This work identifies ragged seedling2 as a maize homolog of Arabidopsis argonaute7 and focuses on the role of small RNA molecules in establishing leaf polarity.
Abstract
Leaves arise from the flank of the shoot apical meristem and are asymmetrical along the adaxial/abaxial plane from inception. Mutations perturbing dorsiventral cell fate acquisition in a variety of species can result in unifacial (radially symmetrical) leaves lacking adaxial/abaxial polarity. However, mutations in maize (Zea mays) ragged seedling2 (rgd2) condition cylindrical leaves that maintain dorsiventral polarity. Positional cloning reveals that rgd2 encodes an ARGONAUTE7 (AGO7)-like protein required to produce ta-siARF, a trans-acting small interfering RNA that targets abaxially located auxin response factor3a (arf3a) transcripts for degradation. Previous studies implicated ta-siARF in dorsiventral patterning of monocot leaves. Here, we show that arf3a transcripts hyperaccumulate but remain abaxialized in rgd2 mutant apices, revealing that ta-siARF function is not required for arf3a polarization. RGD2 also regulates miR390 accumulation and localization in maize shoot apices. Similar to the abaxialized maize mutant leafbladeless1 (lbl1), rgd2 mutants exhibit ectopic accumulation of the abaxial identity factor miR166 in adaxial domains. Thus, hyperaccumulation of arf3a and ectopic accumulation of miR166 are insufficient to condition abaxialized leaf phenotypes in maize. Finally, transcripts of a maize ago1 paralog overaccumulate in lbl1 but not in rgd2 mutants, suggesting that upregulation of ago1 combined with ectopic accumulation of miR166 contribute to abaxialized leaf formation in lbl1. We present a revised model for the role of small RNAs in dorsiventral patterning of maize leaves.
INTRODUCTION
Shoot apical meristems (SAMs) are comprised of a group of indeterminate cells that give rise to all of the aboveground organs of the plant. The angiosperm SAM harbors stem cells that maintain the meristem by replacing cells lost during organogenesis. Leaves are initiated from the flank of the SAM in a position-dependent process whereby cells at the SAM periphery are recruited to become leaf founder cells (Poethig, 1984). As the founder cells are recruited and switch to determinate growth, leaf primordia are elaborated from the SAM along three main axes: the mediolateral axis (medial midrib to lateral margin), the proximodistal axis (proximal sheath to distal blade), and the dorsiventral axis (adaxial/abaxial). Maize (Zea mays) leaves are bifacial (dorsiventrally flattened) and harbor distinct tissue types on their adaxial (upper) and abaxial (lower) surfaces. By contrast, unifacial (radially symmetrical) leaf mutants are deficient in mediolateral development and typically lose either adaxial or abaxial epidermal patterning, with a concomitant conversion of the polarized vascular patterning seen in many wild-type plants (xylem is adaxial and phloem is abaxial) to an abaxialized mutant pattern (phloem surrounds xylem) or an adaxialized mutant vasculature (xylem surrounds phloem; Waites and Hudson, 1995; Timmermans et al., 1998; McConnell et al., 2001; Zhong and Ye, 2004; Eshed et al., 2004). Inspired by the snapdragon (Antirrhinum majus) unifacial leaf mutant phantastica1, a model for dorsiventral patterning was adapted to leaf development whereby the juxtaposition of adaxial and abaxial tissue types is required to organize the mediolateral and proximodistal axes of growth (Koch and Meinhardt, 1994; Waites and Hudson, 1995). Widespread support for this model is provided by phenotypic and molecular analyses of a number of unifacial leaf mutants from Arabidopsis thaliana, rice (Oryza sativa), tobacco (Nicotiana benthamiana), tomato (Solanum lycopersicum), and maize (Zea mays; reviewed in Kidner and Timmermans, 2007; Figure 1).
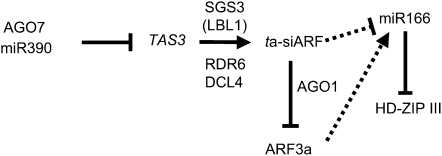
Figure 1.
Proposed Model for the Role of Small Regulatory RNAs in Dorsiventral Patterning.
Model modified from Kidner and Timmermans (2007). Details provided in text. AGO7/RGD2, ARGONAUTE7/RAGGED SEEDLING2; miR390, microRNA 390; SGS3/LBL1, SUPPRESSOR-OF-GENE SILENCING3/LEAFBLADELESS1; RDR6, RNA-DEPENDANT RNA POLYMERASE6; DCL4, DICER-LIKE4; AGO1, ARGONAUTE1; ARF3a, AUXIN RESPONSE FACTOR3a; miR166, microRNA 166; HD-ZIPIII, family of adaxial identity factors.
Unlike any previously described unifacial leaf mutants that are deficient in abaxial or adaxial patterning, the maize mutant ragged seedling2 (rgd2) develops cylindrical leaves that maintain dorsiventral polarity (Henderson et al., 2005). Previous work has suggested that RGD2 function may be required to either interpret or respond to a proposed dorsiventral juxtaposition signal that initiates mediolateral development (Waites and Hudson, 1995; Henderson et al., 2005). Although the mechanisms whereby this leaf patterning response is elaborated are still not fully understood, several microRNAs (miRNAs) and trans-acting small interfering RNAs (ta-siRNAs) are proposed to play important roles in establishing dorsiventral polarity in developing leaves (Figure 1; Nagasaki et al., 2007; Nogueira et al., 2007; reviewed in Husbands et al., 2009).
miRNA biogenesis requires DICER-LIKE1 function to process stem loop miRNA precursors into functional small RNAs (Reinhart et al., 2002). The processed miRNA combines with an ARGONAUTE (AGO) in an RNA-induced silencing complex (RISC) that can then target complementary mRNA sequences for silencing, either via cleavage of the target transcript or by translational inhibition (Fagard et al., 2000; Lanet et al., 2009). miR166 is required to spatially restrict the accumulation of adaxial-determining class III homeodomain-leucine zipper (hd-zipIII) transcripts in both Arabidopsis and maize (McConnell et al., 2001; Juarez et al., 2004). Dominant mutations that disrupt the miR166 binding site of hd-zipIII genes but preserve the protein coding sequence lead to unifacial leaf phenotypes wherein abaxial epidermal characteristics are lost (McConnell et al., 2001; Juarez et al., 2004).
