Abstract
Cytidinediphosphate diacylglycerol synthase (CDS) catalyzes the formation of cytidinediphosphate diacylglycerol, an essential precursor of anionic phosphoglycerolipids like phosphatidylglycerol or -inositol. In plant cells, CDS isozymes are located in plastids, mitochondria, and microsomes. Here, we show that these isozymes are encoded by five genes in Arabidopsis (Arabidopsis thaliana). Alternative translation initiation or alternative splicing of CDS2 and CDS4 transcripts can result in up to 10 isoforms. Most of the cDNAs encoding the various plant isoforms were functionally expressed in yeast and rescued the nonviable phenotype of the mutant strain lacking CDS activity. The closely related genes CDS4 and CDS5 were found to encode plastidial isozymes with similar catalytic properties. Inactivation of both genes was required to obtain Arabidopsis mutant lines with a visible phenotype, suggesting that the genes have redundant functions. Analysis of these Arabidopsis mutants provided further independent evidence for the importance of plastidial phosphatidylglycerol for structure and function of thylakoid membranes and, hence, for photoautotrophic growth.
Cytidinediphosphate diacylglycerol synthase (CDS) catalyzes the transfer of a cytidylyl group from CTP to phosphatidic acid (PA) so that cytidinediphosphate diacylglycerol (CDP-DAG), an important branch point intermediate in glycerolipid biosynthesis, is formed. CDS enzymes are integral membrane proteins that are located in mitochondrial and microsomal membranes of all eukaryotes (Kinney, 1993; Dowhan, 1997; Lykidis and Jackowski, 2001). Within the mitochondria, the enzyme provides the substrate for the biosynthesis of phosphatidylglycerol (PG) and cardiolipin (CL), while the microsomal activity is required for the biosynthesis of phosphatidylinositol and its phosphorylated derivatives. Plants use microsomal CDP-DAG for PG and to a certain extent for phosphatidylserine synthesis as well (Moore, 1982; Delhaize et al., 1999; Babiychuk et al., 2003). Apart from mitochondrial and microsomal CDSs, plants possess plastidial CDS activity in the inner envelope membrane for the formation of plastidial PG (Andrews and Mudd, 1985). Hence, in the different subcellular compartments, CDP-DAG serves as precursor for the biosynthesis of the anionic phosphoglycerolipids, which are minor but indispensable components for essential cellular functions (for review, see Frentzen, 2004; Wada and Murata, 2007; Houtkooper et al., 2009; Xue et al., 2009).
CDSs have been purified from Escherichia coli membranes and yeast mitochondria (Sparrow and Raetz, 1985; Kelley and Carman, 1987), while CDS genes have been functionally characterized from various prokaryotic (Icho et al., 1985; Martin et al., 2000; Sato et al., 2000) and eukaryotic organisms (Wu et al., 1995; Shen et al., 1996; Kopka et al., 1997; Weeks et al., 1997; Volta et al., 1999; Inglis-Broadgate et al., 2005). A comparison ofthe amino acid sequences of the encoded proteins reveals that CDSs have been conserved during evolution. Apart from variable N- and C-terminal extensions, which are present in eukaryotic proteins only and which might be critical for protein targeting and regulation, the sequences are similar to each other, especially within the C-terminal half where the typical CDS motifs are located.
Yeast cells appear to possess a single CDS gene that encodes the total cellular CDS activity associated with both the microsomal and the mitochondrial membranes. Expression of this gene has been shown to be essential for cell viability and spore germination (Shen et al., 1996). In Drosophila melanogaster, the eye-specific CDS isoform, which serves as key regulator of phototransduction, is derived by alternative RNA splicing from the same gene as the constitutively expressed form (Wu et al., 1995). On the other hand, two different CDS genes have been identified in mammals, of which CDS2 encodes the housekeeping enzyme and CDS1 the one specialized for signal transduction in certain tissues (Lykidis and Jackowski, 2001; Inglis-Broadgate et al., 2005). Unlike CDS1 of yeast, the mammalian enzymes encoded by the two genes were found to be associated with the endoplasmic reticulum (ER) only (Inglis-Broadgate et al., 2005). In plants, CDS proteins appear to be encoded by a small gene family that comprises five members in Arabidopsis (Arabidopsis thaliana; Beisson et al., 2003). So far, Kopka et al. (1997) provided evidence that one Arabidopsis gene, namely, CDS1 (At1g62430), encodes a catalytically active CDS protein.
Here, we report the functional characterization of the further four genes of Arabidopsis by heterologous complementation of a cds1-null mutant of yeast and enzymic assays with subcellular fractions of yeast cultures expressing one of the various Arabidopsis cDNAs. We provide evidence that CDS4 and CDS5 encode the plastidial isozymes and show that photoautotrophic growth requires at least one functional CDS4 or CDS5 allele.
