This work shows that chloroplast stromal Hsp70s play a critical role in the step of protein translocation across the chloroplast envelope. It provides evidence that stromal Hsp70 functions in parallel to the Hsp93 system during protein import, making the chloroplast unique among organelles by possessing two simultaneously functioning chaperone/motor systems for protein translocation.
Abstract
Hsp70 family proteins function as motors driving protein translocation into mitochondria and the endoplasmic reticulum. Whether Hsp70 is involved in protein import into chloroplasts has not been resolved. We show here Arabidopsis thaliana knockout mutants of either of the two stromal cpHsc70s, cpHsc70-1 and cpHsc70-2, are defective in protein import into chloroplasts during early developmental stages. Protein import was found to be affected at the step of precursor translocation across the envelope membranes. From solubilized envelope membranes, stromal cpHsc70 was specifically coimmunoprecipitated with importing precursors and stoichiometric amounts of Tic110 and Hsp93. Moreover, in contrast with receptors at the outer envelope membrane, cpHsp70 is important for the import of both photosynthetic and nonphotosynthetic proteins. These data indicate that cpHsc70 is part of the chloroplast translocon for general import and is important for driving translocation into the stroma. We further analyzed the relationship of cpHsc70 with the other suggested motor system, Hsp93/Tic40. Chloroplasts from the cphsc70-1 hsp93-V double mutant had a more severe import defect than did the single mutants, suggesting that the two proteins function in parallel. The cphsc70-1 tic40 double knockout was lethal, further indicating that cpHsc70-1 and Tic40 have an overlapping essential function. In conclusion, our data indicate that chloroplasts have two chaperone systems facilitating protein translocation into the stroma: the cpHsc70 system and the Hsp93/Tic40 system.
INTRODUCTION
Most chloroplast proteins are nucleus encoded and cytosolically synthesized as a precursor form with an N-terminal targeting signal called the transit peptide. Import of these precursor proteins into chloroplasts is mediated by a translocon complex located at the chloroplast envelope. Various translocon components have been assigned functions in the basic steps of the import process. For example, Toc159 (translocon of the outer envelope membrane of chloroplast 159 kD) and Toc34 are the initial receptors for precursors, Toc75 is the protein-translocating channel across the outer membrane, and Tic110 (translocon of the inner envelope membrane of chloroplast 110 kD) most likely functions as the stroma-side receptor for transit peptides during precursor translocation across the inner membrane and as the scaffold for assembling other stromal translocon components (Inaba and Schnell, 2008; Jarvis, 2008). Tic110 has also been shown to form a Ca2+-responsive channel across the inner envelope membrane and therefore may be part of the protein-translocating channel across the inner membrane (Balsera et al., 2009). Several translocon components are encoded by multigene families. In Arabidopsis thaliana, for instance, Toc159 is encoded by a four-gene family: TOC159, TOC132, TOC120, and TOC90, and Toc34 has two family members: TOC33 and TOC34. It has been shown that photosynthetic proteins are preferentially imported through at-Toc159, at-Toc90, and at-Toc33, whereas nonphotosynthetic proteins are preferentially imported through at-Toc132, at-Toc120, and at-Toc34 (Kubis et al., 2003, 2004; Smith et al., 2004; Kessler and Schnell, 2009).
The motors for driving protein import into mitochondria and the endoplasmic reticulum (ER) are Hsp70 family proteins. However, in chloroplasts, it is generally assumed that the most likely motor for driving protein translocation into the stroma is the stromal Hsp93 (also called ClpC). Stromal Hsp93 belongs to the Hsp100 subfamily of the AAA+ (ATPases associated with various cellular activities) family proteins. It is a soluble protein, though a small portion of it associates with the translocon at the inner membrane. Its antibody can specifically precipitate other translocon components together with importing precursors (Akita et al., 1997; Nielsen et al., 1997). Arabidopsis has two genes encoding Hsp93: HSP93-III (ClpC2) and HSP93-V (ClpC1). Knockout mutations in HSP93-III result in no phenotype. Chloroplasts isolated from hsp93-V knockout mutants usually have reduced chloroplast protein import efficiencies (Constan et al., 2004; Kovacheva et al., 2005). However, normal import in the same hsp93-V knockout mutants has also been reported (Sjogren et al., 2004). The reason for the discrepancy is unclear.
Hsp93 is functionally associated with the cochaperone Tic40. Tic40 has an N-terminal membrane anchor followed by a stromal domain composed of a tetratricopeptide repeat domain (Chou et al., 2003) and a C-terminal Hip/Hop (or Sti1) domain that is homologous to the C terminus of cochaperones Sti1p/Hop (Hsp70/Hsp90-organizing protein) and Hip (Hsp70-interacting protein; Stahl et al., 1999; Bedard et al., 2007). It has been shown that the Hip/Hop domain can stimulate ATP hydrolysis by Hsp93 and that the association of Tic40 with Hsp93 is regulated by ATP (Chou et al., 2006). The hsp93-V tic40 double mutant has a similar phenotype as tic40 (Kovacheva et al., 2005), suggesting that these two proteins function together in the same pathway in close association with each other.
It has been suggested that chloroplast stromal cpHsc70 (chloroplast heat shock cognate protein 70 kD) may also be involved in protein translocation into chloroplasts, but the data are inconclusive. In vitro binding assays have shown interactions between cpHsc70 and a recombinant transit peptide of the precursor to the small subunit of RuBP carboxylase (prRBCS; Ivey et al., 2000). However, precursors with altered affinities for Hsp70 in their transit peptides were still efficiently imported into pea (Pisum sativum) chloroplasts (Rial et al., 2003, 2006). Using an antibody against the pea stromal cpHsc70, S78 (also named CSS1; Marshall and Keegstra, 1992; Nielsen et al., 1997), it was shown that no, or very little, importing precursors were precipitated by the anti-S78 antibody from the soluble fraction of solubilized total chloroplast membranes (Akita et al., 1997; Nielsen et al., 1997), suggesting that cpHsc70 was not associated with importing precursors in vivo.
Another chloroplast Hsp70 suggested to be involved in protein import is an intermembrane space Hsp70 named Hsp70-IAP (IAP for import intermediate associated protein) or imsHsp70 (ims for intermembrane space) identified in pea chloroplasts. Hsp70-IAP was identified as an Hsp70 homolog comigrating with Toc75, and it copurified with importing precursors and other translocon components (Schnell et al., 1994; Becker et al., 2004; Chen and Li, 2007). It was suggested to be an outer membrane protein facing the intermembrane space (Marshall et al., 1990; Schnell et al., 1994). However, its identity was shown only on immunoblots by its reactivity to the monoclonal antibody SPA820 raised against human (Homo sapiens) Hsp70. Its encoding gene has not been identified. A recent study further showed that the major protein recognized by SPA820 in pea chloroplasts is located in the stroma (Ratnayake et al., 2008). This result does not exclude the possibility that an intermembrane space Hsp70 recognized by SPA820 still exists, but it does highlight the possibility that some of the data showing interactions of Hsp70-IAP with precursors and the translocon might actually represent the interactions of stromal cpHsc70 with these components. Furthermore, the Arabidopsis Hsp70 gene family has 14 members. Only two of them are localized in chloroplasts, and both have been shown to locate in the stroma (Ratnayake et al., 2008; Su and Li, 2008).
Initial characterizations of the two Arabidopsis stromal cpHsc70s, cpHsc70-1 and cpHsc70-2, have been undertaken (Su and Li, 2008). Knockout mutants of cphsc70-2 have no visible phenotype. Mutants of cphsc70-1 have significantly reduced growth in roots. cpHsc70-1 is also important for thermotolerance of germinating seeds. Nonetheless, double knockout of the two genes is lethal. These results suggest that cpHsc70-1 has some distinct functions in root growth and thermotolerance but that the two cpHsc70s also have some overlapping essential functions. Chloroplasts isolated from 30-d-old cphsc70-1 and cphsc70-2 plants and 7-d-old cphsc70-2 cotyledons exhibit no import defect. Chloroplasts isolated from 7-d-old cphsc70-1 cotyledons, which are variegated with chlorotic lesions, showed reduced precursor association to the chloroplast surface even when no ATP was present in the import system. Under these conditions, precursors have contacts only with the outer surface of the outer envelope membrane (Perry and Keegstra, 1994; Kouranov and Schnell, 1997). Since cpHsc70-1 is an ATPase in the stroma, the reduced precursor association with the chloroplast surface independent of ATP suggests that, in cotyledons, the cphsc70-1 mutation has some secondary effects and caused damage to the surface of chloroplasts. Thus, whether cpHsc70s are directly involved in chloroplast protein import remains to be determined.
Here, we further analyzed the import phenotypes of the cphsc70 mutants. We found that, in 14- to 24-d-old plants, both cphsc70-1 and cphsc70-2 single mutants had significantly reduced protein import efficiencies, suggesting that both cpHsc70s are important for protein import into chloroplasts at these stages. Biochemical data showed that cpHsc70 is a stable part of the translocon and can be coimmunoprecipitated with importing precursors. Double mutant analyses further suggested that cpHsc70 and Hsp93/Tic40 systems most likely function in parallel during chloroplast protein import. We propose that there are two systems driving protein import into the chloroplast stroma: the Hsp93/Tic40 system and the cpHsc70 system.
