Abstract
The Arabidopsis (Arabidopsis thaliana) fatty acid biosynthesis1 (fab1) mutant grows as well as wild type at 22°C, but after transfer to 2°C fab1 plants cannot maintain photosynthetic function and die after 5 to 7 weeks at 2°C. A fab1 suppressor line, S7, was isolated in a screen that identified mutants that remained alive after 16 weeks at 2°C and were able to flower and produce seed after return to 22°C. Relative to wild type, S7 plants had reduced levels of 16:3 fatty acid in leaf galactolipids, indicating reduced synthesis of chloroplast glycerolipids by the prokaryotic pathway of lipid metabolism. The suppressor mutation was identified, by map-based and candidate-gene approaches, as a hypomorphic allele of lysophosphatidic acid acyltransferase1 (lpat1), lpat1-3. LPAT1 encodes the enzyme that catalyzes the second reaction in the prokaryotic pathway. Several lines of evidence indicate that damage and death of fab1 plants at 2°C may be a result of the increased proportion of phosphatidylglycerol (PG) in fab1 that are high-melting-point molecular species (containing only 16:0, 18:0, and 16:1,Δ3-trans fatty acids). Consistent with this proposal, the lpat1-3 mutation strongly affects the fatty acid composition of PG. The proportion of high-melting-point molecular species in PG is reduced from 48.2% in fab1 to 10.7% in fab1 lpat1-3 (S7), a value close to the 7.6% found in wild type.
There are two distinct pathways in higher plants for the synthesis of membrane glycerolipids. Both pathways are initiated by the synthesis of 16:0-acyl carrier protein (16:0-ACP) from acetyl-CoA by plastid acetyl-CoA carboxylase and enzymes of the fatty acid synthase. 16:0-ACP may be elongated by one additional cycle of the fatty acid synthase and then desaturated by the stromal 18:0-ACP desaturase (Lindqvist et al., 1996), so that 16:0-ACP and 18:1-ACP are the primary products of plastid fatty acid synthesis (Ohlrogge and Browse, 1995). The prokaryotic pathway located in the plastid envelope begins with the synthesis of 18:1-lysophosphatidic acid by acyl-ACP:glycerol-3-P acyltransferase. Then 16:0-ACP:lysophosphatidic acid acyltransferase (LPAT) completes the synthesis of phosphatidic acid (PA) that is a key intermediate in glycerolipid synthesis. The PA in the prokaryotic pathway is used for the synthesis of phosphatidylglycerol (PG) and, in some plants, as a precursor via diacylglycerol for the synthesis of monogalactosyldiacylglycerol (MGD), digalactosyldiacylglycerol (DGD), and sulfoquinovosyldiacylglycerol (SQD), the four major glycerolipids of the photosynthetic thylakoid membranes. The alternative eukaryotic pathway begins with the hydrolysis of acyl-ACPs and the synthesis of 16:0-CoA and 18:1-CoA in the plastid envelope for export to the endoplasmic reticulum, where they are used for the synthesis of phosphatidylcholine (PC) and other phospholipids characteristic of the various extrachloroplast membranes of the cell. In addition, the diacylglycerol moiety of PC is returned to the chloroplast envelope—possibly in the form of PA (Benning, 2009)—where it enters the diacylglycerol pool and contributes to the synthesis of all the thylakoid glycerolipids, except PG (Browse et al., 1986; Benning, 2009). On both the prokaryotic and eukaryotic pathways, 16:0 and 18:1 are incorporated into the membrane glycerolipids before membrane-bound desaturases in the chloroplast and endoplasmic reticulum desaturate the acyl groups further to produce the polyunsaturated lipids typical of the different membrane systems of the cell. In many species of higher plants, PG is the only product of the prokaryotic pathway and the remaining chloroplast lipids are synthesized entirely by the eukaryotic pathway. However, in Arabidopsis (Arabidopsis thaliana) and some other species, the two pathways contribute about equally to the synthesis of MGD, DGD, and SQD (Browse et al., 1986; Wallis and Browse, 2010).
Investigations of Arabidopsis mutants deficient in enzymes of these two lipid synthesis pathways have provided considerable information about the relationship between membrane lipid composition and biological function. For example, a number of mutations that reduce the level of membrane unsaturation compromise plant growth at low temperatures (<10°C; Hugly and Somerville, 1992; Wu and Browse, 1995; Murakami et al., 2000; Routaboul et al., 2000). It has been proposed that damage occurring at these temperatures in chilling-sensitive plant species is caused by an Lα to Lβ lipid phase (liquid crystalline to gel phase) transition in cellular membranes (Nishida and Murata, 1996), and, in particular, that molecular species of PG containing only 16:0, 18:0, and 16:1,Δ3-trans (high-melting-point PG) confer chilling sensitivity on plants (Murata, 1983; Murata and Yamaya, 1984). PG purified from chilling-sensitive plants (containing 33%–71% high-melting-point molecular species) entered the gel phase at temperatures approximately 20°C higher than PG from chilling-resistant plants (6%–20% high-melting-point molecular species; Murata and Yamaya, 1984). A role for membrane unsaturation in maintaining plant growth and photosynthesis at low temperatures is also supported by experiments in which transgenic expression of fatty acid desaturases in chilling-sensitive plant species resulted in increased survival of plants at low temperatures (Kodama et al., 1994; Ishizaki-Nishizawa et al., 1996; Murakami et al., 2000).