RESULTS
Arabidopsis CDS Isoforms Are Encoded by Five Genes
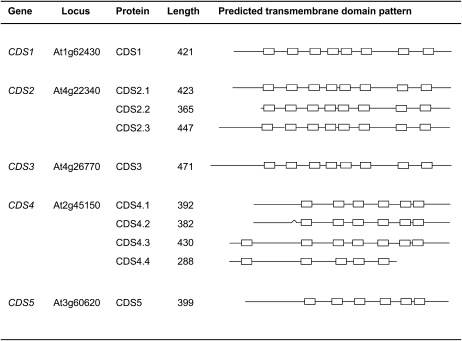
According to predictions derived from EST sequence information (Schwacke et al., 2003), the Arabidopsis genes termed CDS1 to CDS5 might encode 10 different proteins because of the formation of various CDS2 and CDS4 isoforms (Fig. 1). Most of these isoforms can be produced by alternative translation initiation of CDS2 and CDS4 transcripts. This mechanism appears to operate in a number of plant genes and can influence translation regulation and intracellular protein targeting (Mackenzie, 2005; Wamboldt et al., 2009). However, we cannot exclude the possibility that the prediction of some of the CDS isoforms are merely based on incomplete cDNA sequences. With regard to the three CDS2 isoforms, which can be formed by translation initiation at the first, second, and third AUG, this might hold true, but direct evidence for this assumption is currently missing (Fig. 1). Based on the results presented below, the situation appears to be different with regard to the CDS4 isoforms. Translational initiation at the first AUG can result in the formation of CDS4.3 and CDS4.4, while the one at the second AUG causes CDS4.1 and CDS4.2 formation lacking the first predicted transmembrane domain (Fig. 1). In addition, CDS4.2, unlike CDS4.1, has a short deletion of nine amino acids encoded by the 5′ region of exon 2 caused by alternative splicing of CDS4 transcripts. Respective CDS4 transcripts were detected by reverse transcription (RT)-PCR (Fig. 6). Alternative splicing likely causes the formation of CDS4.4 as well. This isoform possesses a deletion in the C-terminal region covering the last two conserved transmembrane domains, suggesting that CDS4.4 represents a catalytically inactive isoform.
Figure 1.
Overview of the predicted CDS proteins encoded by the five genes of Arabidopsis. The transmembrane domains are shown as boxes and based on consensus prediction (Arai et al., 2004).The bend in the structure of CDS4.2 indicates a deletion of nine amino acids due to alternative splicing.
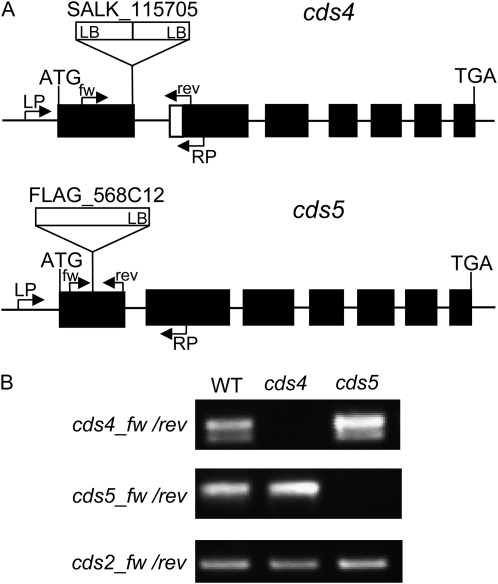
Figure 6.
Analysis of the cds4 and cds5 T-DNA insertion alleles. A, Positions of the T-DNA insertions are indicated relative to the exon-intron structure of CDS genes. Exons are indicated with black bars and introns with thin lines. Primers used for the analysis of the locus (LP and RP) and the respective transcripts (fw and rev) are indicated by arrows. The T-DNAs are shown as white boxes with LB designating the left border. B, Expression of CDS genes in cds4 and cds5 mutants in comparison to the wild type (WT). CDS transcripts were determined by RT-PCR. The lower of the two bands amplified with the CDS4-specific primers is due to the CDS4.2 transcript having a short deletion of 27 nucleotides at the 5′ region of exon 2.
Sequence similarities among the Arabidopsis CDS proteins are reflected in the number and pattern of the predicted transmembrane domains (Fig. 1). CDS1 shares highest sequence identity with the CDS2 proteins (CDS2.1, 78%; CDS2.2, 72%; CDS2.3, 74%), a slightly lower one with CDS3 (63%), and lowest ones with the closely related proteins encoded by CDS4 (CDS4.1/2, 21%; CDS4.3, 18%; CDS4.4, 17%) and CDS5 (20%). The CDS1, CDS2, and CDS3 proteins are more similar to the respective proteins from eukaryotes than from prokaryotes, while the CDS4 and CDS5 proteins show highest sequence similarity to cyanobacterial CDS proteins (Sato et al., 2000). Apart from CDS4.3 and CDS4.4, these proteins are predicted to contain cleavable N-terminal transit peptides for the import into plastids and with a lower probability into mitochondria (Schwacke et al., 2003). These data suggest that CDS4 and CDS5 encode the plastidial isozymes or both the plastidial and mitochondrial ones like PGP1 of Arabidopsis encoding phosphatidylglycerolphosphate (PGP) synthase (Babiychuk et al., 2003). CDS1 and the closely related CDS2 proteins likely represent microsomal isozymes (Kopka et al., 1997). The MultiLoc program (Hoeglund et al., 2006) predicts the proteins to be located in the plasma membrane, but Aramemnon consensus prediction (Schwacke et al., 2003) supports such a subcellular localization with regard to CDS2.2 only. Similar to CDS1, CDS3 contains no typical targeting sequence so that different subcellular localization programs gave inconsistent results.