The production of ta-siRNAs is a plant-specific process that uses components from both miRNA and small interfering (siRNA) biogenesis pathways. Biogenesis of ta-siRNAs begins with RISC-mediated cleavage of a non-protein-coding ta-siRNA precursor (TAS) transcript (Figure 1; Yoshikawa et al., 2005; Allen et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006). Whereas the majority of small RNA-cleaved transcripts are summarily degraded, the TAS cleavage product is stabilized and processed into a double-stranded RNA. The formation of this double-stranded RNA requires SUPPRESSOR-OF-GENE SILENCING3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE (RDR6; Allen et al., 2005; Nogueira et al., 2007). The resultant double-stranded RNA undergoes phased cleavage by DICER-LIKE4 (DCL4) to yield mature 21-nucleotide ta-siRNAs (Allen et al., 2005; Figure 1).
Four TAS gene families (TAS1, TAS2, TAS3, and TAS4) are found in Arabidopsis, whereas tas3 is the only tas family described in maize to date (Nogueira et al., 2007). Posttranscriptional cleavage of TAS3 precursor genes requires a specialized RISC containing miR390 and AGO7/ZIPPY (ZIP). After posttranscriptional cleavage, SGS3-RDR6-DCL4 processes the TAS3 cleavage product into 21-bp ta-siRNAs, a subset of which are called ta-siARFs (Peragine et al., 2004; Allen et al., 2005; Montgomery et al., 2008). As a small RNA substrate bound by AGO1 during posttranscriptional cleavage of the transcription factors AUXIN RESPONSE FACTOR3/ETTIN (ARF3/ETT) and ARF4, ta-siARF regulates vegetative phase change in Arabidopsis shoots (Allen et al., 2005; Hunter et al., 2006). Subsequently, ta-siARF was shown to function redundantly to regulate organ polarity, wherein arf3/ett and arf4 are abaxial determinants (Pekker et al., 2005; Garcia et al., 2006; Fahlgren et al., 2006).
Genetic analyses in maize and rice have demonstrated a primary role for ta-siARF during dorsiventral leaf patterning; mutations in this ta-siRNA pathway generate shootless embryos or filamentous, abaxialized leaf phenotypes (Nagasaki et al., 2007; Nogueira et al., 2007, 2009). By contrast, Arabidopsis mutants in the TAS3 ta-siRNA pathway show no defects in leaf polarity but render a precocious switch from juvenile to adult-staged leaves (Hunter et al., 2003; Peragine et al., 2004). Leafbladeless1 (lbl1) encodes the maize homolog of SGS3 (Nogueira et al., 2007). Macroscopically, lbl1 and rgd2 have very similar mutant phenotypes (Timmermans et al., 1998; Henderson et al., 2005). However, closer inspection reveals that whereas severe lbl1 unifacial leaves lose adaxial epidermal characteristics, rgd2 cylindrical leaves maintain dorsiventral polarity. Recessive alleles of lbl1 exhibit reduced levels of ta-siARF biogenesis and elevated levels of arf3a, which are purported to promote miR166 accumulation and condition an abaxialized phenotype (Figure 1; Nogueira et al., 2007, 2009). Surprisingly, rgd2 lbl1 double mutants have a synergistic shootless phenotype, suggesting that RGD2 and LBL1 operate in overlapping pathways, yet also perform some nonredundant functions (Henderson et al., 2006).
Here, we show that rgd2 encodes a maize AGO7-like protein required for ta-siARF biogenesis and downregulation of arf3a transcripts. Surprisingly, arf3a transcript accumulation remains abaxialized in rgd2 and lbl1 mutants, revealing that ta-siARF function is not required for arf3a polarization. RGD2 function is also required for proper localization and downregulation of miR390 and miR166 in maize shoot apices. Combined with previous genetic and phenotypic analyses of the rgd2-R mutation, these data suggest that arf3a overaccumulation and ectopic accumulation of miR166 are insufficient to confer the loss of dorsiventral polarity in maize leaves. An ago1 homolog hyperaccumulates in lbl1-rgd1 but not in rgd2-R mutants. Taken together, these findings suggest that LBL1 regulates miR166 localization in a separate pathway that does not require RGD2 function, and the combined overaccumulation of ago1 together with ectopic miR166 triggers abaxialized leaf phenotypes that are not observed in rgd2 mutants.
RESULTS
Positional Cloning of rgd2
A mapping population of 189 highly introgressed individuals (see Methods) was used to genetically map the rgd2-R mutation to within a 0.52-centimorgan interval on maize chromosomal bin 1.04. This interval is flanked by the markers IDP1473 and CAP276M21 and is comprised of ∼1.2 megabase pairs on contig 19 of the maize genome (Figure 2A; see Supplemental Table 1 online; Maize Sequence Release 4a.53, October 2009). Among the 40 predicted genes within this interval, a maize homolog (GRMZM2G365589) of Arabidopsis AGO7/ZIP was investigated as a candidate rgd2 gene.
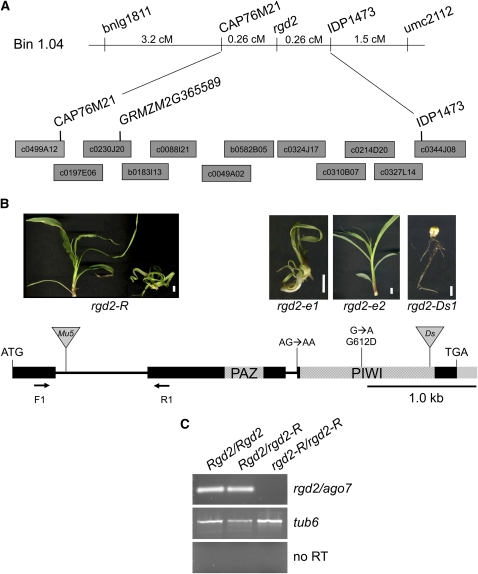
Figure 2.
Positional Cloning and Characterization of rgd2 Alleles.
(A) Positional cloning of rgd2. rgd2-R was mapped to bin 1.04 using simple sequence repeat markers (names) and fine mapped to an interval flanked by CAP76M21 and IDP1473. Candidate gene analysis revealed an ago7-like locus, GRMZM2G365589, on maize BAC c0230J20. Gray blocks indicate BAC clones. cM, centimorgan.
