were collected with an upright laser scanning confocal microscope (TCS SP2; Leica Microsystems). Different fluorochromes were detected sequentially frame by frame with the acousto-optical tunable filter system using 488- and 543-nm laser lines. The images were usually coded green (GFP or Alexa Fluor 488) and red (Alexa Fluor 568), giving yellow colocalization signals in merged images. Before imaging samples, negative control samples were used for microscope settings to detect background signals emitted from 488- and 543-nm laser lines. Negative controls were tsn plants for TSN immunolocalization and plants of the original background line for GFP fusion expressing plants. For cell wall staining, seedlings were incubated for 10 min in 20 mM propidium iodide and rinsed in water for 5 min before imaging. Three independent single copy homozygous lines were imaged for each construct.
For NaCl-triggered relocalization experiments, seedlings were grown for 4 d on agar-containing GM plates and transferred for 2 d in liquid GM. Subsequently, plants were transferred into liquid GM (mock treatment) or liquid GM supplemented with 150 mM NaCl. After 0, 15, or 90 min incubation, seedlings were mounted in their incubation media between slides and cover slips for immediate imaging (within 5 min).
GFP fluorescence patterns at the seedling level were acquired on a Leica DMI6000B or on a Leica Reichert Polyvar (Leica Microsystems), with UV intensities adjusted to produce no signals in non transgenic controls. Fluorescence images were accompanied by visible light acquisitions with Nomarski optics.
Bioinformatics
Oligonucleotides for PCR were designed using Primer3 (http://frodo.wi.mit.edu/primer3/input.htm). Protein localization was predicted using TargetP and SignalP (Emanuelsson et al., 2007) at CBS (http://www.cbs.dtu.dk/services/). Gene sequences identified by transcriptome analysis were screened for common motifs employing the AFGC's Motif Finder at TAIR (http://www.Arabidopsis.org) and MEME (http://meme.sdsc.edu). Arabidopsis transcripts were searched with the identified motifs using Patmatch (TAIR).Coregulation was examined using the Arabidopsis Coexpression Data Mining Tools (http://www.Arabidopsis.leeds.ac.uk/act/) and NetworkDrawer tool at ATTED (http://atted.jp/top_tool.shtml). TSN-dependent transcripts were screened for small RNA targets by interrogation of the ASRP (http://asrp.cgrb.oregonstate.edu/) and PARE (http://mpss.udel.edu) databases. Public transcriptome data was mined and visualized with Genevestigator (https://www.genevestigator.ethz.ch; Hruz et al., 2008).
Accession Numbers
Microarray data from this article can be found in the Gene Expression Omnibus database under the series accession number GSE15559. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TSN1, AT5G07350; TSN2, AT5G61780; TSN-dependent RNA 1 to 15, AT2G43510, AT3G53980, AT2G16005, AT2G37870, AT3G04720, AT1G17860, AT5G42600, AT2G18370, AT2G33790, AT1G12090, AT1G45015, AT3G58550, AT1G73330, AT5G05960, AT2G19970, and AT3G08770; Ps-TSN, BAC06184; Os-TSN, NP_001046985.1; and Hs-TSN, NP_055205.2. Additional Arabidopsis Genome Initiative accession numbers are listed in Supplemental Data Set 1E online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of TSN Proteins.
Supplemental Figure 2. Production of tsn1 tsn2 Mutants and Strategy Employed to Generate Nearly Isogenic Pairs of Complemented and Double Mutant Lines.
Supplemental Figure 3. Cell Elongation Defect in tsn1 tsn2 Roots of 5-d-Old Seedlings.
Supplemental Figure 4. Expression Patterns Driven by the TSN1 and TSN2 Promoters.
Supplemental Figure 5. Organ Distribution of TSN1 and TSN2 Transcripts in Publicly Available Transcriptomic Data.
Supplemental Figure 6. Response of TSN2EC Plants to Long-Term High Salinity Stress.
Supplemental Figure 7. Abiotic Stress and ABA Independence of TSN Levels.
Supplemental Figure 8. tsn1 tsn2 Is Hypersensitive to Mannitol Stress.