According to the Arabidopsis microarray database, CDS3 differs in its expression pattern from the other four genes (Zimmermann et al., 2005). While the latter are constitutively expressed, CDS3 is expressed in certain cell types like pollen tube or trichoblasts only.
Arabidopsis CDS Genes Can Be Functionally Expressed in Yeast
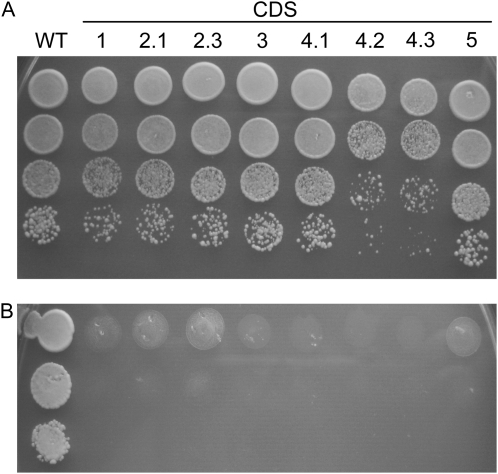
To functionally characterize the various Arabidopsis CDS sequences, the respective cDNAs were cloned into a yeast expression vector under the regulation of the GAL1 promoter and transformed into the heterozygous diploid mutant strain YBR029c. In this mutant strain, one ScCDS1 allele is disrupted by the insertion of a KanMX4 cassette (cds1::KanMX4) via homologous recombination. When sporulation of the transgenic mutant cultures was induced, tetrads were dissected and viability of the spores was analyzed. Each tetrad of the control harboring the empty vector only gave rise to two viable spores, which contained the wild-type ScCDS1 gene as reflected in their kanamycin-sensitive phenotype and confirmed by PCR analysis of genomic DNA with ScCDS1-specific primers. Tetrads of the mutant harboring either the CDS2.2 or the CDS4.4 cDNA sequence contained two viable spores like the tetrads of the control. On the other hand, transformants expressing one of the eight further Arabidopsis sequences formed four viable spores, two of which were kanamycin resistant because they carried the cds1::KanMX4 gene as approved by PCR. As shown in Figure 2, the introduced Arabidopsis genes rescued the nonviable phenotype of the haploid mutant strain only when expression of the introduced genes was induced by Gal in the medium (Fig. 2A). Under these conditions, transformants showed growth rates very similar to the wild type (Fig. 2A), but they stopped growing when expression of the Arabidopsis genes was repressed by Glc (Fig. 2B).
Figure 2.
Functional complementation of Saccharomyces cerevisiae mutant YBR029c lacking CDP-DAG synthase activity by expression of Arabidopsis CDS cDNAs under control of the PGAL promoter. Yeast suspensions of wild-type (WT) and mutant strain containing one of the given Arabidopsis CDS cDNAs were stepwise diluted 10 times and spotted on 2% Gal (A) and 2% Glc (B) and incubated at 28°C for 48 h. cDNAs are termed according to the proteins they encode (see Fig. 1). Unlike wild-type cells, mutant cells were only able to grow when expression of the Arabidopsis gene constructs was induced by Gal.
These data provide direct evidence that not only CDS1 (Kopka et al., 1997) but also the further four members of the Arabidopsis gene family represent CDS genes that were functionally expressed in yeast and thus compensated for the lethal phenotype of the Sccds1-null mutant. Merely CDS2.2 and CDS4.4 failed to restore growth of yeast mutant cells. With regard to CDS4.4, this was expected because the protein lacks part of the active site within the C-terminal region (Fig. 1). This is supported by CDS4.3 having no deletion in its C-terminal region and possessing CDS activity (Figs. 1 and 2). With regard to the CDS2 isoforms, the results suggest that the N-terminal region encoded by exon 1 that is present in CDS2.1 and CDS2.3 but lacking in CDS2.2 is essential for the proteins to acquire a catalytically active conformation. On the other hand, the N-terminal extension only present in CDS2.3 appears to be dispensable for CDS activity (Figs. 1 and 2).