(B) Gene structure of rgd2 wild-type and mutant alleles. The rgd2 open reading frame consists of three exons (black boxes) and shows a high degree of similarity to ago7. Shaded boxes show the approximate positions of the predicted PAZ and PIWI protein domains. Solid gray box represents 3′ untranslated region. Inverted triangles mark the locations of transposon insertions in the rgd2-R and rgd2-Ds1 alleles. Point mutations in the EMS-induced alleles, rgd2-e1 and rgd2-e2, are denoted. Images of representative phenotypes of rgd2 mutant alleles are placed above each corresponding mutation. Bars = 1 cm.
(C) RT-PCR analysis of rgd2 transcripts. Primers F1 and R1, which flank the first intron, were used in rgd2 transcript analyses of seedlings from rgd2-R mutants, wild-type siblings, and heterozygous siblings. No transcripts were detected in rgd2-R mutants after 42 amplification cycles. RT-PCR using tub6 primers served as the control. Three biological replicates comprising one seedling per replicate were performed.
[See online article for color version of this figure.]
The rgd2-R mutation arose in a Mutator (Mu) transposon-mutagenized background (Henderson et al., 2005); the ago7 candidate gene was used as a probe in a DNA gel blot hybridization to search for a gene-specific Mu insertion in rgd2-R mutant plants. Compared with the inbred lines Mo17 and B73, rgd2-R mutants contained an ∼1.6-kb insertion in the maize ago7-like gene (Figure 2B; see Supplemental Figure 1 online). Analyses of genomic PCR products obtained using an ago7-specific primer and a degenerate Mu transposon primer (MuTIR) revealed the presence of a Mu5 (Talbert et al., 1989) transposon located 96 bp into the first intron of the rgd2-R allele of ago7. Attempts to span the Mu5 insertion site using ago7-specific primers (F1 and R1) in RT-PCR of cDNA derived from rgd2-R mutants failed (Figure 2C), suggesting that the Mu5 transposon is improperly spliced from ago7 transcripts of rgd2-R mutants. RT-PCR analyses using an ago7-specific primer (F1) and the MuTIR primer yielded an aberrant ago7 cDNA fragment in which the Mu5 terminus was spliced directly into the first exon of ago7, indicating that rgd2-R is a null mutation.
Identification of three additional mutant alleles verified that rgd2 is an ago7-like maize gene (Figure 2B). Two alleles were obtained via ethyl methanesulfonate (EMS) mutagenesis; rgd2-e1 conditions a severe phenotype and contains a canonical G-to-A transition at the last base pair of the second intron. A predicted null allele, rgd2-e1, produces a mis-spliced transcript containing a premature stop codon that is predicted to eliminate the conserved PIWI domain of RGD2 (described below). A second EMS allele with a relatively mild ragged seedling phenotype, rgd2-e2, contains a G-to-A transition in the third exon that is predicted to cause a Gly to Asp missense substitution at amino acid 612 (Figure 2B). A third mutant allele, rgd2-Ds1, contains a ∼1.5-kb Dissociation (Ds) transposon insertion at base pair 3635 of the third exon. In the W22 inbred genetic background wherein rgd2-Ds1 arose, plants homozygous for this allele are shootless (Figure 2B). Notably, the rgd2-R allele is also shootless when introgressed into W22. The independent origin of these four rgd2 mutations, all of which contain lesions in the ago7-like locus (GRMZM2G365589), verifies that rgd2 encodes an AGO7-like protein in maize.
Like its homologs in Arabidopsis (AGO7/ZIP) and rice (SHOOT ORGANIZATION2/SHOOTLESS4 [SHO2/SHL4]), the rgd2 locus contains three exons and is predicted to encode a protein of 1032 amino acids (Hunter et al., 2003; Nagasaki et al., 2007). RGD2 shares 61% identity/76% similarity with AGO7/ZIP and 76% identity/85% similarity with SHO2/SHL4 (see Supplemental Figure 2 online). RGD2 also contains the highly conserved PAZ domain (87.7% identity with SHO2 and 62.3% identity with AGO7) and the PIWI domain (86.6% identity with SHO2 and 69.0% identity with AGO7). PAZ domains are important for recognizing the 3′ end of small RNAs, while PIWI domains function during endonucleolytic slicing of target transcripts (reviewed in Vaucheret, 2008).
RGD2 Is Required for miR390 Localization
As an AGO7-like protein, RGD2 is predicted to regulate maize dorsiventral patterning via ta-siARF biogenesis (Figure 1; Nogueira et al., 2009). In situ hybridization of seedling apices revealed that rgd2 transcript accumulation is not polarized (Figures 3A and 3B). Unlike the adaxial accumulation observed for AGO7 putative orthologs from Arabidopsis and rice (Itoh et al., 2008; Chitwood et al., 2009), rgd2 transcripts are evenly accumulated throughout the adaxial and abaxial regions of the youngest leaf primordium (P0-P1) and accumulate weakly in the SAM. In older leaf primordia (P2-P6), transcripts are localized to the margins and vascular bundles. No transcript accumulation was detected using an rgd2 sense control (Figure 3C).
Figure 3.
In Situ Hybridization Analysis of rgd2 and miR390 Accumulation in rgd2 Mutant and Wild-Type Sibling Apices.
(A) and (B) Accumulation pattern of rgd2 in the SAM (asterisks) and leaf primordia (P1 and P2).
(A) Transverse wild-type shoot apex.
(B) Longitudinal wild-type shoot apex.
(C) Control in situ hybridization of wild-type shoot apex using a sense rgd2 hybridization probe.
(D) to (G) Accumulation pattern of miR390.
(D) Transverse wild-type shoot apex.
(E) Longitudinal wild-type shoot apex.
(F) Longitudinal rgd2-R shoot apices show ectopic miR390 expression throughout the SAM crown and leaf primordia.
(G) Longitudinal rgd2-e2 shoot apex.
(H) An LNA probe for murine miR124e was used as a sense control for miR390 accumulation.
Bars = 100 μm; arrows indicate areas of transcript accumulation.
[See online article for color version of this figure.]