RESULTS
Chloroplasts from cphsc70 Mutants Had Protein Import Defects at Early Developmental Stages
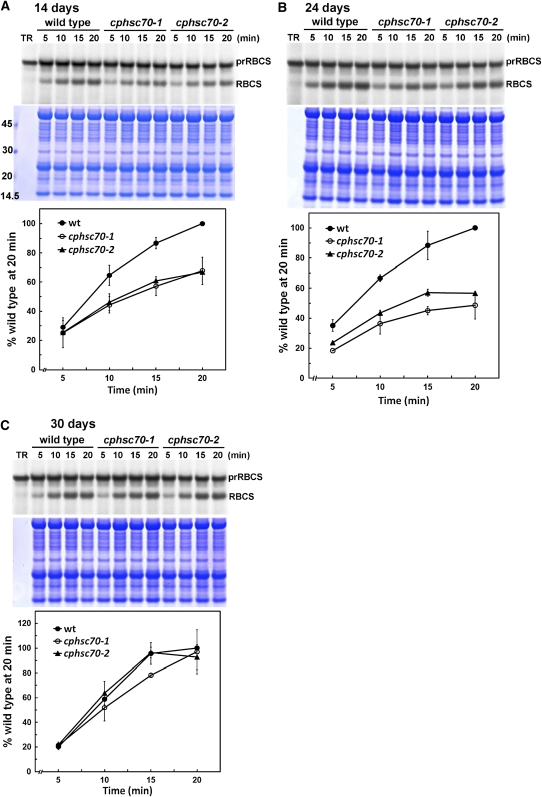
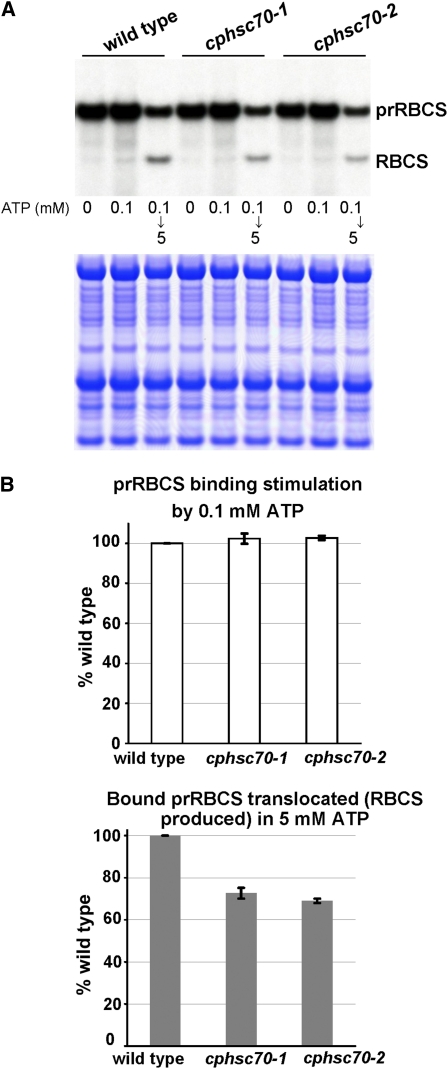
The efficiency of protein import into chloroplasts declines as leaves get older (Dahlin and Cline, 1991), and some mutants show import defects only at younger stages (Jarvis et al., 1998). We therefore decided to analyze further possible import phenotypes of the cphsc70 mutants by isolating chloroplasts from seedlings of different ages. We used prRBCS as the import substrate because it is the best studied precursor protein in terms of import behavior and nearly all translocon components were identified through association with importing prRBCS. As shown in Figure 1A, chloroplasts isolated from both 14-d-old cphsc70-1 and cphsc70-2 plants showed a clear import defect. The defect was still observed in chloroplasts isolated from 24-d-old seedlings (Figure 1B). However, as observed previously (Su and Li, 2008), by 30 d, the import efficiency of the mutant chloroplasts was no longer significantly different from that of wild-type chloroplasts (Figure 1C). Direct comparison of import efficiencies (see Supplemental Figure 1 online) showed that import efficiency in wild-type plants did indeed decline as plants aged from 14 to 30 d. Mutant plants started with a lower import efficiency and did not show further decline from 24 to 30 d. Hence, protein import defects were not observed in our previous study because in 30-d-old plants, import efficiency in the wild type had dropped to the same level as that in the mutants. Import defects were also not observed in chloroplasts isolated from 7-d-old cphsc70-2 mutant plants in our previous studies (Su and Li, 2008). Chloroplasts isolated from 7-d-old seedlings represent mostly chloroplasts from cotyledons that are fully mature. Many studies have suggested that chloroplast biogenesis in cotyledons is different from that in true leaves (Kim and Apel, 2004; Ishizaki et al., 2005; Shimada et al., 2007; Albrecht et al., 2008). Cotyledon chloroplasts may have a lower demand for protein import, either due to their maturity or due to differences in physiology, and the import process is therefore not affected by the loss of any single cpHsc70.
Figure 1.
cphsc70 Mutant Chloroplasts Are Defective in Protein Import.
prRBCS import into chloroplasts isolated from 14-d-old seedlings (A), 24-d-old seedlings (B), or 30-d-old seedlings (C). Chloroplasts were isolated from wild-type (wt) and mutant seedlings of the specified age. In vitro import assays were conducted for 5 to 20 min with [35S]Met-prRBCS. After import, intact chloroplasts were reisolated and analyzed by SDS-PAGE. The gels were stained with Coomassie blue, scanned, and then dried for autoradiography. The region of each stained gel between the endogenous large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase is shown below the autoradiograph to confirm that loading was equal in each lane. Molecular masses of markers in kilodaltons are shown on the left of (A). Imported mature [35S]Met-RBCS was quantified using a phosphor imager and normalized to the amount of endogenous RBCS obtained by scanning the Coomassie-stained gels. The amount of mature RBCS imported in the wild type at 20 min was set as 100%. Data shown in the graphs are means ± sd of two to three independent experiments. TR, in vitro–translated [35S]Met-prRBCS before the import reactions.
[See online article for color version of this figure.]
We have shown previously that both the cphsc70-1 and cphsc70-2 mutant alleles we used contain a T-DNA insertion only in the respective gene. For cphsc70-1, the T-DNA insertion cosegregated with the visible phenotypes (Su and Li, 2008). To provide additional evidence that the import defect observed was not due to a second-site, unrelated insertion, we isolated chloroplasts from cphsc70-1 plants complemented with a 5-kb genomic fragment of cpHsc70-1 (the complemented line is designated as cpHsc70-1 g). This genomic fragment contains only the CPHSC70-1 gene, and previous work has shown that it complemented all the visible phenotypes of cphsc70-1 (Su and Li, 2008). Chloroplasts isolated from cpHsc70-1g plants had a fully recovered import efficiency (see Supplemental Figure 2 online), indicating that the import defect observed in cphsc70-1 was caused by the loss of cpHsc70-1. Furthermore, the observation that both cphsc70-1 and cphsc70-2 have similar import defects also supports that the import defects originate from the loss of cpHsc70 proteins, rather than from T-DNA insertion into other loci in the mutant lines.
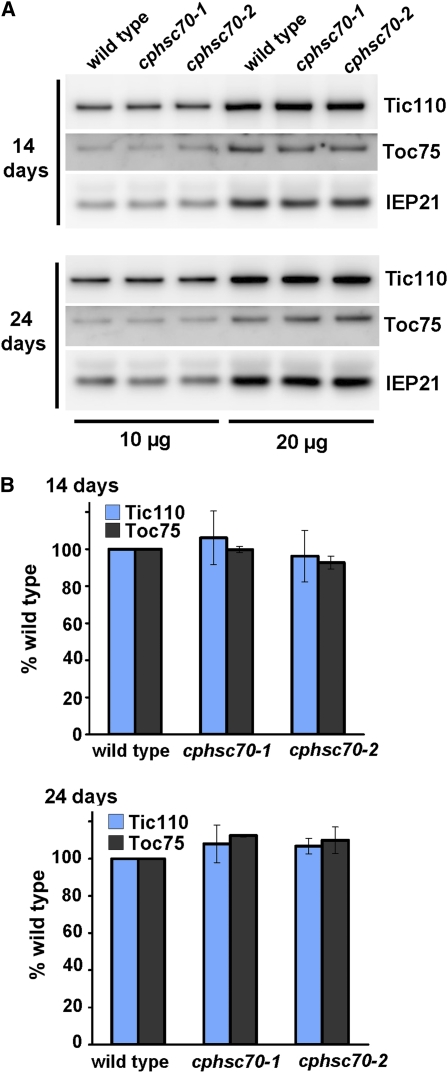
We further analyzed the amounts of major translocon components Toc75 and Tic110 in 14- and 24-d-old plants by immunoblots (Figure 2). The amount of IEP21, an inner membrane protein that is not part of the translocon (Kouranov et al., 1998), was analyzed as a control. The results showed that the amounts of Toc75 and Tic110 were not reduced in the cphsc70 mutants. These data indicate that the observed import defects were not caused by a decrease in the amount of translocon proteins due to secondary effects of the cphsc70 mutations. In addition, because cphsc70-1 and cphsc70-2 had about the same level of reduction in import, the two proteins seem to function equally in supporting import, even though they have other distinct functions in root growth, cotyledon development, and thermotolerance (Su and Li, 2008).
Figure 2.
The Amount of Tic110 and Toc75 Is Not Reduced in the cphsc70 Mutant Chloroplasts.
Total chloroplast proteins from 14- and 24-d-old wild-type, cphsc70-1, and cphsc70-2 seedlings were analyzed by immunoblotting with antibodies against Toc75, Tic110, and the nontranslocon control IEP21. Ten and twenty micrograms of proteins were analyzed. The blots were visualized using a horseradish peroxidase–conjugated secondary antibody chemiluminescent system and the UVP BioSpectrum 600 Image System (A) and quantified with the Image Gauge V4.0 (B). Reading for Toc75 and Tic110 was first normalized to the reading of IEP21 of the same sample and then expressed as percentage of the wild type of the same age. Data from both the 10 and 20 μg analyses were included.
