Abstract
The final size of plant organs, such as leaves, is tightly controlled by environmental and genetic factors that must spatially and temporally coordinate cell expansion and cell cycle activity. However, this regulation of organ growth is still poorly understood. The aim of this study is to gain more insight into the genetic control of leaf size in Arabidopsis (Arabidopsis thaliana) by performing a comparative analysis of transgenic lines that produce enlarged leaves under standardized environmental conditions. To this end, we selected five genes belonging to different functional classes that all positively affect leaf size when overexpressed: AVP1, GRF5, JAW, BRI1, and GA20OX1. We show that the increase in leaf area in these lines depended on leaf position and growth conditions and that all five lines affected leaf size differently; however, in all cases, an increase in cell number was, entirely or predominantly, responsible for the leaf size enlargement. By analyzing hormone levels, transcriptome, and metabolome, we provide deeper insight into the molecular basis of the growth phenotype for the individual lines. A comparative analysis between these data sets indicates that enhanced organ growth is governed by different, seemingly independent pathways. The analysis of transgenic lines simultaneously overexpressing two growth-enhancing genes further supports the concept that multiple pathways independently converge on organ size control in Arabidopsis.
In a fixed environment, the final size of plant organs, such as leaves and flowers, is constant, implying that organ growth is tightly controlled by genetic factors. The two effector systems, cell division and cell expansion, contribute to the final organ size. It is important to note that cell division normally cooccurs with cell expansion to maintain cell size homeostasis (Green, 1976). To avoid ambiguity, we refer to cell proliferation to indicate this combined activity. In Arabidopsis (Arabidopsis thaliana) leaves, proliferation, expansion, and the mature state follow each other in a largely time-dependent fashion (Beemster et al., 2005; Skirycz et al., 2010). After emergence from the shoot apical meristem, the leaf primordium grows mainly through cell proliferation. This phase of growth is progressively replaced, in a distal-proximal manner (Donnelly et al., 1999), by a period of cell expansion associated, in Arabidopsis, with an alternative mode of cell cycle activity, namely, endoreduplication (Beemster et al., 2005). Endoreduplication enhances endopolyploidy linked, in many cases, to increased cell size (Melaragno et al., 1993; Inzé and De Veylder, 2006). The regulation of organ size is poorly understood but must include a complex spatial and temporal coordination of cell expansion and cell cycle activity (Beemster et al., 2003). Any perturbation of one of these processes by modification of the expression of distinct genes, therefore, might affect the final size of an organ. Smaller leaves are produced when the number and/or size of cells are decreased (Horiguchi et al., 2006). For example, down-regulation of the expression of the GROWTH-REGULATING FACTOR5 (GRF5), encoding a putative transcription factor, leads to the production of small leaves containing fewer cells (Horiguchi et al., 2005). Similarly, a reduction in leaf size due to a decrease in cell number without change in cell size is observed in avp1 mutants expressing a lower amount of AVP1, a tonoplast-located, pyrophosphate-dependent H+ pump (Li et al., 2005). More complex cellular behavior occurs when a reduced cell number triggered by inhibiting cell cycle activity is accompanied by an increase in cell size that partly offsets the decrease in cell number (Hemerly et al., 1995; De Veylder et al., 2001). This phenomenon is called compensation (Tsukaya, 2002; Horiguchi et al., 2006) and is also observed in response to the perturbation of more upstream regulators of leaf growth, such as down-regulation of ANGUSTIFOLIA3, encoding a homolog of the human transcription coactivator SYT (Clark et al., 1994; Thaete et al., 1999), and of AINTEGUMENTA, encoding an AP2 transcription factor involved in regulation of the cell division activity (Mizukami and Fischer, 2000).
Conversely, several genes have been described that, when down-regulated or ectopically (over)expressed, increase leaf size (for review, see Gonzalez et al., 2009; Krizek, 2009). For instance, plants overexpressing GRF5 (Horiguchi et al., 2005) produce large organs by increasing the cell number. Similarly, down-regulation of two genes encoding plant-specific putative DNA-binding proteins, PEAPOD1 (PPD1) and PPD2, also enhances cell production, forming enlarged leaves (White, 2006). Degradation of TCP transcripts by overexpression of the microRNA R319a in the jaw-D (JAW) mutant also causes the formation of larger leaves containing more cells, an effect that is partially offset by a reduced cell size (Palatnik et al., 2003; Efroni et al., 2008). Plants with an enhanced expression of AVP1 also produce large leaves due to an increase in the cell number (Li et al., 2005). Besides activation of cell proliferation, enhanced cell expansion can also provoke the formation of larger organs than those of the wild-type, such as in plants overexpressing EXP10, encoding a cell wall-loosening protein, or ARGOS-LIKE, encoding a protein with unknown function (Cho and Cosgrove, 2000; Hu et al., 2006). A few cases are reported in which misexpression of a gene results in an increase of both cell division and cell expansion: ARF2, EBP1, and HRC1 (Okushima et al., 2005; Horváth et al., 2006; Century et al., 2008).
Some of the genes enhancing leaf growth are involved in hormone synthesis or signaling, confirming the important role of phytohormones in plant growth regulation. AVP1 has been suggested to control auxin transport (Li et al., 2005), the transcriptional repressor ARF2 to mediate responses to auxin (Ulmasov et al., 1999; Okushima et al., 2005), and JAW-D to regulate jasmonate biosynthesis (Schommer et al., 2008). Overexpression of the brassinosteroid receptor BRI1 leads to an increase of leaf petiole length (Wang et al., 2001), and GA 20-oxidase1 (GA20OX1), which catalyzes important steps in GA synthesis, causes an enlargement of leaves when ectopically expressed in Arabidopsis (Huang et al., 1998; Coles et al., 1999). Taken together, genes that increase organ size, which we previously designated intrinsic yield genes (IYGs), belong to diverse regulatory pathways, underlining the complexity of growth control (Gonzalez et al., 2009). Although the molecular function of the majority of the IYGs is known, the downstream molecular mechanisms that result in large leaves are not. In addition, the putative connection between these different growth-regulating genes and processes remains elusive.
Moreover, even the cellular basis of the enhanced growth phenotypes is not always known. For some mutants, such as BRI1 and GA20OX1 overexpressors, the cellular nature underlying the increased leaf size phenotype has not been analyzed at all. Although for other lines with enhanced growth characteristics detailed phenotypic information is available, it is impossible to compare results rigorously because different growth parameters are quantified (rosette biomass, leaf biomass, leaf area, etc.) and growth is often measured under different growth conditions.
The aim of this study is to gain more insight into the control of leaf size in Arabidopsis by performing a comparative analysis of transgenic lines that produce enlarged leaves. To this end, five genes belonging to different functional classes were selected that, in our hands, reproducibly induced increased leaf size when overexpressed: AVP1, GRF5, JAW, BRI1, and GA20OX1. The analysis of these lines, when grown under two experimental conditions, showed that the increase in leaf area depends on leaf position and environment and that all five lines affect leaf development in a distinct way. Nevertheless, in all cases, the enlarged leaf size correlated with an increase in cell number. The analysis of the hormone levels, transcriptome, and metabolome of individual lines was consistent with the above cellular analysis in showing that enhanced organ growth in these lines is governed by different, seemingly independent, molecular pathways. To further support the notion of independently converging pathways, an analysis is presented of transgenic lines simultaneously overexpressing several combinations of two growth-enhancing genes in Arabidopsis.
RESULTS
Phenotypic Characterization of Arabidopsis Plants with Enlarged Leaves
Final organ size is determined by environmental parameters as well as intrinsic regulatory mechanisms. Several IYGs, involved in seemingly unrelated pathways, have been shown to produce enlarged organs when overexpressed or mutated (Gonzalez et al., 2009). To unravel the different molecular mechanisms that govern organ size and to identify the potential interrelationship between these regulatory pathways, five transgenic lines overexpressing IYGs (AVP1, BRI1, GRF5, GA20OX1, and JAW) were selected for comparative phenotypic and molecular analyses. Under our experimental conditions, these lines consistently produced significantly enlarged leaves.
In a first step to characterize the phenotype in more detail, plants were grown in two different experimental systems, semihydroponic and in vitro. Leaf areas were measured at individual leaf positions of plants harvested 22 d after stratification (DAS), which is just before bolting occurs in both systems. The growth measurements were performed at this time point to decrease the potential influence of the flowering time on leaf number and/or size.
