Abstract
The Arabidopsis (Arabidopsis thaliana) genome includes eight PIN-FORMED (PIN) members that are molecularly diverged. To comparatively examine their differences in auxin-transporting activity and subcellular behaviors, we expressed seven PIN proteins specifically in Arabidopsis root hairs and analyzed their activities in terms of the degree of PIN-mediated root hair inhibition or enhancement and determined their subcellular localization. Expression of six PINs (PIN1–PIN4, PIN7, and PIN8) in root hair cells greatly inhibited root hair growth, most likely by lowering auxin levels in the root hair cell by their auxin efflux activities. The auxin efflux activity of PIN8, which had not been previously demonstrated, was further confirmed using a tobacco (Nicotiana tabacum) cell assay system. In accordance with these results, those PINs were localized in the plasma membrane, where they likely export auxin to the apoplast and formed internal compartments in response to brefeldin A. These six PINs conferred different degrees of root hair inhibition and sensitivities to auxin or auxin transport inhibitors. Conversely, PIN5 mostly localized to internal compartments, and its expression in root hair cells rather slightly stimulated hair growth, implying that PIN5 enhanced internal auxin availability. These results suggest that different PINs behave differentially in catalyzing auxin transport depending upon their molecular activity and subcellular localization in the root hair cell.
Auxin plays a critical role in plant development and growth by forming local concentration gradients. Local auxin gradients, created by the polar cell-to-cell movement of auxin, are implicated in primary axis formation, root meristem patterning, lateral organ formation, and tropic movements of shoots and roots (for recent review, see Vanneste and Friml, 2009). The cell-to-cell movement of auxin is achieved by auxin influx and efflux transporters such as AUXIN-RESISTANT1 (AUX1)/LIKE-AUX1 for influx and PIN-FORMED (PIN) and the P-glycoprotein (PGP) of ABCB (ATP-binding cassette-type transporter subfamily B) for efflux. Since diffusive efflux of the natural auxin indole-3-acetic acid (IAA; pKa = 4.75) is not favorable and PINs are localized in the plasma membrane in a polar manner, PINs act as rate-limiting factors for cellular auxin efflux and polar auxin transport through the plant body. These PINs' properties explain why representative physiological effects of auxin transport are associated with PINs.
Auxin flows from young aerial parts all the way down to the root tip columella in which an auxin maximum is formed for root stem cell maintenance and moves up toward the root differentiation zone through root epidermal cells, where a part of it travels back to the root tip via cortical cells (Blilou et al., 2005). This directional auxin flow is supported by the polar localization of PINs: PIN1, PIN3, and PIN7 at the basal side of stele cells (Friml et al., 2002a, 2002b; Blilou et al., 2005), PIN4 at the basal side in root stem cells (Friml et al., 2002a), and PIN2 at the upper side of root epidermis and at the basal side of the root cortex (Luschnig et al., 1998; Müller et al., 1998). Another interesting aspect of PIN-mediated auxin transport is the dynamics in directionality of auxin flow due to environmental stimuli-directed changes of subcellular PIN polarity, as exemplified for PIN3, whose subcellular localization changes in response to the gravity vector (Friml et al., 2002b).
An intriguing question is how different PIN proteins have different subcellular polarities, which might be attributable to PIN-specific molecular properties, cell-type-specific factors, or both. The different PIN subcellular polarities in different cell types seemingly indicate that cell-type-specific factors are involved in polarity. In the case of PIN1, however, both classes of factors appear to affect its subcellular localization because when expressed under the PIN2 promoter, PIN1 localizes to the upper or basal side of root epidermal cells, depending on the GFP insertion site of the protein (Wiśniewska et al., 2006). A recent study demonstrated that the polar targeting of PIN proteins is modulated by phosphorylation/dephosphorylation of the central hydrophilic loop of PINs, which is mediated by PINOID (PID; a Ser/Thr protein kinase)/PP2A phosphatase (Michniewicz et al., 2007). The central hydrophilic domain of PINs might provide the molecule-specific cue for PIN polarity, together with as yet unknown cell-specific factors. Different recycling behaviors of PINs, which show variable sensitivities to brefeldin A (BFA), also imply different molecular characters among PIN species. Most PIN1 proteins are internalized by BFA treatment, whereas considerable amounts of PIN2 remain in the plasma membrane in addition to internal accumulation after BFA treatment. Recycling and basal polar targeting of PIN1 is dependent on the BFA-sensitive guanine nucleotide exchange factor for adenosyl ribosylation factors (ARF GEFs), GNOM, which is the major target of BFA. In contrast, apical targeting and recycling of PIN2 is independent of GNOM and controlled by BFA-resistant ARF GEFs (Geldner et al., 2003; Kleine-Vehn and Friml, 2008).
In contrast to their distinct subcellular localizations, the differential auxin-transporting activities of PINs remain to be studied. The divergent primary structures of PIN proteins are not only indicative of differential subcellular polarity, but also would represent their differential catalytic activities for auxin transport. The auxin efflux activities of Arabidopsis (Arabidopsis thaliana) PINs have been demonstrated using Arabidopsis and heterologous systems: PIN1 and PIN5 in Arabidopsis cells (Petrásek et al., 2006; Mravec et al., 2009); PIN2, PIN3, PIN4, PIN6, and PIN7 in tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cells (Lee and Cho, 2006; Petrásek et al., 2006; Mravec et al., 2008); PIN1, PIN2, PIN5, and PIN7 in yeast (Saccharomyces cerevisiae) cells (Petrásek et al., 2006; Blakeslee et al., 2007; Mravec et al., 2009; Yang and Murphy, 2009); and PIN1, PIN2, and PIN7 in HeLa cells (Petrásek et al., 2006; Blakeslee et al., 2007). Among the eight Arabidopsis PIN members, PIN1, PIN2, PIN3, PIN4, PIN6, and PIN7, which share a similar molecular structure in terms of the presence of a long central loop (hereafter called long-looped PINs; Fig. 1A; Supplemental Fig. S1), have been shown to catalyze auxin efflux at the cellular level. On the other hand, PIN5 and PIN8 possess a very short putative central loop (hereafter called short-looped PINs). Although PIN5 was recently shown to be localized in the endoplasmic reticulum (ER) and proposed to transport auxin metabolites into the ER lumen, its cellular function regarding its intracellular auxin-transporting activity has not been shown, and the auxin-transporting activity of PIN8 has yet to be demonstrated. In spite of the same transport directionality (auxin efflux) and similar molecular structures, the long-looped PINs exhibit sequence divergence not only in their central loop, but also in certain residues of the transmembrane domains. This structural divergence of long-looped PINs might be indicative of their differential auxin-transporting activities, which have not yet been quantitatively compared.
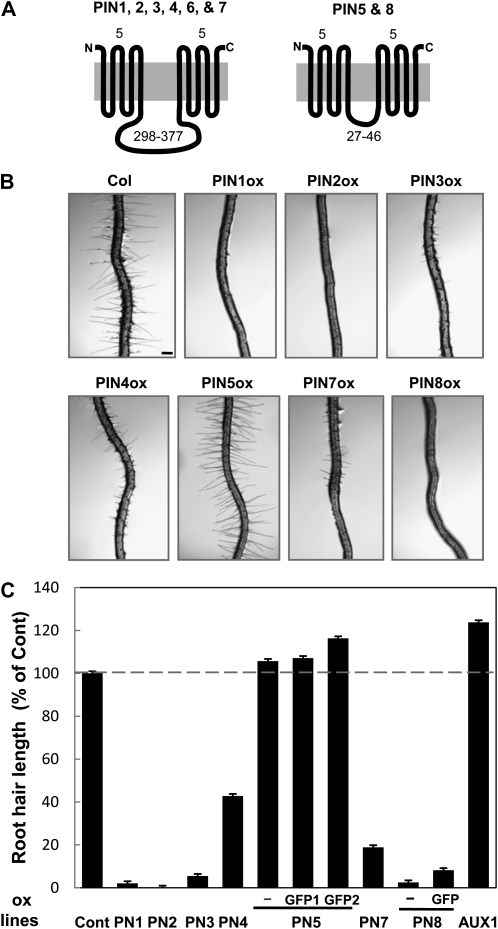
Figure 1.
Differential activities of PINs in the Arabidopsis root hair. A, Two distinctive PIN groups with different central hydrophilic loop sizes. Topology of PIN proteins was predicted by four different programs as described in Supplemental Figure S1. Numbers above indicate the number of transmembrane helices for each N- and C-terminal region, and numbers below indicate the number of amino acid residues of the central hydrophilic domain. B, Representative root images of control (Cont; Columbia-0) and root-hair-specific PIN-overexpressing (PINox; ProE7:PIN-GFP or ProE7:PIN [−]) plants. Bar = 100 μm for all. C, Root hair lengths of control and PINox plants. Six to 12 independent transgenic lines (average = 8.3), and 42 to 243 roots (average = 86.8) and 336 to 2,187 root hairs (average = 727.8) per construct, were observed for the estimation of root hair length. Data represent means ± se. The root hair lengths of PIN5ox lines were significantly longer than those of the control (P = 0.016 for PIN5ox; P < 0.0001 for PIN5-GFP1ox and PIN5-GFP2ox).