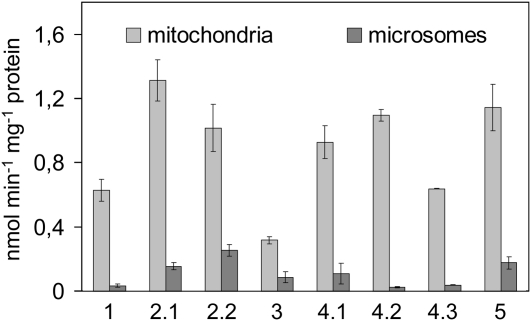
The results of heterologous complementation experiments were supported and extended by enzymic assays using membrane fractions of the Sccds1-null mutant strain expressing an Arabidopsis gene construct. Under assay conditions that allowed protein linear formation rates of CDP-DAG, the eight different CDS proteins clearly displayed CDS activity although their specific activities varied. CDS2.1 gave the highest activities, slightly lower ones were determined with CDS2.2, CDS4.1, CDS4.2, and CDS5, while CDS1, CDS4.3, and CDS3 gave relatively low activities (Fig. 3). Surprisingly, all CDS isozymes displayed appreciably higher activities in the mitochondrial than in the microsomal fractions (Fig. 3). In these fractions, marker enzymes showed typical activity patterns. For instance, phosphatidylethanolamine methyltransferase, a marker enzyme of ER membranes, had about 6-fold higher activities in the microsomal than in the mitochondrial fractions, while the PGP synthase located in the inner mitochondrial membrane had about 20-fold higher activities in the mitochondrial than in the microsomal fraction. Purification of the mitochondria via density gradients provided further evidence that the major part of the CDS activities determined in the crude mitochondria was not caused by microsomal contamination. In addition, a repetition of the CDS assays without detergent gave lower enzyme activities but no distinct alterations in the activity patterns shown in Figure 3. Hence, the unexpectedly low microsomal activities, especially of the CDS1 and CDS2 isozymes, cannot be attributed to a specific inhibition of microsomal activities by detergents as reported for the respective enzymes of castor bean (Ricinus communis; Kleppinger-Sparace and Moore, 1985). Our data rather suggest that not only the plastidial enzymes of Arabidopsis but also those located in extraplastidial membranes can be efficiently imported into yeast mitochondria. In that way, the functional expression studies in yeast clearly verified the identity of the Arabidopsis CDS genes but provided no clues about the intracellular targeting of the CDS isozymes in plant cells.
Figure 3.
Activities and subcellular localization of Arabidopsis CDS isoforms expressed in the yeast mutant strain YBR029c. Incorporation rates of [3H]CTP into CDP-DAG by mitochondrial and microsomal fractions of yeast cells harboring the given CDS isoform are depicted as mean values and sds of three independent preparations.
CDS4 and CDS5 Encode Plastidial Isozymes with Similar Properties
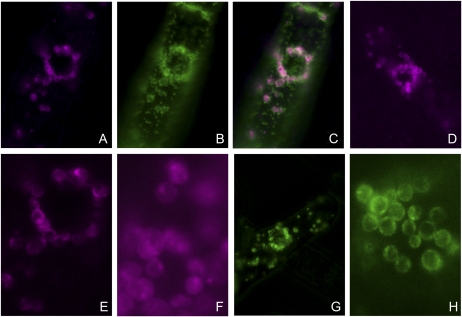
To investigate the intracellular localization of CDS isozymes and their functions in plant lipid metabolism, we focused on isozymes encoded by CDS4 and CDS5 at first. Expression of the four isozymes as fusion proteins with red fluorescent protein (RFP) or GFP at their C termini in tobacco (Nicotiana tabacum) Bright Yellow 2 (BY2) cells gave clear and reproducible results with three isozymes, namely, CDS4.1, CDS4.2, and CDS5. We used fusion proteins of CDS5 as plastidial controls because proteomic studies have detected CDS5 in the envelope membranes of chloroplasts (Froehlich et al., 2003; Heazlewood et al., 2005). As shown in Figure 4, not only the fusion protein of CDS5 but also those of CDS4.1 and CDS4.2 were found to be localized in plastids and appeared to be enriched in the envelope membranes. In contrast to the results with CDS4.1 and CDS4.2, we failed to detect CDS4.3 as fusion protein in BY2 cells. Because the isoforms vary in their N-terminal regions only, the different results must be attributed to the N-terminal hydrophobic region solely present in the CDS4.3 isoform (Fig. 1). This region hardly affected import into yeast mitochondria (Fig. 3) but appears to prevent targeting to plastids and likely causes a rapid degradation of the fusion protein. In vitro import studies with CDS4.3 will perhaps show whether this assumption is correct.
Figure 4.
Subcellular localization of Arabidopsis CDS isozymes expressed as fusion proteins with RFP or GFP in tobacco BY2 cells. A, BY2 cell that expresses CDS4.1-RFP fusion protein was counterstained with Mitotracker green. The RFP signal is shown. B, Same cell as in A, but the Mitotracker green signal is shown. C, Merge of images A and B. D, RFP signal of a BY2 cell expressing CDS5-GFP fusion protein as a positive control because CDS5 has been shown to be located in the chloroplast envelope. E, Enlargement of part of A. F, RFP signal of a BY2 cell expressing CDS4.2-RFP fusion protein. G, GFP signal of a BY2 cell expressing CDS5-GFP fusion protein as positive control. H, GFP signal of plastids within a BY2 cell expressing CDS5-GFP fusion protein. [See online article for color version of this figure.]