[See online article for color version of this figure.]
cpHsc70 Is Part of the Translocon and Is Associated with Importing Precursors
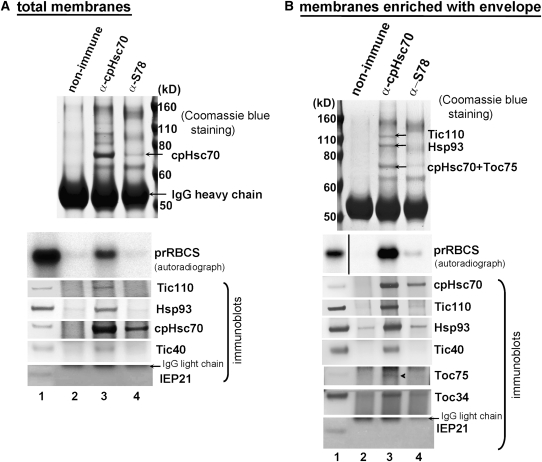
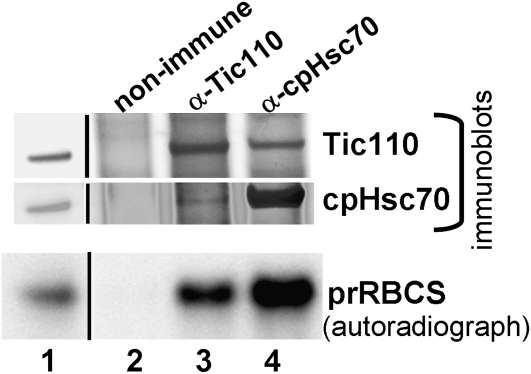
To analyze whether cpHsc70 was associated with importing precursors and the translocon, in vitro–translated [35S]Met-prRBCS was incubated with isolated pea chloroplasts under normal import conditions with 1 mM ATP at room temperature for 5 min. Pilot experiments showed that chloroplasts accumulated the highest amount of precursors en route to the stroma, and a substantial amount of mature RBCS started to appear under these conditions (see Supplemental Figure 3 online). Chloroplasts were reisolated and cross-linked with 1 mM dithiobis-9-succinimidylpropionate (DSP) and quenched by Gly. DSP has been used successfully in identifying translocon complexes (Akita et al., 1997; Chou et al., 2003; Shi and Theg, 2010). Many chloroplasts were already lysed after the DSP and Gly treatments, so a low-speed spin was used to collect the thylakoids and unbroken chloroplasts. The supernatant was further centrifuged at 100,000g for 5 min to prepare a membrane fraction enriched with envelope membranes and importing prRBCS and with less thylakoid membrane contamination (see Supplemental Figure 4 online). The pellet of the low-speed spin was lysed and used to prepare a total-membrane fraction as used previously (Nielsen et al., 1997). Both membrane fractions were solubilized with 1% decylmaltoside (DM), clarified by centrifugation, and used for immunoprecipitation. We used two antibodies for the immunoprecipitation. One is the same anti-S78 antibody used in previous studies (Akita et al., 1997; Nielsen et al., 1997). The other is a commercially available antibody generated to a peptide specific to higher-plant stromal cpHsc70s (Agrisera AS08 348, hereafter referred to as the anti-cpHsc70 antibody). The specificities of both antibodies are shown in Supplemental Figure 5 online. The immunoprecipitates were divided into three portions, and all were first analyzed by SDS-PAGE. The gels were then stained with Coomassie blue to reveal the amount of proteins precipitated, or they were dried and used for autoradiography to reveal the amount of [35S]Met-prRBCS coimmunoprecipitated or used for immunoblotting to detect any coimmunoprecipitated translocon components. As shown in Figure 3A, when solubilized total membranes were used for the immunoprecipitation, the anti-cpHsc70 antibody precipitated a high amount of cpHsc70 (Figure 3A, the Coomassie blue staining panel and the cpHsc70 immunoblot). It also coimmunoprecipitated prRBCS and translocon components Tic110, Hsp93, and Tic40, but not the nontranslocon envelope protein IEP21. In comparison, the anti-S78 antibody precipitated a much lower amount of cpHsc70 and, in agreement with previous data, the amount of prRBCS and translocon components coimmunoprecipitated did not exceed the background level (Figure 3A, lane 4; Akita et al., 1997; Nielsen et al., 1997). These results indicate that previous attempts in detecting the association of cpHsc70 with the translocon and importing precursors were obstructed by the low efficiency of the antibody used.
Figure 3.
cpHsc70 Is Associated with Importing Precursors and Other Translocon Components.
Immunoprecipitation analyses of solubilized total membranes (A) or a membrane fraction enriched with envelope membranes (B). Pea chloroplasts were incubated with prRBCS under import conditions for 5 min, reisolated, and cross-linked with DSP. Supernatant of solubilized total membranes were immunoprecipitated with a nonimmune serum, the anti-cpHsc70 antibody, or the anti-S78 antibody. Immunoprecipitates were divided into three portions and analyzed by SDS-PAGE followed by Coomassie blue staining, autoradiography (prRBCS panel), or immunoblotting with antibodies indicated on the right. Molecular masses of markers in kilodaltons are given for the Coomassie blue–stained gels. Lane 1 of the autoradiographs and immunoblots contained 0.45% of the solubilized membrane supernatant used for the immunoprecipitation. In (A), the positions of cpHsc70 and IgG heavy chain are indicated by arrows. In (B), the three gel bands analyzed by mass spectrometry and the proteins identified are indicated by arrows. The autoradiograph shows two different portions of the same image from the same gel with the same brightness/contrast settings. The arrowhead in the Toc75 panel of the immunoblot indicates the position of Toc75. The antibody cross-reacts with a protein in rabbit sera that migrates right above Toc75. The band migrating above IEP21 in immunoprecipitates (lanes 2 to 4) is the IgG light chain as indicated.
We then analyzed immunoprecipitates obtained from the fraction enriched with envelope membranes. As shown in Figure 3B, the anti-cpHsc70 antibody coimmunoprecipitated with cpHsc70 two proteins the same sizes as Hsp93 and Tic110 in stoichiometric amounts (Figure 3B, arrows in the Coomassie blue staining panel). These three bands were excised and the proteins were identified by mass spectrometry. The top two bands were indeed identified as Tic110 and Hsp93. The cpHsc70 band contained, in addition to cpHsc70, Toc75 (see Supplemental Tables 1 and 2 online).
Autoradiography of immunoprecipitates from the fraction enriched with envelope membranes further revealed that the anti-cpHsc70 antibody coimmunoprecipitated prRBCS. Compared with immunoprecipitation performed with the total membrane fraction shown in Figure 3A, cpHsc70 coimmunoprecipitated a higher percentage of prRBCS in the fraction enriched with envelope membranes. Immunoblots revealed that the immunoprecipitate from the anti-cpHsc70 antibody also contained Tic110, Hsp93, Tic40, and Toc34, but not IEP21. The amount of Toc75 present in the solubilized membranes was very small for unknown reasons, and only a very small amount was immunoprecipitated (arrowhead, Toc75 panel). Similar to the results shown in Figure 3A, the anti-S78 antibody pulled down a much smaller amount of cpHsc70 and very little translocon components. However, a very small amount of prRBCS could now be observed in the immunoprecipitates (Figure 3B, lane 4 of the autoradiograph panel).
cpHsc70 Is Also Important for the Import of the Nonphotosynthetic Precursor Protein prL11
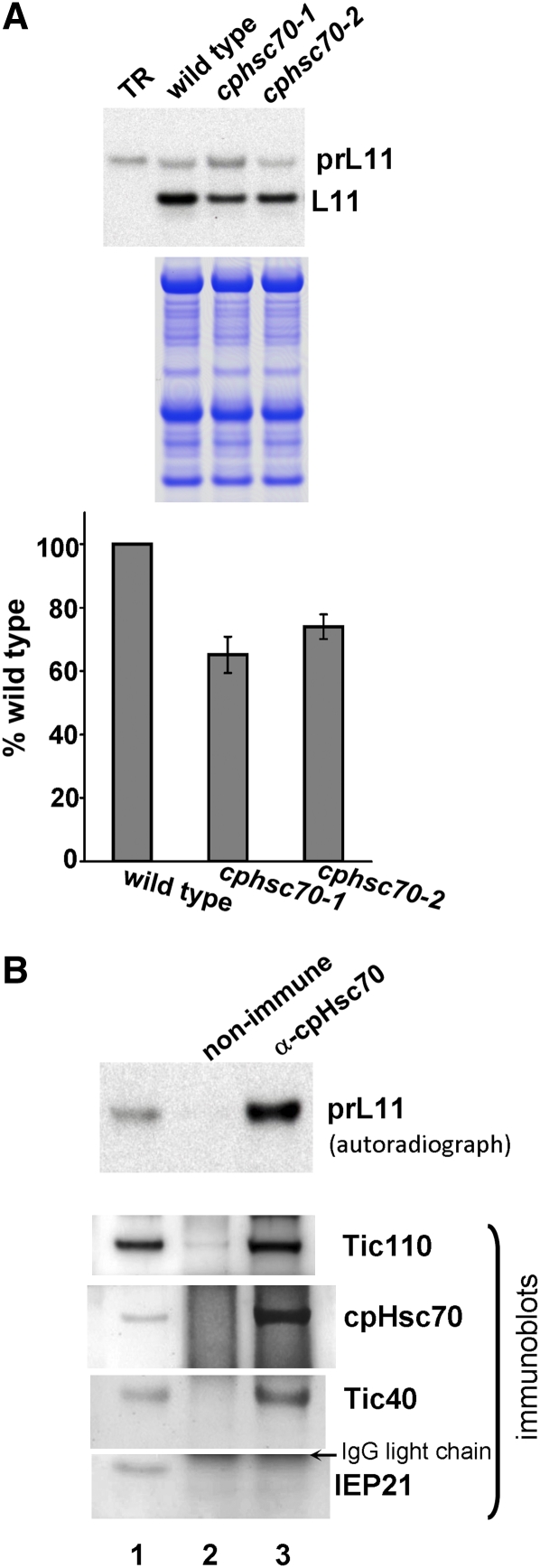
Next, we tested the import of the precursor to the nucleus-encoded chloroplast 50S ribosomal subunit L11, prL11. Whereas prRBCS is used as a representative for photosynthetic proteins that preferentially bind to the receptors at-Toc159 and at-Toc33, prL11 is used as a representative for housekeeping nonphotosynthetic proteins that preferentially bind to the receptors at-Toc132/at-Toc120 and at-Toc34 (Kubis et al., 2003, 2004; Smith et al., 2004). As shown in Figure 4A, cphsc70-1 and cphsc70-2 mutant chloroplasts were defective in importing prL11.
Figure 4.
cpHsc70 Is Important for the Import of the Nonphotosynthetic Precursor Protein prL11.
(A) Chloroplasts were isolated from 21-d-old Arabidopsis plants of various genotypes and incubated with [35S]Met-prL11 under import conditions for 8 min. Intact chloroplasts were reisolated and analyzed by SDS-PAGE. The stained gel is shown below the autoradiograph. Quantification and normalization were performed as described in Figure 1. TR, in vitro–translated [35S]Met-prL11 before the import reactions.