Coimmunoprecipitation analyses in Arabidopsis showed that AGO7 selectively interacts with miR390 (Montgomery et al., 2008). Small RNA gel blot hybridizations were used to analyze the effect of the rgd2-R mutation on miR390 accumulation in maize shoot apices. As shown in Figure 4A, mir390 levels are significantly elevated in rgd2-R mutant apices compared with apices of wild-type seedlings. To determine if the rgd2-R mutation affects the tissue specificity of miR390 accumulation, locked nucleic acid (LNA) probes complementary to miR390 were used for in situ hybridizations of maize seedling shoot apices. As previously described and shown in Figures 3D and 3E, miR390 accumulates in adaxial domains of wild-type maize leaf primordia (Nogueira et al., 2009). By contrast, miR390 accumulation in the crown of the wild-type shoot apex is much lower than that observed in leaf primordia (Figure 3D). Polarized accumulation of miR390 is maintained in rgd2-R mutant leaf primordia; however, marked miR390 overaccumulation is observed in the crown of the rgd2-R mutant SAM compared with wild-type siblings (Figure 3F; see Supplemental Figure 3 online). By contrast, miR390 accumulation in the weakly phenotypic rgd2-e2 mutant is similar to that observed in wild-type apices; very little miR390 is detected within the SAM crown (Figure 3G). An LNA probe complementary to murine miR124e was used as a negative control for all LNA in situ hybridizations; no signal is detected in maize tissues (Figure 3H).
Figure 4.
Accumulation Analyses of Transcripts Involved in the ta-siARF Pathway.
(A) Small RNA gel blot hybridization of miR390, miR166, and ta-siARF in wild-type and rgd2-R shoot apices. All genotypes are introgressed into the Mo17 genetic background.
(B) qRT-PCR analysis of arf3a and tas3a in laser-microdissected leaf primordia and SAMs from wild-type (WT) and rgd2-R mutants. Accumulation was normalized to ubq. Three biological replicates were performed per experiment. Error bars denote 1 se.
(C) qRT-PCR analysis of miR166 precursors in laser-microdissected SAMs from wild-type and rgd2-R mutants. An ago1 putative paralogue (1GenBank accession number AY110984) was analyzed in wild-type, rgd2-R mutant, and lbl1-ref mutant 14-d-old seedlings. Accumulation was normalized to tub6. Each experiment used three biological replicates. Error bars denote 1 se.
RGD2 Is Required for ta-siARF Biogenesis and Regulation of arf3a Transcripts
AGO7 and miR390 are required to cleave the non-protein-coding tas3 transcripts that serve as precursors to ta-siARFs, small RNAs that regulate arf3a transcript accumulation (Figure 1; Allen et al., 2005; Pekker et al., 2005; Hunter et al., 2006; Montgomery et al., 2008). While mature ta-siARF transcripts are shown to accumulate adaxially in maize leaf primordia (Nogueira et al., 2007), the accumulation pattern of the tas3a precursor has not been described. Transcripts of miRNA precursor genes are predicted to form a stem-loop secondary structure that prevents hybridization of probes complementary to the 21-bp miRNA sequence contained within the precursor RNA. Unlike miRNAs, however, the tas3a transcript is not predicted to form a secondary structure that prevents the binding of a ta-siARF LNA probe to two complementary sites found within the tas3a mRNA (Nogueira et al., 2007). Thus, a probe complementary to ta-siARF is also expected to hybridize to tas3a precursor transcripts. As predicted, equivalent signals are detected in the abaxial regions of wild-type leaf primordia following in situ hybridization to either a tas3a antisense probe or to a 16-bp LNA probe that is complementary to the mature ta-siARF (Figures 5A to 5C). Previous studies revealed that precursor tas3a transcripts accumulate to much higher levels than mature ta-siARF (Allen et al., 2005; Lu et al., 2006). Taken together, these data suggest that the signal obtained following in situ hybridization of the ta-siARF LNA probe corresponds to tas3a precursor transcripts, which accumulate in abaxial leaf domains when RGD2 function is intact (Figures 5A to 5C). No accumulation is detected with a sense tas3a hybridization probe (Figure 5E). By contrast, the ta-siARF LNA probe identifies tas3a precursor transcripts in both adaxial and abaxial domains of rgd2-R leaf primordia, wherein RGD2 function is mutated (Figure 5D). These data suggest that tas3a transcripts initially accumulate throughout maize leaf primordia and are preferentially cleaved to generate ta-siARF in adaxial leaf domains by RGD2/miR390 activity, whereupon intact tas3a transcripts persist in abaxial regions.
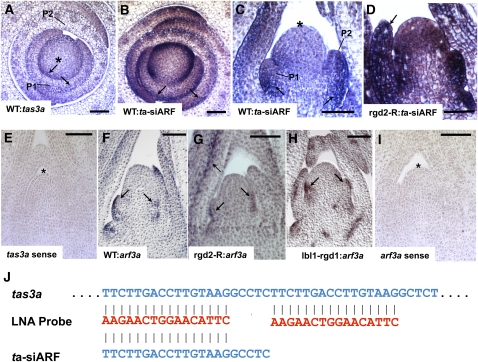
Figure 5.
In Situ Hybridization Analysis of tas3a and arf3a in the Wild Type and in rgd2-R and lbl1-rgd1 Mutants.
(A) Accumulation of tas3a in a wild-type apex transverse section using a tas3a antisense hybridization probe. (B) to (D) Pattern of transcript accumulation obtained using the ta-siARF LNA hybridization probe. (B) Transverse section of a wild-type apex. Note that the pattern of transcript accumulation detected with the ta-siARF LNA probe is equivalent to that observed when using the tas3a probe (A). (C) Longitudinal section of a wild-type SAM (asterisk) and leaf primordia (P1 and P2) (D) Longitudinal section of a rgd2-R mutant apex shows ectopic transcript accumulation in the SAM and the leaf primordia. (E) Control in situ hybridization of wild-type shoot apex using a sense tas3a hybridization probe. (F) to (H) Accumulation pattern of arf3a transcripts. (F) Longitudinal section of a wild-type apex reveals arf3a transcript accumulation in abaxial domains of leaf primordia. (G) and (H) Longitudinal sections of rg2-R (G) and lbl1-rgd1 (H) mutant apices; arf3a transcript accumulation remains abaxialized in both. (I) Control in situ hybridization of wild-type shoot apex using a sense arf3a hybridization probe. (J) The LNA ta-siARF in situ hybridization probe is predicted to hybridize to the mature ta-siARF as well as to two locations in its abundant precursor, the tas3a transcript. Bars = 100 μm. Arrows point to areas of accumulation.
[See online article for color version of this figure.]
Quantitative RT-PCR (qRT-PCR) confirmed that tas3a transcripts overaccumulate by 2.7-fold in rgd2-R mutant apices and by 7.9-fold in rgd2-R leaf primordia (P1-P4) compared with wild-type siblings (Figure 4B). Hyperaccumulation of tas3a transcripts in rgd2-R apices may reflect inefficient processing of tas3a precursors into ta-siARF in the mutant shoots. To test this hypothesis, the accumulation of ta-siARF in rgd2-R and wild-type apices was compared using small RNA gel blot hybridization. A 16-bp LNA probe complementary to ta-siARF detects a small RNA of the expected size in total RNA extracted from wild-type apices. By contrast, ta-siARF hybridization is dramatically reduced in RNA derived from rgd2-R mutant shoot apices (Figure 4A).