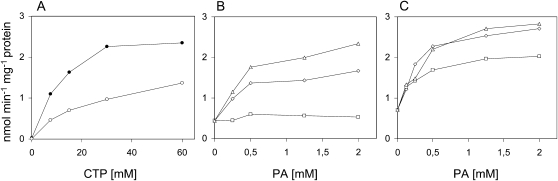
To determine the properties of the plastidial CDS isozymes, mitochondrial membranes of the yeast mutant strains expressing either CDS4.1 or CDS5 were used as enzyme sources. Both isozymes showed a broad pH optimum of about 7.5. They required divalent cations for activity and displayed highest activities with MgCl2 at a final concentration of 20 mm. Addition of Triton X-100 up to 2% (w/v) stimulated the activities about 3-fold. CDS activity as a function of the concentration of the substrates CTP and PA gave saturation kinetics. Highest activities were obtained at about 30 mm CTP and 2 mm PA (Fig. 5). Similar properties have been reported for CDS from different plant species and different subcellular compartments (Andrews and Mudd, 1984; Hanenberg et al., 1993), but CDS activity of pea (Pisum sativum) chloroplast enzymes was inhibited by Triton X-100, unlike the respective Arabidopsis enzymes (Andrews and Mudd, 1984). The Arabidopsis isozymes both showed highest activities with the dioleoyl species of PA, slightly lower ones with the species derived from egg phosphatidylcholine, which predominantly consists of the 1-palmitoyl-2-oleoyl species, and lowest ones with the dipalmitoyl species (Fig. 5). PA species specificity of CDS4.1 was more distinct than that of CDS5, but none of the isozymes preferentially used the prokaryotic PA species carrying an oleoyl group at sn-1 and a palmitoyl group at sn-2 when compared to the 1-palmitoyl-2-oleoyl species. Under optimal assay conditions, CDP-DAG formation rates were constant for at least 20 min, and a protein amount of up to 20 μg and specific enzymic activities of about 2 nmol min−1 mg−1 protein were determined.
Figure 5.
Certain properties of CDS4.1 and CDS5 heterologously expressed in yeast. A, Enzymic activity of CDS4.1 (white circles) and CDS5 (black circles) in dependence on CTP concentrations. B and C, Enzymic activities of CDS4.1 (B) and CDS5 (C) as a function of the concentrations of three different PA species (triangles, dioleoyl; squares, dipalmitoyl species; diamonds, species from egg lecithin).
CDS4 and CDS5 Have Redundant Functions
Arabidopsis mutants available from the Nottingham Arabidopsis Stock Centre and the French National Institute for Agricultural Research (Institut National de la Recherche Agronomique [INRA]) have T-DNA insertions in exon 1 of CDS4 or CDS5, respectively (Fig. 6A). The insertion sites of the respective T-DNA were ascertained by sequencing the PCR products amplified from genomic DNA of the mutant plants with T-DNA and gene-specific primers. These experiments also showed that the SALK_117505 line contains two T-DNAs in form of an inverted repeat flanked by the left border region at both ends (Fig. 6A). According to the analysis of the CDS transcripts of the homozygous mutant plants by RT-PCR, the T-DNA insertions in both CDS4 and CDS5 cause loss-of-function mutations (Fig. 6B). In contrast to the cds4 mutant plants, in which no CDS4 transcripts were detectable, CDS5 transcript regions downstream to the insertion site could be amplified from RNA of the cds5 mutant plants likely because of the aberrant transcripts formed in the mutant. This was supported by sequence analysis of the amplified fragments.
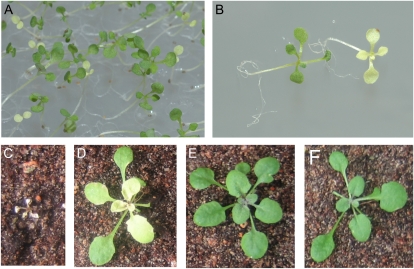
Mutant plants homozygous with regard to the T-DNA insertion in either CDS4 or CDS5, however, showed no obvious phenotype. They grew and developed like wild-type plants (Fig. 7). Because both CDS4 and CDS5 encode plastidial isozymes, these genes most probably have redundant functions.
Figure 7.
Phenotype of Arabidopsis cds4 cds5 mutants. A, Two-week-old seedlings segregating in a progeny of heterozygous mutant plants were cultivated on agar-solidified MSG medium with 3% (w/v) Suc. Homozygous double mutant seedlings are easily recognized by their pale greenish-yellow color and their retarded growth. B, Close-up of a wild-type (left) and mutant seedling (right) shown in A. C to F, Plants grown for 2 weeks on agar with Suc followed by 2 weeks of growth on earth. C, The cds4 cds5 mutant. D, The cds4 cds5 mutant expressing a chimeric CDS4 construct under the control of the 35S promoter that encodes CDS4.1 as a GFP fusion protein. E, The cds4 cds5 mutant expressing the open reading frame of CDS5 under the control of the 35S promoter. F, A wild-type plant. [See online article for color version of this figure.]