One of the Arabidopsis mutants, fatty acid biosynthesis1 (fab1), contains increased levels of the saturated fatty acid, 16:0, as a result of a mutation in the KAS2 gene, which encodes the first step in elongation of 16:0 to 18:0 during fatty acid synthesis (Wu et al., 1994; Carlsson et al., 2002). Although fab1 plants do not exhibit a classic chilling-sensitive phenotype (Wu and Browse, 1995), they are damaged and killed by long-term exposure to low temperatures. During the first 7 to 10 d after transfer from 22°C to 2°C, growth and photosynthetic characteristics of fab1 plants were similar to wild type. However, between 10 and 28 d at 2°C the mutants suffered almost complete loss of photosynthetic function and destruction of chloroplasts, but not other organelles, within leaf cells, and then plants died after 4 to 6 weeks in the cold (Wu et al., 1997). This collapse of photosynthesis and breakdown of chloroplasts in fab1 plants at 2°C suggests that further investigation of this mutant will lead to a better understanding of the relationship between thylakoid lipid structure and photosynthetic function.
We initiated a suppressor screen for mutations that rescue fab1 from death at low temperatures (Barkan et al., 2006). One of the mutants isolated in this screen, S7, showed a reduced level of 16:3 in total leaf lipids, and this altered fatty acid composition cosegregated with the suppressive phenotype as a single, recessive mutation. The low 16:3 in S7 plants indicated a possible block in synthesis of thylakoid lipids by the prokaryotic pathway (Kunst et al., 1988). Reductions in lipid synthesis by the prokaryotic pathway have previously been observed in the gly1 mutant, which is deficient in glycerol-3-P dehydrogenase activity (Miquel et al., 1998), and the act1 mutant, which is deficient in activity of the chloroplast acyl-ACP:glycerol-3-P acyltransferase (Kunst et al., 1988). However, test crosses of S7 with gly1 and act1 produced F1 progeny with levels of leaf 16:3 similar to wild type, indicating that the lipid mutation in S7 is not allelic to either of these previously characterized mutants (Barkan et al., 2006). Here we describe identification of the S7 suppressor, by map-based cloning, as a new, hypomorphic allele of LPAT1 that encodes acyl-ACP:LPAT, the second enzyme in the prokaryotic pathway. Our discovery provides new insight on the cause of the fab1 low-temperature phenotype. In addition, because the lpat1-1 and lpat1-2 null alleles are lethal (Kim and Huang, 2004; Yu et al., 2004), our newly identified allele provides a useful tool to investigate this step in the biochemistry of thylakoid lipid synthesis.
RESULTS
Changes in Leaf Lipids of S7 Plants
Because the overall fatty acid composition of leaf lipids from S7 plants indicated a partial block of the prokaryotic pathway, we separated individual lipids from leaf extracts and analyzed their fatty acid compositions by gas chromatography (GC). The results of this analysis, shown in Table I, confirm that the S7 suppressor line has a reduced flux through the prokaryotic pathway since 16:3, a fatty acid that is confined to the sn-2 position of lipids synthesized by this pathway (Browse et al., 1986), is present in much lower proportions on MGD and DGD of S7 plants. The major chloroplast lipid, MGD, in S7 contained 13.3% 16:3 compared to 33.3% in the parental fab1 line, suggesting that MGD synthesis via the prokaryotic pathway is reduced by approximately 60% in S7. This deficiency appears to be compensated for by increased MGD synthesis by the eukaryotic pathway, since there was a very much smaller drop in total MGD as a percentage of leaf lipids (Table I).
Table I. Fatty acid compositions of leaf lipids from wild-type and mutant Arabidopsis grown at 22°C.
Data are from one of three separate experiments that gave similar results. WT, Wild type; PE, phosphatidylethanolamine; PI, phosphatidylinositol.
| Lipid Class | Genotype | % of Total Polar Lipids | Fatty Acid Composition |
||||||
| 16:0 | 16:1 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 | |||
| % | |||||||||
| MGD | WT | 32.6 | 3.3 | 1.7 | 30.7 | –a | 1.4 | 3.4 | 57.2 |
| fab1 | 35.1 | 5.2 | 3.2 | 33.3 | – | 2.5 | 2.0 | 51.1 | |
| S7 | 30.1 | 9.6 | 3.0 | 13.3 | 1.3 | 3.2 | 2.2 | 65.8 | |
| DGD | WT | 14.2 | 16.8 | – | 2.9 | 2.7 | 1.1 | 4.1 | 71.8 |
| fab1 | 10.4 | 31.4 | 1.2 | 3.2 | 2.1 | 3.5 | 3.4 | 54.5 | |
| S7 | 12.2 | 32.7 | 2.9 | – | 2.2 | 2.8 | 1.7 | 57.2 | |
| PG | WT | 12.9 | 33.0 | 19.8 | – | 2.1 | 5.8 | 12.1 | 27.2 |
| fab1 | 14.8 | 48.2 | 18.4 | – | 1.5 | 5.1 | 7.5 | 18.4 | |
| S7 | 11.5 | 34.5 | 21.3 | – | 1.4 | 5.2 | 11.0 | 25.3 | |
| SQD | WT | 3.4 | 52.7 | 1.4 | – | 5.6 | 3.0 | 5.6 | 31.6 |
| fab1 | 3.5 | 58.3 | 2.0 | 2.9 | 4.2 | 1.8 | 5.8 | 24.4 | |
| S7 | 3.6 | 55.6 | 3.2 | – | 8.2 | 2.4 | 3.0 | 24.9 | |
| PC | WT | 18.7 | 26.6 | – | – | 3.2 | 6.7 | 32.7 | 30.1 |
| fab1 | 18.3 | 34.0 | 2.6 | – | 2.6 | 6.1 | 24.4 | 29.8 | |
| S7 | 20.7 | 38.5 | 1.7 | – | 3.4 | 7.3 | 26.2 | 24.9 | |
| PE | WT | 12.9 | 30.9 | – | – | 2.7 | 4.0 | 34.4 | 28.0 |
| fab1 | 12.9 | 34.2 | 1.1 | – | 1.8 | 5.6 | 26.4 | 29.8 | |
| S7 | 14.7 | 38.1 | – | – | 2.3 | 6.3 | 29.5 | 23.7 | |
| PI | WT | 5.3 | 52.3 | – | – | 5.3 | 2.5 | 19.3 | 20.6 |
| fab1 | 5.0 | 56.1 | – | – | 1.9 | 1.7 | 16.6 | 23.7 | |
| S7 | 7.1 | 55.5 | 1.0 | – | 4.1 | 4.7 | 18.0 | 16.7 | |
<1%.