To comparatively assess the cytological behaviors and molecular activities of different PIN members, it would be favorable to use a single assay system that provides a consistent cellular environment and enables quantitative estimation of PIN activity. In previous studies, we adopted the root hair single cell system to quantitatively assay auxin-transporting or regulatory activities of PINs, PGPs, AUX1, and PID (Lee and Cho, 2006; Cho et al., 2007a). Root hair growth is proportional to internal auxin levels in the root hair cell. Therefore, auxin efflux inhibits and auxin influx enhances root hair growth (Cho et al., 2007b; Lee and Cho, 2008). In addition, the use of a root-hair-specific promoter (Cho and Cosgrove, 2002; Kim et al., 2006) for expression of auxin transporters enables the transporters' biological effect to be pinpointed to only the root hair cell, thus excluding probable non-cell-autonomous effects that could be caused by the general expression of auxin transporters.
In this study, we expressed five long-looped PINs (PIN1, PIN2, PIN3, PIN4, and PIN7) and two short-looped PINs (PIN5 and PIN8) in root hair cells and compared their auxin-transporting activities and cytological dynamics. To directly measure the radiolabeled auxin-transporting activities of PIN5 and PIN8, we used an additional assay system, tobacco suspension cells. Our data revealed that PINs have differential molecular activities and pharmacological responses and that the short-looped and long-looped PINs have different subcellular localizations.
RESULTS
Root Hair Cell-Specific Expression of PINs Reveals Differential Activities in Inhibiting Root Hair Growth
We previously used the root hair cell-specific promoter of EXPA7 (Arabidopsis EXPANSIN A7; Cho and Cosgrove, 2002) to demonstrate that the root-hair-specific expression of PID, PIN3, and PGPs suppresses root hair growth by promoting auxin efflux from the root hair cell (Lee and Cho, 2006; Cho et al., 2007a). To investigate whether the other PINs also have a similar phenotype in the root hair, genomic fragments of PIN genes (PIN1, PIN2, PIN4, PIN5, PIN7, and PIN8) were fused to a reporter gene encoding GFP, and the fusion genes were expressed using the EXPA7 promoter (ProE7) in Arabidopsis. In the case of PIN5, three different transgenes were introduced into Arabidopsis: ProE7:PIN5 (PIN5ox, without GFP), ProE7:PIN5-GFP1 (PIN5-GFP1ox, with GFP after nucleotide 558 [from “A” of the start codon of the genomic sequence]), and ProE7:PIN5-GFP2 (PIN5-GFP2ox, with GFP after nucleotide 1,333). For PIN8, two transgenes were constructed: ProE7:PIN8 (PIN8ox, without GFP) and ProE7:PIN8-GFP (PIN8-GFPox, with GFP after nucleotide 675). For a statistical analysis of the effect of PINox on root hair growth, six to 12 independent transgenic lines per construct were randomly chosen for estimation of root hair length (Fig. 1C; Supplemental Fig. S2).
Root-hair-specific overexpression of the long-looped PINs (PIN1–PIN4 and PIN7; ProE7:PIN-GFP [PINox]) greatly inhibited root hair growth (Fig. 1, B and C; Supplemental Fig. S2), suggesting that these PINs facilitate auxin efflux and consequently decrease internal auxin levels in the hair cell. Overexpression of the short-looped PIN8 also dramatically inhibited root hair growth, as did long-looped PINoxs (Fig. 1, B and C; Supplemental Fig. S2, I and J). However, root-hair-specific overexpression of the auxin influx transporter AUX1 significantly enhanced root hair growth (Fig. 1C), most likely by increasing cellular auxin levels in root hair cells.
Although overexpression of long-looped PINs and PIN8 all significantly decreased root hair growth, the effects varied among the different PIN species. While overexpression of PIN1, PIN2, PIN3, and PIN8 caused inhibition of hair growth by >90%, PIN4ox and PIN7ox mildly inhibited hair growth, with an average of approximately 60% and approximately 80% inhibition (in 9–12 lines), respectively (Fig. 1C). Even among PINs with strong root hair inhibitory effects, slightly different inhibitory strengths were observed. For example, PIN2ox almost completely inhibited hair growth, whereas PIN1ox, PIN3ox, and PIN8ox allowed marginal hair growth (2%–5% of control). Considering the use of the same promoter (ProE7) for PIN expression, the similar root hair inhibitory effect among independent lines for each PINox and the plasma membrane localization of PIN1, PIN2, PIN3, PIN4, and PIN7, it is conceivable that the long-looped PINs have differential auxin efflux activities in the root hair cell.
In contrast to other PINox lines, all three PIN5-overexpressing lines (PIN5ox, PIN5-GFP1ox, and PIN5-GFP2ox) did not inhibit root hair growth, but instead slightly (6% to approximately 16%) enhanced growth (Fig. 1C; Supplemental Fig. S2, E–G). This PIN5ox-mediated increase was marginal but consistent for different PIN5-overexpressing transgenic constructs (P = 0.016 for PIN5ox; P < 0.0001 for PIN5-GFP1ox and PIN5-GFP2ox), suggesting that PIN5 might catalyze cellular auxin influx or enhance auxin availability in the subcellular domains where auxin executes its molecular actions.
Root Epidermal Cell Elongation Is Enhanced by PINox
Because high auxin concentrations inhibit root growth (Evans et al., 1994), we thought that PINox, by lowering cellular auxin levels, might enhance the elongation of root epidermal cells and thus of root growth. Root-hair-specific expression of PINs considerably increased root growth (Fig. 2A), and this effect seems to be at least partly due to enhanced cell elongation. PINoxs increased the length of root-hair-bearing epidermal cells by 8.5% (PIN4ox) to approximately 15% (PIN1ox, PIN2ox, PIN3ox, and PIN8ox; Fig. 2B). The degree of PINox-mediated stimulation of root growth and root hair cell elongation was somewhat proportional to the strength of PINox-mediated root hair inhibition. Among long-looped PINs, while PIN1ox and PIN2ox showed the greatest effect, PIN4ox showed the smallest effect on root growth and root hair cell elongation. The epidermal cell length of PIN5ox plants was similar to that of controls. Together with the result of PINox-mediated root hair inhibition, these data further support the proposal that different PINs have different auxin efflux activities.
Figure 2.
Effect of root hair cell-specific expression of PINs on elongation of roots and root hair epidermal cells. A, Four-day-old seedlings of control (Cont; Columbia-0) and PINox (ProE7:PIN-GFP) lines. Bar = 0.5 cm. B, Root hair epidermal cell length in PINox lines. The length between two consecutive root hairs in the same hair cell file was measured as the root hair cell length for control and PINox lines. Five independent transgenic lines for each transformant were analyzed. Data represent means ± se (n = 350–500). [See online article for color version of this figure.]
Subcellular Localization Pattern of PINs in the Root Hair Cell
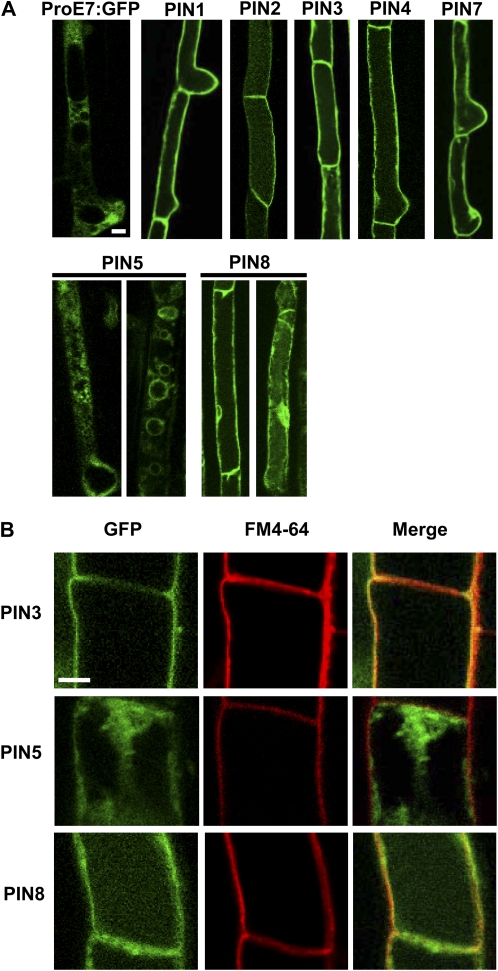
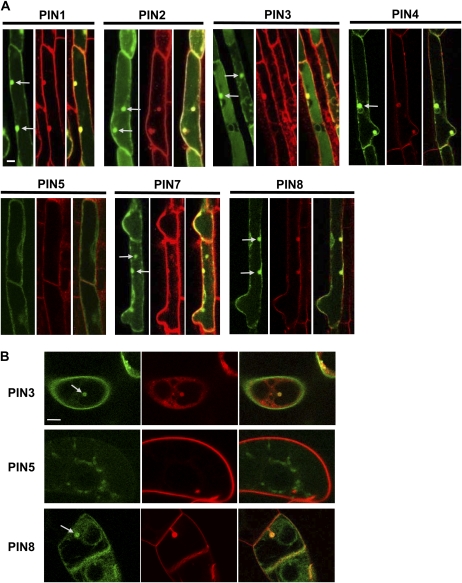
To collectively investigate the subcellular localization of PIN proteins in the root hair cell, we used confocal microscopy to examine PIN-GFP fusion proteins specifically expressed in the root hair. PIN1-, PIN2-, PIN3-, PIN4-, PIN7-, and PIN8-GFP primarily localized to the plasma membrane (Fig. 3). However, while PIN2-GFP consistently showed only plasma membrane localization, other PINs-GFP were sometimes internally localized, such as at the nuclear periphery, in addition to plasma membrane localization. Internal localization was more frequent for PIN8-GFP than for other long-looped PIN-GFPs. Depending on cells, PIN8-GFP was sometimes dominantly internally localized. In contrast to these PINs, PIN5-GFP1 predominantly localized to internal compartments. PIN5-GFP1 was located around variously sized cytoplasmic vesicles, probably at vesicular membranes, and it also formed a network pattern, probably the ER network as demonstrated in tobacco cells (Mravec et al., 2009; Fig. 4A).