Proper biogenesis of ta-siARF is required to regulate arf3a accumulation in maize (Nogueira et al., 2007). Therefore, it was expected that arf3a transcripts would also be elevated in rgd2-R mutants owing to the absence of ta-siARF. Using cDNA prepared from laser microdissected (LM) SAM-P1 and leaf primordial (P1-P4) tissue, the accumulation of arf3a was examined by qRT-PCR. As expected, arf3a transcript accumulation is elevated in rgd2-R mutant SAMs (7.8-fold) and leaf primordia (35.9-fold) (Figure 4B) compared with the corresponding values in wild-type siblings.
Whereas ARF3/ETT transcripts accumulate throughout leaf primordia in Arabidopsis, accumulation of ARF3/ETT protein is only detected in abaxial domains (Pekker et al., 2005; Chitwood et al., 2009). In situ hybridization analyses of wild-type shoot apices confirmed that arf3a transcripts in maize are preferentially abaxialized (Figure 5F), while no accumulation is detected using a sense arf3a probe (Figure 5I). Unexpectedly, arf3a transcript accumulation remains abaxialized in rgd2-R mutant leaves (Figure 5G). An equivalent arf3a accumulation pattern was also observed in lbl1-rgd1 mutants (Figure 5H), suggesting that ta-siARF biogenesis is not required to polarize arf3a accumulation within abaxial domains of maize leaf primordia.
RGD2 Regulates Abaxial Localization of miR166
Previous studies have suggested that miR166 accumulation is negatively regulated by ta-siARF and/or positively regulated by ARF3a (Figure 1; Nogueira et al., 2007, 2009). In light of these findings, the accumulation of miR166 precursor loci was examined in rgd2-R mutant and wild-type SAM-P1 tissue by LM-qRT-PCR. Of the nine miR166 precursor loci identified in the maize genome, four (miR166a, miR166c, miR166h, and miR166i) were previously shown to exhibit abnormal transcript accumulation in lbl1 mutant shoots (Nogueira et al., 2007). However, the accumulation of these miR166 precursor transcripts is not significantly increased in rgd2-R mutant apices compared with wild-type siblings (Figure 4C). By contrast, small RNA gel blot hybridization reveals that the 21-bp mature miR166 accumulates to approximately twofold higher levels in rgd2-R mutant apices (Figure 4A).
The tissue-specific accumulation of mature miR166 was examined by in situ hybridization using an LNA probe. In wild-type apices, miR166 accumulates in abaxial regions of young leaf primordia but is excluded from the SAM and from the pith directly below the SAM (Juarez et al., 2004; Figure 6A). Whereas lbl1 mutants accumulate miR166 throughout the abaxial and adaxial domains of young leaf primordia and below the SAM (Juarez et al., 2004; Nogueira et al., 2007), rgd2-R mutants also exhibit ectopic miR166 accumulation in adaxial leaf domains but do not accumulate miR166 in the pith directly below the SAM (Figure 6B).
Figure 6.
In Situ Hybridization Analysis of miR166 in the Wild Type and rgd2-R Mutants.
(A) and (B) Accumulation of miR166 in shoot apices. Bars = 50 μm.
(A) Longitudinal section of a wild-type apex shows miR166 accumulation in abaxial regions of leaf primordia (indicated by arrows). Asterisk denotes SAM.
(B) Longitudinal section of an rgd2-R mutant apex shows ectopic accumulation of miR166 in adaxial regions of leaf primordia and the crown of the meristem (arrows).
[See online article for color version of this figure.]
Recent studies have shown that SGS3 is required for siRNA-directed cleavage of AGO1 transcripts in Arabidopsis (Mallory and Vaucheret, 2009). Using cDNA from 14-d-old maize seedlings, qRT-PCR was performed in rgd2-R and lbl1-rgd1 mutant backgrounds. In congruence with Arabidopsis, a maize ago1 putative paralog accumulates to nearly sixfold higher levels in lbl1-rgd1 mutants than in the rgd2-R mutant or in wild-type plants (Figure 4C).
DISCUSSION
rgd2 Encodes AGO7 Function in Maize
rgd2-R is a recessively inherited mutant that is defective in mediolateral leaf development. Mutant plants often have cylindrical leaves that, unlike similar unifacial leaf mutants, maintain dorsiventral anatomy. No net loss of adaxial or abaxial characteristics is observed in rgd2-R mutant leaves (Henderson et al., 2005). The rgd2 gene is identified as an ago7-like gene (GRMZM2G365589) using a positional cloning approach; several noncomplementing ago7 mutant alleles were isolated to verify the identity of rgd2 (Figures 2A and 2B). The rgd2-R reference allele harbors a Mu5 transposable element within intron 1, and RT-PCR analyses of rgd2-R mutants fail to amplify wild-type transcripts that span the exon 1–exon 2 boundary (Figure 2C). These data suggest that rgd2-R is a null allele. In Arabidopsis, AGO7 is implicated in the formation of ta-siARFs, which are required to downregulate the abaxial identity factors ARF3 and ARF4 (Allen et al., 2005; Pekker et al., 2005; Hunter et al., 2006). As predicted for a null allele of ago7, rgd2-R mutants are defective in ta-siARF biogenesis (Figure 4A) and show a concomitant increase in arf3a transcript accumulation (Figure 4B).
RGD2 Is Required to Localize miR390 in Maize Shoot Apices
Elegant and detailed coimmunoprecipitation analyses in Arabidopsis demonstrated that AGO7 forms a RISC exclusively with miR390 (Montgomery et al., 2008). Although the accumulation of rgd2 and miR390 transcripts overlaps in young leaf primordia and developing vasculature, miR390 accumulates in adaxial domains (Nogueira et al., 2007; Figures 3D and 3E), whereas rgd2 accumulation is apolar (Figures 3A and 3B). Adaxial accumulation of miR390 is maintained in rgd2-R mutant leaf primordia; however, marked overaccumulation of miR390 is observed within the crown of the mutant SAM (Figures 3F and 4A; see Supplemental Figure 3 online).