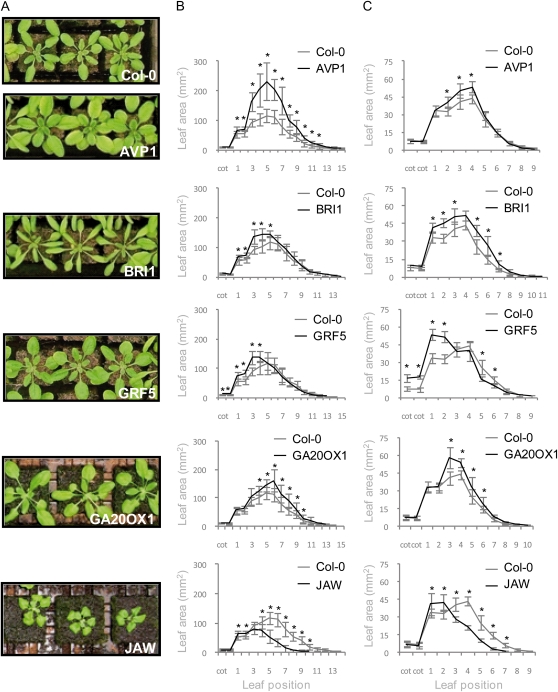
For the semihydroponic conditions, plants were grown in rock wool, an inert porous substrate that transports water and fertilizers to the roots by capillary action. On this substrate, all five IYG lines differed from the wild type and from each other, but all produced larger leaves than those of the control plants (Fig. 1, A and B). However, the effect strongly depended on the position of the leaf in the rosette (Fig. 1B). In the AVP1 line, most leaves were larger than those of the wild-type plants, so that the whole rosette area had significantly increased. Inversely, for JAW, we only observed a slight increase of the size of the first leaves produced, while the later leaves and the rosette as a whole were much reduced. The large leaf size reported previously (Palatnik et al., 2003) only becomes apparent when plants are grown for a prolonged period, well after the time that the Columbia-0 (Col-0) wild type has started flowering. The other lines showed intermediate phenotypes. BRI1 and GRF5 produced enlarged older leaves (leaves 1–5 and 1–4, respectively), while the overexpression of GA20OX1 increased the size of the younger leaves (leaves 4–10) but not the older ones. The extent of the area increase also varied from line to line. The largest increase was observed in AVP1, with a maximum up to 112% compared with the wild type (leaf 7). In JAW, the largest effect observed corresponded to only 14% of increase (leaf 2; Fig. 1B).
Figure 1.
Rosette leaf area of Col-0 and the five IYG lines. Plants were grown under semihydroponic (A and B) or in vitro (C) conditions. A, Plants photographed after 22 DAS. B and C, Area of rosette leaves at different positions. Values are averages ± sd (n = 8–12; * P < 0.05).
When the five IYG lines were grown under in vitro conditions (Fig. 1C), an increase in leaf area was observed as well, although the extent of the increase differed from that under rock wool conditions. As for plants grown on rock wool, in vitro, the increase in area did not occur in all leaves, and in most cases, the leaf age dependence resembled that on rock wool. AVP1-overexpressing plants, in contrast with the large increase in organ size observed on rock wool, only showed a slight enlargement of leaves 2 to 4 compared with the wild type (Fig. 1C). In BRI1, more leaves were larger than on rock wool: leaves 1 to 3 and 5 to 7. Overexpression of GA20OX1 led to a size increase of only the younger leaves. In JAW plants, only the first leaf pair was enlarged, whereas the other leaves remained small. In contrast, in GRF5, the increase in the area of the cotyledon and the first leaves was accompanied by a decrease in size of the other rosette leaves, whereas no such decrease was seen on rock wool.
Taken together, these measurements indicate that the five genes all control organ size in a differentmanner: they increase the size at different leaf positions, and the effect depends on the growth conditions.
Changes in Leaf Length and/or Width
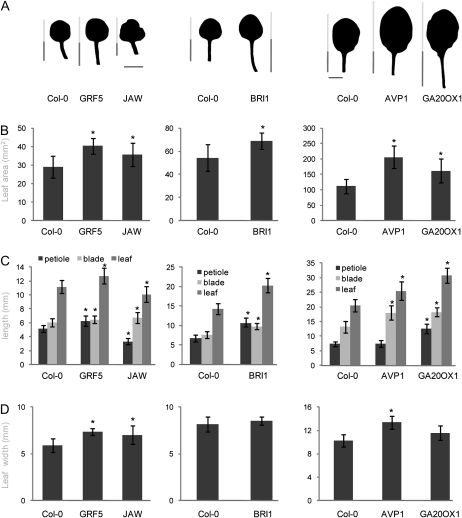
An increase of the total leaf area can be due to a change of the length and/or the width of the leaf. To investigate whether the IYGs affect the leaf size by modifying the same parameters, we measured length (blade and petiole) and width of the leaves at the positions where a large increase in the leaf area had been observed. Thus, whenever individual leaves were analyzed, these were leaves 1 and 2 from in vitro-grown plants for GRF5 and JAW, leaves 1 and 2 from plants grown under semihydroponic conditions for BRI1, and leaf 6 from plants grown under semihydroponic conditions for GA20OX1 and AVP1 (Fig. 2A).
Figure 2.
Leaf size parameters of Col-0 and the five IYG lines. Plants were grown in vitro (GRF5 and JAW) or semihydroponically (BRI1, AVP1, and GA20OX1) for 22 d. Leaves 1 and 2 from GRF5, JAW, and BRI1 and leaf 6 from AVP1 and GA20OX1 (A) were harvested to measure the leaf area (B), blade and petiole lengths (C), and leaf width (D). Values are averages ± se (n = 8–12; * P < 0.05). Bars in A = 5 mm.
In AVP1 and GRF5 plants, enlarged leaves were formed by an increase in size in two directions (i.e. length and width; Fig. 2, C and D), but in AVP1 the increase was similar in both directions (30% in length and 24% in width; Fig. 2, C and D), whereas in GRF5 the increase in width was larger (14% increase in length and 25% in width; Fig. 2, C and D). In AVP1, the increase in leaf length was due to a specific increase of the blade and not of the petiole. In contrast, in the other lines, the increased leaf size was related to an increased growth in only one direction. In the JAW line, the leaves were wider (20%), while in BRI1 and GA20OX1, they were longer (56% and 70% for the petiole and 30% and 38% for the blade, respectively; Fig. 2, C and D), while leaf width did not significantly differ from that of the wild type. Similar changes in length and width were observed for the adjacent leaves of the rosette with an increased area (Supplemental Fig. S1).
From these results, we conclude that the five IYGs have contrasting effects on leaf shape, again indicating that they affect different aspects of leaf growth.
Changes in Leaf Area Occur Late in Development
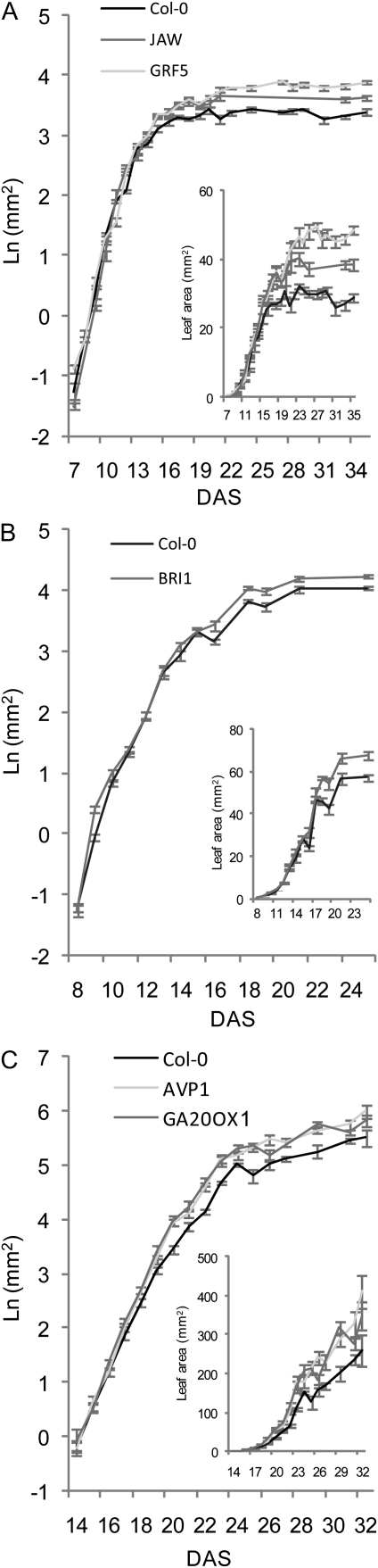
An increase in leaf area can be due to the production of an enlarged primordium during the initiation at the meristem and/or to a progressive increase in size resulting from a faster and/or a prolonged growth period. To identify the developmental stage at which changes in leaf size appear, we measured the evolution of the leaf blade area over time for each IYG and wild-type line (Fig. 3). To this end, leaves 1 and 2 were measured from 7 DAS onward for GRF5, JAW (Fig. 3A), and BRI1 (Fig. 3B), and leaf 6 was measured from 14 DAS onward for AVP1 and GA20OX1 (Fig. 3C).
Figure 3.
Evolution of leaf area of Col-0 and the five IYG lines throughout their development. Plants were grown in vitro (GRF5 and JAW; A) or in soil (BRI1 [B] and AVP1 and GA20OX1 [C]). Leaves 1 and 2 from GRF5, JAW, and BRI1 and leaf 6 from AVP1 and GA20OX1 were harvested on a daily basis to measure the leaf area. Insets, Same data on a linear scale to show absolute differences, particularly at the end of the leaf development. Values are averages ± se (n = 8–10).
During the first days of development, leaf sizes did not differ significantly, although in the case of AVP1 and GA20OX1, variability in size was found at very early time points (14 and 15 DAS). At 14 DAS, the area in the GRF5 and JAW lines became significantly different from that of wild-type plants (Fig. 3A), but in BRI1, this occurred at day 16. Interestingly, after 19 DAS, the leaf area in the wild type remained unchanged, whereas there was a further slow growth in JAW and a marked and prolonged growth in GRF5 until 25 DAS. The area of the first leaves in BRI1 and the wild type did not change after 21 d. In AVP1 and GA20OX1 lines, the increase of area became significant after 16 to 17 DAS.
