Abstract
Nck is a functionally versatile multidomain adaptor protein consisting of one SH2 and three SH3 domains. In most cases, the SH2 domain mediates binding to tyrosine-phosphorylated receptors or cytosolic proteins, which leads to the formation of larger protein complexes via the SH3 domains. Nck plays a pivotal role in T-cell receptor mediated reorganization of the actin cytoskeleton as well as in the formation of the immunological synapses. The modular domain structure and the functionality of the individual domains suggest that they might act independently. Here we report an interesting intramolecular interaction within Nck that occurs between a non-canonical yet conserved (K/R)x(K/R)RxxS sequence in the linker between the 1st and 2nd SH3 domain (SH3.1/SH3.2) and the 2nd SH3 domain (SH3.2). Since this interaction masks the proline-rich sequence binding site of the SH3.2 domain, the intramolecular interaction is self-inhibitory. This intramolecular interaction could, at least partially, explain the remarkable specificity of Nck toward proteins with proline rich sequences. It may prevent non-specific low-affinity binding while keeping the site available for high affinity bivalent ligands that can bind multiple sites in Nck. This indicates that Nck does not simply adopt a “beads on a string” architecture but incorporates a higher order organization for improved specificity and functionality.
Non-catalytic tyrosine kinase (Nck) is a 47-kDa adaptor protein, consisting of three N-terminal SH3 domains and a single SH2 domain at the C-terminus (1–3). There are two human proteins that share the designation Nck : Nck1/Nckα and Nck2/Nckβ (also known as Grb4). The Nck1 and Nck2 isoforms exhibit a high degree of sequence homology, being 68% identical and 79% similar over the entire protein sequences. The modular architecture of the Nck protein provides a scaffold for a variety of protein-protein interactions, and more than 60 interaction partners have been reported so far. In most of the cases, the SH2 domain mediates binding to tyrosine phosphorylated receptors or cytosolic proteins, which leads to the formation of larger protein complexes via SH3 domains (1, 2).
Nck is functionally quite versatile. For instance, Nck is known to be involved in cellular activation, reorganization of the actin cytoskeleton, cell movement, as well as responses to cell stresses. In T cells, Nck plays a pivotal role in T-cell receptor (TCR) mediated reorganization of the actin cytoskeleton as well as in the formation of immunological synapses (1, 2). Upon activation of T cells, Nck is recruited to a membrane proximal site via interaction with tyrosine-phosphorylated SLP76, providing a scaffold for the Wiskott-Aldrich Syndrome Protein (WASP) dependent actin remodeling machinery (2–5). Nck is also directly recruited to the CD3ε component of TCR in an activation dependent manner (6). However, the roles of the CD3ε/Nck interaction remain disputed (1, 6–9).
Each SH3 domain exhibits remarkable specificities where identified interaction partners having clear preferences for paticular SH3 domains in Nck (1, 2, 10). These specificities allow Nck to have multiple simultaneous interactions with different binding partners. For example, WASP binding occurs through the 3rd SH3 domain (SH3.3) (5), whereas the WASP interacting protein (WIP) (11) and p21-activated kinase (Pak1) (12, 13) bind to the 2nd SH3 domain (SH3.2) (2–5). The 1st SH3 domain (SH3.1) interacts with CD3ε in a non-canonical fashion (6, 7, 9). Recent determinations of the structures of isolated Nck domains (14–19) as well as the structures of the Nck/CD3ε and Nck/PINCH1 complexes (9, 20) provide insights into the ligand binding specificities of individual domains. However, the functions and specificities of the multi-domain adaptor protein might not be fully elucidated by the sum of those of the individual domains. Recent interactome analysis showed that a significant number of proteins that showed interaction with individual isolated SH3/SH2 domains were not identified in the analysis using full-length Nck (10). On the other hand, it is also known that a Nck construct which has all SH3 domains showed stronger binding than isolated individual domains for some of the binding partners (21). Still, the basis of the specificity, in the context of the full-length protein, has not been addressed.
Here we report an interesting intramolecular interaction within the Nck2 molecule, identified by NMR structural analysis of full length Nck and its various deletion mutants. The analysis revealed that an intramolecular interaction occurs between a conserved non-canonical (K/R)x(K/R)RxxS sequence in the SH3.1/3.2 linker and the SH3.2 domain of Nck2. This interaction masks the proline-rich sequence (PRS) binding site of the SH3.2 domain and thus works as a self-inhibitory mechanism. The PRS of two physiologically identified binding partners, SOS1 (22, 23) and Pak1 (12, 13), exhibited much lower affinities (by ~5 folds) in the presence of the intramolecular interaction. Thus, the intramolecular interaction might work as a safety switch keeping the site available only for high affinity stable interactions that can overcome the intramolecular interaction. Our identification of this higher-order structure in Nck might help understanding the functional versatility of this multidomain adaptor protein.