Figure 3.
Subcellular localization of PIN-GFP proteins in the Arabidopsis root hair cell. A and B, Root hair specifically expressed PIN-GFP (ProE7:PIN-GFP; PIN5 = ProE7:PIN5-GFP1) fusion proteins were visualized by confocal microscopy. Root hair cells were visualized by only GFP signal (A) or by both GFP and FM4-64 dye signals (B). GFP and FM4-64 signals at the plasma membrane were merged for PIN3 and PIN8, whereas they were not for PIN5 (B). Bars = 10 μm for A and 5 μm for B.
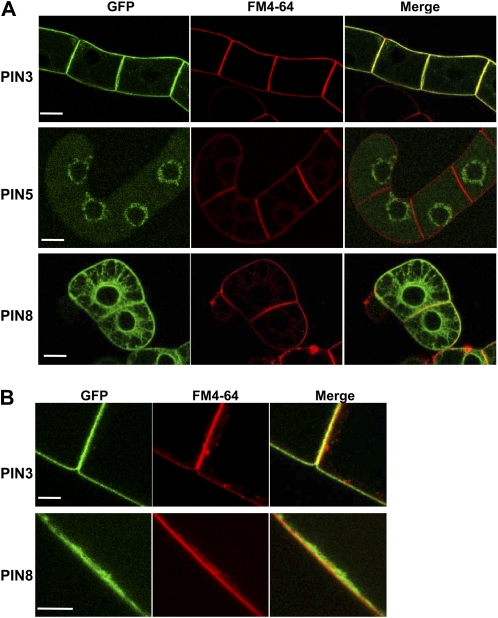
Figure 4.
Subcellular localization of short-looped PINs in tobacco cells. A and B, PIN-GFP (ProTA:PIN-GFP; PIN5 = ProTA:PIN5-GFP1) fusion proteins were visualized by confocal microscopy. Expression of the fusion proteins was induced by 0.01 mm Dex for 24 h. Cells were visualized by GFP and FM4-64 dye signals. GFP and FM4-64 signals at the plasma membrane were merged for PIN3 and PIN8, whereas they were not for PIN5. Bars = 20 μm for A and 10 μm for B.
To further support the idea that the PIN8-localized cell boundary is the plasma membrane, we examined whether PIN8-GFP signals overlap with the FM4-64-stained (<3 min) plasma membrane. High-magnification confocal images showed that PIN8-GFP signals, as did PIN3-GFP signals, overlapped with FM4-64 signals (Fig. 3B). In contrast, PIN5-GFP signals did not overlap with FM4-64 signals. This observation suggests that PIN8 at least partly localizes at the plasma membrane but PIN5 does not.
PIN8 Shows Auxin Efflux Activity in Tobacco BY-2 Suspension Cells
We previously demonstrated that auxin transporters, such as PIN3 and PGP4, localize to the plasma membrane and act as efflux transporters for 1-naphthalene acetic acid (NAA) in tobacco BY-2 cells (Lee and Cho, 2006; Cho et al., 2007a). PIN4, PIN6, PIN7, and PGP19 were also shown to catalyze auxin efflux in tobacco BY-2 cells (Petrásek et al., 2006). Recently, it was reported that PIN5 has an auxin efflux activity in the yeast plasma membrane and presumably an intracellular transporting activity in Arabidopsis cells (Mravec et al., 2009). In this study, to directly measure the cellular auxin-transporting activity of short-looped PINs (PIN5 and PIN8) in plant cells, we expressed their GFP-tagged versions using the dexamethasone (Dex)-inducible promoter (ProTA; Aoyama and Chua, 1997) in tobacco BY-2 cells.
Treatment with Dex resulted in considerable induction of the PIN5-GFP1 and PIN8-GFP fusion proteins (Fig. 4A). As seen in Arabidopsis root hair cells, the PIN5- and PIN8-GFP fusion proteins showed clear internal localization, revealing a typical ER network distribution pattern. However, while PIN5-GFP1 predominantly located in internal compartments, PIN8-GFP additionally continued along the cell boundary. In colocalization analyses of PIN-GFP and FM4-64, PIN8-GFP signals in the cell boundary overlapped with FM4-64 signals as did PIN3-GFP signals, whereas PIN5-GFP1 signals scarcely overlapped with the FM4-64-stained plasma membrane (Fig. 4). These data suggest that PIN8 localizes to both the plasma membrane and the ER network in tobacco cells, as observed in Arabidopsis root hair cells (Fig. 3).
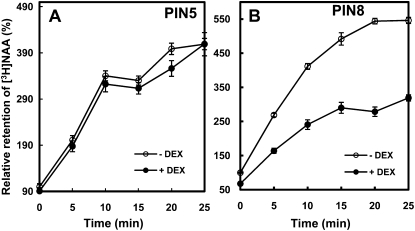
For the cellular auxin-transporting assay, transgenic tobacco cells were treated with [3H]NAA, and the retention of cellular [3H]NAA was measured at specific time intervals. NAA can easily enter the cell by diffusion but needs active auxin efflux transporters to be exported to the apoplast (Delbarre et al., 1996). Induction of PIN8 significantly decreased the retention of [3H]NAA compared to uninduced control cells (Fig. 5B). After incubation with [3H]NAA for 20 min, PIN8ox cells retained approximately 60% [3H]NAA compared with control cells, indicating that PIN8 mediated a net auxin efflux from tobacco cells. On the other hand, induction of PIN5 did not significantly alter [3H]NAA retention levels in tobacco cells compared to control cells. The inability to detect a net flux of auxin in PIN5-expressing cells could be due to its intracellular auxin-transporting activity, rather than to intercellular auxin transport.
Figure 5.
Relative retention of [3H]NAA in PINox BY-2 tobacco cells. A and B, Relative accumulation of [3H]NAA by PIN5ox (ProTA:PIN5-GFP1; A) and PIN8ox (ProTA:PIN8-GFP; B). Expression of transgenes was induced by 0.01 mm Dex for 24 h (−Dex = noninduced). Data represent means ± se from eight replicates.
Exogenous Auxin Restores the Root Hair Growth of PINox Transformants
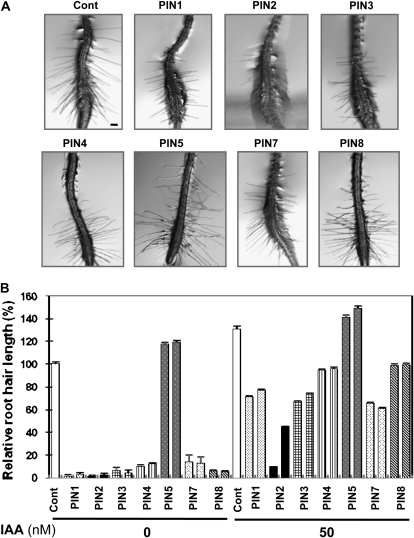
If PINoxs inhibit root hair growth by lowering auxin levels in the root hair cell, application of exogenous auxin should restore root hair growth in these transformants. While 50 nm IAA enhanced the root hair growth of control plants by 30% and of PIN5ox transformants by approximately 20%, it stimulated the root hair growth of other PINox transformants by 5- to 44-fold (average 15-fold) when compared with that of untreated transformants (Fig. 6). The auxin-mediated root hair restoration level in PINoxs was on average 82% of the root hair length of untreated control plants (100%). This result indicates that the diffusive and AUX1-facilitated auxin influx rate surpasses the PINox-mediated auxin efflux rate until a certain equilibrium point is reached. Although exogenous IAA overall greatly increased the root hair growth of PINox transformants, the degree of auxin-mediated root hair restoration varied among PINoxs. PIN2ox plants showed the least hair restoration by IAA, implying that PIN2 has the strongest auxin efflux activity in the root hair cell.
Figure 6.