Thus, in all five lines, the initial size and growth of the leaf was unaltered, but growth at later stages was faster, and at least in some cases (GRF5, BRI1, and JAW) the duration was also prolonged.
Changes in Leaf Area Are Mainly Due to an Increase in Cell Number
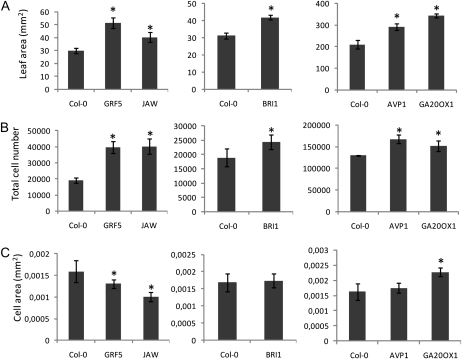
Final organ size is the result of the action of two processes, cell division and cell expansion. To analyze the extent to which cell proliferation and/or cell expansion contribute to enlarged leaves, the leaf epidermal cell number and size were analyzed in the five IYG lines. The measurements were carried out at 22 DAS for BRI1 and 30 DAS for GRF5, JAW, AVP1, and GA20OX1. The cell number in all transgenic lines was higher than that of the wild-type controls (Fig. 4B). The increase was particularly high in GRF5 and JAW (more than 2-fold). The final cell size, compared with that of control plants, increased in GA20OX1 (27%), remained unchanged in BRI1 and AVP1, and decreased in JAW (29%) and GRF5 (20%; Fig. 4C).
Figure 4.
Cellular basis of size differences between Col-0 and the five IYG lines. Plants were grown in vitro (GRF5 and JAW) or in soil (BRI1, AVP1, and GA20OX1). Leaves 1 and 2 from GRF5, JAW, and BRI1 and leaf 6 from AVP1 and GA20OX1 were harvested 22 d (BRI1) or 30 d (AVP1, GA20OX1, GRF5, and JAW) after sowing to measure the leaf area (A), epidermal cell number (B), and cell area (C). Values are averages ± sd (n = 5; * P < 0.05).
To investigate whether endoreduplication was related to leaf and cell size differences (Melaragno et al., 1993), the ploidy level of cells was analyzed in the mature leaves of the five IYG lines. Curiously, an increase in ploidy level was observed only in GRF5 that formed reduced cells (Supplemental Fig. S2).
In conclusion, although the analyzed genes affect leaf growth differently, a common mechanism, cell proliferation, drives the size change in all five lines. The increase in leaf area was entirely or almost entirely due to an increased cell number in GRF5, JAW, BRI1, and AVP1. Indeed, in the former two, the cell number actually increased at the expense of the cell size. The GA20OX1 line differed from the other lines in also having an increase in cell size.
Hormone Measurement
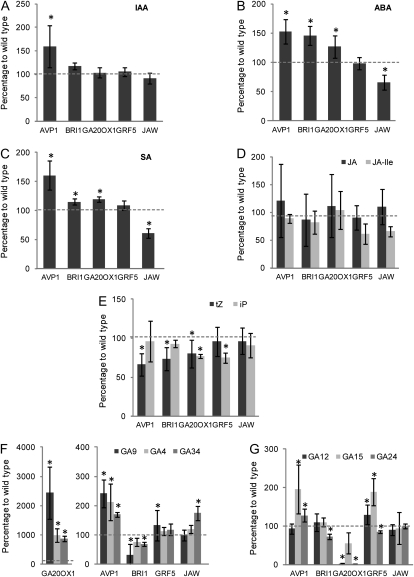
Plant hormones play a key role in growth regulation. For four of the five IYGs, there is evidence for a link with the action of a specific hormone: BRI1 to brassinosteroid perception (Wang et al., 2001), GA20OX1 to GA biosynthesis (Coles et al., 1999), AVP1 to auxin transport (Li et al., 2005), and JAW to jasmonate biosynthesis (Schommer et al., 2008). In a first step toward unraveling the mechanisms by which the five analyzed genes affect leaf size, we investigated the level of several hormones: indole acetic acid (IAA), abscisic acid (ABA), salicylic acid (SA), cytokinin (isopentenyladenine [iP] and trans-zeatin [tZ] forms), jasmonate (jasmonic acid [JA] and JA-Ile), and GAs, from young seedlings grown in vitro and harvested at stage 1.03, when the third rosette leaf is approximately 1 mm in length, approximately 14 d after sowing (Fig. 5).
Figure 5.
Hormonal content in Col-0 and the five IYG lines. Levels of IAA (A), ABA (B), SA (C), cytokinin (iP and tZ forms; D), jasmonate (JA and JA-Ile; E), and GA (F and G) were determined from the aerial parts of seedlings grown in vitro and harvested at stage 1.03. The graphs represent the percentage of hormone content compared with wild-type plants. Values are averages ± sd (n = 4; * P < 0.05).
Auxin levels were not significantly affected in the transgenic lines, with the exception of AVP1, in which an increase of 50% was observed (Fig. 5A). ABA levels increased in AVP1, BRI1, and, to some extent, GA20OX1, remained unaltered in GRF5, and decreased in JAW (Fig. 5B). The response of SA had some similarities to that of ABA, with an increase of 50% in AVP1 and to a lower extent in BRI1 and GA20OX1, a decrease in JAW, but no change in GRF5 (Fig. 5C). JA levels did not fluctuate in any lines, but the active form JA-Ile decreased by approximately 30% in JAW and GRF5, although not significantly (Fig. 5D), confirming previous measurements in the JAW mutants (Schommer et al., 2008). The tZ form of cytokinin decreased by approximately 30% in AVP1 and BRI1 and by 20% in GA20OX1 (Fig. 5E). The iP form decreased slightly (20%) in GA20OX1 and GRF5 (Fig. 5E). Several GAs were measured: the biologically active compound, GA4, and its inactive metabolite form, GA34, as well as other precursors and intermediates (GA9, GA12, GA15, and GA24). In GA20OX1, the level of the bioactive form GA4 increased dramatically, but also that of GA34 and GA9 (Fig. 5F; note the scale difference for GA20OX1). The concentrations of GA12, GA15, and GA24, which are precursors of GA located upstream of the action site of GA20OX1 in the GA synthesis pathway, decreased (Fig. 5G). An increase, albeit to a much lower level, in GA4, GA34, and GA9 was also observed in AVP1, no change was seen in JAW and GRF5, and a decrease in GA9 occurred mainly in BRI1. Levels of GA15 increased in AVP1 and GRF5, and those of GA34 increased in JAW.
In conclusion, hormone levels are affected in all IYG lines in a unique manner. Striking examples are the GA20OX1 and BRI1 lines, with massively increased and strongly decreased levels of GA9, respectively, and the AVP1 and JAW lines with increased IAA, ABA, and SA in the former and decreased levels of these three hormones in the latter.
Metabolite Measurements
Vegetative plant growth depends on the ability to produce, store, and use carbon and nitrogen sources. To analyze whether changes in metabolite content were common in the five lines, the mutant and wild-type plants were grown in vitro. The vegetative aboveground part of the seedlings was harvested at stage 1.03, the biomass was determined, and 65 metabolites were quantitatively characterized with a combination of robotized enzymatic and colorimetric assays, namely gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS; Supplemental Table S1). As many metabolites are diurnally regulated (Gibon et al., 2006), the plants were harvested and analyzed in the middle and at the end of the light period. In these two sets of samples, the total aboveground biomass increased by 5% and 7% in AVP1, 9% and 5% in GA20OX1, 17% and 10% in BRI1, 27% and 28% in JAW, and 29% and 24% in GRF5.
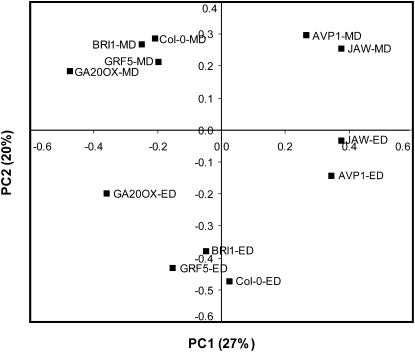
To obtain an overview of these large data sets, a principal component analysis (PCA) was carried out (Fig. 6). The first principal component (PC1) accounted for 27% of the variation and separated the genotypes, while the second principal component (PC2) accounted for 20% of the variation and separated samples according to the time of day. The separation of genotypes in PC1 was similar at both times but was not obviously related to leaf size (see above) or biomass. The BRI1 and GRF5 lines were close to the wild-type Col-0, GA20OX1 was clearly separated and isolated, and AVP1 and JAW were also separated from Col-0, but in the other direction to that of GA20OX1. Inspection of the weightings in PC1 (Supplemental Table S2) revealed that variation in the levels of raffinose, galactinol (an intermediate in the synthesis of raffinose), Asn (an amino acid that also typically accumulates in carbon starvation; Brouquisse et al., 1998), tocopherol, maltotriose, and gentiobiose (a metabolite possibly related to cellulose synthesis) largely contributed to the separation of the genotypes. Metabolites with a high weighting in PC2 included starch, the sugar signal metabolite trehalose-6-phosphate (T6P), and Phe. Thus, from the PCA, no simple or general relation can be concluded between the metabolite profile and plant or leaf sizes.