Experimental Procedures
All chemicals were purchased from Sigma (St. Louis, MO) unless otherwise noted. All stable-isotope-labeled materials were acquired from Cambridge Isotope laboratories (Cambridge, MA). Nck2_71-90 (NH2-71LKDTLGLGKTRRKTSARDAS90-CONH2), SOS1 (NH2-1146EVPVPPPVPPRR1157-CONH2), and Pak1 (NH2-9QDKPPAPPMRN19-CONH2) peptides were purchased from Tufts University Core Facility (Boston, MA)
Expression and purification of Nck proteins
The human Nck2 gene was purchased from Open Biosystems (Huntsville, AL). The primers used to amplify the genes are listed in the Supplemental material. The PCR products were ligated to a modified pET28b vector, pET28P, or pET30b vector at NdeI and XhoI sites following the manufacturer’s instruction. The pET28P vector has His10-tag and the PreScission Protease (GE Healthcare Biosciences, Pittsburgh, PA) sequences between the NcoI and NdeI sites. The pET30 vector was used to make C-terminal poly-histidine tag constructs. The pET28P vector was used to make N-terminal poly-histidine tagged constructs, in which the poly-histidine tag can be cleaved by the PreScission Protease. The C-terminal deletion constructs are produced from the N-terminal poly-histidine tagged full-length Nck2 construct by introducing a stop codon (TGA). QuikChange Site-Directed Mutagenesis (Stratagene, La Jolla, CA) was used to make the C-terminal deletion constructs as well as point mutants of Nck2. Either BL21(DE3) or Rosetta2(DE3) were used as host strains for expression. All the clones were verified by DNA sequencing.
All proteins were expressed in E. coli by induction with IPTG at the final concentration of 1 mM. Overexpressed proteins in the soluble fraction were then purified using Ni-NTA affinity chromatography using standard procedures. All the proteins were then purified using Ni-NTA affinity chromatography using standard procedures. N-terminal poly-histidine tagged constructs were then cleaved by the PreScission Protease. All the proteins were subjected to a gel filtration chromatography using Superdex-75 column (GE Healthcare Biosciences, Pittsburgh, PA). For the uniformly 15N13C-labeled samples, the cells were cultured in 15N-13C M9 media containing 8.5 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 2mM MgCl2, 0.1 mM CaCl2, which is supplemented with 1 g/L of 15NH4Cl, 2 g/L of 13C glucose as nitrogen and carbon sources.
NMR experiments
All experiments were performed on a Bruker Avance 500 spectrometer equipped with a cryogenic probe at 25 C° unless otherwise indicated. All the Nck protein samples were prepared in 10 mM Na phosphate buffer (pH 6.8) with 100 mM NaCl and 1 mM DTT. Spectra were processed using XWINNMR (Bruker) and analyzed with Sparky(24). The resonance assignments for individual domains as well as deletion constructs of Nck (Δ169 and 70–168) were accomplished by HNcocaN and HNcocanH experiments (25), and standard triple resonance experiments (26).
Results
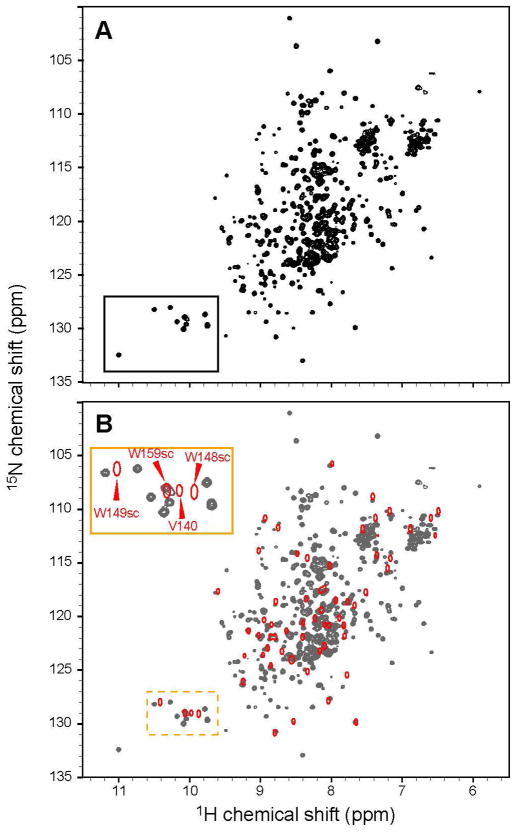
Although the modular domain structure of Nck protein suggests that its individual domains might be independent, there is some evidence of functional cross-talk between structural elements in the full-length Nck2 (Nck2-FL) molecule (10, 21). In exploring the possibility of domain-domain or domain-linker interactions in Nck, the 1H15N HSQC spectrum of Nck2-FL was compared with those of the isolated individual SH3 domains under the same experimental conditions (Figure 1 and Figure S1). As there are ~100 residues of linker regions in the Nck2-FL protein, the central part of the spectrum did not show many isolated peaks (Figure 1A). Nevertheless, it was possible to compare the peak positions of the well-isolated peaks and those of the tryptophan sidechains. As shown in the expanded spectrum in Figure S1, the peaks of the Trp indole sidechains in isolated SH3.1 and SH3.3 are not much different from those in the full-length protein. However, the tryptophan sidechain resonances from SH3.2 have quite different positions from those in Nck2-FL (Figure 1B). Since the Trp148 in SH3.2 is a critical residue for the binding to a PRS, the SH3.2 domain seems to have different environment when it is in the full-length protein as compared to the isolated state.
Figure 1.
Comparison of 1H15N HSQC spectra of Nck-FL and isolated SH3.2. (A) 1H15N HSQC spectrum of 100 μM [ul-2H15N] Nck-FL. (B) 1H15N HSQC spectrum of 100 μM [ul-15N] Nck SH3.2 (Red contour) was overlaid to that of [ul-2H15N] Nck-FL (Gray). The region containing the tryptophan indole signals is enlarged in the insert and the assignments are indicated (“sc” stands for Trp sidechain indole signals).