Consistent with previous results (Wu et al., 1994; Barkan et al., 2006), all of the lipids in fab1 leaves had increased 16:0 compared with wild type. Three of the major membrane lipids, MGD, DGD, and PC, from S7 plants all showed a further increase in 16:0 relative to fab1. The only substantial decrease in 16:0 was seen in PG; for this lipid 16:0 was decreased from 48.2% in fab1 to 34.5% in S7—close to the value for wild-type PG of 33.0%. This change in fatty acid composition of PG is significant because it likely reflects a decrease in high-melting-point species of PG (Roughan, 1985). We have previously proposed that high-melting-point PG contributes to the photosynthetic decline and death of fab1 at low temperature (Wu et al., 1997; Barkan et al., 2006). To test whether S7 plants contain substantially lower levels of high-melting-point PG than fab1, we submitted wild-type, fab1, and S7 leaf lipids for lipidomics analysis by mass spectrometry (Esch et al., 2007). The results obtained (Table II) indicate that high-melting-point molecular species account for 7.6% of PG in wild type and 48.2% in fab1; these values agree with results obtained earlier using HPLC techniques (Wu and Browse, 1995). In S7, high-melting-point molecular species make up only 10.7% of PG. Thus the lipid mutation in S7 provides a molecular species composition in PG that is closer to that of wild type than to that of the fab1 progenitor.
Table II. Molecular species composition of leaf PG from wild-type and mutant Arabidopsis.
Analyses were conducted by collision-induced dissociation time-of-flight mass spectrometry as described under “Materials and Methods.” Data are normalized from the means of five replicates with ses < 10% of the means. WT, Wild type.
| Fragment Mass | Molecular Species | Percentage of Total PG |
||
| WT | fab1 | S7 | ||
| 760.5 | 18:3/16:1 | 40.3 | 21.6 | 37.4 |
| 762.5 | 18:3/16:0 + 18:2/16:1 | 30.0 | 18.5 | 29.2 |
| 764.5 | 18:2/16:0 + 18:1/16:1 | 13.5 | 7.3 | 14.8 |
| 766.5 | 18:1:16:0 | 8.1 | 4.3 | 7.8 |
| – | Other unsaturated species | 0.5 | 0.1 | 0.1 |
| 768.6 | 18:0/16:0 | 0.6 | 0.3 | 0.1 |
| 738.5 | 16:0/16:1 | 3.6 | 26.1 | 6.5 |
| 740.5 | 16:0/16:0 | 3.4 | 21.8 | 4.1 |
| Total high-melting-point species | 7.6% | 48.2% | 10.7% | |
The S7 Line Contains a Mutation in LPAT1
To identify the suppressor locus in S7, we first crossed this line to Landsberg erecta, selfed the F1 progeny, and grew F2 plants. Based on the expected Mendelian segregation, 25% of the F2 would be fab1 homozygotes and 50% of these would be heterozygous for the recessive suppressor mutation. We identified six fab1 plants by GC of leaf fatty acids, grew them to maturity, and collected seed from each plant separately. We chose one of the plants whose progeny segregated for the suppressor phenotype (approximately one-quarter surviving at 2°C) as the parent of an F3 mapping population. A total of 800 F3 seeds were planted and the resulting plants grown at 2°C for 6 weeks. DNA was prepared from leaf samples of surviving plants. Bulk-segregant analysis (Lukowitz et al., 2000), using DNA pooled from 50 individuals, indicated Columbia DNA associated with marker NGA111 that is on the lower arm of chromosome1 close to the fab1 locus. Columbia DNA was also associated with marker NGA1107 on chromosome 4, indicating that the suppressor mutation lay close to this marker. Analysis of DNA from individual plants indicated that the suppressor mutation was approximately 10 centimorgans from the marker. Because NGA1107 is close to the end of chromosome 4, this degree of linkage suggested a map window centered around 15,500 kb on the physical map. The 1,200 kb region around this position contains approximately 350 genes including 12 that have known or putative functions in lipid metabolism (Beisson et al., 2003). One of these, LPAT1 (At4g30580), was a particularly appealing candidate since it encodes the second enzyme of the prokaryotic pathway. Null mutants at this locus, lpat1 (=ats2), are embryo lethal (Kim and Huang, 2004; Yu et al., 2004), but the presence of 13% 16:3 in MGD from S7 leaves would be consistent with a hypomorphic lpat1 allele.