Figure 6.
PCA of metabolites in Col-0 and the five IYG lines. PC1 accounted for 27% of the variation and separated the genotypes, while PC2 accounted for 20% of the variation and separated samples according to the time of day. ED, End of the day; MD, middle of the day.
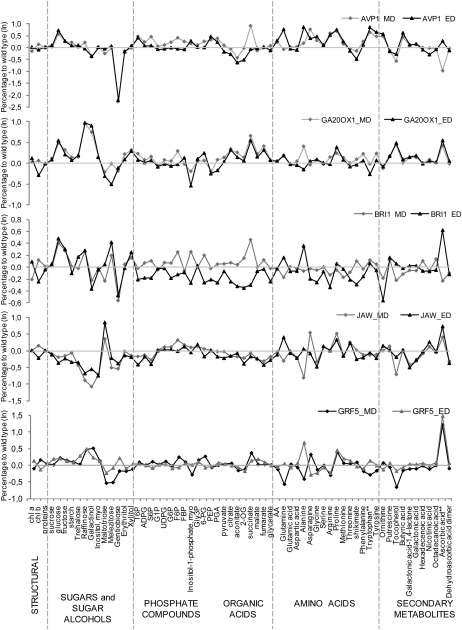
To allow a more detailed exploration of the data, we calculated the relative change and its significance for each metabolite compared with the wild-type control for each line and harvest time (Supplemental Table S3). The resulting ratios are plotted in Figure 7. All metabolite levels were normalized at a given time point on the average value in the wild-type control at that time and combined for both harvest times to provide a combined data set with 10 samples per genotype (Supplemental Table S3).
Figure 7.
Metabolite levels in the five IYG lines compared with Col-0. The graphs represent the percentage difference in metabolite content in AVP1, GA20OX1, BRI1, JAW, and GRF5 lines relative to wild-type levels. ED, End of the day; MD, middle of the day.
Many metabolites had a very diverse response between the five lines, but some were quite consistent. Ascorbic acid increased in all five IYG lines at the end of the day, and in three of the five at midday it was also often high. Glc, Fru, and succinate increased in four of the five IYG lines (in each case, JAW was the exception). Fru-6-P increased in all the IYG lines at midday but not at the end of the day. In the five lines, some metabolite levels, such as those of starch, protein, and Suc, resembled those in wild-type plants. These metabolites correlated highly significantly and negatively with the biomass in a population of more than 90 genetically diverse Arabidopsis accessions (Sulpice et al., 2009).
We compared the breadth of the response in the five lines by investigating for each line how many metabolites showed significant differences (P < 0.05) from the wild type. This comparison was made for the data sets for the middle and the end of the day and for the combined data set (Supplemental Table S3). The number of metabolites showing significant changes was the largest in AVP1 (25, 16, and 28) and JAW (22, 22, and 32) and smaller in GA20OX1 (16, eight, and 26), GRF5 (nine, 11, and 15), and BRI1 (five, 10, and 12). In AVP1 and GA20OX1, most of the significant changes in the middle of the day (19 of 25 and 16 of 16 metabolites, respectively) and at the end of the day (12 of 16 and six of eight metabolites, respectively) involved an increase of a metabolite, but in JAW and BRI1 in the middle of the day (18 of 22 and three of five metabolites, respectively) and the end of the day (26 of 32 and eight of 12 metabolites, respectively), these changes correspond to a decrease. In GRF5, many of the significant changes in the middle of the day correlated with a decrease of metabolites (six of nine), while most at the end of the day with an increase (10 of 11). Visual inspection of Figure 7 also revealed that the most marked and widespread changes occurred in AVP1, a rising trend to reduced metabolite levels in several of the lines in the evening data set and a general and marked trend to low levels of metabolites at both time points in JAW.
Many metabolite levels changed between the two harvest times in the six genotypes (Supplemental Tables S1 and S4). Most of these time-dependent changes were retained in the transgenic lines (Supplemental Tables S1 and S4), as was apparent from the PCA (Fig. 6). For example, starch increased, Glc and Fru decreased, T6P and Phe increased, and Orn, citrate, and aconitate decreased in the wild type between the middle and end of the light period as well as in all five transgenic lines. However, some metabolites had different time-dependent changes. For example, in BRI1 and, to a lesser extent, in AVP1, Gln, Ala, and succinate increased markedly in the middle of the day but were unaltered or decreased at the end of the day compared with the wild-type control. In these genotypes, several other organic acids did not change at the middle of the day but decreased at the end of the day (e.g. pyruvate and 2-oxoglurarate; Supplemental Table S3), while ascorbic acid decreased in the middle of the day but increased at the end of the day. This general shift to low levels of metabolites in BRI1 at the end of the day was also visible in Figure 7.
The spectrum of the changes in the compounds in the different IYG lines was remarkably diverse, but some patterns emerged. In AVP1, many of the 28 metabolites with significant differences were amino acids. Out of the 16 amino acids that were analyzed, 11 increased in AVP1 (Fig. 7; Supplemental Table S3). Other patterns in AVP1 included an increase in the levels of many phosphorylated intermediate compounds involved in glycolysis (almost all between Glc-6-P and phosphoenolpyruvate) in the middle of the day and a strong decrease in organic acids (pyruvate, citrate, aconitate, and 2-oxoglutarate) at the end of the day. Although not significant at the individual time points, the sugar signal metabolite T6P (Lunn et al., 2006; Smeekens et al., 2010) increased by 50% to 60%. Overall, the metabolite profile in AVP1 points to a shift toward nitrogen metabolism and changes in carbon signaling.
In GA20OX1, the level of many sugars and organic acids was elevated. There was also a greater than 2-fold increase in raffinose and galactinol and a significant increase in myoinositol at both time points, all three metabolites involved in raffinose metabolism. In GRF5, in addition to a small increase of raffinose and galactinol, there was a remarkably large increase in ascorbic acid (Fig. 7; Supplemental Table S3). In BRI1, many phosphorylated intermediates and organic acids decreased at the end of the day (Fig. 7; Supplemental Table S3). In JAW, maltotriose and ascorbate increased but many other metabolites decreased, including Glc, raffinose, galactinol, and myoinositol (Fig. 7; Supplemental Table S3). There was also a consistent decrease at both harvest times of Suc-6-P and ADP-Glc, the dedicated precursors for Suc and starch synthesis, and the sugar-signaling metabolite T6P. This general decrease of metabolites in JAW indicates that growth may be outstripping the supply of resources, which may be relevant for understanding the transient nature of the growth stimulation in this line.
In summary, the five lines show very diverse changes in metabolite composition, both with respect to breadth and spectrum of the changes. Nevertheless, a few metabolites behaved in a very similar manner in all or most of the lines when compared with the wild type, such as ascorbic acid, which increased in all five mutants at the end of the day and in three of them at midday, and Glc, Fru, and succinate, which increased in four of the five lines. Some classes of metabolites, such as those involved in raffinose metabolism (raffinose, galactinol, and myoinositol), changed in several of the transgenic lines, but with responses depending on the line.
Transcript Profiling
To obtain more insight into the molecular changes associated with the overexpression of the five IYGs, we extracted RNA from the vegetative part of seedlings at stage 1.03 for microarray transcript profiling. The number of genes found to be differentially expressed (fold change > 1.5, false discovery rate < 0.05) differed strongly between the lines. In AVP1 and JAW, the number of genes showing changes in expression compared with the wild type was relatively high: 455 and 509 and 237 and 259 up-regulated and down-regulated genes, respectively. In contrast, in GA20OX1 and BRI1, 29 and 26 genes were differentially expressed, respectively, compared with the wild type. The GRF5 line showed intermediate changes, with 72 up-regulated and 113 down-regulated genes.
Differential transcripts were investigated with PageMan to calculate the functional overrepresentation of MapMan categories (Usadel et al., 2006) and BiNGO, a cytoscape plugin, to analyze the overrepresentation of Gene Ontology (GO) categories (Maere et al., 2005) and were compared with publicly available data sets corresponding to hormonal treatment experiments (Nemhauser et al., 2006). The five data sets obtained were also compared with the Bio-Array Resource Venn SuperSelector tool (http://www.bar.utoronto.ca/) to find potential overlaps.
AVP1 Affects a Large Diversity of Transcripts
Among the 964 differentially expressed genes in AVP1, several functional classes were overrepresented, including hormone metabolism and signaling, stress response, cell wall modification, and kinase activity (Supplemental Table S5). In addition, by using BiNGO to analyze overrepresentation of cellular components (Maere et al., 2005), we found that 245 of the differentially expressed genes encoded proteins for which the cellular localization is the membrane (data not shown).
The genes from the hormone class were not related to one specific hormone, which is in agreement with the changed content of several of these regulatory molecules in the AVP1 line (Fig. 5), but the most prominent class of hormone-regulated genes are responsive to ABA (Supplemental Table S6). Indeed, many genes followed the same variation in the AVP1 data set as after ABA addition (Nemhauser et al., 2006), which is consistent with the marked increase of ABA in AVP1. For example, among the down-regulated genes in AVP1, we found several DUF-26 class genes whose expression also decreased after ABA treatment. DUF-26 proteins, found to be overrepresented with the PageMan tool, constitute a class of receptor kinases with a still unknown function. Biotic stress-responsive genes were also enriched in the AVP1 data set, most of which were down-regulated (31 out of 45 genes). Among these 31 genes, 10 were also down-regulated after ABA treatment. Several RECOGNITION OF PERONOSPORA PARASITICA and Toll/interleukin-1 receptor-nucleotide-binding site proteins, involved in disease signaling, were highly down-regulated.