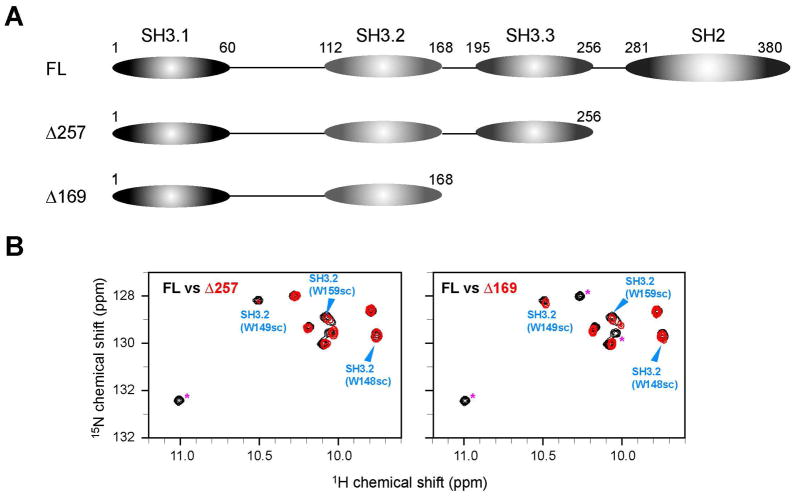
One immediate possible explanation would be the presence of the intramolecular interaction between SH3.2 and an internal PRS. Although the sequence of the full-length Nck2 protein was analyzed for such an element, no region was discovered that has the canonical PxxP motif in the protein. Therefore, we conducted a thorough search of the site for intramolecular SH3.2 interaction by means of a series of deletion mutants. As the first step to identify the intramolecular binding site, we examined the possibility that Nck2 SH3.2 interacts with the C-terminal half of the Nck2 protein. Two different C- terminal deletion constructs, Δ169 and Δ257, in which C-terminal side of the protein is deleted at the C- terminal boarder of the 2nd and 3rd SH3 domains, were made, (Figure 2A). The presence of intramolecular interaction was examined by signal distribution in the Trp indole regions. However, even the shortest construct, Δ169, which has only the SH3.1 and SH3.2 domains, showed almost the same chemical shifts as intact Nck for the Trp residues of SH3.2, indicating that no SH3.2 interaction exists in the C-terminal half of Nck (Figure 2B).
Figure 2.
C-terminal deletion constructs of Nck2 (A) Schematic representation of C-terminal deletion constructs of Nck2 (B) Comparison of 1H15N HSQC spectrum between 100 μM [ul-2H15N] Nck-FL and 100 μM [ul-2H15N] Nck Δ257 (left, red contour) or [ul-2H15N] Nck Δ169 (right, red contour). The region containing the tryptophan indole signals is shown. Arrowheads with assignments indicate the tryptophan indole signals from SH3.2 domains. Signals from SH2 domain are indicated with asterisk in Δ257 spectrum. In Δ169 spectrum SH2 and SH3.2 domain signals are indicated with asterisk.
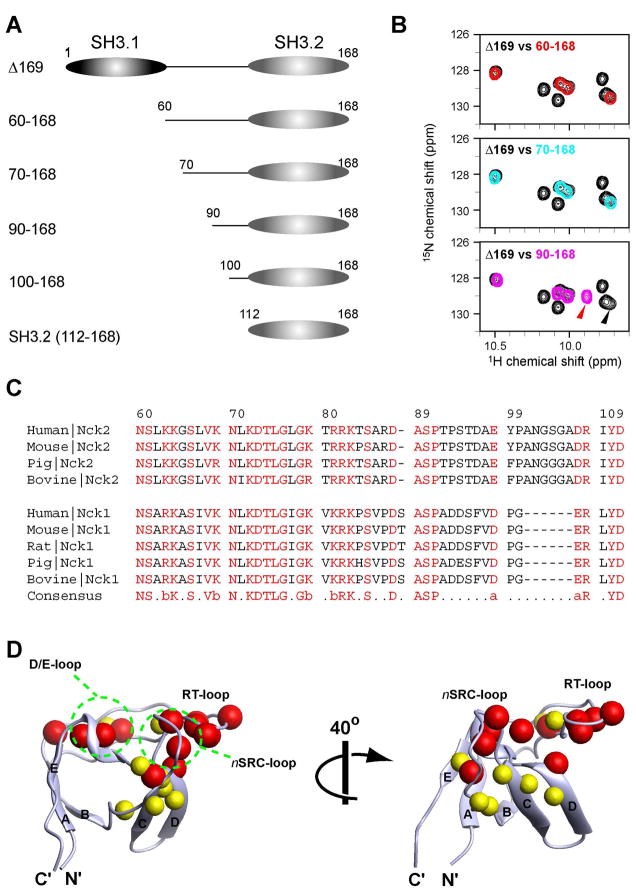
Subsequently, we investigated possible interaction sites in the SH3.1 domain and the SH3.1/SH3.2 linker by deleting the N-terminus of Nck2 Δ169 (Figure 3A). The deletion of the SH3.1 domain (Nck2_60–168) still yields the same spectra (Figure 3B, top), excluding the involvement of SH3.1 in the domain-domain interaction. It rather indicates that the SH3.2 binding site would have to be in the 41 a.a. SH3.1/SH3.2 linker (residues 60–110). Comparison of the sequence of human Nck2 with other Nck proteins showed that the N-terminal half of the SH3.1/SH3.2 linker is highly conserved (residues 60–91, 61% identity among Nck proteins), which implies the possible importance of this region in Nck proteins (Figure 3C). Although deletion of the first 10 a.a. (Nck2_70–168) did not affect the signals from the SH3.2 domain residues (Figure 3B, middle), deletion of the conserved region (Nck_90–168) caused significant changes in the SH3.2 signals (Figure 3B, bottom).