We prepared RNA from wild-type, fab1, and S7 plants and amplified cDNAs of LPAT1 from each. Sequencing showed that the wild-type and fab1 cDNAs corresponded with the sequence in The Arabidopsis Information Resource database (www.arabidopsis.org), while the S7 allele contained a G to A change in the seventh exon of the gene. This change is predicted to encode an Ala-290Thr mutation in the LPAT1 protein (Fig. 1). The Ala-290 residue is located in one of the three domains (D1–D3 in Fig. 1) that are conserved in LPAT proteins from plants, animals, and microbes (Eberhardt et al., 1997), although Phe or Ser are found in place of Ala in some sequences. We designate this new mutant allele lpat1-3.
Figure 1.
Sequence alignment of LPAT1 homologs. The Arabidopsis amino acid sequence was aligned with LPATs from Escherichia coli (PlsC), yeast (Saccharomyces cerevisiae; Slc1p), and human (Hs LPAAT; GenBank AAC51649). The three conserved domains (D1–D3) are shown and the site of the lpat1-3 mutation, A290T, is indicated.
LPAT1 Expression Complements the Lipid and Suppressor Phenotypes of S7 Plants
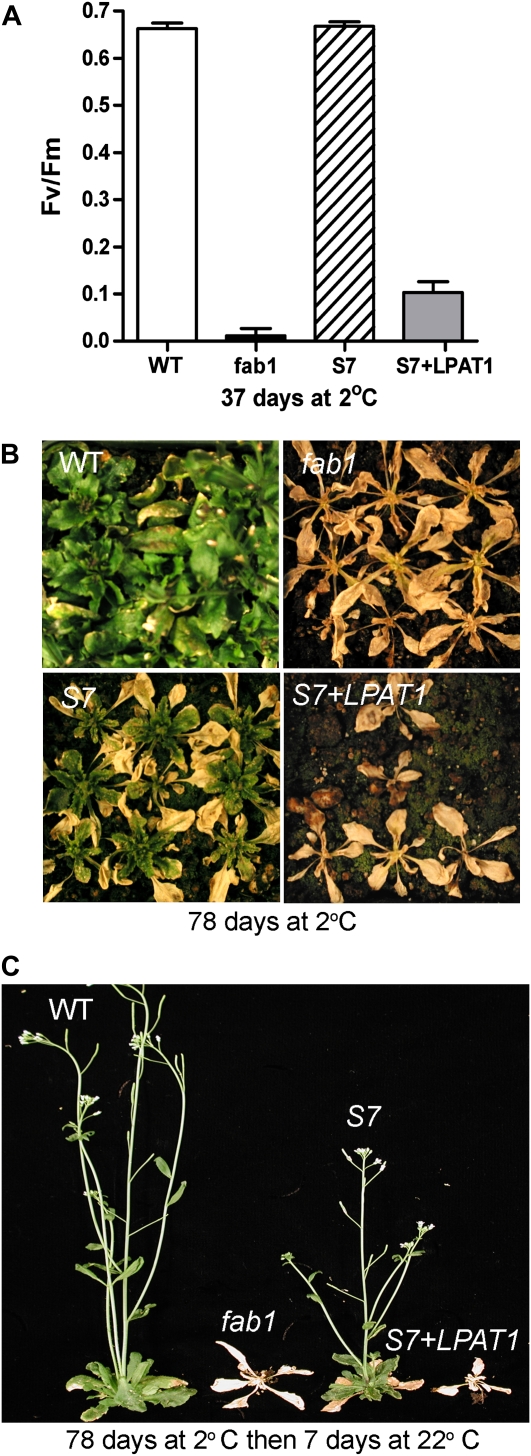
To test whether the lpat1-3 mutation is the basis of the altered lipid composition and the suppressor allowing growth of S7 plants at 2°C, we transformed S7 plants with an LPAT1 cDNA under control of the cauliflower mosaic virus 35S promoter. Eight independent T1 transgenic plants had levels of 16:3 in total leaf lipids that were increased to those of fab1 controls, indicating that the altered lipid composition of S7 plants is indeed the result of reduced LPAT1 activity. A line containing a single insert of the 35S:LPAT1 transgene was identified by a 3:1 segregation pattern in the T2 generation, and a homozygous T2 individual was identified by T3 pedigree analysis. Homozygous S7 35S:LPAT1 plants were grown together with wild-type, fab1, and S7 controls at 22°C for 25 d and then transferred to 2°C. After 36 d at 2°C wild-type and S7 plants were healthy while fab1 and S7 35S:LPAT1 plants showed symptoms of chlorosis and poor growth similar to those reported previously for fab1 (Wu and Browse, 1995; Wu et al., 1997). Measurements of the potential quantum yield of PSII, Fv/Fm, showed that wild-type and S7 plants maintained Fv/Fm close to 0.7, while the values for fab1 and S7 35S:LPAT1 plants were less than 0.1 (Fig. 2A). After 78 d at 2°C, the older leaves on S7 plants were senescent but new green leaves had been produced (Fig. 2B), and the plants quickly recovered and set seed after being returned to 22°C (Fig. 2C). In contrast, fab1 and S7 35S:LPAT1 plants were completely dead as indicated by their failure to recover after transfer to 22°C (Fig. 2C). These experiments establish lpat1-3 as the suppressor mutation in S7, and we now refer to this line as fab1 lpat1-3.
Figure 2.