Another functional category significantly enriched in the AVP1 data set corresponded to the cell wall class, with 37 genes differentially expressed. Among these genes, four arabinogalactan proteins, six pectate lyases, three expansins, and several other genes involved in cell wall modification were transcriptionally modified compared with the wild type. Although the functional classes of enzymes implicated in major or minor carbohydrate metabolism were not significantly overrepresented, several changes in individual gene expression correlated with the metabolite profiling. For example, the expression of APL4, encoding one of the large subunits of the ADP-Glc pyrophosphorylase catalyzing the first and limiting step in starch biosynthesis and being probably a regulatory subunit (Smith et al., 2004; Crevillén et al., 2005; Ventriglia et al., 2008), increased. In addition, the Glc-6-P translocator 2 gene, involved in the transport of Glc-6-P and suggested to be important for starch synthesis, also increased. The latter is known to be sugar induced, which would match the increase of Glc in AVP1. These changes could explain the slight increase in starch measured in AVP1.
This gene expression profiling shows that overexpression of AVP1 affects many pathways and that numerous differentially expressed genes have a membrane-associated GO annotation or are related to ABA response.
JAW Transcription Profiling: Jasmonate But Also ABA Response, and Inositol Metabolism
The 496 genes differentially expressed in JAW were first submitted to overrepresentation of functional class analysis with PageMan. The categories “hormone metabolism,” “abiotic stress response,” “RNA transcription regulation,” “minor carbohydrate metabolism,” and “transport” were enriched in this data set (Supplemental Table S5).
As several genes involved in hormone metabolism were overrepresented in the JAW microarray data, we compared the list of differentially expressed genes (Supplemental Table S7) with publicly available hormone-related data sets (Nemhauser et al., 2006). As described previously (Schommer et al., 2008), genes responding to methyl jasmonate (MeJA) treatment were overrepresented in the JAW data set. Numerous genes normally induced by MeJA treatment, such as LIPOXYGENASE2 and ALLENE OXIDE SYNTHASE involved in jasmonate synthesis, but also JASMONATE-ZIM-DOMAIN PROTEIN1 involved in jasmonate signaling, were down-regulated. These data were consistent with the small decrease of JA-Ile observed in the JAW transgenic plants (Fig. 5). In addition to genes responding to MeJA, an enrichment of genes responding to ABA treatment was found in the JAW data set. Many genes induced by ABA were down-regulated in JAW, and genes down-regulated after ABA treatment were induced. This effect can be explained by the large decrease in ABA content in JAW. In addition, as shown previously (Schommer et al., 2008), TCP-type transcription factors were down-regulated in JAW plants. Interestingly, several other classes of transcription factors were overrepresented in the JAW transcriptome, such as members of the AUX/IAA family or factors belonging to the Basic Helix-Loop-Helix family of transcription factors (Supplemental Tables S5 and S7).
The minor carbohydrate metabolism GO class was also enriched in the JAW data set with several genes involved in raffinose and galactinol metabolism. For example, the decreased expression of myoinositol-1-phosphate synthase (MIPS1), involved in the first step of myoinositol synthesis, could be linked to the strong reduction of myoinositol content in JAW. In antisense StIPS-1 transformants of potato (Solanum tuberosum), low levels of myoinositol, galactinol, and raffinose were found (Keller et al., 1998). Recently, a combination of comparative expression profiling, resequencing, and trait association mapping allowed the identification of MIPS1 as a candidate gene that may contribute to the determination of biomass in Arabidopsis (Sulpice et al., 2009). Consistently, we found that GALACTINOL SYNTHASE2 (GOLS2) and GOLS3, coding for key enzymes in the synthesis of raffinose oligosaccharides, were down-regulated in JAW. In addition, the increased expression of the trehalase gene could explain the reduction in trehalose content. Inhibition of trehalase activity leads to trehalose accumulation (Goddijn et al., 1997).
Taken together, the JAW expression profiling data show, in addition to the down-regulation of the expression of TCP genes, a change in expression of genes related to MeJA and ABA and in myoinositol and raffinose metabolism.
Repression of BR Signaling in BRI1
Overexpression of BRI1 under its own promoter increased 6.7-fold the mRNA levels, but remarkably, only a few genes were differentially regulated (16 up-regulated and nine down-regulated; Supplemental Table S8), especially when considering that the external application of brassinolide to plants affects the expression of large sets of genes involved in cell wall and long-chain fatty acid biosynthesis, cytoskeleton organization, light response, transcription regulation, and hormone response (Vert et al., 2005). We postulate that continuous activation of the brassinosteroid pathway causes the establishment of a new steady state in which only a few genes are sufficiently differentially expressed to identify them by microarrays.
In agreement with this view, the BRI1 transcriptome revealed that overexpression of the BRI1 gene affected brassinosteroid signaling pathways. The BR-degrading BAS1 was up-regulated and the BR biosynthesis genes CPD, DWF4, and BR6ox2 were down-regulated, suggesting a negative feedback loop on BR biosynthesis. In addition, GA3ox1, encoding an enzyme involved in the biosynthesis of GA, was up-regulated. This increased expression could be a consequence of the decreased GA4 content in BRI1 (Fig. 5), because transcripts of GA3ox1 accumulated to a higher level in plants treated with uniconazole P, an inhibitor of GA biosynthesis (Matsushita et al., 2007).
Two genes encoding cell wall-modifying enzymes were up-regulated: the xyloglucan endotransglucosylase/hydroxylase XTH19, belonging to the same family as BRU1, the expression of which is induced by BL treatment in soybean (Glycine max; Zurek and Clouse, 1994), and the cellulose synthase AtCSLG3. Most interestingly, BRI1 overexpression down-regulated PPD2, a transcriptional repressor previously implicated in growth control. Deletion of both PPD1 and PPD2 in the Landsberg erecta ecotype produced enlarged leaves (White, 2006).
An artificial microRNA construct targeting the two PPD genes was introduced into the wild-type Col-0 plants. When the transgenic plants with reduced PPD1 and PPD2 expression were grown next to BRI1 plants, the similarity in phenotype between these two lines was striking: the petiole elongation described for BRI1 (Wang et al., 2001) and the epinastic dome-shaped blade described in ppd mutants (White, 2006) were observed, suggesting that down-regulation of PPD is an integral part of the BRI1-induced phenotypes (Supplemental Fig. S3).
No Specific Pathway Modified in GA20OX1 or GRF5
Overexpression of GA20OX1 led to a large increase in GA content (Fig. 5), but only 14 genes were up-regulated and 14 were down-regulated (Supplemental Table S9). As in the BRI1 line, this is in contrast to the large set of genes that were differentially regulated by external GA treatment. The down-regulation of GA3ox1 found in our data set confirmed the negative feedback loop exerted by GA on its biosynthesis. The other genes differentially expressed in GA20OX1 did not belong to a particular functional class or did not respond to specific hormone treatments, not even GA.
With PageMan to identify the overrepresentation of functional categories in the GRF5 data set, no particular class of proteins was enriched significantly. The comparison of this data set with hormone-related microarrays showed that several genes up-regulated after MeJA treatment were down-regulated in GRF5 (Supplemental Table S10), probably linked to the decrease in MeJA. No other specific genes responding to a particular hormone were found.
Comparative Analysis of Transcript Profiling
In all five IYG lines studied, an increase in cell number contributed to the enlargement of the leaf area. With the exception of GA20OX1, this increased cell number was the major or only reason for the enhanced leaf size. Probably, common regulators might be involved in the regulation of cell division. To investigate the possible relationship between the five IYG lines, we analyzed the overlap between the different data sets with the Bio-Array Resource Venn SuperSelector (http://www.bar.utoronto.ca/; Supplemental Table S11).
Surprisingly, not a single gene was significantly affected in the five data sets. An overlap between differentially expressed genes was found between a maximum of three data sets. For example, in the up-regulated genes, AT5G22460, encoding a thioesterase family protein with unknown precise function, was found in the AVP1, GA20OX1, and JAW data sets. Four genes (AT1G23130, Bet v I allergen family protein; AT2G29490, glutathione transferase 19; AT3G05890, rare-cold-inducible 2B; and AT5G24660, similar to an unknown protein) were down-regulated in AVP1 as well as in JAW and GRF5. For down-regulated genes, the highest overlap was found between the GRF5 and JAW data sets, with 12% of common down-regulated genes but without specific functional classes in the overlap.
AVP1 and JAW had a large number of differentially expressed genes in common: 15% corresponding to 113 genes, with 18 and 35 genes with increased and decreased expression, respectively, in both data sets. Surprisingly, 35 and 25 genes down-regulated and up-regulated in JAW, respectively, were up-regulated and down-regulated in AVP1. Among these genes, 14 were differentially expressed after ABA treatment, a hormone with opposite changes in AVP1 and JAW.
In summary, comparison of the transcriptome data sets from the five IYGs was consistent with the view that they all impinge on distinct pathways when overexpressed.