Layer-specific LM-qRT-PCR has shown that maize mir390 precursor transcripts are expressed in the outer cell layer (L1) of the SAM but not in the L2 (Nogueira et al., 2009). Intriguingly, in situ hybridizations detect mature miR390 accumulation in both the L1 and L2 (Nogueira et al., 2009; Figure 3E). One possible explanation for this apparent discrepancy is that mir390 precursor transcripts are processed more rapidly in the L2 than the L1; a second hypothesis is that mature miR390 is mobile and traffics from the L1 to the L2. Although ta-siRNAs are shown to act non-cell-autonomously (Chitwood et al., 2009), several studies examining the localization of artificial miRNAs and miRNAs with non-native promoters concluded that miRNAs act cell autonomously (Alvarez et al., 2006; Schwab et al., 2006; Tretter et al., 2008). Still other studies have suggested that miR166 and miR390 may traffic within the shoot apex (Juarez et al., 2004; Kidner and Martienssen, 2004; Nogueira et al., 2009). Our analyses of differential rgd2 mutant alleles support a scenario wherein miR390 acts non-cell-autonomously, and RGD2 functions to restrict its mobility into the crown of the maize shoot apex.
The conserved PAZ domains of AGO proteins recognize and bind the 3′ ends of small RNAs (Lingel et al., 2004; Ma et al., 2004; Hutvagner and Simard, 2008). The unspliced Mu insertion in rgd2-R mutant transcripts is expected to disrupt or eliminate the PAZ domain from RGD2, thereby preventing its binding of miR390, which overaccumulates ectopically in the rgd2-R mutant SAM crown (Figure 3F). By contrast, rgd2-e2 mutants harbor a point mutation in the PIWI domain that causes a missense amino acid substitution; the PAZ domain is predicted to be unaltered (Figure 2B). Unlike rgd2-R mutants, rgd2-e2 mutants do not show ectopic miR390 overaccumulation within the SAM crown (Figure 3G). Likewise, miR390 accumulation is unchanged in lbl1 mutants (Nogueira et al., 2009), indicating that ta-siARF function is not required for proper localization of miR390. Taken together, these data suggest that an intact PAZ domain is correlated with normal miR390 accumulation in the maize SAM. We speculate that RGD2 is required to bind miR390 and restrict it from trafficking into the crown of the SAM (Figure 7).
Figure 7.
Revised Model for the Role of Small Regulatory RNAs in Dorsiventral Patterning.
rgd2 is the maize homolog of Arabidopsis ago7 and is required to regulate miR390 accumulation and localization in the maize shoot apex. RGD2 and LBL1/SGS3 are both required to regulate miR166 accumulation levels. Like LBL1, RGD2 is required to properly localize miR166, suggesting that miR166 polarization requires ta-siARF function. Although rgd2 mutants overaccumulate miR166 and arf3a, miR166 is properly localized, resulting in no net loss of dorsiventral patterning in rgd2 mutants. However, in lbl1 mutants, the ectopic accumulation of miR166 combined with the hyperaccumulation of ago1 may account for the abaxialized mutant leaf phenotype. Abbreviations are as described in Figure 1.
[See online article for color version of this figure.]
Polarized Degradation of tas3a Is Patterned by Localization of miR390
An in situ hybridization probe that recognizes tas3a precursor transcripts was used to determine their accumulation pattern in wild-type shoot apices (Figure 5). Wild-type apices showed tas3a accumulation only in abaxial regions of leaf primordia, which is consistent with the biogenesis and function of ta-siARF in adaxial domains. An equivalent in situ hybridization signal is detected using a 16-bp LNA probe, which contains two complementary sites in the tas3a transcript (Figures 5B to 5D and 5J). In light of previous analyses revealing that ta-siARF accumulates to lower levels than most miRNAs (Allen et al., 2005; Lu et al., 2006), these data strongly suggest that the signals obtained with the ta-siARF LNA probe are derived from tas3a transcript accumulation and not ta-siARF accumulation.
Unlike wild-type siblings of rgd2-R mutants, ta-siARF LNA in situ hybridization analyses of rgd2-R mutants detect tas3a accumulation throughout the abaxial and adaxial domains of mutant leaf primordia (Figure 5D). These data suggest that the failure to process tas3a precursor transcripts into mature ta-siARF results in the ectopic accumulation of intact tas3a transcripts in rgd2-R mutant primordia. Both the overabundance of tas3a transcripts and the failure to detect ta-siARF in rgd2 mutant shoots support this hypothesis (Figures 4A and 4B). AGO7 and miR390 are both required to cleave tas3a and begin the process of ta-siARF biogenesis (Montgomery et al., 2008). Although rgd2 transcript accumulation is not polarized, miR390 is restricted to adaxial domains of maize leaf primordia (Figures 3A to 3D). These data indicate that the abaxial accumulation pattern of tas3a transcripts in wild-type leaves is patterned by the adaxial expression of miR390, not by RGD2 per se. By the same token, the ectopic tas3a accumulation observed in adaxial domains of rgd2-R mutant leaf primordia is likely the result of failure to incorporate previously adaxialized miR390 into a functional RISC that normally converts tas3a into ta-siARF.
Upregulation of arf3a and Ectopic miR166 Are Insufficient to Confer an Abaxialized Leaf Phenotype in Maize
ARF3/ETT is an abaxial identity factor in Arabidopsis (Pekker et al., 2005); however, the absence of arf3 mutants and/or ta-siARF insensitive lines has hindered analyses of ARF3 function in maize. arf3a transcript accumulation is detected in abaxial regions of wild-type maize (Figure 5F). Although the rgd2-R mutation confers marked overaccumulation of arf3a transcripts in the SAM and in young leaf primordia (Figure 4B), this accumulation pattern remains abaxialized (Figure 5G), and dorsiventral polarity is maintained in mutant cylindrical leaves (Henderson et al., 2005). A similar pattern of abaxialized arf3a transcript overaccumulation is observed in lbl1 rgd1 double mutants, which are also defective in ta-siARF biogenesis (Figure 5H). Therefore, like the adaxialized expression of miR390, the abaxial accumulation of arf3a transcripts is regulated independently of ta-siARF biogenesis. Moreover, rgd2-R mutants retain dorsiventral leaf polarity (Henderson et al., 2005), suggesting that ta-siARF biogenesis is not a requirement for leaf polarity in maize.