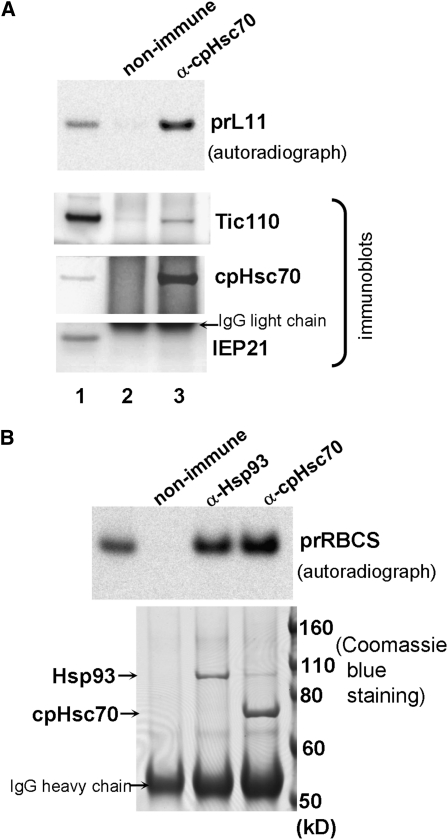
(B) Isolated pea chloroplasts were incubated with [35S]Met-prL11 under import conditions for 5 min, reisolated, and cross-linked with DSP. Supernatant of solubilized crude envelope membranes were immunoprecipitated with a nonimmune serum or the anti-cpHsc70 antibody. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with antibodies indicated on the right. The polyvinylidene fluoride membrane was further exposed to x-ray film for autoradiography to reveal the amount of [35S]Met-prL11. Lane 1 contained 0.45% of the solubilized membrane supernatant used for the immunoprecipitation.
[See online article for color version of this figure.]
We further confirmed that cpHsc70 is associated with importing prL11. Using the same import conditions as those for prRBCS, chloroplasts with importing prL11 were treated with cross-linkers and lysed. The lysed chloroplasts were centrifuged at 3000g to remove most of the thylakoid membranes. The supernatant was then centrifuged at 100,000g to prepare a crude envelope membrane fraction similar to that described (Keegstra and Yousif, 1988). The crude envelope membrane was solubilized with 1% DM and used for immunoprecipitation. The immunoprecipitates were analyzed by immunoblotting and autoradiography. As shown in Figure 4B, the anti-cpHsc70 antibody could indeed immunoprecipitate prL11. The immunoprecipitate also contained translocon components Tic110 and Tic40, but not the nontranslocon component IEP21. Control experiments in which precursors were added after the chloroplasts were lysed showed no precursor proteins immunoprecipitated by the anti-cpHsc70 antibody (see Supplemental Figure 6 online). These results indicate that cpHsc70 participates in the import of prL11. Therefore, similar to other Tic components (Kovacheva et al., 2005; Teng et al., 2006), but different from the two Toc receptor families, cpHsc70 is important for the import of both photosynthetic and nonphotosynthetic proteins.
cpHsc70 Coprecipitated with a Population of Tic110 Enriched with Importing Precursors
To confirm the association of cpHsc70 with other translocon components, we performed reciprocal immunoprecipitations to see if Tic110 could also coimmunoprecipitate cpHsc70. Using the same import and cross-linking conditions, chloroplasts with importing prRBCS were lysed and used to prepare the crude envelope membrane fraction. Solubilized crude envelope membranes were immunoprecipitated with an anti-Tic110 antibody or the anti-cpHsc70 antibody. The immunoprecipitates were divided into two portions and analyzed by immunoblotting and autoradiography. As shown in Figure 5, the anti-Tic110 antibody could indeed immunoprecipitate cpHsc70, supporting that cpHsc70 is part of the translocon. Interestingly, although the anti-cpHsc70 antibody precipitated a lower amount of Tic110 than did the anti-Tic110 antibody (Figure 5, Tic110 panel, compare lane 4 to lane 3), the amount of prRBCS precipitated by the anti-cpHsc70 antibody was higher (prRBCS panel). Since Tic110 is part of the inner membrane channel (Balsera et al., 2009) and the stroma-side receptor for transit peptides (Inaba et al., 2003, 2005) and precursors is processed to mature proteins while still bound to the translocon (Chen and Li, 2007), all precursors crossing the inner membrane should be associated with Tic110. Thus, the result in Figure 5 suggests that those Tic110 molecules associated with cpHsc70 are enriched with importing precursors.
Figure 5.
cpHsc70 Copurified with a Population of Tic110 Enriched with Precursors.
Pea chloroplasts were incubated with prRBCS under import conditions for 5 min, reisolated, and cross-linked with DSP. Solubilized crude envelope membranes were immunoprecipitated with a nonimmune serum, the anti-cpHsc70 antibody, or the anti-Tic110 antibody. Immunoprecipitates were divided into two portions and analyzed by SDS-PAGE followed by autoradiography (prRBCS panel) or immunoblotting with antibodies indicated on the right. Lane 1 contained 0.45% of the solubilized membrane supernatant used for the immunoprecipitation. Pictures of lane 1 were from a different portion of the same image of the same gel as lanes 2 to 4 of the same panel with the same brightness/contrast settings.
It has been shown that a portion of several translocon components preassembles into a Toc-Tic supercomplex even in the absence of importing precursors (Nielsen et al., 1997; Kouranov et al., 1998). We therefore further investigated whether cpHsc70 was also preassembled with the complex or cpHsc70 was recruited to the translocon only during precursor translocation. Isolated chloroplasts without incubating with precursors were directly treated with DSP and then lysed. The crude envelope fraction was solubilized and used for immunoprecipitation with the anti-cpHsc70 antibody (see Supplemental Figure 7 online). cpHsc70 still coprecipitated with translocon components Tic110, Tic40, Toc34, and Toc75, but not with the nontranslocon protein IEP21. The same result was obtained when the immunoprecipitation was performed with the anti-Tic110 antibody (see Supplemental Figure 7 online). This result indicates that, like other translocon components, a portion of the cpHsc70 protein complement is assembled with the Toc-Tic supercomplex even in the absence of importing precursors.
The cphsc70 Mutant Chloroplasts Were Defective in the Translocation Step of the Import Process
Hsp70 family proteins are ATPases. Protein import into chloroplasts involves at least two ATP-dependent steps (Jarvis, 2008). The first step is a binding or early import intermediate step, which requires a low amount of ATP (<0.1 mM; Olsen et al., 1989). At this step, transit peptides penetrate deep into the translocon and reach as far as Tic110 (Inaba et al., 2003) but are still inaccessible to the processing peptidase and the precursors are still largely exposed on the chloroplast surface. The second step is a translocation step, which requires >1 mM ATP (Theg et al., 1989). During this step, the precursors are fully translocated across the inner membrane into the stroma and processed to their mature size. To gain further insight into the function of cpHsc70 in the import process, we investigated which of these steps requires the participation of cpHsc70. Energy-depleted chloroplasts isolated from the wild type and the two cphsc70 mutants were incubated with energy-depleted [35S]Met-prRBCS in the dark and in the absence or presence of 0.1 mM ATP to analyze the binding step. Chloroplasts were then reisolated and half of the chloroplasts that had been incubated with 0.1 mM ATP were further incubated with 5 mM ATP to analyze the translocation step. As shown previously, Arabidopsis chloroplasts usually have a high amount of precursors associated with the chloroplast surface even when no ATP is present in the import system, and the reason for this high background is not clear (Figure 6A, 0 mM ATP lanes; Fitzpatrick and Keegstra, 2001; Aronsson and Jarvis, 2002). Nonetheless, when 0.1 mM ATP was added, the amount of precursors bound was further increased. The level of increase was similar in the wild type and the mutants (Figure 6A, 0.1 mM ATP lanes, and Figure 6B, top graph). However, when chloroplasts were reisolated to separate the chloroplasts from the unbound precursors, and the bound precursors on the chloroplasts were allowed to import by the addition of 5 mM ATP, the amount of protein that was translocated and processed into mature protein was reduced by 30% in both mutants compared with the wild type (Figure 6A, 0.1 mM then 5 mM ATP lanes, and Figure 6B, bottom graph). This result indicates that cpHsc70 is involved in the step of precursor translocation across the envelope membranes, which is consistent with the proposed function of Hsp70 as a motor driving protein translocation into the stroma.
Figure 6.
cphsc70 Mutants Are Defective in the Translocation Step of the Import Process.
Energy-depleted [35S]Met-prRBCS and chloroplasts were incubated with chloroplasts in the absence or present of 0.1 mM ATP in the dark at room temperature for 5 min. Chloroplasts were pelleted, and half of the chloroplasts that had been incubated with 0.1 mM ATP were further incubated with 5 mM ATP at room temperature for 15 min.
(A) Samples were analyzed by SDS-PAGE. The gel was first stained with Coomassie blue, scanned, and then dried and used for autoradiography. A picture of the Coomassie-stained gel is shown below the autoradiograph.
(B) Quantification of binding stimulation and translocation. The percentage of prRBCS binding increase stimulated by 0.1 mM ATP was calculated for each sample and then normalized to the wild type (top graph). The percentage of bound prRBCS that was translocated and processed into mature RBCS was calculated for each sample and then normalized to the wild type (bottom graph). Data shown in the graphs are means ± sd of two duplicate experiments. All values have been corrected for the number of Met residues in prRBCS and RBCS and for the amount of proteins loaded using the amount of endogenous RBCS obtained from scanning the Coomassie blue–stained gels.
[See online article for color version of this figure.]
cpHsc70 and Hsp93 Were Associated with Importing Precursors and Other Translocon Components Even in the Absence of Cross-Linkers
To investigate further whether cpHsc70 directly participates in precursor import or is only peripherally associated with other translocon components to assist their folding or assembly, chloroplasts were again incubated with prL11 under import conditions and reisolated. However, this time chloroplasts were lysed directly without cross-linker treatment. Omitting the cross-linkers can also confirm that the coimmunoprecipitation results we observed were not due to nonspecific association within the spacer arm length of the cross-linkers. Analyses of the immunoprecipitates from the solubilized crude envelope membrane showed that, even in the absence of cross-linkers, cpHsc70 still specifically associated with importing prL11 and Tic110 but not the nontranslocon envelope protein IEP21 (Figure 7A). Compared with experiments with cross-linker treatment (Figure 4B), cpHsc70 pulled down a similar amount of prL11 but a lower amount of Tic110 in the absence of cross-linkers, suggesting that the association of cpHsc70 with precursors is more stable than is the association of cpHsc70 with Tic110. Specific association of cpHsc70 with other translocon components in the absence of cross-linkers also suggested that cpHsc70 is a stable part of the translocon.