Expression of an LPAT1 cDNA complements the S7 suppressor phenotype. A, After 37 d at 2°C, wild-type (WT) and S7 plants maintain high Fv/Fm, while S7 plants transformed with a 35S:LPAT1 transgene (S7 + LPAT1) and fab1 plants have low Fv/Fm (data are mean ± se for 10 plants of each line). B, After 78 d at 2°C, only wild-type and S7 plants remain green. C, When returned to 22°C, wild-type and S7 plants flower and set seed, while fab1 and S7 + LPAT1 plants fail to recover.
To investigate the biological consequences of the lpat1-3 mutation independently of fab1, we crossed fab1 lpat1-3 to wild type and isolated a homozygous lpat1-3 mutant. Interestingly, the leaf fatty acid composition of this new line contained only 1.8% 16:3 (Table III), considerably less than the 5.6% 16:3 measured in fab1 lpat1-3 (S7) leaves (Barkan et al., 2006). We interpret this difference as indicating that the reduced KASII activity caused by the fab1 mutation (Wu et al., 1994) results in an increase in 16:0-ACP, the substrate for LPAT1, which, in turn, increases synthesis of PA and prokaryotic lipids by the mutant LPAT1 enzyme encoded by lpat1-3.
Table III. Overall leaf fatty acid compositions of wild-type Arabidopsis and the lpat1-3, act1, and act1 lpat1-3 mutants.
Data are the means of five replicates with ses < 10% of the means. WT, Wild type.
| Fatty Acid | Percentage of Total |
|||
| WT | lpat1-3 | act1 | act1 lpat1-3 | |
| 16:0 | 15.7 | 13.1 | 14.4 | 11.9 |
| 16:1,Δ3-trans | 2.2 | 1.8 | 2.2 | 1.5 |
| 16:3 | 13.3 | 1.8 | 1.1 | 0.8 |
| 18:0 | 0.9 | 1.5 | 1.5 | 1.5 |
| 18:1 | 2.5 | 9.9 | 7.2 | 15.4 |
| 18:2 | 12.4 | 17.1 | 17.5 | 19.6 |
| 18:3 | 51.4 | 54.4 | 55.3 | 48.8 |
Plants of the fab1 lpat1-3 line are smaller than those of fab1 and wild type (Barkan et al., 2006), and lpat1-3 plants were also observed to be small and slow to develop. However, the act1 mutants, which are also defective in prokaryotic lipid synthesis, are similar in growth and development to wild type (Kunst et al., 1988; Xu et al., 2006). We transformed lpat1-3 with the 35S:LPAT1 transgene and complemented both the lipid and growth defects of the mutant (data not shown), thus demonstrating that LPAT1 deficiency causes the mutant phenotype. Next, we crossed the act1-1 and lpat1-3 mutants and produced a double-mutant line. act1-1, lpat1-3, and act1-1 lpat1-3 plants, along with wild-type controls, were grown side by side. The leaf fatty acid compositions of these plants (Table III) indicate that all three mutant lines have very low flux through the prokaryotic pathway, as judged by the very low levels of 16:3—a fatty acid that is only present in galactolipids synthesized by the prokaryotic pathway. The proportion of total fatty acids lost from 16:3 is distributed among the 18-carbon unsaturated fatty acids. The levels of 16:3 found in act1-1 lpat1-3 and act1 leaves suggest that prokaryotic flux in these mutants is less than one-tenth of that in wild type, while the lpat1-3 mutant has a slightly higher flux (Table III). After 33 d of growth, wild-type and act1-1 plants had well-developed bolts with flowers and filled siliques (Fig. 3). By contrast, the lpat1-3 plants were mainly maturing rosettes that had only just initiated reproductive growth. Plants of the act1-1 lpat1-3 line were noticeably smaller rosettes that had not yet bolted. The mutations in lpat1-3 and act1-1 lpat1-3 primarily result in a reduced growth rate. Mutant plants had a number of rosette leaves (10–12) similar to wild-type and act1 plants at the time of floral initiation, indicating that developmental processes were substantially unaffected. The growth phenotype of the act1-1 lpat1-3 double mutant is similar to that of act1-1 (ats1-1) plants in which LPAT1 expression was reduced by RNA interference (Xu et al., 2006). Thus, the lpat1-3 mutant causes reduced growth and development in wild-type and act1-1 genetic backgrounds, as well as in the fab1 background (Barkan et al., 2006).
Figure 3.
Growth and development of mutants deficient in chloroplast acyltransferases. Wild type (WT), act1, lpat1-3, and act1 lpat1-3 were grown for 33 d at 22°C.
DISCUSSION
The Arabidopsis fab1 mutation causes a Leu-337Phe substitution in KASII (At1g74960), the 3-ketoacyl-ACP synthase responsible for elongation of 16:0-ACP during fatty acid synthesis (Carlsson et al., 2002). The amino acid substitution likely results in structural instability of the KASII protein due to the difficulty of accommodating the imidazole ring of the mutant Phe-337 residue. The result is a 40% reduction in KASII activity in extracts from fab1 leaves relative to wild type and increased proportions of 16:0 in all the major leaf glycerolipids of fab1 plants (Wu et al., 1994). Of particular interest is the change in fatty acid composition of PG, the major chloroplast phospholipid. The PG from fab1 leaves has a 50% increase in the proportion of 16:0 compared with PG from wild-type leaves (Table I; Wu et al., 1994, 1997). Although fab1 plants contain 43% to 48% high-melting-point PG (Table II; Wu and Browse, 1995), this mutant does not show classic symptoms of chilling sensitivity, based on established tests that quickly lead to damage and death of chilling-sensitive plants (Wu and Browse, 1995). Instead, fab1 plants show a decline in photosynthetic function (as indicated by the fluorescence parameter, Fv/Fm) starting 10 d after transfer to 2°C (Wu et al., 1997).