Combining Different IYG Lines
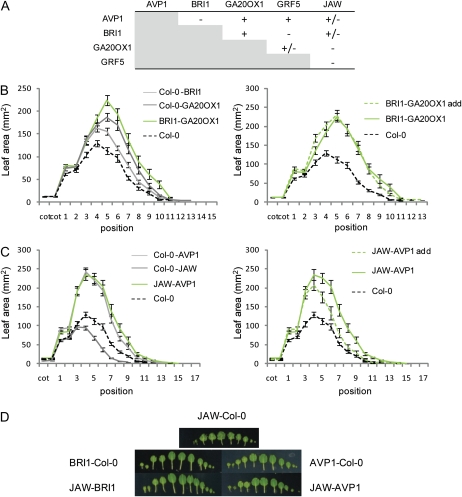
The phenotypic and molecular analyses suggest that the five IYGs work in different pathways. Further support for this conclusion comes from the analysis of plants expressing combinations of two IYGs. To obtain these double transgenic plants, homozygous lines expressing the respective IYG were crossed with each other and the F1 progenies were analyzed. As controls, crosses were made with a Col-0 wild type. Leaves were harvested from plants grown on soil, and their areas were measured. An overview of the results of all crosses is shown in Figure 8A. In most cases, the lines heterozygous for the transgene had a phenotype intermediate between the wild type and the homozygous line or were similar to the homozygous parent. The combination of the different IYG lines led to various effects. For example, for BRI1×GA20OX1 or AVP1×JAW, the phenotype of the progeny can be explained by distinct interactions between the transgenes. Indeed, when BRI1 and GA20OX1 were overexpressed in the same plant, the final leaf size corresponded to the sum of the effects of the heterozygous parents (Fig. 8B), suggesting that these genes autonomously affect organ size. Other combinations (AVP1×GA20OX1 and AVP1×GRF5) also resulted in plants with this additive phenotype, corresponding to the sum of the effects of the heterozygous parents (Supplemental Fig. S4). Conversely, in the combination AVP1×JAW, the leaf size was clearly similar to that of plants overexpressing AVP1 heterozygously only (Fig. 8C), suggesting that in an AVP1 background, the effect of overexpression of JAW on leaf size is repressed. This phenomenon was also observed when BRI1 and JAW were combined (Fig. 8D). Interestingly, besides this inhibition of the effect of JAW up-regulation on leaf size, the strong leaf serration in the homozygous JAW plants was also lost in the AVP1 or BRI1 background (Fig. 8D) but not in the GRF5 or GA20OX1 background.
Figure 8.
Effect of combined pairs of two IYGs on leaf area. Leaf series were harvested from 22-d-old plants grown in soil, and the area of individual leaves was measured. A, Interpretation based on individual rosette leaf area: + corresponds to an additive effect where the crossed plants show a phenotype corresponding to the sum of the effect of the heterozygous parents; − corresponds to an effect where the crossed plants have a phenotype smaller than the sum of the effect of the heterozygous parents; +/− corresponds to an intermediate effect. B, Left, Leaf area of Col-0, Col-0-BRI1, Col-0-GA20OX1, and BRI1-GA20OX1. Right, Individual leaf area of Col-0 and BRI1-GA20OX1 compared with the leaf area expected in case of an additive effect (BRI1-GA20OX1 add). C, Left, Individual leaf area of Col-0, Col-0-AVP1, Col-0-JAW, and AVP1-JAW. Right, Individual leaf area of Col-0 and AVP1-JAW compared with the leaf area expected in case of an additive effect (AVP1-JAW add). D, Photographs of representative leaf series made with JAW-Col-0, BRI1-Col-0, AVP1-Col-0, BRI1-JAW, and AVP1-JAW. For a given cross, the leaf area expected in case of an additive effect was calculated by adding the areas of the two heterozygous parents and subtracting the area from the control Col-0. Values are averages ± se (n = 10–14).
These results provide genetic evidence that AVP1 and GA20OX1 or GA20OX1 and GRF5 control leaf size in an independent manner, whereas AVP1 and JAW or BRI1 and JAW work in pathways that are somehow interconnected. Interestingly, the three lines that show an increase in leaf area due to an increase in cell number accumulated hormones differently: AVP1 and BRI1 contained more ABA and SA and JAW less of both hormones.
DISCUSSION
To gain insight into the molecular mechanisms that control leaf size, we performed a comparative analysis of five Arabidopsis transgenic lines that had previously been described to display increased leaf growth when compared with the wild type (Huang et al., 1998; Wang et al., 2001; Palatnik et al., 2003; Horiguchi et al., 2005; Li et al., 2005). As the published information on these lines is not comparable because of different growth conditions, we quantitatively measured leaf growth parameters, cell area, cell number, and physiological and molecular traits for the five lines grown under identical conditions and excluding “seed” effects. Thus, our data allow a comprehensive comparative analysis of five genes that positively affect the leaf size.
Leaf Size Increase, Leaf Position, and Environmental Effect
By analyzing the individual areas of all rosette leaves, we confirmed previous findings showing that overexpression of AVP1, BRI1, GRF5, GA20OX1, and JAW produce enlarged leaves, but, as detailed here, in surprisingly different ways, depending on leaf age and growth conditions. The different lines can be classified into three classes, based on which leaves are affected. One class includes the GRF5, BRI1, and JAW lines that mainly enhance the size of the first leaves produced. In GRF5, as described previously (Horiguchi et al., 2005), enlarged first leaves are formed, and in BRI1, the petiole length increases also (Wang et al., 2001) and the leaves are elongated. A similar increase in petiole and blade length has also been described for plants overexpressing DWF4, encoding an enzyme of the BR biosynthesis pathway (Choe et al., 2001). In JAW, only the first leaves are larger and the others are smaller, possibly because of the slow maturation occurring in these plants (Efroni et al., 2008). Another class includes GA20OX1, previously described to promote fast growth (Huang et al., 1998; Coles et al., 1999), that has little effect on the size of the first leaves but a major one on that of the subsequent leaves. Finally, the AVP1 line represents a third class in which the size of almost all leaves is enhanced. The measurements of the area of all rosette leaves were done before the plants started bolting to decrease the potential influence of flowering time on leaf growth. At this time point, an increase in leaf size is already observed in the five lines when compared with wild-type plants. In addition, the analysis of the leaf area evolution over time revealed that, for some lines, such as GRF5, the analyzed leaves keep on growing after 22 d, the time at which the leaf series were made. This observation suggests that some of the rosette leaves might show a further increase in size when plants are mature.
The analysis of these lines under two growth conditions also revealed that these genes are influenced by environmental factors. Indeed, when AVP1 is grown on rock wool and JAW is grown in vitro, the enhanced leaf size is much more pronounced than under the other growth condition.
Proliferation Is a Key Driver to Increase Plant Organ Size
The final size of a leaf requires the contribution of two essential processes, cell division and cell expansion. Analysis of cell number and cell area of mature leaves revealed that an increase in cell number is the sole cause of the increase in size for four of the five IYG lines, except for GA20OX1, in which cell division and cell expansion play a role. These measurements confirm the previously described effect of AVP1, JAW, and GRF5 on cell number (Horiguchi et al., 2005; Li et al., 2005; Efroni et al., 2008). Interestingly, for GRF5 and JAW, the increase in cell number is partially counteracted by a reduced cell size. In AVP1, such a phenomenon is not observed.
The lines BRI1 and GA20OX1 stimulate BR signaling and increase levels of bioactive GA, respectively. Both brassinosteroids and GAs are required for developmental processes related to cell expansion (Hooley, 1994; Szekeres et al., 1996; Clouse and Sasse, 1998; Pérez-Pérez et al., 2002). In our experiments, GA20OX1 overexpression, in contrast to the other lines, affects cell expansion, but, unexpectedly, both the BRI1 and GA20OX1 lines considerably enhance the cell number. The effect of BRI1 on the cell number is in agreement with studies showing that applied BR increases cell number in det2 mutants (Nakaya et al., 2002). Recently, GAs also have been implicated in the control of cell proliferation. Indeed, in the quadruple DELLA mutant, in which these growth repressors in GA signaling are down-regulated, cell proliferation and cell expansion rates increase (Achard et al., 2009).
Analysis of leaf area evolution in all five IYG lines revealed that the increment in leaf size appears late during leaf development, implying that the initial growth associated with cell proliferation is similar in the IYG lines and in the wild type and that growth at later stages is faster in the transgenic plants. Together with the changes in the final cell number, this finding suggests that the five IYG lines are involved in the control of the proliferation rate or of the length of the proliferation period during leaf development. For GRF5, BRI1, and JAW, the question remains whether a prolonged cell proliferation period occurs at the transition zone between cell division and cell expansion or as an effect on the activity of the dispersed meristemoid cells. In JAW, the retraction of the cell proliferation gradient (Donnelly et al., 1999) is delayed. In addition, the shape of this cell cycle front arrest differs also, suggesting an extended division period in the marginal regions (Nath et al., 2003; Efroni et al., 2008). The down-regulation of PPD2 in BRI1 and the similarity in the leaf phenotype between BRI1 and a double ppd1 ppd2 mutant indicate that the increased blade area in BRI1 could result from prolonged cell division of dispersed meristemoid cells, as described for ppd1 ppd2 Landsberg erecta mutants (White, 2006). Taken together, our data suggest that the transition between cell proliferation and cell expansion is tightly genetically controlled and plays a crucial role in the achievement of the final leaf size.