Figure 7.
cpHsc70 and Hsp93 Were Associated with Importing Precursors in the Absence of Cross-Linkers.
(A) Chloroplasts were incubated with [35S]Met-prL11 under import conditions for 5 min, reisolated, and lysed directly without adding cross-linkers. Supernatant of solubilized crude envelope membranes were immunoprecipitated with a nonimmune serum or the anti-cpHsc70. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with antibodies indicated on the right. The PVDF membrane was further exposed to x-ray film for autoradiography to reveal the amount of [35S]Met-prL11 precipitated. Lane 1 contained 0.9% of the solubilized membrane supernatant used for the immunoprecipitations.
(B) Same as in (A) except [35S]Met-prRBCS was used. In addition, the anti-Hsp93 antibody was also used for the immunoprecipitation. Immunoprecipitates were divided into two portions analyzed by SDS-PAGE followed by autoradiography or Coommassie blue staining.
We further confirmed the cpHsc70-precursor association with the import of another precursor, prRBCS. We also compared the association with the other chaperone/motor protein Hsp93. Using the same import conditions, solubilized crude envelope membrane containing prRBCS without cross-linker treatment was immunoprecipitated with the anti-Hsp93 or the anti-cpHsc70 antibody. The immunoprecipitates were analyzed by SDS-PAGE followed by autoradiography or Coomassie blue staining. As shown in Figure 7B, the anti-cpHsc70 antibody precipitated a slightly higher amount of prRBCS than did the anti-Hsp93 antibody. In comparison, Coomassie blue staining revealed that the anti-Hsp93 antibody precipitated a much higher amount of Hsp93 than did the anti-cpHsc70 antibody. Immunoblotting confirmed that the major proteins precipitated by the two antibodies were indeed Hsp93 and cpHsc70, respectively (see Supplemental Figure 8 online). There may be two possible explanations for this result. It is possible that precursors are associated with Hsp93 and cpHsc70 in the same complex and cpHsc70 has pulled down a population of Hsp93 highly enriched with precursors. The second possibility is that Hsp93 and cpHsc70 can independently associate with precursors.
The cphsc70-1 hsp93-V Double Mutant Showed an Additive Import Defect
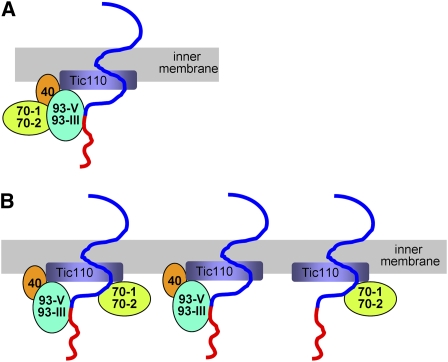
Our data in Figures 1 to 7 indicate that cpHsc70 is part of the translocon and is important for protein translocation across the chloroplast envelope. Hsp70 family proteins function as motors driving protein translocation into mitochondria and the ER. The motor system driving protein translation into chloroplasts has been assumed to be the Hsp93/Tic40 system. Therefore, an important next step is to clarify the functional relationship between the cpHsc70 system and the Hsp93/Tic40 system. Assuming that they provide the major driving force for translocation like in other systems, the two systems may function together as a single motor if, for example, cpHsc70 functions in facilitating the assembly of the Hsp93/Tic40 system (Figure 8A). Alternatively, the two systems may function as independent motors and the precursor could be associated with either one or both chaperones at any given time (Figure 8B). In the first scenario, double mutations in the two systems should result in no additional defects, similar to the finding that the tic40 hsp93-V double mutant has a similar phenotype to that of the tic40 single mutant (Kovacheva et al., 2005). On the other hand, if the two systems operate in parallel as depicted in Figure 8B, double mutations in the two systems should result in loss of both motor systems and have severe consequences.
Figure 8.
Two Possible Models for the Relationship between the cpHsc70 and the Hsp93/Tic40 Systems.
(A) The cpHsc70 and Hsp93/Tic40 systems may function as one complex, for example, with cpHsc70 assisting the assembly of the Hsp93/Tic40 system.
(B) The cpHsc70 and Hsp93/Tic40 systems may function in parallel, for example, by both independently associating and facilitating the translocation of precursors across the inner membrane. Therefore, at any given time, a precursor could be associated with either one, or both, chaperones as depicted in the figure. The red and blue regions of the line indicate the transit peptide and mature regions of a precursor protein, respectively. Since it is not known how Hsp93 and cpHsc70 bind to a precursor, they are placed arbitrarily on the precursor.
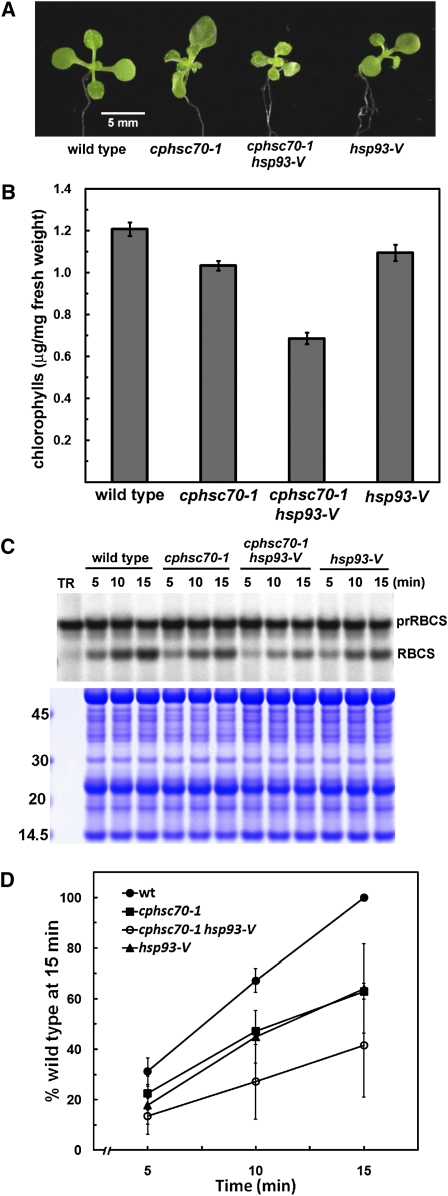
Arabidopsis Hsp93 is encoded by two genes: HSP93-III and HSP93-V. Overexpression of Hsp93-III can complement the phenotypes of hsp93-V (Kovacheva et al., 2007), indicating functional redundancy between the two proteins. However, phenotype analyses of individual mutants suggest that Hsp93-V is the major functional form (Constan et al., 2004; Sjogren et al., 2004; Kovacheva et al., 2005). We therefore first generated a double mutant of cphsc70-1 hsp93-V. The hsp93-V mutant used (Salk_014058) was the same null allele reported previously (clpC1-1 in Sjogren et al., 2004 and hsp93-V-2 in Kovacheva et al., 2005). Compared with the wild type, both the cphsc70-1 and the hsp93-V single mutants had only slightly reduced chlorophyll contents (Figures 9A and 9B). However, the double mutant showed a much stronger defect with a 50% reduction in chlorophyll. Chloroplasts were isolated from 14-d-old mutants and the wild type and analyzed for their import efficiencies. While chloroplasts from both cphsc70-1 and hsp93-V had an import efficiency ∼60% of that of the wild type, chloroplasts from the cphsc70-1 hsp93-V double mutant had an import efficiency <40% of that of the wild type (Figures 9C and 9D). This result suggests that cpHsc70-1 and Hsp93-V have a redundant function in chloroplast protein import. However, because of the presence of the second gene, HSP93-III, the result can also be interpreted as reduced assembly of the remaining Hsp93-III/Tic40 system due to the cphsc70-1 mutation. We have not been able to obtain the cphsc70-2 hsp93-V double mutant because the two loci are only 409 kb apart on chromosome V.
Figure 9.
The cphsc70-1 hsp93-V Double Mutant Has More Severe Phenotypes.
(A) Plants of the indicated genotypes were grown on MS medium for 14 d and photographed.
(B) Leaf chlorophyll contents of seedlings as shown in (A) were determined. Data are means ± sd of three replicates.
(C) Chloroplasts were isolated from 14-d-old seedlings of the indicated genotypes and used to perform import time-course assays with prRBCS. After import, intact chloroplasts were reisolated and analyzed by SDS-PAGE. An equal amount of protein was loaded in each lane. The gel was stained with Coomassie blue and dried for autoradiography. The region of the gel between the endogenous large and small subunits of ribulose-1,5-bis-phosphate carboxylase/oxygenase is shown to confirm that loading was equal in each lane. Molecular masses of makers in kilodaltons are shown on the left.