Supplemental Figure 9. Growth and Fitness of tsn1 tsn2 and TSN1EC Plants in the Greenhouse.
Supplemental Figure 10. Complementation of the Root Elongation and Germination Phenotypes of tsn1 tsn2 by TSNpro:GFP:TSN Constructs.
Supplemental Figure 11. Response of the tsn1 tsn2 Mutant to Short-Term NaCl Stress.
Supplemental Figure 12. RISC-Directed mRNA Cleavage Activity in tsn1 tsn2.
Supplemental Figure 13. tsn1 tsn2 Downregulated Transcripts Are Targets of the Unfolded Protein Response.
Supplemental Figure 14. Salt Responses in tsn1 tsn2 Seedlings.
Supplemental Table 1. Partial Lethality of the tsn1 tsn2 Mutation.
Supplemental Data Set 1A. Transcripts Differentially Expressed (P < 0.05) in a Microarray Experiment Comparing tsn1 tsn2 with TSN1EC.
Supplemental Data Set 1B. Transcripts Differentially Expressed (P < 0.05) in a Microarray Experiment Comparing tsn1 tsn2 with TSN2EC.
Supplemental Data Set 1C. Transcripts Differentially Expressed (P < 0.05) in a Microarray Experiment Comparing tsn1 tsn2 with TSN1EC on NaCl Supplemented Media.
Supplemental Data Set 1D. Expression Levels of Known miRNA Targets in the CATMA Microarray Experiments.
Supplemental Data Set 1E. Oligonucleotides Employed for Cloning and qRT-PCR.
Acknowledgments
35S:HDEL-GFP and 35S:freeGFP were kindly provided by Jim Haseloff and Woo Sik Chung, respectively. Anti-NMT1 and α-tubulin antibodies were a gift of Carmela Giglione and Béatrice Satiat-Jeunemaitre, respectively. This work has benefited from the facilities and expertise of the Imagif Cell Biology Unit of the Gif campus (www.imagif.cnrs.fr), which is supported by the Conseil Général de l'Essonne. The support of Nicole Vartanian and of the Scientific Council of the University of Paris 12 to M.W.B. and the skilled technical help of Tessa Fredriksen in the initial phases of this project are gratefully acknowledged.
References
- Abe S., Sakai M., Yagi K., Hagino T., Ochi K., Shibata K., Davies E. (2003). A Tudor protein with multiple SNc domains from pea seedlings: Cellular localization, partial characterization, sequence analysis, and phylogenetic relationships. J. Exp. Bot. 54: 971–983 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bensmihen S., Giraudat J., Parcy F. (2005). Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 56: 597–603 [DOI] [PubMed] [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Broadhurst M.K., Lee R.S.F., Hawkins S., Wheeler T.T. (2005). The p100 EBNA-2 coactivator: a highly conserved protein found in a range of exocrine and endocrine cells and tissues in cattle. Biochim. Biophys. Acta 1681: 126–133 [DOI] [PubMed] [Google Scholar]
- Broadhurst M.K., Wheeler T.T. (2001). The p100 coactivator is present in the nuclei of mammary epithelial cells and its abundance is increased in response to prolactin in culture and in mammary tissue during lactation. J. Endocrinol. 171: 329–337 [DOI] [PubMed] [Google Scholar]
- Caudy A.A., Ketting R.F., Hammond S.M., Denli A.M., Bathoorn A.M., Tops B.B., Silva J.M., Myers M.M., Hannon G.J., Plasterk R.H. (2003). A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414 [DOI] [PubMed] [Google Scholar]
- Charrier B., Champion A., Henry Y., Kreis M. (2002). Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong S.D.X., Good A.G., Taylor G.J., Freeman M.C., Moorhead G.B.G., Muench D.G. (2004). Large-scale identification of tubulin-binding proteins provides insight on subcellular trafficking, metabolic channeling, and signaling in plant cells. Mol. Cell. Proteomics 3: 970–983 [DOI] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash A.B., Orrico F.C., Ness S.A. (1996). The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 10: 1858–1869 [DOI] [PubMed] [Google Scholar]
- Decottignies A., Sanchez-Perez I., Nurse P. (2003). Schizosaccharomyces pombe essential genes: A pilot study. Genome Res. 13: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S., Anrather D., Roitinger E., Djamei A., Hufnagl T., Barta A., Csaszar E., Dohnal I., Lecourieux D., Hirt H. (2006). Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res. 34: 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri S., Swarup R., Mockaitis K., Dharmasiri N., Singh S.K., Kowalchyk M., Marchant A., Mills S., Sandberg G., Bennett M.J., Estelle M. (2006). AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312: 1218–1220 [DOI] [PubMed] [Google Scholar]