Ultrastructural studies revealed that the decline in photosynthetic capacity is accompanied by disruption of granal and stromal thylakoids, and the development of small vacuoles (0.5–1.0 μm) in close association with the chloroplasts. This process continues with degradation and complete dismantling of chloroplasts in leaf mesophyll cells. Other organelles and cell structural features appear to be maintained during this period of chloroplast breakdown, so that affected leaf tissue is still alive but very chlorotic at the end of a 3-week cold treatment (Wu et al., 1997). fab1 plants kept at 2°C for up to 5 weeks are able to recover and set seed after a return to 22°C (Wu et al., 1997). We originally suggested that this chloroplast degradation at low temperature was a form of autophagy; however, recent reports of vacuole formation and chloroplast degradation during natural or dark-induced senescence provide alternative models for the process that occurs following the collapse of photosynthesis in fab1 plants at 2°C (Otegui et al., 2005; Ishida et al., 2008).
Based on these results, we have proposed that the levels of high-melting-point PG found in fab1 plants (and chilling-sensitive species) disrupt long-term maintenance of the photosynthetic machinery at low temperatures. In this model, altered PG composition would be one of several traits that evolved in tropical and subtropical plants (perhaps because they confer a selective advantage) but are incompatible with growth in a cold climate (Wu and Browse, 1995). However, it is important to note that this characterization of the fab1 mutant does not establish increased high-melting-point PG as the cause of low-temperature damage. It could be argued that increased 16:0 in another membrane lipid (DGD, for example), or the general increase in the proportion of 16:0 in membrane lipids, is the key determinant of damage.
Our characterization of the lpat1-3 mutation as a suppressor of the fab1 low-temperature phenotype addresses these possible explanations. It therefore provides additional support for the proposal that high-melting-point PG is a barrier to growth and survival of fab1 plants at low temperatures. Compared with fab1, the fab1 lpat1-3 (S7) line contains a higher proportion of 16:0 in the total leaf lipids (25.5% versus 23.3%; Barkan et al., 2006). There are also increases in the level of 16:0 in all of the major leaf glycerolipids except PG and SQD (Table I). The decrease of 16:0 in SQD is relatively small (from 58.3% in fab1 to 55.6% in fab1 lpat1-3) and is accompanied by a small increase in 18:0, the other saturated fatty acid. By contrast, 16:0 in PG decreases from 48.2% in fab1 to 34.5% in fab1 lpat1-3. More importantly, the percentage of high-melting-point molecular species in leaf PG of fab1 lpat1-3 is 10.7%, compared with 48.2% in fab1 and 7.6% in wild type (Table II). Given the evidence linking PG molecular species composition with low-temperature damage in plants (Nishida and Murata, 1996), it is reasonable to conclude that the lpat1-3 acts as a suppressor through its effect on reducing the proportion of high-melting-point PG. However, this is not the only alteration in thylakoid lipid composition that can suppress the fab1 phenotype. Mutations at the fad5 locus that reduce desaturation of 16:0 to 16:3 in MGD also result in fab1 fad5 plants that survive at 2°C. We have proposed that the change in shape of MGD molecules caused by the fad5 mutations may compensate for disruptive changes in the shape and packing of PG molecules induced by the fab1 mutation (Barkan et al., 2006).
Our identification of the S7 suppressor as a hypomorphic lpat1 allele is additionally useful because two, previously characterized, null lpat1 alleles are not viable (Kim and Huang, 2004; Yu et al., 2004). Interestingly, these null alleles are embryo lethal, causing arrest of embryos between the globular and torpedo stages of development. Evidently, LPAT1 activity is not essential for growth and development of either the male or female gametophytes, nor for the earliest stages of embryo development. Furthermore, homozygous lpat1 embryos were produced in the predicted Mendelian ratio (Kim and Huang, 2004; Yu et al., 2004), and this indicates that development and function of lpat1-1 and lpat1-2 sperm cells are sufficient for them to bring about fertilization as effectively as wild-type sperm cells.
LPAT1 is required for operation of the prokaryotic pathway that produces MGD, DGD, PG, and SQD for thylakoid biogenesis (Wallis and Browse, 2010), so it is reasonable to conclude that synthesis of one or more of these lipids is essential for embryo (and plant) development. In the act1 mutant, which is blocked in acyl-ACP:glycerol-3-P acyltransferase—the step preceding LPAT1—accumulation of prokaryotic MDG + DGD + SQD is reduced by >98% (Kunst et al., 1988; Xu et al., 2006), making it unlikely that synthesis of any of these three lipids by the prokaryotic pathway is essential. Leaves of act1 plants contain levels of MGD, DGD, and SQD that are similar to wild type due to increased flux through the eukaryotic pathway, and growth and development of the mutants are very similar to wild type (Fig. 3; Kunst et al., 1988, 1989). Chloroplast PG is not synthesized by the eukaryotic pathway, but the rate of PG synthesis by the prokaryotic pathway appears to be preferentially maintained in the act1 mutant at 50% of the wild-type rate. There appears also to be reduced turnover of PG in act1 so that PG is present in act1 leaves in amounts that are approximately 70% of wild type (Kunst et al., 1988; Xu et al., 2006). These observations suggested the possibility that lpat1 null mutants are embryo lethal because they are unable to synthesize chloroplast PG. However, a pgp1 null mutant that is also deficient in chloroplast PG synthesis is not embryo lethal, but instead is able to germinate and grow on Suc media, although it is not autotrophic (Hagio et al., 2002). Characterization of this pgp1 null mutant, and other evidence (Nussberger et al., 1993; Liu et al., 2004), indicate that PG is a component of the photosystem light-harvesting complexes and required for photosynthetic functions, but this does not provide an explanation for the embryo-lethal phenotype of lpat1 null mutants.