Overexpression of a ta-siARF-insensitive arf3/ett transcript recapitulates the zip-2/ago7 mutant phenotype in Arabidopsis (Hunter et al., 2006), which might suggest that the rgd2-R mutant phenotype is conditioned entirely by arf3a overaccumulation. However, mutations in putatively orthologous ta-siARF biogenesis genes condition disparate phenotypes in maize and Arabidopsis (Timmermans et al., 1998; Mourrain et al., 2000; Hunter et al., 2003; Peragine et al., 2004; Henderson et al., 2005), and in the absence of a maize ta-siARF insensitive line, this hypothesis remains quite speculative. On the other hand, the lack of abaxialized leaf phenotypes in rgd2 mutants indicates that arf3a overaccumulation alone is not sufficient to disrupt dorsiventral leaf patterning in maize.
LBL1 encodes the maize SGS3 protein (Nogueira et al., 2007) and is thereby predicted to function directly downstream of RGD2 during ta-siARF biogenesis (Figure 1). Unlike rgd2 mutants, however, lbl1 mutations confer severe disruptions in dorsiventral patterning and abaxialized, unifacial leaf phenotypes (Timmermans et al., 1998; reviewed in Chitwood et al., 2007). Furthermore, instead of epistasis, rgd2 lbl1 double mutants display synergistic shootless phenotypes, which suggest that RGD2 and/or LBL1 perform nonoverlapping functions outside of ta-siARF biogenesis (Henderson et al., 2006). Similar to lbl1 and the rice ago7 mutant sho2, rgd2-R mutant shoot apices exhibit ectopic overaccumulation of miR166 in adaxial leaf domains, although lbl1 also accumulates ectopic miR166 at the base of the SAM (Nagasaki et al., 2007; Nogueira et al., 2007; Figures 5B and 6B). As such, our findings are consistent with suggested models wherein ta-siARF negatively regulates the expression of the abaxial-determinant miR166 (Figure 1; Nagasaki et al., 2007; Nogueira et al., 2007, 2009). Thus, despite the fact that rgd2 and lbl1 mutants both overexpress arf3a and ectopically accumulate miR166, they condition disparate leaf phenotypes. These data, when considered in light of the synergistic rgd2 lbl1 double mutant phenotype, suggest that LBL1 performs additional functions contributing to maize leaf polarity, outside of the ta-siARF biogenesis pathway.
In addition to its role during ta-siARF biogenesis, Arabidopsis SGS3 is also required for production of AGO1 siRNAs that downregulate the accumulation of AGO1 mRNA in conjunction with miR168 (Mallory and Vaucheret, 2009). Unlike AGO7, AGO1 binds numerous miRNA species, including miR166. Our data demonstrate that lbl1-ref mutants overaccumulate transcripts of a putative paralog of ago1, whereas rgd2-R mutants do not (Figure 4C). Recent studies have shown that a maize ago1-like transcript accumulates in a gradient within leaf primordia, with higher accumulation in adaxial domains (Brooks et al., 2009). Intriguingly, miR166 accumulates ectopically in adaxial leaf domains of lbl1 mutants, in a pattern that overlaps with ago1 accumulation. We propose that upregulation of ago1 in lbl1 mutants (Figure 4C), in conjunction with the ectopic overaccumulation of miR166, may result in enhanced posttranscriptional degradation of adaxial identity factor hd-zipIII genes. This increase in miR166 activity may explain why lbl1 mutants lose adaxial identity while rgd2 mutants do not. Furthermore, Arabidopsis SGS3 functions during viral defense and in additional siRNA pathways besides ta-siRNA biosynthesis (Mourrain et al., 2000; Peragine et al., 2004; Borsani et al., 2005). We propose a revised model (Figure 7) wherein additional functions of LBL1 outside of ta-siARF biogenesis contribute to the abaxialized phenotypes observed in lbl1 mutants and to the synergistic, shootless phenotype seen in rgd2 lbl1 double mutants (Henderson et al., 2006).
The interplay of several small RNAs is critical to the dorsiventral patterning of maize leaves (Chitwood et al., 2007; reviewed in Kidner and Timmermans, 2007). Future studies probing the mechanisms of miR390 polarization and its RGD2-mediated localization in the shoot apex will enhance our understanding of the role of regulatory RNAs during leaf development. In addition, identification and analyses of arf3a mutants in maize may help decipher the convergent and divergent mechanisms by which ta-siARF regulates the development of monocot and eudicot shoots.
METHODS
Plant Materials
The rgd2-R mutation arose in a Mutator transposon line (Henderson et al., 2005) and was introgressed into the maize (Zea mays) inbred Mo17 genetic background for at least four to five generations to generate the mapping population used in this study. Primers for novel molecular markers developed during this study are listed in Supplemental Table 1 online.
EMS-mutagenized Mo17 pollen was crossed to rgd2-R heterozygous individuals to obtain additional mutant alleles of rgd2 as described (Neuffer, 1993). One EMS allele, rgd2-e1, was identified in a noncomplementation screen of ∼6200 F1 plants. However, the rgd2-R/rgd2-e1 heterozygous plant was seedling lethal, and the lesion was not recoverable for further genetic analyses. An additional mutant allele of rgd2, rgd2-e2, was obtained via screens of ∼3000 M2 EMS-mutagenized families in the B73 inbred background. An M2 family segregated a filamentous leaf phenotype and was mapped to bin 1.04 using bulk segregant analyses. The rgd2-Ds1allele was identified in a large-scale transposon mutagenesis program using the transposons Activator and Dissociation (Ahern et al., 2009). Analyses of the lbl1-rgd1 allele were performed in the Mo17 genetic background.
In Situ Hybridizations
Samples from 14-d-old seedlings were fixed, processed, sectioned, and hybridized to gene-specific probes as described (Jackson, 1991) with modifications as described (Juarez et al., 2004). Double-digoxigenin (DIG)–labeled LNA probes complementary to miR390, miR166, and ta-siARF were purchased from Exiqon. Five picomoles of LNA probe were used per slide; hybridizations were performed at 50°C, and washes were performed at 55°C. The rgd2 and tas3a probes (see Supplemental Table 1 online) were DIG labeled (Roche) according to the manufacturer's instructions. All samples were imaged on a Zeiss Axio Imager Z1m microscope using a 5-megapixel Zeiss AxioCam MRc5 and AxioVision Release 4.6 software.