- Dorner S., Lum L., Kim M., Paro R., Beachy P.A., Green R. (2006). A genomewide screen for components of the RNAi pathway in Drosophila cultured cells. Proc. Natl. Acad. Sci. USA 103: 11880–11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Lecellier C.H., Parizotto E.A., Himber C., Voinnet O. (2004). Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl. Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Friberg A., Corsini L., Mourao A., Sattler M. (2009). Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J. Mol. Biol. 387: 921–934 [DOI] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.-P., Martin-Magniette M.-L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.-P., Lecharny A., Brunaud V. (2008). CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucl. Acids Res. 36: D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Grishok A., Sinskey J.L., Sharp P.A. (2005). Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 19: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M.J., Finch-Savage W.E., Grappin P., Job D. (2008). Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 13: 7–13 [DOI] [PubMed] [Google Scholar]
- Hong B., Ichida A., Wang Y., Gens J.S., Pickard B.G., Harper J.F. (1999). Identification of a calmodulin-regulated Ca2+-ATPase in the endoplasmic reticulum. Plant Physiol. 119: 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008 (online), doi/10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Jammes F., Lecomte P., de Almeida-Engler J., Bitton F., Martin-Magniette M.L., Renou J.P., Abad P., Favery B. (2005). Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 44: 447–458 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yang B., Harris N.S., Deyholos M.K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58: 3591–3607 [DOI] [PubMed] [Google Scholar]
- Johnson M.A., Perez-Amador M.A., Lidder P., Green P.J. (2000). Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc. Natl. Acad. Sci. USA 97: 13991–13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan T.W., Winter S., Rackwitz H.R., Heid H.W. (2000). Nuclear coactivator protein p100 is present in endoplasmic reticulum and lipid droplets of milk secreting cells. Biochim. Biophys. Acta, Gen. Subj. 1523: 84–90 [DOI] [PubMed] [Google Scholar]
- Kim J.K., Gabel H.W., Kamath R.S., Tewari M., Pasquinelli A., Rual J.F., Kennedy S., Dybbs M., Bertin N., Kaplan J.M., Vidal M., Ruvkun G. (2005). Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Kim Y.O., Pan S., Jung C.H., Kang H. (2007). A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 48: 1170–1181 [DOI] [PubMed] [Google Scholar]
- Kuhn J.M., Breton G., Schroeder J.I. (2007). mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 50: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Kuhn J.M., Hugouvieux V., Schroeder J.I. (2008). mRNA cap binding proteins: Effects on abscisic acid signal transduction, mRNA processing, and microarray analyses. Curr. Top. Microbiol. Immunol. 326: 139–150 [DOI] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., Van Der Straeten D., Verbelen J.P. (2001). In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiol. 125: 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Kim H.S., Han H.J., Moon B.C., Kim C.Y., Harper J.F., Chung W.S. (2007). Identification of a calmodulin-regulated autoinhibited Ca2+-ATPase (ACA11) that is localized to vacuole membranes in Arabidopsis. FEBS Lett. 581: 3943–3949 [DOI] [PubMed] [Google Scholar]
- Leverson J.D., Koskinen P.J., Orrico F.C., Rainio E.M., Jalkanen K.J., Dash A.B., Eisenman R.N., Ness S.A. (1998). Pim-1 kinase and p100 cooperate to enhance c-myb activity. Mol. Cell 2: 417–425 [DOI] [PubMed] [Google Scholar]
- Li C.L., Yang W.Z., Chen Y.P., Yuan H.S. (2008). Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 36: 3579–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic Z.J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14: 229–236 [DOI] [PubMed] [Google Scholar]
- Low S.H., Vasanth S., Larson C.H., Mukherjee S., Sharma N., Kinter M.T., Kane M.E., Obara T., Weimbs T. (2006). Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev. Cell 10: 57–69 [DOI] [PubMed] [Google Scholar]
- Ma S., Gong Q., Bohnert H.J. (2006). Dissecting salt stress pathways. J. Exp. Bot. 57: 1097–1107 [DOI] [PubMed] [Google Scholar]