IAA restores root hair growth of PINox transformants. A, Representative root images of control (Cont; Columbia-0) and root-hair-specific PIN-overexpressing (PINox; ProE7:PIN-GFP) plants. Seedlings were treated with 50 nm IAA for 1 d. Bar = 100 μm for all. B, Root hair lengths of control and PINox plants. Two independent homozygous lines for each transformant were analyzed. Data represent means ± se (n = 56–112; average = 103).
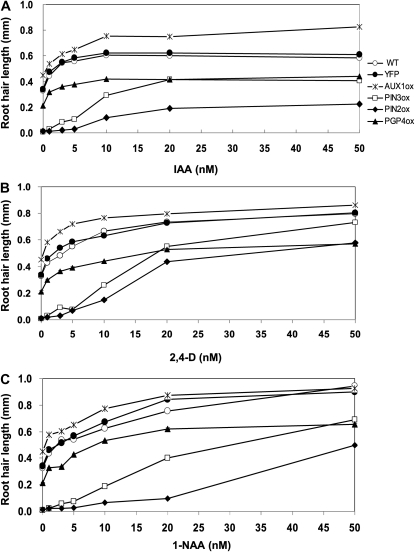
To obtain insights into the different molecular properties of influx and efflux transporters and also among different PINs, we examined the auxin-mediated root hair restoration kinetics of auxin transporter-overexpressing lines. Two control lines (the wild type and ProE7:YFP) and AUX1ox (ProE7:AUX1-YFP), PIN3ox (ProE7:PIN3-GFP), PIN2ox (ProE7:PIN2-GFP), and PGP4ox (ProE7:PGP4-YFP) lines were treated with 0 to 50 nm of three different auxins, IAA, 2,4-dichlorophenoxyacetic acid (2,4-D), and NAA, which have different membrane permeabilities (Fig. 7). The two control lines showed similar kinetics of increased root hair length in response to auxin. An intriguing difference in the kinetics is that while control, AUX1ox, and PGP4ox lines showed a hyperbolic pattern of root hair restoration with increasing exogenous auxin concentrations, PIN2ox and PIN3ox lines showed a sigmoidal pattern. This might be due to strong auxin efflux activities by PIN2 and PIN3, leading to a poor root hair restoration capacity of these overexpressing transformants at low concentrations of exogenous auxin. PIN2ox showed a stronger resistance to auxin-mediated root hair restoration than did PIN3ox for the auxin concentrations tested.
Figure 7.
Different root-hair-restoring behaviors of auxin transporter-expressing transformants in response to different auxin species. A, IAA effect on root hair growth of wild-type (WT), ProE7:YFP (YFP), ProE7:AUX1-YFP (AUX1ox), ProE7:PIN2-GFP (PIN2ox), ProE7:PIN3-GFP (PIN3ox), and ProE7:PGP4-YFP (PGP4ox) plants. B, The effect of 2,4-D on root hair growth. C, NAA effect on root hair growth. Root hair length was estimated from five independent experiments, each including 50 to 100 root hairs from five to 10 roots. Data points are means ± se (n = 320–430 [average = 380] for A; n = 270–430 [average = 370] for B; n = 310–430 [average = 380] for C).
Auxin transporter-expressing transformants had slightly variable responses to different auxin species. The increase in root hair length in control and transporter-expressing lines was saturated at 10 to 20 nm IAA, whereas root hair length continued to increase even at levels above 20 nm 2,4-D or NAA, probably reflecting different membrane-permeating capabilities among auxin species. This continuous increase of root hair length was more obvious with NAA, which has the highest membrane permeability among the three auxins.
Another interesting observation is that after IAA treatment, the root hair restoration curves of different transgenic lines did not tend to converge; rather, they continued to differ even at higher IAA concentrations (Fig. 7A). In contrast, with increasing concentrations of 2,4-D and NAA, the differences in root hair length among different lines were reduced (Fig. 7, B and C). In particular, the root hair lengths of AUX1ox and control plants were almost identical in 20 to 50 nm NAA. This experiment also demonstrated how much auxin is required for an auxin efflux transporter-expressing line to restore the control level of root hair length (i.e. that of wild-type plants without exogenous auxin). IAA of 1 to 3 nm was enough in PGP4ox plants to restore the control level, and approximately 20 nm IAA was required in PIN3ox lines to restore the control level (Fig. 7A). However, the control level was not restored in PIN2ox plants, even at 50 nm IAA. The 2,4-D and NAA also had similar effects except that the control level was restored in PIN2ox plants at higher auxin concentrations (Fig. 7, B and C). The root hair restoration of PIN2ox plants by high concentrations of 2,4-D and NAA could be because 2,4-D tends to accumulate high levels in the cytosol and NAA has a high diffusive influx capacity (Delbarre et al., 1996). These data again demonstrate different auxin-transporting behaviors between different auxin transporters.
Inhibition of Auxin Efflux Transporters Restores Root Hair Growth of PINox Transformants
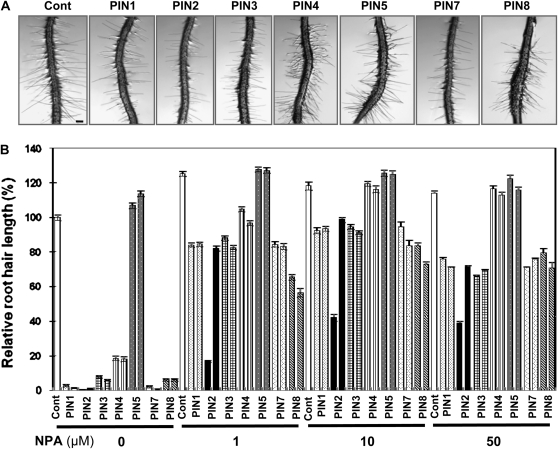
We reasoned that the inhibition of PINox-mediated auxin efflux activity should result in a restoration of root hair growth in PINox lines. PINox seedlings were treated with varying concentrations (1–50 μm) of naphthylphthalamic acid (NPA), an auxin efflux inhibitor. Root hair growth in most PINox lines (PIN1ox, PIN2ox, PIN3ox, PIN7ox, and PIN8ox) was significantly restored by even 1 μm NPA (Fig. 8). Although NPA slightly enhanced the root hair lengths of control and PIN5ox plants (10%–18%), the hair restoration effect by NPA was most prominent in the short root-haired PINox lines. The lengths of PIN1ox, PIN2ox, PIN3ox, PIN4ox, PIN7ox, and PIN8ox root hairs treated with 10 μm NPA were on average 62-fold longer than those untreated with NPA. Most dramatically, PIN1ox, PIN2ox, and PIN7ox root hairs showed an average 113-fold increase in length when treated with 10 μm NPA. Considering only the final restored root hair length, PINox plants had slightly different sensitivities to NPA. PIN4ox showed the greatest restoration, and PIN2ox showed the least. Interestingly, a few PIN2ox lines were strongly resistant to NPA and to IAA (Figs. 6B and 8B). These results collectively reflect slightly different NPA sensitivities and different auxin efflux activities of separate PIN species.
Figure 8.
NPA restores root hair growth of PINox transformants. A, Representative root images of control (Cont; Columbia-0) and root-hair-specific PINox (ProE7:PIN-GFP) plants treated with 1 μm NPA for 1 d. Bar = 100 μm for all. B, Root hair lengths of control and PINox plants treated with 0, 1, 10, and 50 μm NPA. Two independent homozygous lines for each transformant were analyzed. Data represent means ± se for each line (n = 154–234; average = 191).
Effects of BFA on the Subcellular Localization and Activity of PINs
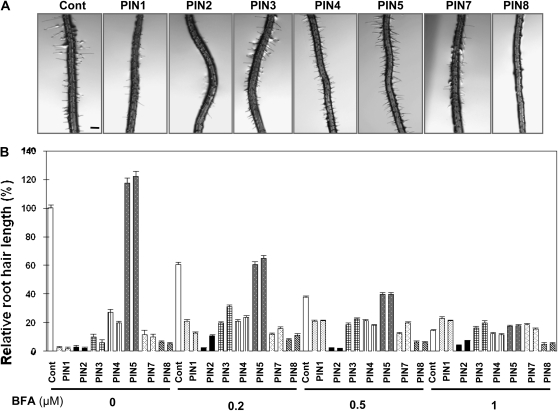
BFA, a blocker of PIN trafficking to the plasma membrane, restores root hair growth in the PIN3ox line, most likely by lowering the concentration of PINs in the plasma membrane and thus increasing cellular auxin levels (Lee and Cho, 2006). To determine whether BFA exhibits a similar effect on the root hairs of other PINox lines, transgenic seedlings were treated with BFA (0, 0.2, 0.5, and 1 μm) for 24 h. Increasing BFA concentrations significantly inhibited the root hair growth of long root-haired control and PIN5ox plants in a dose-dependent manner. However, most other PINox lines showed a slight restoration of root hair growth in response to BFA (Fig. 9). When compared with each BFA-treated control line, short-haired PINox lines (PIN1ox–PIN4ox, PIN7ox, and PIN8ox) showed significant root hair restoration by BFA. The average root hair length of these short-haired PINox lines was 8.9% of the control value (0 μm BFA). This restoration frequency increased to 26.1, 38.2, and 92.8% of the control value, respectively, after treatment with 0.2, 0.5, and 1 μm BFA (Fig. 9; Supplemental Table S1). Among different short-haired PINox lines, BFA-mediated root hair restoration of PIN2ox and PIN8ox lines was much less effective than for other PINox lines. In particular, PIN2ox lines showed the greatest resistance to BFA-mediated root hair restoration. The differential root hair responses of PINox lines to BFA might be indicative of different sensitivity to BFA in their intracellular trafficking.