The cds4 cds5 Double Mutant Requires Suc for Growth
To provide direct evidence for CDS4 and CDS5 having redundant functions, homozygous single (cds4 and cds5) mutant plants were crossed and the progenies were analyzed. Mutant plants being homozygous with regard to only one of the two CDS genes displayed a phenotype very similar to wild-type plants (Fig. 7). On the other hand, homozygous cds4 cds5 double mutant plants were easily distinguishable from wild-type plants. Seedlings of the double mutant were not viable in soil, grew more slowly on agar-solidified medium with Suc than wild-type plants, and developed pale yellow-green leaves only (Fig. 7). Total chlorophyll content was reduced by at least 80%, while chlorophyll a/b ratio was appreciably increased.
We transformed heterozygous double mutant plants with a CDS5 cDNA under the control of the 35S promoter. Among the progenies of the selfed transgenic plants, homozygous double mutant plants expressing the chimeric gene construct were identified, which developed and grew like wild-type plants (Fig. 7). These data demonstrate that disruption of both CDS4 and CDS5 is responsible for the mutant phenotype, which can be compensated for by expression of a functional allele. Respective data obtained with a chimeric gene encoding CDS4.1 as a fusion protein with GFP support the conclusion but also showed that expression of the fusion protein could largely but not completely compensate for the phenotype of the double mutants. These transgenic plants were able to grow on soil and to produce fertile seeds, but they grew slower and contained lower pigment levels than wild-type plants (Fig. 7). Expression of an unmodified CDS4.1 isozyme in double mutant plants will show whether GFP at the C terminus of the CDS isozyme interferes with its catalytic activity or stability or whether CDS4.1 must be coexpressed with CDS4.2 to achieve a complete restoration of the wild-type phenotype.
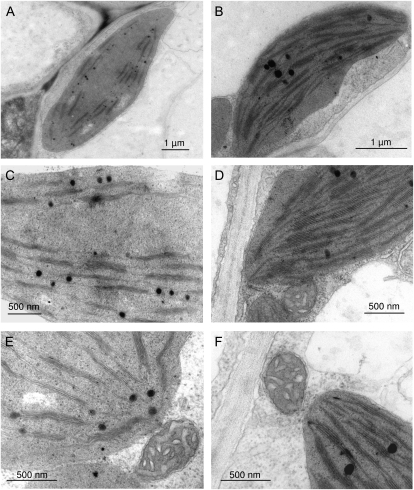
The ultrastructure of mesophyll cells from leaves of the double cds4 cds5 mutant was analyzed by transmission electron microscopy in comparison to respective cells from wild-type plants. As illustrated in Figure 8, mutant plastids were similar in size to those of wild-type organelles but showed severe defects in thylakoid structure. The surface areas of these membranes were drastically reduced so that mutant plastids contained relatively large stroma regions lacking thylakoid structure. On the other hand, the number of membranes per granum was only slightly affected. Unlike plastids, the other organelles of cds4 cds5 showed no obvious morphological differences between mutant and wild-type cells (Fig. 8).
Figure 8.
Ultrastructure of chloroplasts of cds4 cds5 mutant (A, C, and E) and wild-type plants (B, D, and F). A and B, Chloroplast of mesophyll cells. C and D, Thylakoid membranes of the chloroplast. E and F, Mitochondrium and part of the chloroplast.
PG Biosynthesis Is Impaired in the cds4 cds5 Double Mutant
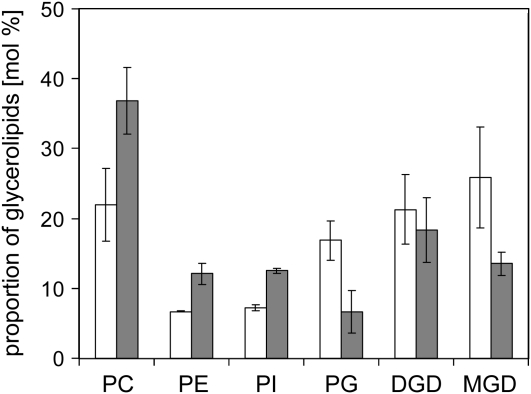
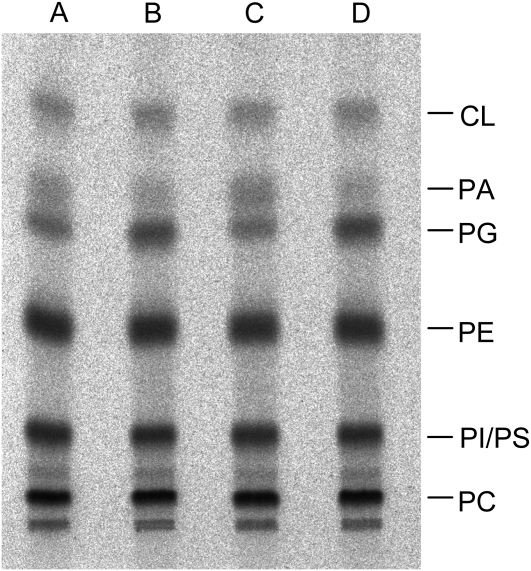
To investigate the effect of CDS4 and CDS5 gene disruption on glycerolipid metabolism, we analyzed the glycerolipid composition of the double mutant plants in comparison to wild-type plants that were cultivated on agar-solidified Murashige and Skoog medium containing Suc. As shown in Figure 9, the mutant plants had lower levels of galactolipids than wild-type plants and correspondingly higher levels of phospholipids apart from PG, the level of which was reduced to about 40% of that of the wild-type level. In addition, the fatty acid composition of PG from the mutant plants unlike that from the wild-type plants lacked 16:13t, which is specifically esterified at the sn-2 position of PG from chloroplasts. In vivo phosphate labeling experiments with cds4 cds5 double mutant and wild-type plants also showed that PG biosynthesis is compromised in the mutant plants. After 