- Martinez I.M., Chrispeels M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Hayashi Y., Yamada K., Shimada T., Nishimura M., Hara-Nishimura I. (2003). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44: 661–666 [DOI] [PubMed] [Google Scholar]
- Morel J.B., Godon C., Mourrain P., Beclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Palacios L., Ochoa B., Gomez-Lechon M.J., Castell J.V., Fresnedo O. (2006). Overexpression of SND p102, a rat homologue of p100 coactivator, promotes the secretion of lipoprotein phospholipids in primary hepatocytes. Biochim. Biophys. Acta 1761: 698–708 [DOI] [PubMed] [Google Scholar]
- Paukku K., Kalkkinen N., Silvennoinen O., Kontula K.K., Lehtonen J.Y. (2008). p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res. 36: 4474–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham J.W., Pellino J.L., Lee Y.S., Carthew R.W., Sontheimer E.J. (2004). A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94 [DOI] [PubMed] [Google Scholar]
- Pierre M., Traverso J.A., Boisson B., Domenichini S., Bouchez D., Giglione C., Meinnel T. (2007). N-myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19: 2804–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L., Gallardo K., Debeaujon I., Vandekerckhove J., Job C., Job D. (2004). The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge R.W., Uozumi Y., Plazinski J., Hurley U.A., Williamson R.E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Sami-Subbu R., Choi S.B., Wu Y., Wang C., Okita T.W. (2001). Identification of a cytoskeleton-associated 120 kDa RNA-binding protein in developing rice seeds. Plant Mol. Biol. 46: 79–88 [DOI] [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benkova E., Friml J. (2006). Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Scadden A.D. (2005). The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 12: 489–496 [DOI] [PubMed] [Google Scholar]
- Scadden A.D.J. (2008). Gene expression is reduced in trans by inosine-containing dsRNA. Biochem. Soc. Trans. 36: 534–536 [DOI] [PubMed] [Google Scholar]
- Schauer S.E., Jacobsen S.E., Meinke D.W., Ray A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sclep G., Allemeersch J., Liechti R., De Meyer B., Beynon J., Bhalerao R., Moreau Y., Nietfeld W., Renou J.P., Reymond P., Kuiper M.T., Hilson P. (2007). CATMA, a comprehensive genome-scale resource for silencing and transcript profiling of Arabidopsis genes. BMC Bioinformatics 8: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Silverstein K.A., Moskal W.A., Jr., Wu H.C., Underwood B.A., Graham M.A., Town C.D., VandenBosch K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51: 262–280 [DOI] [PubMed] [Google Scholar]
- Siomi H., Siomi M.C. (2009). On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Steven J., Clough A.F.B. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Sundström J.F., et al. (2009). Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Sussman M.R., Amasino R.M., Young J.C., Krysan P.J., Austin-Phillips S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Drapkin R., Yalamanchili R., Mosialos G., Kieff E. (1995). The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15: 4735–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian N., Marcotte L., Giraudat J. (1994). Drought rhizogenesis in Arabidopsis thaliana (differential responses of hormonal mutants). Plant Physiol. 104: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Boston R.S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Wang C., Washida H., Crofts A.J., Hamada S., Katsube-Tanaka T., Kim D., Choi S.B., Modi M., Singh S., Okita T.W. (2008). The cytoplasmic-localized, cytoskeletal-associated RNA binding protein OsTudor-SN: Evidence for an essential role in storage protein RNA transport and localization. Plant J. 55: 443–454 [DOI] [PubMed] [Google Scholar]
- Yang J., Aittomaki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O. (2002). Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21: 4950–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. (2006). Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.F., Yuan L.J., Shao Y., Du W., Yan D.W., Lu Y.T. (2008). The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 31: 562–574 [DOI] [PubMed] [Google Scholar]