Figure 9.
BFA restores root hair growth of PINox transformants. A, Representative root images of control (Cont; Columbia-0) and root-hair-specific PINox (ProE7:PIN-GFP) plants treated with 0.2 μm BFA for 1 d. Bar = 100 μm for all. B, Root hair length of control and PINox plants treated with 0, 0.2, 0.5, and 1 μm BFA. Two independent homozygous lines for each transformant were analyzed. Data represent means ± se for each line (n = 118–303; average = 184).
BFA inhibits the exocytic trafficking of PIN proteins from endosomes to the plasma membrane and drives PINs to localize to so-called BFA compartments (Boutté et al., 2006). Thus far, five PIN species (PIN1, PIN2, PIN3, PIN4, and PIN7) have been shown to be in BFA compartments (Geldner et al., 2001, 2003; Friml et al., 2002b). In this study, we examined the cytological effect of BFA on seven PIN proteins in root hair cells for two purposes: the first was to determine whether the two short-looped PINs (PIN5 and PIN8) are also targeted to the BFA compartment, and the second was to demonstrate that the BFA-mediated root hair restoration of PINox lines correlates with the internalization of PIN proteins in BFA compartments. When the fluorescence of PIN-GFP proteins in PINox root hair cells was analyzed after 2 h incubation in liquid medium containing 50 μm BFA, we detected fluorescent PIN proteins in round intracellular compartments in most PIN-GFP lines, including the short-looped PIN8-GFP, but not PIN5-GFP (Fig. 10A). The BFA compartments were colabeled with FM4-64 after a 10-min incubation with the dye. Concurrently, BFA treatment seemed to considerably decrease PIN-GFP signals in the plasma membrane. Similar results were obtained with tobacco BY-2 cells where PIN5-GFP was not but PIN8-GFP was integrated into the BFA compartment (Fig. 10B). These results suggest that the BFA-mediated root hair restoration of PINox lines is caused by a decreased auxin efflux at the plasma membrane and that PIN8 in the BFA compartment originates from the plasma membrane.
Figure 10.
The BFA effect on PIN trafficking in Arabidopsis root hair and tobacco cells. A, Confocal microscopy images of root hair cells from Arabidopsis transformant (ProE7:PIN-GFP) seedlings. B, Confocal microscopy images of transgenic (ProTA:PIN-GFP) tobacco cells. Seedlings and cells were treated with 50 μm BFA for 2 h and with 2 μm FM4-64 for 15 min before observation. GFP signals (left) and FM4-64 signals (middle) were merged in the right panels for each PIN. BFA compartments are indicated by arrows. Bars = 10 μm for all.
PIN8 Localizes in the Cell Plate, Which Originates from the Plasma Membrane
To further support the idea that PIN8 exists in the plasma membrane, we observed the presence of PIN proteins in the cell plate during cytokinesis of tobacco BY-2 cells. Plasma-membrane-originated PIN1 and PIN2 are localized in the cell plate of dividing BY-2 cells (Dhonukshe et al., 2006). Therefore, if PIN8 is only localized in the internal organelles, such as ER, it would not be found in the cell plate. In our experiment, BY-2 cells were treated with BFA to block probable PIN trafficking to the cell plate via the ER-Golgi secretory pathway (Dhonukshe et al., 2006). PIN8-GFP clearly targeted to the cell plate, as did PIN3-GFP, and PIN8- or PIN3-located cell plates were completely colocalized with FM4-64 signals, which also originated from the plasma membrane (Fig. 11). In contrast, PIN5-GFP was not found in the cell plate. These data suggest again that PIN8 exists in the plasma membrane, whereas PIN5 resides only in the internal compartment, such as the ER.
Figure 11.
PIN8 localizes in the cell plate. Confocal microscopy images of transgenic tobacco cells (ProTA:PIN3-GFP, ProTA:PIN5-GFP1, and ProTA:PIN8-GFP). Cells were treated with 50 μm BFA for 2 h and with 2 μm FM4-64 for 15 min before observation. GFP signals (left) and FM4-64 signals (middle) were merged in the right panels for each PIN. The arrows indicate PIN-residing cell plates (for PIN3 and PIN8, but not for PIN5). Bar = 10 μm for all.
DISCUSSION
The Root Hair Cell Provides a Dual Biological Marker System for PIN Activities
In this study, we adopted the Arabidopsis root hair single cell system as the primary biological marker to estimate the auxin-transporting activity of PIN proteins. Numerous genetic and physiological studies have indicated that auxin positively regulates root hair growth (Masucci and Schiefelbein, 1994, 1996; Okada and Shimura, 1994; Leyser et al., 1996; Pitts et al., 1998; Reed, 2001). Our previous study with this root hair assay system demonstrated that the root-hair-specific expression of PIN3 and PID caused a reduction of root hair growth, and the auxin efflux inhibitor NPA restored root hair growth in these transgenic seedlings (Lee and Cho, 2006). Conversely, the root-hair-specific expression of the auxin influx transporter AUX1 enhanced root hair growth (Cho et al., 2007a). These data strongly suggest that the PIN-mediated inhibition of root hair growth resulted from PIN-mediated reduction of intracellular auxin levels in the hair cell. The root hair system, in a certain aspect, is a robust assay system against cellular metabolic inhibitors, such as BFA and kinase inhibitors, which, however, are useful for assessing PIN trafficking behaviors. Although these inhibitors are general toxins for cellular processes, root hair growth was restored in PIN- or PID-overexpressing transformants by the application of these chemicals (Lee and Cho, 2006). Another advantage of using the root hair system for assaying auxin transporters is that it provides a reasonable quantitative system. Statistical analyses of root hair length from multiple independent transgenic lines enabled a quantitative comparison of cellular auxin-transporting activities by different auxin transporters. Although many different PINs were assayed in the tobacco suspension cell system, it would be difficult to perform a statistical auxin transport assay with many independent cell lines.
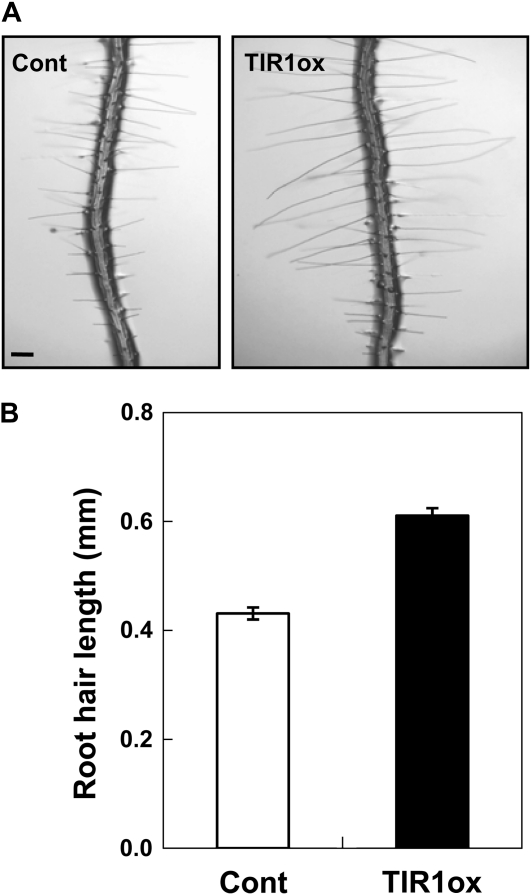
Although we could quantitatively compare auxin-transporting activities among different PIN proteins by taking advantage of the root hair system, an important point is that PIN-mediated auxin-transporting activity should be properly interpreted in terms of the degree of root hair inhibition. Auxin-promoted root hair growth is attributable primarily to the result of nuclear events. Auxin receptors (TIR1 and related F-box proteins) are localized in the nucleus, and their loss of function results in defects in root hair growth (Dharmasiri et al., 2005). In contrast, root-hair-specific expression of TIR1 enhances root hair growth (Fig. 12; Supplemental Fig. S3). Furthermore, root-hair-specific expression of the nondegradable dominant mutants of AXR2/IAA7 and AXR3/IAA17 (Aux/IAA transcriptional repressors) almost completely blocks root hair formation (Lee and Cho, 2006; Li et al., 2009; Won et al., 2009) whereas an activator version of AXR3/IAA17 (ProE7:VP16-IAA17mImII) greatly enhanced root hair growth (Li et al., 2009). These data indicate that TIR1- and Aux/IAA-modulated nuclear transcription is the primary regulatory point for root hair growth, leading to the conclusion that auxin levels in the nucleus could be important for root hair growth. Interestingly, among seven different PINs tested, the short-looped PIN5 and PIN8 were intracellularly localized, while others predominantly localized in the plasma membrane (Figs. 3 and 4). In particular, immunogold-labeled PIN5-GFP was found in the ER around the nucleus (Mravec et al., 2009), suggesting that PIN5 might supply auxin to the nucleus so as to enhance transcriptional activity for root hair growth (Supplemental Fig. S4). Other plasma-membrane-localized PINs are thought to transport auxin from the cytosol to the apoplast, leading to root hair inhibition. On the other hand, PIN8, localized in both the plasma membrane and internal compartments, might catalyze auxin sequestration into certain intracellular vesicles or auxin export from the cytosol, all of which would reduce the availability of auxin for the transcription of root hair morphogenetic genes and thus inhibit root hair growth. In summary, the different subcellular localization of PINs and their different effects on root hair growth inspire a model illustrating specific subcellular roles for individual PINs.