(D) Imported mature RBCS was quantified using a phosphor imager. The amount of mature RBCS imported into wild-type (wt) chloroplasts at 15 min was set as 100%. Data are means ± sd of three independent experiments.
cpHsc70-1 Has a Vital Function in Common with Tic40
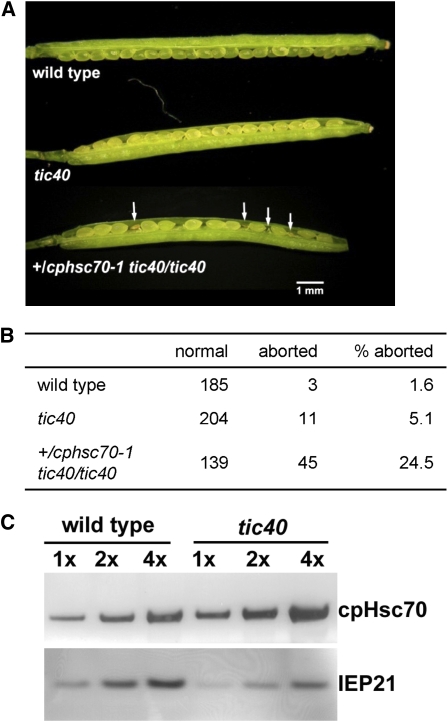
Although Arabidopsis Hsp93 is encoded by two genes, their cochaperone Tic40 is encoded by a single gene, which makes genetic interaction analyses less complicated. We therefore crossed the cphsc70-1 mutant with the tic40 mutant. The tic40 mutant used is the tic40-2 null allele previously reported (Chou et al., 2003). In the progenies of plants homozygous for cphsc70-1 and heterozygous for tic40 (genotype cphsc70-1/cphsc70-1 +/tci40), or heterozygous for cphsc70-1 and homozygous for tic40 (+/cphsc70-1 tic40/tic40), the double mutant was not observed (Table 1). Meanwhile, approximately one-quarter of the seeds from the +/cphsc70-1 tic40/tic40 plants were aborted (Figures 10A and 10B), indicating that the cphsc70-1 tic40 double mutation is lethal. This result indicates that cpHsc70-1 and Tic40 have an overlapping, essential function. Because Tic40 is a cochaperone, not a chaperone, and it is an integral inner envelope membrane protein associated with the translocon, the most plausible explanation for the lethality is that cpHsc70-1 and Tic40/Hsp93 perform the same function during protein import. It is possible that in the tic40 mutant, both Hsp93-III and Hsp93-V can no longer function efficiently in driving import and that the tic40 mutant remains viable because of the presence of a second motor system, the cpHsc70 system. In agreement with this hypothesis, we observed an increase in the amount of cpHsc70 in tic40 chloroplasts (Figure 10C). In comparison, the amount of nontranslocon envelope protein IEP21 was reduced. Furthermore, according to publicly available microarray data (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Tissue_Specific), cpHsc70-1 is essentially the only cpHsc70 present during the globular stage of embryogenesis because cpHsc70-2 is barely expressed at that stage. Therefore, it is possible that the cphsc70-1 tic40 double mutant embryos basically had both motor systems knocked out and could no longer develop. We also performed reciprocal crosses of the +/cphsc70-1 tic40/tic40 plants with the wild type. Genotype analyses of F1 seedlings revealed that the double mutation caused reduced viability, but not lethality, in both male and female gametes (see Supplemental Table 3 online). The presence of cpHsc70-2 may have sustained some plastid functions in the gametes.
Table 1.
Genotypes of Progenies from cphsc70-1/cphsc70-1 +/tic40 and +/cphsc70-1 tic40/tic40 Plants
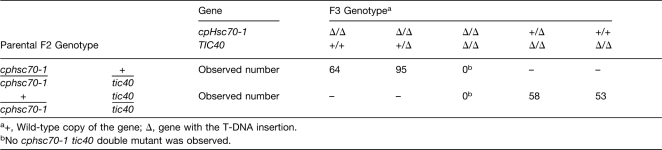 |
Figure 10.
cpHsc70-1 Has a Vital Function in Common with Tic40.
(A) The cphsc70-1 tic40 double mutation was lethal. Typical and fully elongated siliques from wild-type, tic40, and +/cphsc70-1 tic40/tic40 plants are shown. Arrows indicate aborted seeds.
(B) Scoring of the number of seeds aborted from plants of the three genotypes shown in (A).
(C) The amount of the cpHsp70 was increased in the tic40 mutant. Chloroplasts from wild-type and tic40-mutant plants were isolated and analyzed by SDS-PAGE and immunoblotting with antibodies indicated on the right. A serial dilution of proteins was analyzed. 1x = 4.5 μg.
[See online article for color version of this figure.]
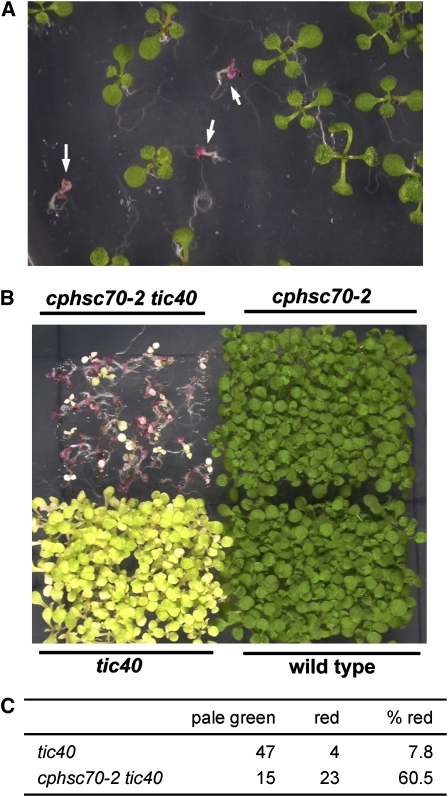
We also generated the cphsc70-2 tic40 double mutant. Slightly less than one-quarter of progenies from cphsc70-2/cphsc70-2 +/tic40 plants were double mutants (16 out of 86 plants), indicating that the double mutation did not severely affect plant viability. However, during germination, a high percentage of double mutant seedlings grew much more slowly and accumulated high levels of anthocyanins (Figure 11A). These plants eventually developed, and the mature plants were indistinguishable from the tic40 single mutant (see Supplemental Figure 9 online). When seeds of double mutants were germinated, 60.5% of seedlings showed the slow growth and anthocyanin accumulation phenotype. In comparison, only 7.8% of tic40 single-mutant seedlings showed such a phenotype (Figures 11B and 11C). These results again indicate that, in the tic40 mutant, the amount of cpHsc70 became critical for plant growth, especially in early developmental stages.
Figure 11.
A Higher Percentage of cphsc70-2 tic40 Double Mutant Seedlings Grew More Slowly and Accumulated Anthocyanins.
(A) Seeds from cphsc70-2/cphsc70-2 +/tic40 plants were germinated and grown on MS medium for 8 d. Arrows indicate cphsc70-2 tic40 double mutants.
(B) Seeds from the wild type, cphsc70-2, tic40, and cphsc70-2 tic40 were germinated and grown on MS medium for 9 d.
(C) Scoring of the percentage of seedlings with the slow growth and anthocyanin accumulation (red) phenotype in tic40 and cphsc70-2 tic40 mutants.
DISCUSSION
It has been shown recently in the moss Physcomitrella patens that a stromal Hsp70 system functions in protein import into chloroplasts (Shi and Theg, 2010). Protein import is impaired in heat-shocked chloroplasts isolated from temperature-sensitive stromal hsp70 mutants, or in chloroplasts from a knockdown mutant of GrpE, the nucleotide exchange factor for Hsp70. We show here that we observed the same import defects in mutants of Arabidopsis cpHsc70s. These results indicate that the function of Hsp70 in protein translocation may be conserved from moss to higher plant chloroplasts, and from the ER, mitochondria to chloroplasts. We have further provided biochemical data showing cpHsc70 is a stable part of the translocon and dissected the import process to show that cpHsc70 is involved in the translocation step of the import process. Moreover, because the generally assumed motor system for chloroplast protein import has been the Hsp93/Tic40 system, we further analyzed the relationship between the cpHsc70 and the Hsp93/Tic40 systems. Using double mutant analyses, our data suggest that the two systems function in parallel.
It has been difficult to discern biochemically whether stromal cpHsc70 was involved in protein import because the cpHsc70-precursor complex was difficult to immunoprecipitate from solubilized total chloroplast membranes (Akita et al., 1997; Nielsen et al., 1997). In Physcomitrella chloroplasts, demonstration of precursor-Hsp70 association was achieved only when the precursor of GrpE, which may have a tendency to associate with Hsp70, was used or when a very high amount of prRBCS afforded by Escherichia coli overexpressed recombinant prRBCS was used (Shi and Theg, 2010). However, we found that if a membrane fraction enriched with envelope membranes and a better antibody were used, the cpHsc70-precursor complex could be solubilized and specifically immunoprecipitated with other translocon components in stoichiometric amounts. We further showed that the complex between cpHsc70 and importing precursors was stable even in the absence of cross-linkers. Therefore, we identified the reasons for previous technical difficulties and provided clear evidence to show that cpHsc70 is part of the chloroplast translocon.
Because cpHsc70 may have other functions as a general chaperone, the observed import defects in the cphsc70 mutants may be caused by secondary effects of cpHsc70 affecting the folding of other translocon components. However, data from Physcomitrella showing that import was further impaired in heat-shocked chloroplasts isolated from temperature-sensitive stromal hsp70 mutants suggest that cpHsc70 is directly involved in import (Shi and Theg, 2010). We provide several additional lines of evidence to support that cpHsc70 plays an active role in the import process. First, if the primary function of cpHsc70 is to assist the folding or assembly of other translocon components, it is not necessary for cpHsc70 to associate with Tic110 and Hsp93 in the inner membrane in stoichiometric amounts. Second, cpHsc70 could be specifically coimmunoprecipitated with importing precursors and Tic110 even when no cross-linker was added. This result indicates that cpHsc70 is a stable part of the translocon. Third, the Tic110 molecules associated with cpHsc70 are enriched with importing precursors, suggesting that assembly of Tic110 with cpHsc70 is an active part of the translocation process. Finally, the lethality of the cphsc70-1 tic40 double mutant indicates that cpHsc70-1 and Tic40 have an overlapping essential function. If the function of cpHsc70 was to assist the assembly or function of Hsp93 or Tic40, then the cphsc70-1 tic40 double mutant should have the same phenotype as the tic40 single mutant. Indeed, in comparison, the phenotype of the hsp93-V tic40 double mutant is similar to tic40, or even slightly healthier than tic40 (Kovacheva et al., 2005). If the import defects of the cphsc70 mutants were the results of a lower population of properly folded Tic110, then the cphsc70-1 tic40 double mutant should have the additive phenotypes of cphsc70-1 plus tic40, rather than complete lethality.