Figure 3.
N, C-terminal deletion constructs of Nck2 (A) Schematic representation of the N, C-terminal deletion constructs of Nck2 (B) Comparison of 1H15N HSQC spectrum between 100 μM [ul-15N] Nck2 Δ169 and 100 μM [ul-15N] Nck2_60–168 (top, red contour), [ul-15N] Nck2_70–168 (middle, cyan contour), or [ul-15N] Nck2_90–168 (middle, pink contour). The region containing the tryptophane indole signals is shown. The peak positions of W148 indole, which shifted significantly between Nck2 Δ169 and Nck2_90–168, are indicated with arrowheads that have same color with its spectrum. (C) Sequence alignment of human, mouse, rat, pig and bovine Nck proteins at the SH3.1/SH3.2 linker regions. (D) Mapping of the residues that are shifted by the deletion of the residues 70–89 (red) or 90–110 (yellow). Residues that are shifted >0.05 ppm in Nck2_70–168 with respect to 90–168 are indicated as red spheres. Yellow spheres are for the residues shifted >0.05 ppm in Nck2_90–168 with respect to isolated SH3.2. Residues shifted >0.05 ppm in both cases are indicated with red spheres. Chemical shift changes are normalized by the equation :((δN/5)2+(δH)2)1/2, where δN and δH are chemical shit changes in nitrogen and proton dimensions. Although Asn112 was also shifted >0.05 ppm in Nck2_90–168 with respect to isolated SH3.2, the residue is not shown in Figure 4D since it is not defined in the determined structure. The solution structure of the isolated SH3.2 domain, which contains residues 114–170 (2FRW), was used for mapping (16).
Figure 3D shows the location of the residues that were shifted with the deletion of the conserved 70–89 region (red spheres). Interestingly, the shifted residues are found at the binding site for PRS. A PRS usually binds to the surface formed by the RT and n-SRC loops, and it perfectly overlaps with the binding residues for the 70–89 region. It is also worth noting that this intramolecular interaction sequence has no Pro residues or other known SH3 binding motifs. Thus, the interaction might represent a new mode of interaction involving SH3 domains. Although there are some differences in chemical shifts between Nck2_90–168 and the isolated SH3.2 domain, those residues are mostly in proximity to the N-terminus of the domain (Figure 3D, yellow spheres), which might reflect the local environmental change caused by the loss of N-terminal proximal sequences. This assumption is further corroborated by the fact that Nck2_90–168 showed the same chemical shifts as Nck2_100–168 (data not shown).
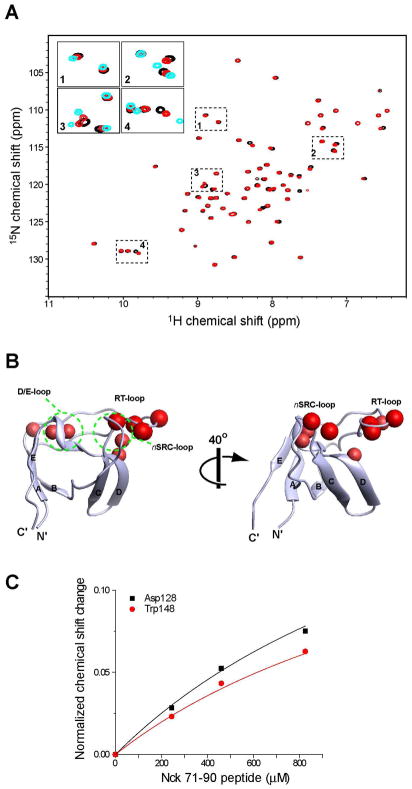
To confirm that the chemical shift changes observed at the PRS binding site actually reflect direct interaction between the 70–89 sequence and the SH3.2 domain, we then synthesized a peptide corresponding to the 71–90 residues of Nck2 and titrated it to the isolated SH3.2 domain. As shown in Figure 4A, titration of the 71–90 peptide to the SH3.2 domain induced chemical shift changes, most of the titrated signals having moved toward the chemical shifts of Nck2_70–168. This indicates that the external peptide can reproduce the intramolecular interaction. The residues perturbed by the 71–90 peptide overlaps well with those shifted by the deletion of 70–89 (Figure 4B). The result confirms that the intramolecular binding site for SH3.2 consists of the residues 71–90 in Nck2. The KD for this trans interaction is very weak (~1.7 mM), as estimated from the concentration dependence of the chemical shift changes (Fig. 4C).
Figure 4.