Figure 12.
TIR1-mediated nuclear events are implicated in root hair growth. A, Representative root images of control (Cont; ProE7:GFP) and root-hair-specific TIR1ox (ProE7:TIR1) transformants. Bar = 100 μm for both. B, Root hair lengths of control and TIR1ox plants. The root hair length value of TIR1ox is a mean of values from five independent homozygous lines (Supplemental Fig. S3). Data represent means ± se (n = 200 for control and 1,000 for TIR1ox).
In addition to root hair growth, the root hair cell provides another assay parameter for PIN activity: root hair epidermal cell elongation. Auxin (IAA) concentrations higher than 10−10 M inhibit Arabidopsis root growth by suppressing cell elongation (Evans et al., 1994). The Cholodny-Went model and recent experiments with auxin-sensitive promoter:reporter systems during root gravitropism (Ottenschläger et al., 2003) further support the observation that high endogenous auxin concentrations inhibit root cell elongation. Based on these previous studies, we thought that PINox proteins, by lowering cellular auxin levels, could enhance the elongation of root epidermal cells. Root-hair-specific expression of PINs considerably increased root growth, and this seems to be at least partly due to the enhancement of cell elongation (Fig. 2). This result suggests several points. First, the elongation of certain epidermal cell files (in this study, root hair cell files) can affect overall root elongation. Second, lowering endogenous auxin levels enhances root cell elongation. Third, in the root hair cell, auxin shows dual and opposing functions in epidermal cell elongation and root hair elongation: high auxin concentrations inhibit root hair epidermal cell elongation but simultaneously enhance hair tip growth.
Long-Looped PINs and PIN8 Show Differential Auxin-Exporting Activities in the Root Hair Cell
The expression of plasma-membrane-localized long-looped PINs (PIN1–PIN4 and PIN7) and PIN8 in the root hair cell inhibited root hair growth but differentially. These PINs can be classified into three groups depending on their strength in root hair inhibition: very strong (PIN2), strong (PIN1, PIN3, and PIN8), and weak (PIN4 and PIN7; Fig. 1C). In this study, we expressed these PIN proteins using the same root-hair-specific EXPA7 promoter (Fig. 3). In addition, we took statistical analyses for the root hair measurement with multiple independent transgenic lines for each PINox construct. Unless we suppose the existence of elusive PIN-specific regulators in the root hair cell, we consider that this differential PIN-mediated root hair inhibition basically results from their different auxin efflux activities due to different molecular properties.
We still do not know how auxin is conveyed through the PIN protein and which residues in the protein are critical for the auxin-transporting catalytic activity. The central loop domain of PINs seems to be the phosphorylation target for their proper trafficking to the plasma membrane (Michniewicz et al., 2007), and the transmembrane domains may be responsible for auxin-transporting activity. When only the transmembrane domains are aligned, PIN1 and PIN2 share 70% to 80% amino acid identity with each other and with other long-looped PINs, and PIN3, PIN4, and PIN7 show over 90% identity, forming a tight cluster (Supplemental Fig. S1, A and B). Conversely, PIN5 and PIN8 show 40% to 60% amino acid identity with other PINs. Alignment of the central loop regions also revealed a similar phylogenetic relationship between PINs (Supplemental Fig. S1C). The strength of PIN-mediated root hair inhibition appears to correlate somewhat with this phylogenetic relationship, as the PIN3 paralog group showed generally weaker root hair inhibition than did PIN1 and PIN2. However, it is notable that, in spite of the high similarity of their primary structures, PIN4 induced a much weaker root hair inhibition on average than did PIN3. This indicates that only a small number of different amino acid residues (about 10% difference in the amino acid sequences, for example, between PIN3 and PIN4) can make a big difference in auxin-transporting activity, which is in contrast to the small activity difference between PIN1 and PIN3 despite their relatively greater sequence divergence. In the case of PIN3 and PIN4, our results imply that some critical residues for auxin-transporting activity could be included in that small number of nonidentical amino acid residues.
The different responses of the PINox lines to exogenous auxin are also indicative of differential auxin efflux activity. In particular, PIN2ox lines showed a very strong resistance to exogenous auxin-mediated root hair restoration (Fig. 6). The strong auxin-transporting activity by PIN2 is also reflected in hair restoration kinetics after treatment with different auxin concentrations, in which PIN2 very effectively facilitates auxin efflux at low concentrations, between 1 and 20 nm (Fig. 7). The root hair restoration responses of PINox lines to BFA also showed diversity among PINs. Root hair growth was restored in most PINox lines in response to BFA treatment, most likely due to BFA-mediated inhibition of PIN trafficking to the plasma membrane (Paciorek et al., 2005) and, thus, because of reduced auxin efflux and increased cellular auxin levels (Fig. 9; Supplemental Table S1). However, PIN2ox and PIN8ox lines showed resistance to the BFA effect, where PIN2ox lines showed much stronger resistance. This could be because of the different sensitivities of PIN trafficking in response to BFA. BFA caused the formation of internal compartments for all PINs, except PIN5, in the root hair cell (Fig. 10), indicating that trafficking of these PINs is basically affected by the BFA-sensitive trafficking machinery. However, it is known that while PIN1 trafficking is mediated by BFA-sensitive ARF GEF, PIN2 trafficking is somewhat resistant to BFA (Geldner et al., 2003; Kleine-Vehn and Friml, 2008). We think that the poor root hair restoration of PIN2ox transformants in response to BFA was due to this resistance. The resistance of PIN8ox lines to BFA could be either because the trafficking pathway of PIN8 to the plasma membrane is resistant to BFA or because the internally localized PIN8 still allows cellular auxin efflux in an unknown manner. In contrast to BFA, NPA largely caused a significant restoration of root hair growth for most PINox lines (Fig. 8), suggesting that NPA is a general inhibitor of auxin efflux transporter PINs.
Short-Looped PINs Show Different Subcellular Localization and Different Auxin-Transporting Activities
Two short-looped PINs, PIN5 and PIN8, localize in internal compartments that are probably ER-related organelles (Mravec et al., 2009). However, in this study, we propose that PIN8, in addition to its internal localization, also localizes in the plasma membrane. Several lines of evidence support this notion. First, PIN8 expression in the tobacco cell decreased auxin retention in the cell. This result indicates that PIN8 catalyzes auxin efflux from the cell. A parsimonious explanation for this is that PIN8 facilitates auxin efflux in the plasma membrane, although it is also possible that PIN8 promotes auxin accumulation into secretory vesicles, which is reminiscent of neurotransmitter secretion (Baluška et al., 2003; Supplemental Fig. S4). Second, the PIN8-GFP signal overlaps with FM4-64-labeled plasma membrane in Arabidopsis root hair and tobacco BY-2 cells, while the PIN5-GFP signal does not (Figs. 3B and 4). Third, as observed for other well-defined plasma-membrane-localized PINs, BFA caused the formation of PIN8-containing internal compartments both in Arabidopsis root hair and tobacco cells, which were colabeled with the endocytic tracer FM4-64 (Fig. 10). BFA blocks the recycling pathway of PINs, albeit leaving the endocytosis process operational, and this causes internal compartment formation of endocytosed PINs (Geldner et al., 2001, 2003). Therefore, PIN8-labeled BFA compartments suggest that the PIN8 molecules originate from the plasma membrane. This contrasts with the case for PIN5, which was predominantly internally localized and did not form BFA compartments (Fig. 10). Fourth, PIN8 targets to the cell plate in dividing cells. Plasma-membrane-localized PINs were known to traffic to the cell plate (Dhonukshe et al., 2006 for PIN1 and 2; Fig. 11 for PIN3). While ER-localized PIN5-GFP did not target to the cell plate, PIN8-GFP clearly targeted to the cell plate, which completely overlapped with the plasma-membrane-originated FM4-64 tracer (Fig. 11).