The lpat1-3 mutant is comparable to act1 in the level of PG and of 16:3 in the leaf lipids (Table III; data not shown); however, lpat1-3 plants grow more slowly than act1 and wild type (Fig. 3). We considered the possibility that accumulation of lyso-PA, one of the substrates of the LPAT1 enzyme, might contribute to the phenotypes of the hypomorphic and null lpat mutants. To test this possibility, we crossed the lpat1-3 mutant to act1-1. Chloroplast lyso-PA synthesis is blocked by the act1-1 mutation (Kunst et al., 1988), but this does not result in improved growth of act1 lpat1-3 plants, instead these double mutants grow more slowly than lpat1-3 plants (Fig. 3; Xu et al., 2006). It is unlikely that accumulation of 16:0-ACP, the other LPAT1 substrate, is related to the growth phenotype since the levels of this fatty acid synthase product are expected to be less in lpat1-3 than in fab1 lpat1-3 or fab1 chloroplasts. Although these observations do not identify the mechanism through which reduced LPAT1 activity affects plant growth (and embryo viability), the lpat1-3 allele that we have identified from our suppressor screen provides a new tool to investigate the biochemical and biology roles of the chloroplast LPAT1 enzyme.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type in this study. Mutant plants lines fab1, act1, S7, and lpat1-3 are in the Columbia-0 genetic background. Wild-type and mutant plants were grown at 22°C under 16 or 24 h of illumination, 120 μmol quanta m−2 s−1. For cold-resistant test, plants were grown at 2°C for up to 12 weeks.
Mapping the S7 Suppressor Locus
To map the suppressor, an S7 plant was crossed to the Landsberg erecta ecotype and the resulting F1 plants were allowed to self. F2 plants homozygous for the fab1 mutation were selected and their progeny tested for segregation of the recessive suppressor phenotype. An F2 plant that was homozygous fab1 and heterozygous for the suppressor was used as the parent of an F3 mapping population. Eight-hundred F3 seeds were sown on soil and the resulting plants tested for survival at 2°C. DNA was prepared from leaves of individual surviving plants. Samples of 50 DNA preparations were pooled and used for bulk-segregant analysis. Subsequently, DNA from 170 individuals was tested using marker NGA1107 to identify an approximate position for the suppressor locus.
Cloning and Sequencing of LPAT1 Alleles
LPAT1 cDNAs were amplified from first-strand cDNA synthesized from total RNA of wild-type, fab1, and S7 plants using reverse transcription (RT)-PCR. To amplify LPAT1 cDNAs from different mutant plant lines, gene-specific primers (5′-GTTTTTGCTCCAGATTCCGCC-3′ for N terminus and 5′-AAGACAACTTCTGAGATCAGC-3′ for C terminus) were designed to sequences in 5′-untranslated and 3′-untranslated regions of the LPAT1 coding region. RNA samples from eight plants of each line were used to amplify RT-PCR products that were sequenced to identify the gene mutation.
Gene Cloning and Gene Construction
An LPAT1 cDNA containing full-length open reading frame was amplified using RT-PCR with gene-specific primers, 5′-CACCATGGATGTCGCTTCTGCTCGGAGC-3′ and 5′-TTAGAGATCCATTGATTCTGCAAT-3′ from first-strand cDNA from wild-type plants, and cloned into pENTR gateway entry vector using Topoisomerase ligation. The cDNAs were confirmed by DNA sequencing and inserted into Gateway plant expression vector, pB2GW7.0 (Karimi et al., 2002) under control of the cauliflower mosaic virus 35S promoter, by LR Clonase (Invitrogen) reaction.
S7 + LPAT1 and lpat1-3 + LPAT1 transgenic plants were generated by the floral-dip method (Clough and Bent, 1998) with use of Agrobacterium tumefaciens strain GV3101 containing the 35S:LPAT1 construct. Plants resistant to BASTA were examined by PCR for genotype, and lipid composition using GC.
Extraction and Analysis of Lipids
The overall fatty acid compositions of leaf tissues were determined as previously described (Wu et al., 1994) with the exception that 1.5 mL water and 0.5 mL hexane were used to extract fatty acids into the organic phase. Samples (1 μL) of the organic phase were analyzed by GC on a 30 m × 0.53 mm Alltech Econo-cap column containing a 1.2 μm EC-WAX phase (Alltech Associates, Inc.). The GC was programmed for an initial temperature of 160°C for 1 min, followed by an increase of 20°C/min to 190°C and a second increase of 4.5°C/min to the final temperature of 203°C.
The more detailed analyses of lipid and fatty acid composition were performed as described previously (Wu et al., 1994). Aliquots of the lipid extract were separated by one-dimensional thin-layer chromatography on (NH4)2SO4-impregnated silica gel G (Wu et al., 1994) using acetone:benzene:water (30:10:2.7 v/v; Khan and Williams, 1997). To determine the fatty acid composition and the relative amounts of individual lipids, the silica gel from each spot was transferred to a screw-capped tube and fatty acid methyl esters were prepared and analyzed as described above. A known amount of 17:0 PC was added as internal standard prior to derivitization.