If Hsp93/Tic40 and cpHsc70 really function as motors, the dual chaperone/motor system setup seems to be unique to chloroplasts. The Hsp70 family proteins are the motors driving protein translocation into the ER and mitochondria. AAA+ proteins, of which Hsp93 is a member, have been shown to be the motors driving protein translocation for ER-associated degradation and for unwinding and delivering polypeptide chains through the central pore of several proteases (Tucker and Sallai, 2007). In yeast mitochondria, Hsp78, an Hsp100 family member, partially substitutes for the matrix Hsp70 in protein import only when the Hsp70 system is defective (Schmitt et al., 1995). In chloroplasts, however, hsp93-V and the cphsc70 single mutants already show import defects, indicating that both systems are functioning under normal conditions. Thus, chloroplasts have employed both systems to drive protein translocation into the stroma. If cpHsc70 and Hsp93/Tic40 really function as motors, it is interesting to speculate why both systems have been recruited by chloroplasts while other organelles seem to have only adopted one system. Plastids need to import a wide array of proteins through different developmental stages. Two motor systems, each encoded by two genes, may provide the versatility required. Although data obtained by us and others (Kovacheva et al., 2005) indicate that the cpHsc70 and Hsp93/Tic40 systems do not seem to distinguish among photosynthetic and nonphotosynthetic proteins, it is still possible that the two systems have slightly different binding preferences with regard to polypeptide position or hydrophobicity. It is also possible that each system is more important in certain developmental stages or in certain cell types. This possibility may explain why import defects of the hsp93-V mutant have not been consistently observed (Sjogren et al., 2004). Different laboratories may have used mutant plants of different developmental stages. Our data also showed that import defects of cphsc70 mutants are observed only in young true leaves. The presence of two systems may also explain why precursors with altered affinities for Hsp70 in their transit peptides are still efficiently imported into chloroplasts (Rial et al., 2003, 2006).
We cannot rule out the possibility that cpHsc70 and Hsp93/Tic40 function together at different steps of the transport process, similar to the ClpB/Hsp104 and DnaK/Hsp70 bichaperone system for ATP-dependent protein disaggregation in the cytosol (Glover and Lindquist, 1998). Indeed, Arabidopsis mutants entirely lacking Hsp93 or cpHsc70 (i.e., the hsp93-III hsp93-V and the cphsc70-1 cphsc70-2 double mutants) are not viable (Kovacheva et al., 2007; Su and Li, 2008). However, our double mutant analyses suggest that cpHsc70 and Hsp93/Tic40 are not epistatic to each other, but rather are more likely to function in parallel. It should be noted that both cpHsc70 and Hsp93 have other functions in chloroplast biogenesis. cpHsc70 may assist protein folding under stressed or normal conditions, and at least cpHsc70-1 is important for thermotolerance of germinating seeds (Su and Li, 2008). In Chlamydomonas, stromal Hsp70B regulates the assembly state of VIPP1 (vesicle inducing protein in plastid 1), which may be important for thylakoid biogenesis or maintenance (Liu et al., 2005, 2007). Hsp93 in the stroma is suggested to function primarily as the regulatory chaperone for the ClpP proteases and/or as an independent chaperone for general protein folding/unfolding and assembly (Adam and Clarke, 2002). Hsp93 has also been proposed to be involved in photosystem biogenesis (Sjogren et al., 2004) and in insertion of Tic110 from the stroma (Vojta et al., 2007). The lethality of the hsp93-III hsp93-V and the cphsc70-1 cphsc70-2 double mutants could be caused by failure to carry out these functions. Furthermore, we cannot exclude the possibility that Tic40 may also interact with cpHsc70s, similar to the interactions of Hip and Hop with cytosolic Hsp70s.
An Hsp70 from pea chloroplasts, named Hsp70-IAP, has been shown to associate with importing precursors and to be part of the translocon complex (Schnell et al., 1994; Chen and Li, 2007). Hsp70-IAP comigrated with Toc75. Although Hsp70-IAP was suggested to localize in the intermembrane space of the envelope, recent data showed that it is most likely a stromal protein (Ratnayake et al., 2008). Our immunoprecipitation data (Figure 3B) showed that stromal cpHsc70 not only coimmunoprecipitated Toc75, but also these two proteins comigrated as one band in SDS-PAGE. Taken together, these data suggest that the long sought after Hsp70-IAP may be the stromal cpHsc70 (Ratnayake et al., 2008).
Solid evidence for direct contacts with precursors has not been obtained for either Hsp93 or cpHsc70 in vivo. Furthermore, elucidating the molecular mechanism of the translocation process will also require a great deal of future research. For example, it is important to understand the energetics and the conformational changes involved in the translocation process. It also remains to be established whether the stromal cpHsc70 requires any cochaperones, such as the J domain–containing proteins or membrane tethers like the mitochondrial Tim44, for its function in chloroplast protein import.
METHODS
Plant Materials and Growth Conditions
Wild-type and mutant Arabidopsis thaliana plants used in this study were of the ecotype Columbia. The knockout mutants of cphsc70-1 (Salk_140810) and cphsc70-2 (Salk_095715) were identified as described (Su and Li, 2008). Genotypes of mutants were verified by PCR amplification of genomic DNA using specific primers (see Supplemental Table 4 online). The cpHsc70-1g complemented line was produced as described (Su and Li, 2008). Briefly, a 5-kb genomic fragment containing the promoter and coding regions of cpHsc70-1 was amplified by PCR from the wild type and subcloned into the binary vector pCambia1390 (CAMBIA), and the construct was used to transform the cphsc70-1 mutant. The hsp93-V mutant used (Salk_014058) was the same null allele reported previously (clpC1-1 in Sjogren et al., 2004 and hsp93-V-2 in Kovacheva et al., 2005). The tic40 mutant used is the tic40-2 null allele previously reported (Chou et al., 2003).
For chloroplast isolation, sterilized Arabidopsis seeds were plated on 0.3% Gelrite-solidified 1× Murashige and Skoog (MS) medium containing Gamborg's B5 vitamin and 0.5% sucrose, except for mutants containing the tic40 mutation, for which 2% sucrose was used. To facilitate harvesting 14-d-old seedlings for chloroplast isolation, seeds were plated on a piece of 3MM filter paper placed on the MS medium. After a 3-d cold stratification, seeds were grown in growth chambers under a 16-h photoperiod at 22°C with a light intensity of ∼80 μmol m−2 s−1. For growing pea seedlings (Pisum sativum cv Little Marvel; De Bruyn Seed Store), imbibed seeds were grown on vermiculite for 9 to12 d under a 12-h photoperiod at 20°C with a light intensity of ∼150 μmol m−2 s−1. Total chlorophyll contents of Arabidopsis seedlings were determined as described (Lichtenthaler, 1987).
In Vitro Translation and Protein Import and Postimport Analyses
[35S]Met-prRBCS and [35S]Met-prL11 synthesis, pea and Arabidopsis chloroplast isolation, and protein import assays were conducted as described (Perry et al., 1991) with the following modifications. In vitro transcription and translation were performed either by first synthesizing RNA using in vitro transcription (Perry et al., 1991) followed by in vitro translation using the wheat germ lysate (Promega) or by the TNT-wheat germ lysate-coupled transcription/translation system (Promega). For chloroplast isolation, a house blender was used instead of the Polytron, and the grinding buffer for Arabidopsis chloroplast isolation was modified to 50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol, 2 mM EDTA, and 0.5% BSA. After import, samples were resolved by SDS-PAGE using the NuPAGE gel system (Invitrogen) with the MES running buffer. Quantification of gel bands was performed using the Fuji FLA5000 phosphor imager (Fuji Photo Film). Immunoblots were performed by electroblotting of proteins onto polyvinylidene difluoride membranes, probing membranes using specific primary antibodies and an alkaline phosphatase-conjugated secondary antibody, and visualized by the NBT-BCIP colorimetric system. The data shown in Figure 2 were obtained using horseradish peroxidase–conjugated secondary antibodies and the Immobilon Western Chemiluminescent HRP system (Millipore), visualized with the UVP BioSpectrum 600 Image System (Ultra Violet Products), and quantified with Image Gauge V4.0 (Fuji Photo Film).
Separation of the import process into the binding and translocation steps was performed as described (Olsen et al., 1989; Teng et al., 2006). Briefly, isolated chloroplasts were kept on ice in the dark for 2 h to deplete internal ATP. Precursors used were desalted using a Sephadex G-25 gel filtration column to remove ATP from the in vitro translation system. Chloroplasts were incubated with precursors first in the absence or presence 0.1 mM ATP in the dark at room temperature for 5 min. The chloroplasts were pelleted by centrifugation. Half of the chloroplasts that had been incubated with 0.1 mM ATP were further resuspended in import buffer containing 5 mM ATP and incubated at room temperature for another 15 min. All the import steps were performed under a green safe light.
Immunoprecipitation and Antibodies
To analyze the association of cpHsc70 with importing precursors, [35S]Met-prRBCS and [35S]Met-prL11 were incubated with isolated pea chloroplasts in 1 mM ATP at room temperature for 5 min. Chloroplasts were reisolated and cross-linked with 1 mM DSP at 4°C for 15 min except for the experiments shown in Figure 7, in which no cross-linking nor subsequent Gly quenching was performed. The reaction was terminated by adding Gly to a final concentration of 50 mM and further incubated at 4°C for 15 min to quench the free DSP. To prepare a fraction enriched with envelope, the reaction mixture was centrifuged at 1000g. The supernatant was collected and further centrifuged at 100,000g for 5 min to obtain the membrane fraction enriched envelope membranes. To prepare a crude envelope membrane fraction, chloroplasts after cross-linking were pelleted and resuspended in import buffer, lysed by one to two freeze-thaw cycles, and further centrifuged at 3000g for 5 min to remove most of the thylakoid membranes. The supernatant was further centrifuged for 100,000g for 15 to 30 min to obtain the crude envelope membrane fractions. For a total membrane fraction, the crude envelope membrane fraction was combined with the thylakoid membrane fraction from the 3000g 5-min spin. All membrane fractions were solubilized in an IP buffer containing 50 mM HEPES-KOH, pH 8.0, 150 mM NaCl, 4 mM MgCl2, 1% DM, and 10% glycerol. The solubilized membranes were clarified by centrifugation at 100,000g for 45 min. For immunoprecipitation, antibodies were added to the clarified supernatant and incubated for 4 h at 4°C and then precipitated by Protein-A-agarose. After washing with the IP buffer for at least three times, samples were eluted by SDS-PAGE sample buffer and analyzed by SDS-PAGE. The antibody against cpHsc70 was purchased from Agrisera. The anti-S78 antibody used was as described (Akita et al., 1997; Nielsen et al., 1997). Antibodies against Tic and Toc proteins were prepared as described (Tu et al., 2004; Chou et al., 2006). In brief, the polypeptide region (the first residue of the precursor protein is designated as residue 1) and the animal species used to generate each antibody were as follows: full-length pea Toc75 precursor, pea Toc34 (1 to 258), pea Hsp93 (86 to 332), and Arabidopsis Tic110 (431 to 1016) in rabbits and Arabidopsis Tic40 (227 to 447) in mice. The rat anti-Tic110 and anti-Toc75 antibodies were generated for this study. The antigens used were pea Toc75 (132 to 809) and Arabidopsis Tic110 (431 to 1016). The corresponding cDNA was subcloned into pET21a and pET21d vectors, respectively, with C-terminal His tags. The proteins were overexpressed in Escherichia coli and purified by affinity chromatography. The rat anti-Tic110 and anti-Toc75 antibodies were used in a 1:1000 dilution for Figures 4 and 7 and Supplemental Figure 7 online. The anti-IEP21 used was as described (Kouranov et al., 1998). Mass spectrometry identification of proteins from excised gel bands was performed by the University of California Davis Genome Center Proteomics Core Facility using in-gel trypsin digestion followed by liquid chromatography–tandem mass spectrometry.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or The Arabidopsis Information Resource data libraries under the following accession numbers: Arabidopsis cpHsc70-1 (At4g24280), cpHsc70-2 (At5g49910), Hsp93-III (ClpC2, At3g48870), Hsp93-V (ClpC1, At5g50920), Tic40 (At5g16620), and pea stromal cpHsc70 S78 cDNA (L03299) and protein (Q02028).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Chloroplast Protein Import Efficiency Declines as Plants Become Older.