Small RNA Gel Blot Hybridization
miRNA gel blots were performed as described (Allen et al., 2004). Fifteen hand-dissected apices (shoot apex plus six leaf primordia) were pooled for each wild-type and rgd2-R mutant sample; RNA was extracted using the Trizol lysis method and prepared for RNA gel blot transfer as described (Fu et al., 2002). Approximately 20 μg of total RNA was loaded per lane. Twenty-one base pair oligonucleotide probes complementary to miR390 and miR166 were end labeled with 32P. Double-DIG-labeled LNA ta-siARF probes were as described above. Hybridizations and washes were performed at 42°C.
qRT-PCR
SAM or leaf primordia (P1-P4) tissue was harvested by LM, and cDNA was synthesized from amplified RNA as described by Zhang et al. (2007). Gene-specific primers were designed (see Supplemental Table 1 online) for use with SYBR-Green (Quanta) in qRT-PCR as described (Zhang et al., 2007). Three biological replicates were examined, and samples were normalized to beta-6 tubulin or ubiquitin expression as described using Bio-Rad iQ5 Version 1.0 software (Livak and Schmittgen, 2001; Zhang et al., 2007).
Accession Numbers
Sequence data can be found in the GenBank data library under the following accession numbers: BAC c0230J20 (AC206196), rgd2 (GQ918490), Zm ago1 (AY110984), tubulin (L10633), and ubiquitin (S94464).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Analysis of Maize ago7
Supplemental Figure 2. Amino Acid Alignment of RGD2, AGO7, and SHO2.
Supplemental Figure 3. Additional in Situ Hybridization Analyses of miR390 Accumulation in rgd2-R Mutant Apices.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Marja Timmermans and Dan Chitwood for assistance with small RNA in situ hybridizations; Xiaolan Zhang donated LM SAM cDNA, and Dave Henderson provided helpful discussions of the data. We thank Gary Muehlbauer for identification of rgd2 mutant alleles and Erik Vollbrecht, Tom Brutnell, and Kevin Ahern for identifying the rgd2-Ds1 allele. Scott Tingey and Amanda Jones provided physical mapping information that expedited the positional cloning of rgd2, and Taiowa Montgomery offered technical advice on small RNA gel blot hybridizations. This work was supported by the National Science Foundation (Grants IOS-0649810 and IOS-0820610 to M.J.S.).
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasiciolli V., Vaucheret H. (2006). DRB4-dependant TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 18: 758–762 [DOI] [PubMed] [Google Scholar]
- Ahern K.R., Deewatthanawong P., Schares J., Muszynski M., Weeks R., Vollbrecht E., Duvick J., Brendel V.P., Brutnell T.P. (2009). Regional mutagenesis using Dissociation in maize. Methods 49: 248–254 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNAs-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L.B., III, et al. (2009). Microdissection of shoot meristem functional domains. PLoS Genet. 5: e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Guo M., Nogueira F.T., Timmermans M.C. (2007). Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development. 134: 813–823 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T.S., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C.P. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Fagard M., Boutet S., Morel J.B., Bellini C., Vaucheret H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97: 11650–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T., Howell M., Allen E., Dvorak S., Alexander A., Carrington J. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Fu S., Meeley R., Scanlon M.J. (2002). Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Henderson D.C., Muehlbauer G.J., Scanlon M.J. (2005). Radial leaves of the maize mutant ragged seedling2 retain dorsiventral anatomy. Dev. Biol. 282: 455–466 [DOI] [PubMed] [Google Scholar]
- Henderson D.C., Zhang X., Brooks L., Scanlon M.J. (2006). RAGGED SEEDLING2 is required for normal expression of KANADI2 and REVOLUTA homologues in the maize shoot apex. Genesis 44: 372–382 [DOI] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig S.R. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutierrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A.Y., Chitwood D.H., Plavskin Y., Timmermans M.C. (2009). Signals and prepatterns: new insights into organ polarity in plants. Genes Dev. 23: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J. (2008). Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Itoh J., Sato Y., Nagato Y. (2008). The SHOOT ORGANIZATION2 gene coordinates leaf domain development along the central-marginal axis in rice. Plant Cell Physiol. 49: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Jackson D. (1991). In situ hybridization in plants. Molecular Plant Pathology: A Practical Approach, Bowles D.J., Gurr S.J., McPherson M., (Oxford, UK: Oxford University Press; ), pp. 163–174 [Google Scholar]
- Juarez M.T., Kui J., Thomas J., Heller B., Timmermans M.C.P. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Koch A.J., Meinhardt H. (1994). Biological pattern-formation – From basic mechanisms to complex structures. Rev. Mod. Phys. 66: 1481–1507 [Google Scholar]
- Kidner C.A., Martienssen R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Timmermans M.C.P. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant Biol. 10: 13–20 [DOI] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crete P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A., Simon B., Izaurralde E., Sattler M. (2004). Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 11: 576–577 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Meyers B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.B., Ye K., Patel D.J. (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Vaucheret H. (2009). ARGONAUTE1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 10: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONATUE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Mourrain P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Nagasaki H., Itoh J., Hayashi K., Hibara K., Satoh-Nagasawa N., Nosaka M., Mukouhata M., Ashikari M., Kitano H., Matsuoka M., Nagato Y., Sato Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M.G. (1993). Mutagenesis. The Maize Handbook, Freeling M., Walbot V., (New York: Springer; ), pp. 212–219 [Google Scholar]
- Nogueira F.T., Chitwood D.H., Madi S., Ohtsu K., Schnable P.S., Scanlon M.J., Timmermans M.C. (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 5: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F.T., Madi S., Chitwood D.H., Juarez M.T., Timmermans M.C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J., Eshed Y. (2005). AUXIN RESPONSE FACTORS mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (1984). Cellular parameters of leaf morphogenesis in maize and tobacco. Contemporary Problems of Plant Anatomy, White R.A., Dickinson W.C., (New York: Academic Press; ), pp. 235–238 [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert L.E., Patterson G.I., Chandler V.L. (1989). Mu transposable elements are structurally diverse and distributed throughout the genus Zea. J. Mol. Evol. 29: 28–39 [DOI] [PubMed] [Google Scholar]
- Timmermans M.C., Schultes N.P., Jankovsky J.P., Nelson T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125: 2813–2823 [DOI] [PubMed] [Google Scholar]
- Tretter E.M., Alvarez J.P., Eshed Y., Bowman J.L. (2008). Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol. 147: 58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Madi S., Borsuk L., Nettleton D., Elshire R.J., Buckner B., Janick-Buckner D., Beck J., Timmermans M., Schnable P.S., Scanlon M.J. (2007). Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (2004). Amphivasal vascular bundle1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45: 369–385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.