Quantification of PG molecular species was obtained by lipidomics analysis of lipid extracts performed at the Kansas State University Lipidomics Facility (Esch et al., 2007).
Measurements of Chlorophyll Fluorescence
Chlorophyll fluorescence from leaf tissues was measured using a PAM fluorometer (Walz). The ratio of variable fluorescence to maximal fluorescence (Fv/Fm), representing the potential quantum yield of PSII photochemistry, was measured in dark-adapted leaf tissues described by Barkan et al. (2006).
Acknowledgments
We thank Deirdre Fahy for assistance in preparing the figures.
References
- Barkan L, Vijayan P, Carlsson AS, Mekhedov S, Browse J. (2006) A suppressor of fab1 challenges hypotheses on the role of thylakoid unsaturation in photosynthetic function. Plant Physiol 141: 1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al. (2003) Arabidopsis genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol 25: 71–91 [DOI] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem J 235: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AS, LaBrie ST, Kinney AJ, von Wettstein-Knowles P, Browse J. (2002) A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J 29: 761–770 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eberhardt C, Gray PW, Tjoelker LW. (1997) Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J Biol Chem 272: 20299–20305 [DOI] [PubMed] [Google Scholar]
- Esch SW, Tamura P, Sparks AA, Roth MR, Devaiah SP, Heinz E, Wang XM, Williams TD, Welti R. (2007) Rapid characterization of the fatty acyl composition of complex lipids by collision-induced dissociation time-of-flight mass spectrometry. J Lipid Res 48: 235–241 [DOI] [PubMed] [Google Scholar]
- Hagio M, Sakurai I, Sato S, Kato T, Tabata S, Wada H. (2002) Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol 43: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Hugly S, Somerville C. (1992) A role for membrane lipid polyunsaturation in chloroplast biogenesis at low temperature. Plant Physiol 99: 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki-Nishizawa O, Fujii T, Azuma M, Sekiguchi K, Murata N, Ohtani T, Toguri T. (1996) Low-temperature resistance of higher plants is significantly enhanced by a nonspecific cyanobacterial desaturase. Nat Biotechnol 14: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Khan MU, Williams JP. (1997) Improved thin-layer chromatographic method for the separation of major phospholipids and glycolipids from plant lipid extracts and phosphatidylglycerols and bis(monoacylglyceryl) phosphate from animal lipid extracts. J Chromatogr 140: 179–185 [DOI] [PubMed] [Google Scholar]
- Kim HU, Huang AHC. (2004) Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol 134: 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Hamada T, Horiguchi G, Nishimura M, Iba K. (1994) Genetic enhancement of cold tolerance by expression of a gene for chloroplast omega-3 fatty acid desaturase in transgenic tobacco. Plant Physiol 105: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C. (1988) Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc Natl Acad Sci USA 85: 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C. (1989) Altered chloroplast structure and function in a mutant of Arabidopsis deficient in plastid glycerol-3-phosphate acyltransferase activity. Plant Physiol 90: 846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y, Huang W, Schneider G, Shanklin J. (1996) Crystal structure of delta9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J 15: 4081–4092 [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Cassagne C, Browse J. (1998) A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol 117: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Tsuyama M, Kobayashi Y, Kodama H, Iba K. (2000) Trienoic fatty acids and plant tolerance of high temperature. Science 287: 476–479 [DOI] [PubMed] [Google Scholar]
- Murata N. (1983) Molecular species composition of phosphatidylglycerols from chilling-sensitive and chilling-resistant plants. Plant Cell Physiol 24: 81–86 [Google Scholar]
- Murata N, Yamaya J. (1984) Temperature-dependent phase behavior of phosphatidylglycerols from chilling-sensitive and chilling-resistant plants. Plant Physiol 74: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida I, Murata N. (1996) Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu Rev Plant Physiol Plant Mol Biol 47: 541–568 [DOI] [PubMed] [Google Scholar]
- Nussberger S, Dorr K, Wang DN, Kuhlbrandt W. (1993) Lipid-protein interactions in crystals of plant light-harvesting complex. J Mol Biol 234: 347–356 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Noh YS, Martinez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ. (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41: 831–844 [DOI] [PubMed] [Google Scholar]
- Roughan PG. (1985) Phosphatidylglycerol and chilling sensitivity in plants. Plant Physiol 77: 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul JM, Fischer SF, Browse J. (2000) Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol 124: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JG, Browse J. (2010) Lipid biochemists salute the genome. Plant J 16: 1092–1106 [DOI] [PubMed] [Google Scholar]
- Wu J, James DW, Jr, Dooner HK, Browse J. (1994) A mutant of Arabidopsis deficient in the elongation of palmitic acid. Plant Physiol 106: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, Browse J. (1995) Elevated levels of high-melting-point phosphatidylglycerols do not induce chilling sensitivity in an Arabidopsis mutant. Plant Cell 7: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, Lightner J, Warwick N, Browse J. (1997) Low-temperature damage and subsequent recovery of fab1 mutant Arabidopsis exposed to 2°C. Plant Physiol 113: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yu B, Cornish AJ, Froehlich JE, Benning C. (2006) Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant J 47: 296–309 [DOI] [PubMed] [Google Scholar]
- Yu B, Wakao S, Fan J, Benning C. (2004) Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol 45: 503–510 [DOI] [PubMed] [Google Scholar]