Titration of Nck_71–90 peptide to the isolated SH3.2 domain. (A) Comparison of the 1H15N HSQC spectra of 100 μM [ul-15N] Nck SH3.2 with (red) or without (black) 800 μM of the Nck2_70–90 peptide. Four regions in the HSQC spectrum are enlarged as inserts. In the inserted figure, 1H15N HSQC spectrum of 100 μM [ul-15N] Nck2_70–168 is also shown in cyan. (B) Mapping of the residues that are shifted by titration of Nck_71–90. Residues that are shifted >0.2 ppm with titration 800 μM of the 71–90 peptide are indicated as red spheres, Chemical shift changes are normalized by the equation :((δN/5)2+(δH)2)1/2, where δN and δH are chemical shit changes in nitrogen and proton dimensions. The structure and orientation of the molecules are same as in Figure 3. (C) Concentration dependent chemical shift changes induced by the Nck2_71–90 peptide. Chemical shift changes for Asp128 and Trp148 with fast exchanges are shown. The maximum chemical shift change are assumed to be the difference between Nck2_70–168 and isolated SH3.2.
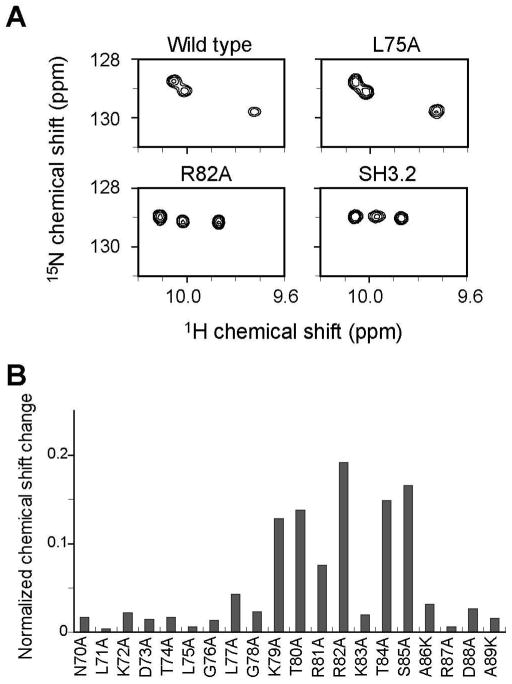
To further identify the hot spot of this interaction, we made mutants of Nck2_70–168 at every position in the 70–89 region. Some of these exhibited intact intramolecular interaction and gave very similar spectra to that of the wild type, as with L75A (Fig. 5A, top). However, as respresented by R82A, several residues showed disrupted interaction exhibiting similar peak patterns as isolated SH3.2 (Fig. 5A, bottom). The analysis identified a stretch of 7 residues containing a non-canonical KxRRKxS sequence in the 1st and 2nd SH3 domain (SH3.1/SH3.2) linker as the hot spot of this intramolecular interaction (Figure 5B). In addition, the D128A mutation in acidic RT-loop and the W148A mutation to the Trp residue that are known to be important for SH3-PRS interaction disrupted the intramolecular interaction (Supplement figure S3). This further supports the notion that the basic KxRRKxS sequence binds to acidic RT-loop with the overlapping with the PRS recognition site.
Figure 5.
Mutation analysis of Nck2_70–168. (A) Comparison of 1H15N HSQC spectra. Tryptophan indole regions of Nck2_70–168 wild type (upper, left), L75A (upper, right), R82A (lower, left), and isolateed SH3.2 domain (lower, right) are shown. (B) Normalized chemical shift changes of Trp148 indole signals in the Nck2_70–168 mutants. Chemical shift changes are normalized as in Figure 4.
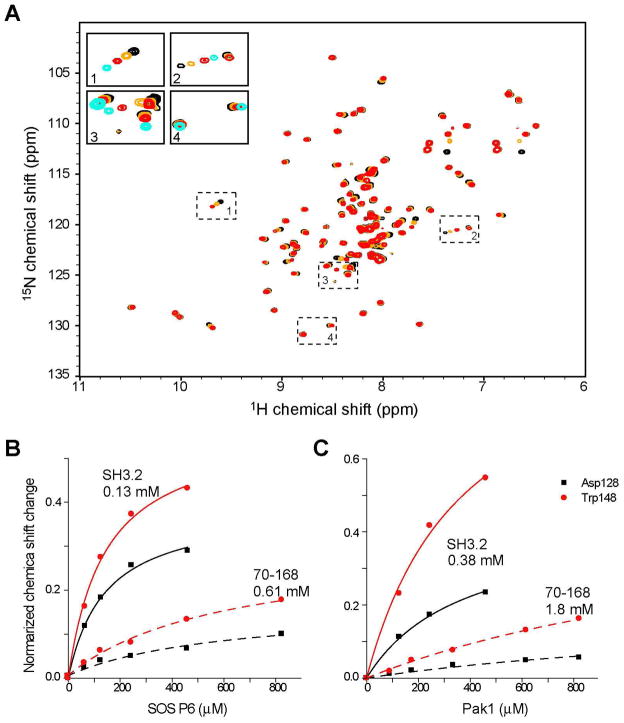
As mentioned above, the intramolecular interaction occurs at the PRS recognition site in the Nck2 SH3.2 domain. Thus, we next tested if the intramolecular interaction masks the PRS recognition site in the SH3.2 domain and, thereby, inhibits the interaction to its ligands. SOS1 (23) and Pak1 (12, 13) are both known to bind to the Nck SH3.2 domain. We synthesized two class II PRS peptides from the sequences of SOS1 (1146-EVPVPPPVPPRR-1157) and Pak1 (9-QDKPPAPPMRN-19) and compared their binding to the isolated SH3.2 domain and Nck2_70–168. The SH3.2 domain showed chemical shift perturbation patterns with fast-to-intermediate exchange kinetics upon binding to the SOS1 (Fig. 6A) and Pak1 peptides (data not shown), and sub mM affinities (Fig. 6B and 6C). As expected, the two PRS peptides bind to the typical canonical PRS recognition site in the SH3.2 (112–168) domain (data not shown). In comparison, the Kd values of these peptides to Nck2_70–168 were greatly increased by 4.7 fold in both cases (Kd values are shown in Fig. 6B and 6C). These results show that the intramolecular interaction is inhibitory to the ligand binding. Both peptides were still able to bind to Nck2_70–168 at much higher concentrations (Figure 6A and Supplemental Figure S3). In addition, the direction of the chemical shift changes with the SOS1 peptide was perfectly matched with that of the isolated SH3.2 domain with a saturating peptide concentrations (Figure 6A, insert panel). This indicates that the PRS peptides can replace the intramolecular interaction.