As briefly mentioned earlier, the dual localization of PIN8 in both the plasma membrane and the endomembrane system opens the possibility of several PIN8-mediated auxin transport processes, although they are currently speculative (Supplemental Fig. S4). One is cellular auxin efflux, either in the plasma membrane or via secretory vesicles, which coincides with PIN8-mediated auxin efflux in tobacco cells (Fig. 5). The other possibility is relevant to the availability of intracellular auxin for its signaling. The internally localized PIN8 might sequester auxin in certain storage compartments, which may result in reduced auxin concentrations in the nucleus and, thus, reduced auxin signaling. The inhibition of auxin signaling-dependent root hair growth by PIN8ox supports both ideas of the intracellular auxin transport and the plasma-membrane-level auxin transport.
Root hair expression of PIN5 revealed internal localization of the protein, which, unlike other PINs, did not shorten but rather enhanced root hair growth (Fig. 1C; Supplemental Fig. S2, E–G). This result may indicate that PIN5 facilitates cellular auxin influx. However, because PIN5 is located in internal compartments, at least in the ER, it is not likely to facilitate auxin import from the apoplast to the cytosol. Therefore, a plausible mechanism for PIN5 in promoting root hair elongation, which should involve the activation of root hair morphogenetic genes, is that PIN5 imports auxin into the nucleus through the nearby ER, thereby enhancing nuclear auxin availability (Supplemental Fig. S4). The insignificant cellular auxin transport activity of PIN5 in tobacco suspension cells (Fig. 5) also supports the idea that PIN5-mediated auxin transport does not occur intercellularly but is probably intracellular.
Previous studies and this study suggest that different PIN members have differential biological roles that are specified by differential polarity in the plasma membrane, differential organellar localization, and differential catalytic activity. The differential polarity of PIN localization in the plasma membrane contributes to the directional flow of auxin, local auxin gradient formation, and organogenesis and tropic movements (Kleine-Vehn and Friml, 2008). Together with the recent PIN5 study (Mravec et al., 2009), our study indicates that internally localized PINs have distinctive functions, such as internal transport of auxin conjugates (Mravec et al., 2009) and probably an increase (for PIN5) or decrease (for PIN8) of nuclear auxin availability for auxin signaling and auxin-mediated root hair growth (this study). On the other hand, the biological significance of differential auxin-transporting activities, which was demonstrated here, remains to be explored. Hence, we are tempted to speculate that different PINs with differential catalytic strengths, together with differential subcellular polarities, may have been selected in a tissue-specific manner so as to fine-tune local auxin concentrations for precise auxin responses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana), Columbia ecotype, was used as the wild-type plant in this study. All seeds were grown on agarose plates containing 4.3 g/L Murashige and Skoog (MS) nutrient mix (Sigma-Aldrich), 1% Suc, 0.5 g/L MES (pH 5.7), KOH, and 0.8% agarose. All seeds were cold treated (4°C) for 3 d and germinated at 23°C under 16-h-light/8-h-dark photoperiods. Transformed plants were selected on hygromycin-containing plates (10 μg/mL). For all pharmacological experiments, 3-d-old seedlings of homozygous transformants were transferred to new plates and grown for an additional day, after which root hairs were observed.
Transgene Constructs
The Arabidopsis EXPA7 promoter ProE7 (Cho and Cosgrove, 2002) was used for cloning all PIN transgenes. The binary vector pCAMBIA1300-NOS was used as the cloning vector. ProE7:PIN3-GFP, ProE7:YFP, ProE7:GFP, ProE7:AUX1-YFP, and ProE7:PGP4-YFP were as previously described (Lee and Cho, 2006; Cho et al., 2007a). The ProE7:PIN1-GFP, ProE7:PIN2-GFP, ProE7:PIN4-GFP, and ProE7:PIN7-GFP constructs were generated by connecting the ProE7 promoter to PIN-GFP coding regions from ProPIN1:PIN1-GFP (Benková et al., 2003), ProPIN2:PIN2-GFP (Xu and Scheres, 2006), ProPIN4:PIN4-GFP (Vieten et al., 2005), and ProPIN7:PIN7-GFP (Blilou et al., 2005) in the binary vector pCAMBIA1300-NOS (Hyg+). ProE7:PIN2-GFP was as described by Cho et al. (2007a), and ProE7:PIN4-GFP was generated by replacing the PIN2 region with the genomic PIN4 fragment in the same vector. For ProE7:PIN1-GFP, the PIN1-GFP fragment was generated by PCR of genomic DNA from the ProPIN1:PIN1-GFP transformant using the following primer sets: 5′-TCTCCGTCGACCAAAAGATGATTACGGC-3′ (containing the SalI site) and 5′-TCACGGGATCCATGATGTATAGTTCATCC-3′ (containing the BamHI site) for the first fragment of PIN1-GFP, 5′-CATTATCAACAAAATACTCCAATTGGCG-3′ (containing the BamHI site) and 5′-TTTCAGAGCTCATCATCCCACAAAAGAA-3′ (containing the SacI site) for the second fragment of PIN1-GFP, and 5′-TCGCCCCCGGGGTGAGCAAGGGCG-3′ (containing the XmaI site) and 5′-CGGCCCCCGGGCTTGTACAGCTCGTCC-3′ (containing the XmaI site) for the GFP region. ProE7:PIN7-GFP was similarly generated by PCR of genomic DNA from the ProPIN7:PIN7-GFP transformant using the following primer sets: 5′-ACTTTGTCGACTCCGGCGAACAACAA-3′ (containing the SalI site) and 5′-CCGTTGGATCCCCAAACAAACATATG-3′ (containing a BamHI site) for the first fragment of PIN7-GFP, and 5′-CATTATCAACAAAATACTCCAATTGGCG-3′ (containing the BamHI site) and 5′-TCTCTGGTACCTCTTCAGCCAAGCAG-3′ (containing the KpnI site) for the second fragment. The third fragment of ProE7:PIN7-GFP was cloned by first cutting out a part of PIN7 along with the whole GFP region with BamHI from ProPIN7:PIN7-GFP and inserting it between the first and second fragments of the PIN7 gene of the BamHI-digested vector. The PIN5 coding region (−6 to +1,744 relative to ATG) of ProE7:PIN5 was generated by PCR of Arabidopsis genomic DNA using the following primer sets: 5′-ATTTTCTCGAGAAAACCATGATAAATTGTG-3′ (containing the XhoI site) and 5′-TAAAATCTAGACTTACGCTGTGCTTAGAAC-3′ (containing the XbaI site). The PIN8 coding region (−6 to +1,780 relative to ATG) of ProE7:PIN8 was generated by PCR of Arabidopsis genomic DNA using the following primer sets: 5′-TACAACTCGAGAAAAACATGATCTCCTGG-3′ (containing the XhoI site) and 5′-AACATTCTAGATTCATAGGTCCAATAGAAA-3′ (containing the XbaI site). The PIN5 coding region of ProE7:PIN5-GFP1 was generated by PCR of Arabidopsis genomic DNA using the following primer sets: 5′-ATTTTCTCGAGAAAACCATGATAAATTGTG-3′ (containing the XhoI site) and 5′-AGGAACCCGGGCTCTCCCACAACCAC-3′ (containing the XmaI site) for the first fragment, and 5′-TTGTGCCTAGGAAGTCGTTCCTTGAGGTC-3′ (containing the AvrII site) and 5′-TAAAATCTAGACTTACGCTGTGCTTAGAAC-3′ (containing the XbaI site) for the second fragment. The PIN5 coding region of ProE7:PIN5-GFP2 was generated by PCR of Arabidopsis genomic DNA using the following primer sets: 5′-ATTTTCTCGAGAAAACCATGATAAATTGTG-3′ (containing the XhoI site) and 5′-TATATCCCGGGGTATTCACTGTTATCTC-3′ (containing the XmaI site) for the first fragment, and 5′-ACAGTCCTAGGATGCATATATACGCAGG-3′ (containing the AvrII site) and 5′-TAAAATCTAGACTTACGCTGTGCTTAGAAC-3′ (containing the XbaI site) for the second fragment. The PIN8 coding region of ProE7:PIN8-GFP was generated by PCR of Arabidopsis genomic DNA using the following primer sets: 5′-TACAACTCGAGAAAAACATGATCTCCTGG-3′ (containing XhoI site) and 5′-GATCTCCCGGGCACTATTGCTACTTCTT-3′ (containing the XmaI site) for the first fragment and 5′-TAGCACCTAGGAGAACAAGATCTGTTGG-3′ (containing the AvrII site) and 5′-AACATTCTAGATTCATAGGTCCAATAGAAA-3′ (containing the XbaI site) for the second fragment. For cloning the GFP region in ProE7:PIN5-GFP1, ProE7:PIN5-GFP2, and ProE7:PIN8-GFP, the GFP fragment was generated using the primers 5′-TCGCCCCCGGGGTGAGCAAGGGCG-3′ (containing an XmaI site) and 5′-CGGCCCCTAGGCTTGTACAGCTCGTCC-3′ (containing an AvrII site). For ProTA:PIN5-GFP1 and ProTA:PIN8-GFP constructs, the PIN-GFP coding regions were inserted into XhoI-SpeI sites in the modified ProTA-7001 vector (Aoyama and Chua, 1997) after PCR amplifying them from their ProE7:PIN-GFP versions using the following primer sets: 5′-ATTTTCTCGAGAAAACCATGATAAATTGTG-3′ (containing the XhoI site) and 5′-TAAAATCTAGACTTACGCTGTGCTTAGAAC-3′ (containing the XbaI site) for PIN5-GFP1, and 5′-TACAACTCGAGAAAAACATGATCTCCTGG-3′ (containing the XhoI site) and 5′-AACATTCTAGATTCATAGGTCCAATAGAAA-3′ (containing the XbaI site) for PIN8-GFP. The ProE7:TIR1 (TIR1ox) construct was designed on the ProE7-containing pCAMBIA1300-NOS vector. The genomic fragment of TIR1 was amplified by PCR using primers, 5′-GAGATGTCGACGTTATTTGGCCGCAATG-3′ (containing the SalI site) and 5′-TCGTTCCCGGGTCTTATAATCCGTTAGTA-3′ (containing the XmaI site), and this PCR product was inserted between SalI and XmaI sites of the vector. All constructs were confirmed by nucleotide sequencing. The constructs were transformed into Arabidopsis (ecotype Columbia) or tobacco (Nicotiana tabacum) BY-2 cells by Agrobacterium tumefaciens (strain C58C1)-mediated infiltration (Bechtold and Pelletier, 1998). Transgene insertion in Arabidopsis transformants was checked by PCR analysis of the genomic DNA.