Molecular Profiling: Multifactorial Control of Organ Size
The hormone analysis has demonstrated that different hormones are affected in the five IYG lines. In AVP1, SA as well as auxin levels are increased, supporting the suggested involvement of AVP1 in auxin transport (Li et al., 2005). Somewhat unexpectedly, in AVP1, BRI1, and GA20OX1, the ABA levels are enhanced. ABA is most often associated with stress responses, but little evidence hints at a possible positive role in shoot growth (Sharp et al., 2000; Cheng et al., 2002). The amounts of the bioactive GA4, GA9, and GA34 drastically increased in GA20OX1, whereas GA9 and GA34 and, to a lesser extent, GA4 decreased in BRI1. Whether the observed changes in hormone levels in the different lines play a role in organ size control remains unclear. The elevated levels of GAs in GA20OX1 had previously been shown to be responsible for stem elongation and early flowering (Huang et al., 1998). We found that BRI1 and GA20OX1 show an opposite accumulation of GA9 and GA34 and, to a lesser extent, of GA4, but they present similarly enhanced leaf growth characteristics (increase in petiole and blade lengths but no change in blade width). In agreement, the cellular basis of the observed increased leaf size differs in the two lines: in GA20OX1, an increase in cell number and size was measured, but in BRI1, only the cell number had increased. In addition, the leaf shape of the lines is different, with BRI1 having a dome-shaped leaf morphology, in contrast to that of GA20OX1. The overproduction of bioactive GA in GA20OX1 might also be responsible for the increased leaf size. In GRF5, a few changes in hormone content are observed, suggesting that the effect on growth is not achieved through hormonal control or, at least, not through one of the analyzed hormones.
Metabolite profiling revealed a rather diverse response. Several metabolites increased in four (Glc, Fru, and succinate) or all (ascorbic acid) IYG lines. Increased Glc and Fru might be a sensitive indicator of the carbon supply, and ascorbate, one of the metabolites of the Halliwell-Asada cycle responsible for redox control in plants, is a key antioxidant and is implicated in the cell cycle control in plants (Arrigoni, 1994). However, other features of the metabolic response in the IYG lines are very diverse. In some of the IYG lines, the changes in the metabolite profile are broad (especially in AVP1) or more limited (in BRI1 and GRF5). The widespread changes in AVP1 differ from those seen after overexpression of a soluble pyrophosphatase in the cytosol (Jelitto et al., 1992), indicating that the metabolite and growth phenotypes are due to the function of AVP1 as a protein pump rather than to depletion of pyrophosphate. In this line, amino acids and the signal metabolite T6P increase and organic acids decrease, pointing to a change in carbon and nitrogen metabolisms. The JAW line and, to a lesser extent, BRI1 lines show a strong trend toward decreased levels of metabolites, indicating that they might promote growth in excess of the rate of resource acquisition.
A significant finding is that all five IYGs affect the transcriptome in very different ways, with remarkably few processes and genes shared between two or more lines. Nevertheless, in all five lines, cell proliferation increases during leaf development (see below). Some transgenes, such as AVP1, JAW, and GRF5, affect the expression of hundreds of genes, but only a few genes are affected in plants overexpressing the two genes involved in hormone signaling (BRI1) or biosynthesis (GA20OX1), some of which are involved in feedback inhibition. The latter is even more remarkable because BR and GA treatments of plants affect hundreds of genes (Nemhauser et al., 2006), suggesting that when the hormone levels are kept constitutively high (such as 7-fold more bioactive GA in GA20OX1 plants), a new steady state is established in which only a few transcripts are changed. This finding suggests that transient hormone treatments push the systems to a new stable state, which triggers feedback mechanisms that in time lessen this effect.
When hormone, metabolite, and transcript profiling data are combined, AVP1 and JAW change massively the transcripts and metabolites and extensively the hormones, including increased IAA, ABA, SA, and GA4 in AVP1 and decreased IAA, ABA, and SA in JAW. The other lines showed moderate to small changes of transcripts and metabolites (GRF5) or hardly any changes in transcripts and only small changes in metabolites (GA20OX1 and, to a lesser extent, BRI1). Interestingly, in the two lines in which the genetic intervention involved a perturbation of a phytohormone, the change in growth was accompanied by rather restricted changes in metabolites and transcripts.
Thus, transcript, metabolite, and hormone profiling support the notion that all five genes affect different processes. Nevertheless, all five genes enhance leaf organ size primarily by increasing cell number and cell proliferation during leaf development. Hence, we speculate that the cell cycle machinery is likely a direct or indirect target of these five genes.
Multiple Largely Independent Pathways Control Leaf Growth
We have shown that different pathways regulate cell proliferation during leaf development. These pathways can work independently and not influence each other or, alternatively, can be interconnected in a kind of growth-regulating network. The differences found in the phenotypes as well as the low overlap between the different data sets suggest that the five analyzed genes work independently. The analysis of plants expressing two IYGs, obtained by crosses, confirmed this independency between several of these genes. For three combinations (AVP1×GA20OX1, AVP1×GRF5, and BRI1×GA20OX1), the size of the progeny corresponds to the sum of the effect of the individual heterozygous parents, showing that the two combined genes probably do not influence each other. This finding suggests that AVP1, by acting on the auxin level and/or by modifying the carbohydrate and amino acid metabolism, works in a way seemingly independent from GA20OX1, which influences the GA content, or from GRF5, for which a few significant changes were observed in the produced data sets. Furthermore, although a link between BR and GA has already been proposed (Wang et al., 2009), the overexpression of BRI1 does not influence the effect on the leaf size of GA20OX1 and inversely. Interestingly, Arabidopsis mutants with enlarged leaves obtained by screening mutant populations, known as grandifolia-D, all have a duplication of the bottom part of chromosome 4. Genetic evidence suggests that this phenotype is caused by the enhanced expression of at least two genes (Horiguchi et al., 2009).
In other cases, genes seem to be interconnected in a growth-regulatory network, and in some cases, the overexpression of two genes in the same plant partly masks the phenotype of one of the genes. For example, when AVP1 or BRI1 is overexpressed in the same plant together with JAW, the effect of overexpression of JAW on leaf area and leaf serration is completely abolished. This inhibition might be explained by the different hormone contents observed in these lines. Indeed, ABA and SA increased in AVP1 and BRI1 but decreased in JAW. The level of IAA is increased in AVP1 and slightly decreased in JAW. Also, the genome-wide transcript analysis revealed that AVP1 and JAW had a large number of differentially expressed genes in common (113), among which 35 and 25 genes down-regulated and up-regulated in JAW, respectively, were up-regulated and down-regulated in AVP1. Among these genes, 14 were differentially expressed after ABA treatment. This opposite accumulation of ABA and the expression profile of genes responding to this hormone might explain the inhibition of the JAW phenotype when combined with AVP1. However, a GO search on these genes did not reveal any enrichment for growth-regulating genes. Further functional analysis is required to decipher the potential role of these hormones and genes in the observed phenotypes. JAW and AVP1 group together in PC1 of the PCA of the metabolite profiles and share a decrease in raffinose metabolism-related metabolites, which points to common features in the manner in which they regulate growth. JAW and BRI1 are separated in PC1. Nevertheless, both show a trend toward decreased levels of many, although not always the same, metabolites, especially at the end of the day. In other words, growth might outstrip resource availability in these two lines and might contribute to the lack of additivity when these genes are combined.
CONCLUSION
This comparative analysis of genes that enhance leaf size reveals that multiple pathways control organ growth in a largely independent manner and provides evidence that combining overexpression of some of these genes leads to additive or near-additive gain in leaf size. Many more IYG lines have been described (Gonzalez et al., 2009; Krizek, 2009), and comparative phenotypic and molecular characterization of these lines will allow the identification of downstream molecular mechanisms that trigger increased organ size but will also bring more insight into all the parallel or common pathways controlling organ growth. Combination of these different genes should allow the building of molecular networks of organ growth regulation for better engineering of crops with superior biomass production.
MATERIALS AND METHODS
Plant Material
Seeds of the Arabidopsis (Arabidopsis thaliana) AVP1-overexpressing line (AT1G15690) were a kind gift of Dr. Roberto A. Gaxiola (University of Connecticut; Li et al., 2005), the GRF5-overexpressing line (AT3G13960) was from Dr. Hirokazu Tsukaya (University of Tokyo; Horiguchi et al., 2005), seeds from the pBRI1:BRI1 line (BRI1; AT4G39400) were from Dr. Joanne Chory (Salk Institute; Wang et al., 2001), and the GA20OX1-overexpressing line (AT4G25420) was a kind gift of Professor Peter Hedden (Rothamsted Research; Coles et al., 1999).
Artificial microRNA (5′-TGATACTTTTCTGTTCGCGTG-3′) that targets both PPD1 and PPD2 (AT4G14713 and AT4G14720, respectively) was designed with the WMD2 tool (http://wmd2.weigelworld.org) and engineered into the MIR319a stem loop as described previously (Schwab et al., 2006). Quantitative reverse transcription-PCR was done with the primer sets reported previously (White, 2006) that identified lines with both down-regulated PPD1 and PPD2 transcript levels. A homozygous line of the T3 generation had transcript levels of PPD1 and PPD2 down-regulated to 13% and 40% of the wild-type levels, respectively (Supplemental Fig. S3). Seeds from all the transgenic lines were in the Col-0 background.