16 h of incubation with labeled phosphate, PG labeling of the mutant plants amounted to 40% of that of wild-type plants, while PA labeling was higher in the mutant than in the wild-type plants, but comprised 2% of the total labeling at most (Fig. 10). On the other hand, inactivation of both cds4 and cds5 genes had no effect on CL labeling. Hence, these data are in line with the subcellular localization of CDS4 and CDS5 isozymes within plastids. They provide evidence that inactivation of the plastidial CDS isozymes prevents plastidial PG biosynthesis and inhibits thylakoid membrane development. In that way, the double mutant plants showed a phenotype similar to that of Arabidopsis mutants in which the PGP1 gene encoding both the plastidial and mitochondrial PGP synthase was disrupted (Hagio et al., 2002; Babiychuk et al., 2003), although the biogenesis of thylakoid membranes was less severely compromised in the cds double mutant than in pgp1 loss-of-function mutant plants.
Figure 9.
Membrane lipid composition of wild-type (white bars) and cds4 cds5 (gray bars) mutant plants. After a 5-week cultivation on agar-solidified MSG medium with Suc, lipids were extracted from the plants, separated by thin-layer chromatography, and quantified by gas-liquid chromatography of fatty acid methyl esters derived from the individual glycerolipods. Three replicates were analyzed and sds are indicated. PC, Phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; DGD, digalactosyldiacylglycerol; MGD, monogalactosyldiacylglycerol.
Figure 10.
In vivo [33P]phosphate labeling pattern of phosphoglycerolipids of the cds4 cds5 mutant and the wild type. After an 18-h incubation with labeled phosphate, lipids were extracted from the mutant (lanes A and C) and wild-type plants (B and D) and separated by thin-layer chromatography, and radioactively labeled lipids were visualized with a Bioimager and identified by cochromatography with authentic lipids (PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine).
DISCUSSION
In this study, we functionally characterized CDS genes of Arabidopsis and provided direct evidence that not only CDS1 but also the further four genes encode CDS isozymes located in different subcellular compartments of plant cells. A comparison of the genes and encoded proteins suggest that CDS2 and CDS3 derive from the same ancestral gene as CDS1 and code for proteins with features typical of CDS from other eukaryotes (Kopka et al., 1997). On the other hand, CDS4 and CDS5 probably evolved via gene duplication from a respective gene of the photosynthetic active endosymbiont. Here, we demonstrated that CDS4 and CDS5 encode preproteins that are targeted to plastids. With regard to CDS5, these results are consistent with proteomic studies, which detected CDS5 in the envelope membranes of chloroplasts (Froehlich et al., 2003; Heazlewood et al., 2005). In contrast to CDS4.1 and CDS4.2, it is unlikely that CDS4.3 represents a further plastidial CDS4 isozyme because the hydrophobic N-terminal extension appears to mask the typical plastidial transit peptide of CDS4.1 and CDS4.2. Perhaps in plants translation of CDS4 transcripts predominantly starts at the second AUG so that the formation of CDS4.3 and CDS4.4 is largely prevented, but this needs further investigation. Inactivation of both CDS4 and CDS5 genes was required to obtain a mutant phenotype clearly different from that of wild-type plants (Fig. 7). Hence, the different phenotypes of the single and double mutants provide compelling evidence that CDS4 and CDS5 have redundant functions. This was supported by the phenotype of transgenic double mutant plants, in which expression of a functional CDS4 or CDS5 gene restores photoautotrophic growth (Fig. 7), as well as by the enzyme properties of CDS5 being similar to those of CDS4 (Fig. 5). It is also in line with the rice genome containing one gene only (Os02g034460) closely related to the two Arabidopsis genes.