Measurement of Root Hair and Root Hair Cell Length
Root hair lengths were measured as described by Lee and Cho (2008) with modifications. For estimation of root hair length, digital photographs of roots were taken under a stereomicroscope (Leica MZ FLIII) at 40× to 50× magnifications. Hairs in the hair maturation region (approximately 0.78 mm from the tip) were counted. To measure hair length, four to five consecutive hairs protruding perpendicularly from each side of the root, for a total of eight to 10 hairs from both sides of the root, were measured. Short hair bulges were considered arrested at the early bulge stage and were not counted. Root epidermal cell length was estimated by measuring the distance (gap) between two consecutive root hairs or bulges in the same root hair file along the long axis of the root. Fifteen to 20 cells were observed per root.
Reporter Gene Expression and Microscopy Observation
Fluorescence from GFP (green) and FM4-64 (red) were observed using an LSM 510 confocal laser scanning microscope (Carl Zeiss). Green and red fluorescence were detected using 488/505- to 530-nm and 587/615-nm excitation/emission filter sets, respectively. Localization of transporter reporter (PIN-GFP) fusion proteins was observed from 3- or 4-d-old seedlings. For observation of subcellular localization of PIN-GFP after BFA treatment, seedlings were incubated in half-strength liquid MS medium for the indicated time periods. The control liquid medium included the same amount of the solvent dimethylsulfoxide, which was used to dissolve BFA. The PIN5-GFP1 and PIN8-GFP localization in transgenic tobacco BY-2 cells was also observed after 24-h treatment of cells with 10 μm Dex. Unless specially mentioned, Arabidopsis roots and tobacco BY-2 cells were treated with BFA (50 μm) for 2 h and with FM4-64 (2 μm) for 3 min (FM4-64 alone) or for 15 min (when cotreated with BFA) before observation.
Culture and Transformation of Tobacco BY-2 Cells
Tobacco BY-2 cells (Nagata et al., 1992) were cultured in darkness at 23°C with shaking (150 rpm). The liquid culture medium included 3% (w/v) Suc, 4.3 g/L MS salts, 100 mg/L inositol, 1 mg/L thiamine, 0.2 mg/L 2,4-D, and 200 mg/L KH2PO4, pH 5.8. The suspension cells were subcultured weekly. Stock BY-2 calli were maintained on solid medium with 0.8% (w/v) agar and subcultured monthly. Transgenic cells and calli were maintained on the same medium supplemented with 150 mg/L hygromycin. For transformation of tobacco BY-2 cells, 50 mL of a 3-d-old culture was cocultivated with 5 mL of A. tumefaciens harboring ProTA:PIN5-GFP1 and ProTA:PIN8-GFP in a petri dish in the dark for 3 d at 23°C. Inoculated cells were washed three times and transferred onto solid medium supplemented with 150 mg/L hygromycin. After 5 weeks, positive calli were selected and transferred into new selection media. Individual calli were then maintained either on solid media or in liquid media according to requirements.
Auxin Retention Assay in Tobacco BY-2 Cells
The auxin retention assay was performed as described by Delbarre et al. (1996) with modifications. After incubation with or without 10 μm Dex for 24 h, the transgenic BY-2 cells in the exponential growth phase were filtered, and 1 to 1.5 g of cell cake was resuspended and equilibrated in uptake buffer (20 mm MES, 40 mm Suc, and 0.5 mm CaSO4, pH adjusted to 5.7 by KOH) for 45 min with continuous orbital shaking. Equilibrated cells were collected by filtration, resuspended in fresh uptake buffer, and incubated in the orbital shaker for 1.5 h in darkness at 23°C. [3H]NAA was mixed with 5 mL of the cell suspension for a final concentration of 2 nm [3H]NAA, and 0.5-mL aliquots were rapidly filtered by vacuum on GF/C glass fiber filters (Whatman) at different time intervals (0, 5, 10, 15, 20, and 25 min). Cell cakes were washed in 3 mL ice-cold distilled water twice, transferred to scintillation vials, and extracted with 0.5 mL ethanol for 30 min, and radioactivity was determined by liquid scintillation counting. Four independent experiments with two replicates for each experiment were conducted.
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At1G12560 (EXPA7), At1G73590 (PIN1), At5G57090 (PIN2), At1G70940 (PIN3), At2G01420 (PIN4), At5G16530 (PIN5), At1G77110 (PIN6), At1G23080 (PIN7), At5G15100 (PIN8), At2G47000 (PGP4), and At2G38120 (AUX1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Predicted membrane-spanning helices and phylogenies of Arabidopsis PIN proteins.
Supplemental Figure S2. Differential effects of root hair-specific expression of PINs on root hair growth.
Supplemental Figure S3. Effect of root hair-specific expression of TIR1 on root hair growth.
Supplemental Figure S4. A model illustrating different subcellular PIN localizations and physiological effects in the root hair assay system.
Supplemental Table S1. Relative root hair restoration of PINox lines in response to BFA.
Supplementary Material
Acknowledgments
We thank Zee-Won Lee at the Korea Basic Science Institute for help with microscopy imaging analyses.
References
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 606–612 [DOI] [PubMed] [Google Scholar]
- Baluška F, Šamaj J, Menzel D. (2003) Polar transport of auxin: carrier-mediated flux across the plasma membrane or neurotransmitter-like secretion? Trends Cell Biol 13: 282–285 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Martinez-Zapater JM, Salinas J, , Arabidopsis Protocols. Humana, Totowa, NJ, pp 259–266 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Crosnier M, Carraro N, Traas J, Satiat-Jeunemaitre B. (2006) The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. (2002) The regulation of Arabidopsis root hair initiation and expansin gene expression. Plant Cell 14: 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Lee OR, Ganguly A, Cho HT. (2007b) Auxin-signaling: short and long. J Plant Biol 50: 79–89 [Google Scholar]
- Cho M, Lee SH, Cho HT. (2007a) P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J. (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Baluška F, Schlicht M, Hlavacka A, Šamaj J, Friml J, Gadella TWJ. (2006) Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev Cell 10: 137–150 [DOI] [PubMed] [Google Scholar]
- Evans ML, Ishikawa H, Estelle M. (1994) Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194: 215–222 [Google Scholar]
- Friml J, Benková E, Blilou I, Wiśniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT. (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Friml J. (2008) Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho HT. (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18: 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Cho HT. (2008) Auxin and root hair morphogenesis. Emons AMC, Ketelaar T , Root Hairs (Plant Cell Monographs Vol 12). Springer, Berlin, pp 45–64 [Google Scholar]
- Leyser HM, Pickett FB, Dharmasiri S, Estelle M. (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Li H, Cheng Y, Murphy A, Hagen G, Guilfoyle TJ. (2009) Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol 149: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Mravec J, Kubeš M, Bielach A, Gaykova V, Petrášek J, Skůpa P, Chand S, Benková E, Zažímalová E, Friml J. (2008) Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135: 3345–3354 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof Y, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1142 [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. (1992) Tobacco BY2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. Int Rev Cytol 13: 1–30 [Google Scholar]
- Okada K, Shimura Y. (1994) Modulation of root growth by physical stimuli. Meyerowitz EM, Somerville CR, , Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 665–684 [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazimalová E, Ruthardt N, Petrásek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Juergens G, Geldner N, et al. (2005) Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wiśniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Wiśniewska J, Xu J, Seifertová D, Brewer PB, Růžička K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. (2009) cis-Element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B. (2006) Polar auxin transport and patterning: grow with the flow. Genes Dev 20: 922–926 [DOI] [PubMed] [Google Scholar]
- Yang H, Murphy AS. (2009) Functional expression and characterization of Arabidopsis ABCB, AUX1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J 59: 179–191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.