Growth Conditions
Environmental conditions during seed formation as well as seed storage duration can affect seed vigor. Therefore, all experiments were conducted with wild-type and transgenic seeds that had been harvested from plants grown side by side on the same tray. Plants were grown in vitro in half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% Suc at 21°C under a 16-h-day/8-h-night regime. For the in vitro culture, plants were grown at a density of one plant per 4 cm2. Plants were also grown in rock wool, a semihydroponic inert porous substrate that transports water and fertilizers to the roots by capillary action, and in soil. They were watered daily with 0.5 g L−1 Hyponex under the same daylength regime.
Growth Analysis
For the rosette leaf area measurements, eight to 12 seedlings were grown on rock wool or under in vitro conditions for 22 d. Individual leaves (cotyledons and rosette leaves) were dissected, and their area was measured with the ImageJ software (http://rsb.info.nih.gov/ij/).
For the leaf area analysis, leaves were harvested daily from eight to 10 plants grown in vitro (or in soil). The leaves were cleared with 100% ethanol, mounted in lactic acid on microscope slides, and photographed. The leaf area was determined with the ImageJ software (http://rsb.info.nih.gov/ij/).
Abaxial epidermal cells (40–100 cells) were drawn for four to five leaves with a DMLB microscope (Leica) fitted with a drawing tube and a differential interference contrast objective. Photographs of leaves and drawings were used to measure the leaf area and to calculate the average cell area, respectively, with the ImageJ software (http://rsb.info.nih.gov/ij/). Leaf and cell areas were subsequently used to calculate cell numbers.
Ploidy Analysis
Leaves were frozen and then chopped with a razor blade in 400 μL of buffer (45 mm MgCl2, 30 mm sodium citrate, 20 mm MOPS, pH 7, and 1% Triton X-100; Galbraith et al., 1991), filtered over a 30-μm mesh, and 1 μL of 1 mg mL−1 4,6-diamidino-2-phenylindole was added. The distribution of the nuclear DNA content was analyzed using a CytoFlow ML flow cytometer and FLOMAX software (Partec).
Microarray Experiment
Total RNA was extracted with a RNeasy plant mini kit (Qiagen) from the aerial part of in vitro-grown seedlings at stage 1.03 (Boyes et al., 2001). RNA from three biological repeats was hybridized to the ATH1 array (Affymetrix). Expression data were processed with the robust multichip average (background correction, normalization, and summarization) in BioConductor (Irizarry et al., 2003a, 2003b; Gentleman et al., 2004). An improved chip description file was used, in which each probe was assigned uniquely to one transcript and was excluded when it aligns to a different transcript with 24 or more perfect matches (Casneuf et al., 2007). Differentially expressed genes were identified with the BioConductor package “limma” (Smyth, 2004). P values were corrected for multiple testing (Benjamini and Hochberg, 1995).
Differential transcripts were investigated with PageMan (Usadel et al., 2006) and BiNGO (Maere et al., 2005) to calculate the functional overrepresentation of MapMan and GO categories. Public microarray data corresponding to various hormone treatment experiments (Nemhauser et al., 2006) were used for comparison.
Hormone Analysis
The aboveground part of seedlings grown in vitro until stage 1.03 was harvested in the middle of the day from four independent experiments and frozen in liquid nitrogen. The phytohormones IAA, ABA, SA, cytokinin (iP and tZ forms), jasmonate (JA and JA-Ile), and GAs were extracted and quantified as described previously (Yoshimoto et al., 2009).
Determination of Structural Components and Metabolites
The aboveground part of seedlings at stage 1.03 was harvested in the middle or the end of the day from three to five independent experiments. Chlorophyll, total protein, starch, Glc, Fru, Suc, and total amino acids were assayed as described (Cross et al., 2006), and malate and fumarate were assessed as described (Nunes-Nesi et al., 2007). Metabolite extraction for GC-MS was carried out as described previously (Schauer et al., 2006). Fifty milligrams of plant material was homogenized using a ball mill precooled with liquid nitrogen. Derivatization and GC-MS analysis were done as described previously (Lisec et al., 2006). Metabolites determined by LC-MS were assayed as described (Lunn et al., 2006). Supplemental Table S12 lists the metabolites analyzed by GC-MS or LC-MS. PCA was performed using the statistical software StatistiXL (Broadway) implemented in Microsoft Excel.
Microarray data were deposited in the Gene Expression Omnibus database under accession number GSE20458.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rosette leaf length and width of Col-0 and the five IYG lines.
Supplemental Figure S2. Ploidy levels in Col-0 and the five IYG lines.
Supplemental Figure S3. Rosette phenotypes of BRI1 and PPD lines.
Supplemental Figure S4. Effects of combined pairs of two IYGs on leaf area.
Supplemental Table S1. Metabolite content in Col-0 and the five IYG lines.
Supplemental Table S2. PCA.
Supplemental Table S3. Differences in metabolite content in the five IYG lines compared with the wild type.
Supplemental Table S4. Differences in metabolite content between middle and end of day in Col-0 and the five IYG lines.
Supplemental Table S5. GO categories overrepresented in AVP1 and JAW microarray data sets.
Supplemental Table S6. Genes differentially expressed in AVP1 and comparison with data sets corresponding to hormone treatment experiments.
Supplemental Table S7. Genes differentially expressed in JAW and comparison with data sets corresponding to hormone treatment experiments.
Supplemental Table S8. Genes differentially expressed in BRI1 and comparison with data sets corresponding to hormone treatment experiments.
Supplemental Table S9. Genes differentially expressed in GA20OX1 and comparison with data sets corresponding to hormone treatment experiments.
Supplemental Table S10. Genes differentially expressed in GRF5 and comparison with data sets corresponding to hormone treatment experiments.
Supplemental Table S11. Overlap between the five microarray data sets.
Supplemental Table S12. Metabolites analyzed by GC-MS or LC-MS.
Supplementary Material
Acknowledgments
We thank our colleagues of the System Biology of Yield group for fruitful discussions and suggestions. We also thank Regina Feil and Dr. John Lunn for the LC-MS analyses and Dr. Sandra Trenkamp and Dr. Alisdair Fernie for the GC-MS analyses.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GTS, Genschik P. (2009) Gibberellin signaling controls cell proliferation in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Arrigoni O. (1994) Ascorbate system in plant development. J Bioenerg Biomembr 26: 407–419 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Fiorani F, Inzé D. (2003) Cell cycle: the key to plant growth control? Trends Plant Sci 8: 154–158 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, Gaudillère JP, Raymond P. (1998) Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol 117: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, Van de Peer Y, Huber W. (2007) In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinformatics 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. (2008) Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol 147: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AML, Gusterson BA, Cooper CS. (1994) Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 7: 502–508 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P. (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Ventriglia T, Pinto F, Orea A, Mérida Á, Romero JM. (2005) Differential pattern of expression and sugar regulation of Arabidopsis thaliana ADP-glucose pyrophosphorylase-encoding genes. J Biol Chem 280: 8143–8149 [DOI] [PubMed] [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M. (2006) Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol 142: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S. (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) BioConductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn OJM, Verwoerd TC, Voogd E, Krutwagen RWHH, de Graaf PTHM, Poels J, van Dun K, Ponstein AS, Damm B, Pen J. (1997) Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol 113: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Beemster GTS, Inzé D. (2009) David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol 12: 157–164 [DOI] [PubMed] [Google Scholar]
- Green PB. (1976) Growth and cell pattern formation on an axis: critique of concepts, terminology, and modes of study. Bot Gaz 137: 187–202 [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley R. (1994) Gibberellins: perception, transduction and responses. Plant Mol Biol 26: 1529–1555 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. (2006) Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J Plant Res 119: 37–42 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Gonzalez N, Beemster GTS, Inzé D, Tsukaya H. (2009) Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant J 60: 122–133 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RGF, Bachem CWB, Bögre L. (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua NH. (2006) The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47: 1–9 [DOI] [PubMed] [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. (1998) Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jelitto T, Sonnewald U, Willmitzer L, Hajirezeai M, Stitt M. (1992) Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Keller R, Brearley CA, Trethewey RN, Müller-Röber B. (1998) Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1d-myo-inositol 3-phosphate synthase. Plant J 16: 403–410 [Google Scholar]
- Krizek BA. (2009) Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y. (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakaya M, Tsukaya H, Murakami N, Kato M. (2002) Brassinosteroids control the proliferation in leaf cells of Arabidopsis thaliana. Plant Cell Physiol 43: 239–244 [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E. (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A. (2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J 43: 29–46 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL. (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: 1991–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME, Else MA, Thorne ET, Gherardi F. (2000) Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: evidence for an interaction with ethylene. J Exp Bot 51: 1575–1584 [DOI] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Conceição Piques M, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner E, Cooper CS, Goodwin G. (1999) Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet 8: 585–591 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2002) Interpretation of mutants in leaf morphology: genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between cell and organismal theories. Int Rev Cytol 217: 1–39 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventriglia T, Kuhn ML, Ruiz MT, Ribeiro-Pedro M, Valverde F, Ballicora MA, Preiss J, Romero JM. (2008) Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol 148: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K. (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57: 498–510 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- White DWR. (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. (1994) Molecular cloning and characterization of a brassinosteriod-regulated gene from elongating soyabean (Glycine max L.) epicotyls. Plant Physiol 104: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.