Figure 6.
Titration of the PRS peptides to isolated SH3.2 domain. (A) Titration of the SOS1 peptide to 130 μM [ul-15N] Nck 70–168. The 1H15N HSQC of free state (black), as well as with 130 μM (orange) and 830 μM of (red) SOS peptide. The four dotted regions in the HSQC spectrum are enlarged and shown in inserted panels. In the inserted figures, 1H15N HSQC spectrum of 130 μM [ul-15N] Nck SH3.2 with 830 μM of SOS peptide is also shown in cyan. (B and C) Concentration dependent chemical shift changes induced by (B) SOS and (C) Pak1 peptides. Chemical shifts are normalized as in Figure 4. Chemical shift changes for Asp128 and Trp148 with fast exchanges are shown. Kd values that were calculated from the concentration dependent chemical shift changes are also shown in the figure.
Discussion
Using NMR structural analysis of the full length Nck2 as well as a variety of its deletion mutants, we identified an unusual intramolecular interaction within the Nck2 molecule. It occurs between a non-canonical 79-KTRRKTS-85 sequence in the SH3.1/3.2 linker and SH3.2 of Nck2. The stretch of the sequence is mostly conserved except for Thr80 (Val in Nck1) and Thr84 (Pro or His in Nck1 as well as Pro in mouse Nck2). Considering the high homology between the SH3.2 domains, this intramolecular interaction appears to be a common characteristic shared by Nck family proteins. However, as a simple mutation of Thr84 in Nck2 to Pro (Nck1 type mutation) or Ala disrupted the interaction, we cannot exclude the possibility that the intramolecular interaction is a unique feature of the Nck2 molecule (Fig. 5).
The interaction occurs at the PRS binding site in SH3.2 and is self-inhibitory. Attempts to determine the structure of Nck2_70–168 were not successful, due to the fact that 1H-15N correlations were not observed in a HSQC spectrum for the residues 83–87. However, we were able to identify NOEs between the intramolecular interaction site and the Trp148 indole (see Supplemental Figure S5). Since the crosspeaks perfectly match the NOEs that are observed for the mainchain amide resonance of R82, which forms a part of intramolecular interaction motif, it is likely that Trp149 Hε1 is in close proximity to the amide proton of R82. We tried to find conditions under which those signals can be observed, by varying the temperature (10–35 C°) and/or lowering the salt concentration (the NaCl concentration was lowered to 0 mM from 100 mM), which turned out to be unsuccessful. In addition, residues 79–82 showed, on an average, only 30% signal intensities compared with the SH3.2 domain signals. This indicates the dynamic nature of the intramolecular interaction. This conclusion is also supported by the very weak (~2 mM) affinity of the 71–90 peptide to the isolated SH3.2 domain. It is also possible the intramolecular interaction site adopts multiple conformations. The dynamic intramolecular interaction allows the two physiological binding partners containing PRS, SOS and Pak1, to replace the inhibitory intramolecular interaction at higher concentrations. Intramolecular interactions between SH3 domains and PRS have been reported for several proteins and been proved to be functionally important (27–30). Still, to our knowledge, the current report is the first case of an SH3 intramolecular interaction with a non-canonical sequence devoid of prolines, which are generally required for SH3 domain interaction. It is worth noting that the sequence also lacks an obvious hydrophobic sequence patterns, such as are known to be important for PRS interaction as well as basic interactions that are supposed to mimic the PRS interaction. Nevertheless, it would be difficult to detect this kind of interaction by sequential analysis.
Besides the PRS sequences, the Nck1 SH3.2 domain is known to bind to the 45-residue juxtamembrane (JM) domain of EGFR, which is not a typical PxxP proline-rich motif (17). More specifically, NMR analysis identified that a basic region extending from Arg647 to Thr654 (647-RHIVRKRT-654) of EGFR binds to Nck1 SH3.2 with a KD value of 80 μM. Although the EGFR JM domain peptide is not identical to the intramolecular interaction site of Nck2, these peptides share a highly basic nature characterized by a cluster of three consecutive basic residues. This indicates, along with a very acidic electrostatic potential on the SH3 domain surface, the importance of electrostatic interaction in the intramolecular interaction.