Supplemental Figure 2. Genomic Fragment of CPHSC70-1 Complemented the Import Defect of the cphsc70-1 Mutant.
Supplemental Figure 3. Import Time Course Experiment to Determine a Better Time Point for Immunoprecipitation.
Supplemental Figure 4. The Fraction Enriched with Envelope Membranes Contained More Tic110 and prRBCS and Less Chlorophyll a/b Binding Protein Than the Total Membrane Fraction.
Supplemental Figure 5. Specificity of Anti-cpHsc70 Antibody.
Supplemental Figure 6. No Precursor Was Precipitated by the Anti-cpHsc70 or AntiHsp93 Antibody if Precursors Were Added after Chloroplast Lysis.
Supplemental Figure 7. cpHsc70 Was Associated with the Toc-Tic Translocon Complex in the Absence of Importing Precursors.
Supplemental Figure 8. Immunoblots of the Samples Shown in Figure 7.
Supplemental Figure 9. Phenotypes of tic40 and cphsc70-2 tic40 Mutants.
Supplemental Table 1. Identification of Coimmunoprecipitated Proteins by LC/MS/MS.
Supplemental Table 2. Peptide Report of MS/MS Spectra.
Supplemental Table 3. Genotypes of F1 Plants from Reciprocal Crosses of +/cphsc70-1 tic40/tic40 with the Wild Type.
Supplemental Table 4. Primers Used for Mutant Verification.
Acknowledgments
We thank Miranda Jane Loney and Heiko Kuhn for English editing, Kentaro Inoue for critical reading of the manuscript, Ming-Jung Liu for assistance in analyzing the gametophytic effects of the cphsc70-1 tic40 double mutation, Danny Schnell for the anti-IEP21 antibody and the construct for production of pea Toc75 (132-809), Ken Keegstra and John Froehlich for the anti-S78 antibody, Neil Hoffman for the anti-CAB antibody, and Rudy Alvarado for assistance in protein identification with mass spectrometry. This work was supported by the National Science Council (Grant NSC-98-2321-B-001-033) and the Academia Sinica of Taiwan.
References
- Adam Z., Clarke A.K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7: 451–456 [DOI] [PubMed] [Google Scholar]
- Akita M., Nielsen E., Keegstra K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V., Ingenfeld A., Apel K. (2008). Snowy cotyledon 2: The identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol. Biol. 66: 599–608 [DOI] [PubMed] [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Balsera M., Goetze T.A., Kovacs-Bogdan E., Schurmann P., Wagner R., Buchanan B.B., Soll J., Bolter B. (2009). Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J. Biol. Chem. 284: 2603–2616 [DOI] [PubMed] [Google Scholar]
- Becker T., Hritz J., Vogel M., Caliebe A., Bukau B., Soll J., Schleiff E. (2004). Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol. Biol. Cell 15: 5130–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard J., Kubis S., Bimanadham S., Jarvis P. (2007). Functional similarity between the chloroplast translocon component, Tic40, and the human co-chaperone, Hsp70-interacting protein (Hip). J. Biol. Chem. 282: 21404–21414 [DOI] [PubMed] [Google Scholar]
- Chen K.Y., Li H. (2007). Precursor binding to an 880-kD Toc complex as an early step during active protein import into chloroplasts. Plant J. 49: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Chu C.C., Chen L.J., Akita M., Li H. (2006). Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Fitzpatrick L.M., Tu S.L., Budziszewski G., Potter-Lewis S., Akita M., Levin J.Z., Keegstra K., Li H. (2003). Tic40, a membrane-anchored co-chaperone homologue in the chloroplast protein translocon. EMBO J. 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D., Froehlich J., Rangarajan S., Keegstra K. (2004). A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136: 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin C., Cline K. (1991). Developmental regulation of the plastid protein import apparatus. Plant Cell 3: 1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick L.M., Keegstra K. (2001). A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 27: 59–65 [DOI] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S. (1998). Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Inaba T., Alvarez-Huerta M., Li M., Bauer J., Ewers C., Kessler F., Schnell D.J. (2005). atTic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17: 1482–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Li M., Alvarez-Huerta M., Kessler F., Schnell D.J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278: 38617–38627 [DOI] [PubMed] [Google Scholar]
- Inaba T., Schnell D.J. (2008). Protein trafficking to plastids: one theme, many variations. Biochem. J. 413: 15–28 [DOI] [PubMed] [Google Scholar]
- Ishizaki Y., Tsunoyama Y., Hatano K., Ando K., Kato K., Shinmyo A., Kobori M., Takeba G., Nakahira Y., Shiina T. (2005). A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Ivey R.A., III, Subramani S., Bruce B.D. (2000). Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol. 122: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Keegstra K., Yousif A.E. (1988). Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 118: 316–325 [Google Scholar]
- Kessler F., Schnell D. (2009). Chloroplast biogenesis: Diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 21: 494–500 [DOI] [PubMed] [Google Scholar]
- Kim C., Apel K. (2004). Substrate-dependent and organ-specific chloroplast protein import in planta. Plant Cell 16: 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Chen X., Fuks B., Schnell D.J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Schnell D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S., Bedard J., Patel R., Dudley P., Twell D., Rios G., Koncz C., Jarvis P. (2005). In vivo studies of the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bedard J., Wardle A., Patel R., Jarvis P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., Kurth J., Leister D., Jarvis P. (2003). The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bedard J., Kovacheva S., Lilley K., Biehl A., Leister D., Rios G., Koncz C., Jarvis P. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Liu C., Willmund F., Golecki J.R., Cacace S., Hess B., Markert C., Schroda M. (2007). The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50: 265–277 [DOI] [PubMed] [Google Scholar]
- Liu C., Willmund F., Whitelegge J.P., Hawat S., Knapp B., Lodha M., Schroda M. (2005). J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J., DeRocher A., Keegstra K., Vierling E. (1990). Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl. Acad. Sci. USA 87: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.S., Keegstra K. (1992). Isolation and characterization of a cDNA clone encoding the major Hsp70 of the pea chloroplastic stroma. Plant Physiol. 100: 1048–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L.J., Theg S.M., Selman B.R., Keegstra K. (1989). ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 246: 6724–6729 [PubMed] [Google Scholar]
- Perry S.E., Keegstra K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E., Li H., Keegstra K. (1991). In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol. 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Ratnayake R.M., Inoue H., Nonami H., Akita M. (2008). Alternative processing of Arabidopsis Hsp70 precursors during protein import into chloroplasts. Biosci. Biotechnol. Biochem. 72: 2926–2935 [DOI] [PubMed] [Google Scholar]
- Rial D., Arakaki A., Almará A., Orellano E., Ceccarelli E. (2006). Chloroplast Hsp70s are not involved in the import of ferredoxin-NADP+ reductase precursor. Physiol. Plant. 128: 618–632 [Google Scholar]
- Rial D.V., Ottado J., Ceccarelli E.A. (2003). Precursors with altered affinity for Hsp70 in their transit peptides are efficiently imported into chloroplasts. J. Biol. Chem. 278: 46473–46481 [DOI] [PubMed] [Google Scholar]
- Schmitt M., Neupert W., Langer T. (1995). Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 14: 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Mochizuki M., Ogura K., Froehlich J.E., Osteryoung K.W., Shirano Y., Shibata D., Masuda S., Mori K., Takamiya K. (2007). Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19: 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren L.L., MacDonald T.M., Sutinen S., Clarke A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.D., Rounds C.M., Wang F., Chen K., Afitlhile M., Schnell D.J. (2004). atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 165: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl T., Glockmann C., Soll J., Heins L. (1999). Tic40, a new “old” subunit of the chloroplast protein import translocon. J. Biol. Chem. 274: 37467–37472 [DOI] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2008). Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.S., Su Y.S., Chen L.J., Lee Y.J., Hwang I., Li H.M. (2006). Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18: 2247–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg S.M., Bauerle C., Olsen L.J., Selman B.R., Keegstra K. (1989). Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264: 6730–6736 [PubMed] [Google Scholar]
- Tu S.L., Chen L.J., Smith M., Su Y.S., Schnell D., Li H.M. (2004). Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P.A., Sallai L. (2007). The AAA+ superfamily–A myriad of motions. Curr. Opin. Struct. Biol. 17: 641–652 [DOI] [PubMed] [Google Scholar]
- Vojta L., Soll J., Bolter B. (2007). Requirements for a conservative protein translocation pathway in chloroplasts. FEBS Lett. 581: 2621–2624 [DOI] [PubMed] [Google Scholar]