Although not extensively analyzed in this paper, the SH2 domain also seems to be in a different environment in intact Nck as compared with the isolated condition (Supplemental Figure S6A). Since the deletion of the first two SH3 domains did not interfere with the SH2 domain signals and isolation of SH3.3 did not change the distribution of those signals, it can be assumed that the SH2 domain is most likely to interact with the SH3.3/SH2 linker (Supplemental Figure S6B). It is quite interesting to point out that some of the SH2 residues that show different chemical shifts between the excised domain and Nck-FL are situated in the AB loop, which is responsible for the recognition of phosphorylated Tyr (Supplemental Figure S6C). Thus, similar to SH3.2, the SH2 domain might also have intramolecular interaction that can modulate ligand binding. The presence of intramolecular modulation in the SH2 domain has also been pointed out in the screening analysis, aimed at identifying the binding partners of the individual domains using mass spectroscopy (10). However, by the time of the present study, the region responsible for the intramolecular interaction had yet to be identified.
In Tec family kinases, PRS is involved in an intramolecular interaction with an adjacent SH3 domain, which prevents the Itk SH3 domain and the proline-rich region from interacting with their respective protein ligands, Sam68 and Grb-2 (27). As indicated in the paper, with the presence of an intramolecular interaction, the ligand binding affinity can be specifically and temporally regulated by post-translational modification such as phosphorylation. The conserved Ser85 residue of Nck at the intramolecular interaction site is predicted to be a phosphorylation site for 90 kDa ribosomal protein S6 kinase (p90-RSK) and/or protein kinase C (PKC) (31). Since both of the kinases become up-regulated by activation of the TCR signaling pathway, Nck function can also be regulated by phosphorylation of Ser85 at the intramolecular interaction site. Since the electrostatic interaction between the basic intramolecular interaction site and the acidic surface of SH3.2 is considered to be the driving force of the interaction, phosphorylation at the interaction site is most likely to be energetically disfavoring the intramolecular masking, thus opening up SH3.2 to its ligands. Indeed, the Ser85Glu mutation disrupted the intramolecular interaction as judged from the 1H15N HSQC spectrum of the mutant (Fig. S3).
It is also possible that this intramolecular interaction with the linker region improves the specificity of the SH3.2 domain and the Nck molecule per se. A protein, which only has the SH3.2 interaction site, would not be favored by Nck molecule, since it requires additional energy to replace intramolecular masking in trans. On the other hand, a bivalent SH3.2 ligand, which has an additional binding site in other domains in Nck or outside of the PRS recognition site in the SH3.2 domain, could overcome the intramolecular interaction easier in cis fashion, using the additional interaction as an anchor. It is worth noting that some of the Nck ligands, including Pak1 and SOS1, are known to bind to Nck-FL much more tightly than the individual SH3 domain of Nck, which suggests that a cooperative interaction is necessary for tight complex formation (10, 12, 21–23). For example, Pak1 can bind both SH3.2 and SH3.3 domains. It is likely that Pak1 bind first with SH3.3 domain which is already available and replace SH3.2 masking for tighter binding. On the other hand, an unwanted nonspecific ligand with low-affinity would not be able to access the SH3.2 domain.
In conclusion, we provide evidence for the presence of a higher order structure in Nck. The intramolecular interaction occurs between the basic (K/R)x(K/R)RxxS sequence in the SH3.1/3.2 linker and the acidic SH3.2 domain of Nck. Although the function of the intramolecular interaction has to be addressed in vivo, its dynamic nature as well as the presence of a conserved Ser residue shows new features of Nck in improving its specificity and functional maneuverability.
Supplementary Material
Acknowledgments
Financial supports: This work was supported by the NIH (grants AI037581 and GM47467 and EB002026), and the Korea Research Foundation Grant funded by the Korean Government (to SP) (MOEHRD, Basic Research Promotion Fund) (KRF-2007-313-C00439)
Abbreviations
- Nck
Non-catalytic tyrosine kinase
- Nck-FL
Full-length Nck
- SH3.1
1st SH3 domain
- SH3.2
2nd SH3 domain
- SH3.3
3rd SH3 domain
- TCR
T-cell receptor
- WASP
Wiskott-Aldrich Syndrome Protein
- WIP
Wasp interacting protein
- Pak1
p21-activated kinase
- PRS
proline-rich sequence
- JM
juxtamembrane
Footnotes
Supporting information available: List of primer for used to amplify the Nck2 genes, Comparison of 1H15N HSQC spectra of Nck2-FL and isolated domains of Nck2 (SH3.1, SH3.3), additional mutation data, titration of the Pak1 peptide to Nck2_70–168 and evidence for intermolecular NOEs are shown in supplemental figures. These materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lettau M, Pieper J, Janssen O. Nck adapter proteins: functional versatility in T cells. Cell Communication and Signaling. 2009;7:1. doi: 10.1186/1478-811X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buday L, Wunderlich L, Tamas P. The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal. 2002;14:723–731. doi: 10.1016/s0898-6568(02)00027-x. [DOI] [PubMed] [Google Scholar]
- 3.McCarty JH. The Nck SH2/SH3 adaptor protein: a regulator of multiple intracellular signal transduction events. Bioessays. 1998;20:913–921. doi: 10.1002/(SICI)1521-1878(199811)20:11<913::AID-BIES6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Yablonski D, Kane LP, Qian D, Weiss A. A Nck-Pak1 signaling module is required for T-cell receptor-mediated activation of NFAT, but not of JNK. Embo J. 1998;17:5647–5657. doi: 10.1093/emboj/17.19.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivero-Lezcano OM, Marcilla A, Sameshima JH, Robbins KC. Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol Cell Biol. 1995;15:5725–5731. doi: 10.1128/mcb.15.10.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 7.Kesti T, Ruppelt A, Wang JH, Liss M, Wagner R, Tasken K, Saksela K. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3epsilon. J Immunol. 2007;179:878–885. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- 8.Szymczak AL, Workman CJ, Gil D, Dilioglou S, Vignali KM, Palmer E, Vignali DA. The CD3epsilon proline-rich sequence, and its interaction with Nck, is not required for T cell development and function. J Immunol. 2005;175:270–275. doi: 10.4049/jimmunol.175.1.270. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi K, Yang H, Ng E, Park SY, Sun ZY, Reinherz EL, Wagner G. Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity. J Mol Biol. 2008;380:704–716. doi: 10.1016/j.jmb.2008.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lettau M, Pieper J, Gerneth A, Lengl-Janssen B, Voss M, Linkermann A, Schmidt H, Gelhaus C, Leippe M, Kabelitz D, Janssen O. The adapter protein Nck: Role of individual SH3 and SH2 binding modules for protein interactions in T lymphocytes. Protein Sci. 2010 doi: 10.1002/pro.334. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anton IM, Lu W, Mayer BJ, Ramesh N, Geha RS. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adaptor protein Nck. J Biol Chem. 1998;273:20992–20995. doi: 10.1074/jbc.273.33.20992. [DOI] [PubMed] [Google Scholar]
- 12.Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 13.Galisteo ML, Chernoff J, Su YC, Skolnik EY, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 14.Frese S, Schubert WD, Findeis AC, Marquardt T, Roske YS, Stradal TE, Heinz DW. The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J Biol Chem. 2006;281:18236–18245. doi: 10.1074/jbc.M512917200. [DOI] [PubMed] [Google Scholar]
- 15.Ran X, Song J. Structural insight into the binding diversity between the Tyr-phosphorylated human ephrinBs and Nck2 SH2 domain. J Biol Chem. 2005;280:19205–19212. doi: 10.1074/jbc.M500330200. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Li M, Ran X, Fan JS, Song J. Structural insight into the binding diversity between the human Nck2 SH3 domains and proline-rich proteins. Biochemistry. 2006;45:7171–7184. doi: 10.1021/bi060091y. [DOI] [PubMed] [Google Scholar]
- 17.Hake MJ, Choowongkomon K, Kostenko O, Carlin CR, Sonnichsen FD. Specificity determinants of a novel Nck interaction with the juxtamembrane domain of the epidermal growth factor receptor. Biochemistry. 2008;47:3096–3108. doi: 10.1021/bi701549a. [DOI] [PubMed] [Google Scholar]
- 18.Santiveri CM, Borroto A, Simon L, Rico M, Alarcon B, Jimenez MA. Interaction between the N-terminal SH3 domain of Nck-alpha and CD3-epsilon-derived peptides: non-canonical and canonical recognition motifs. Biochim Biophys Acta. 2009;1794:110–117. doi: 10.1016/j.bbapap.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Park S, Takeuchi K, Wagner G. Solution structure of the first SRC homology 3 domain of human Nck2. J Biomol NMR. 2006;34:203–208. doi: 10.1007/s10858-006-0019-5. [DOI] [PubMed] [Google Scholar]
- 20.Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–523. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Wunderlich L, Goher A, Farago A, Downward J, Buday L. Requirement of multiple SH3 domains of Nck for ligand binding. Cell Signal. 1999;11:253–262. doi: 10.1016/s0898-6568(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 22.Wunderlich L, Farago A, Buday L. Characterization of Interactions of Nck with Sos and Dynamin. Cell Signal. 1999;11:25–29. doi: 10.1016/s0898-6568(98)00027-8. [DOI] [PubMed] [Google Scholar]
- 23.Hu Q, Milfay D, Williams LT. Binding of NCK to SOS and activation of ras-dependent gene expression. Mol Cell Biol. 1995;15:1169–1174. doi: 10.1128/mcb.15.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goddard TD, Kneller DG. University of California; San Francisco: 2006. [Google Scholar]
- 25.Sun ZY, Frueh DP, Selenko P, Hoch JC, Wagner G. Fast assignment of 15 N-HSQC peaks using high-resolution 3D HNcocaNH experiments with non-uniform sampling. J Biomol NMR. 2005;33:43–50. doi: 10.1007/s10858-005-1284-4. [DOI] [PubMed] [Google Scholar]
- 26.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 27.Andreotti AH, Bunnell SC, Feng S, Berg LJ, Schreiber SL. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 28.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 29.Groemping Y, Lapouge K, Smerdon SJ, Rittinger K. Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell. 2003;113:343–355. doi: 10.1016/s0092-8674(03)00314-3. [DOI] [PubMed] [Google Scholar]
- 30.Yuzawa S, Ogura K, Horiuchi M, Suzuki NN, Fujioka Y, Kataoka M, Sumimoto H, Inagaki F. Solution structure of the tandem Src homology 3 domains of p47phox in an autoinhibited form. J Biol Chem. 2004;279:29752–29760. doi: 10.1074/jbc.M401457200. [DOI] [PubMed] [Google Scholar]
- 31.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.