Analysis of membrane lipid composition of the double mutant suggests that the inactivation of both CDS4 and CDS5 inhibits the formation of plastidial PG but has no effect on the mitochondrial and microsomal biosynthesis of anionic phospholipids as reflected in the levels of CL and phosphatidylinositol in the mutant in comparison to the wild type (Fig. 8). These data are in line with biochemical studies showing that plastidial CDP-DAG is channeled into the biosynthesis of plastidial PG via PGP synthase and phosphatase (Andrews and Mudd, 1984, 1985). The diminished pool of plastidial PG in the double mutant prevents proper development of thylakoid membranes (Figs. 9 and 10) because of a massive decrease in the total number of photosystem reaction centers and an even stronger one in light-harvesting antenna complexes. This was expressed by a drastic reduction in the total chlorophyll content associated with an increase in the chlorophyll a/b ratio of the pale greenish yellow leaves of cds4 cds5 and the inability of the mutant plants to grow photoautotrophically (Fig. 7). In that way, the cds double mutant resembled Arabidopsis PG mutants defective or deficient in other steps of the plastidial PG biosynthetic pathway like those catalyzed by lysophosphatidic acid acyltransferase (ATS2) or PGP synthase (PGP1; Hagio et al., 2002; Xu et al., 2002, 2006; Babiychuk et al., 2003). These mutants clearly differ from the ats1 mutants deficient in plastidial glycerol-3-phosphate acyltransferase (ATS1) catalyzing the first step of de novo glycerolipid biosynthesis (Kunst et al., 1988; Xu et al., 2006). In ats1 mutants, in which ATS1 expression was further inhibited by RNA interference, plant growth was severely inhibited, while PG level stayed largely constant, so that very small but green mutant plants developed (Xu et al., 2006). In contrast to ats1 mutant, all other mutants with defects in downstream steps of the plastidial PG pathway showed a reduction not only in growth but also in PG content so that the mutant plants were pale greenish to yellow as reported here for the cds double mutant (Fig. 7). Based on the phenotype of cyanobacteria mutants, this holds true with regard to defects in phosphatidylglycerol phosphatase catalyzing the last step of the pathway as well (Wu et al., 2006).
The cds double mutant showed more severe defects in PG biosynthesis and thylakoid structure and function than the pgp1-1 mutant carrying a point mutation in its PGP1 gene (Xu et al., 2002) or the ats1-1 mutants in which ATS2 gene expression was reduced by RNA interference (Xu et al., 2006). The defects of cds4 cds5 were, however, less pronounced than pgp1 mutants lacking both plastidial and mitochondrial PGP synthase activities (Hagio et al., 2002; Babiychuk et al., 2003). While the PG level of the cds double mutant was reduced to 40% of the wild-type level (Fig. 9), that of the pgp1 mutants comprised 12% to 15% of the wild-type content only. This difference might be merely caused by higher levels of extraplastidial PG in cds4 cds5 than in pgp1 lacking not only the plastidial but also the mitochondrial isozyme (Babiychuk et al., 2003).
The less severe defects in thylakoid biogenesis and structure of cds4 cds5 than of pgp1 (Hagio et al., 2002; Babiychuk et al., 2003) might suggest that the double mutant, unlike pgp1, still had the ability to produce low but significant amounts of PG within their plastids. This could be due to leakiness of the cds mutations, especially cds5, of which aberrant transcripts were detectable in the mutant leaves, or to a further not yet identified plastidial isozyme, such as CDS3, which might be targeted to plastids, although the probability of this prediction is relatively low (Schwacke et al., 2003) and expression of the CDS3 gene is barely detectable in leaves (Zimmermann et al., 2005). In addition, CDP-DAG imported from the ER or mitochondria might be used for the biosynthesis of plastidial PG in the double mutant plants. In vitro experiments showed that mitochondria of mammalian cells can import CDP-DAG from the ER and incorporate its phosphatidyl group into PGP and PG (Daum and Vance, 1997).
According to the different phenotype of Synechocystis mutant strains lacking either CDS (Sato et al., 2000) or PGP synthase activity (Hagio et al., 2000), however, it appears more likely that the less severe defects in thylakoid ultrastructure of cds4 cds5 from Arabidopsis than of pgp1 were not primarily caused by plastidial PG but by different intermediates formed within the plastids of the different mutants. In both mutant strains of Synechocystis, PG starvation results in defects in growth and photosynthesis. The defects in photosynthesis were, however, less severe in the ΔcdsA than in the ΔpgsA mutant strain. PG deprivation in ΔcdsA mutant cells caused reductions in cellular chlorophyll content and, thus, in a decrease of CO2-dependent photosynthesis rates, while photosynthesis rate on a chlorophyll basis was only slightly lower in the mutant than in the wild-type cells (Sato et al., 2000). On the other hand, reduction of PG in ΔpgsA mutant cells caused distinctly lower photosynthesis rates not only on a cell but also on a chlorophyll basis (Hagio et al., 2000). According to these data, it is likely that CDP-DAG enhances the defects caused by PG deprivation or that PA can partly substitute PG. To elucidate which of the various assumptions hold true with regard to the cds double mutant of Arabidopsis will be a challenging goal of future experiments.
MATERIALS AND METHODS
Computer Analysis of CDS Sequences
Information about gene models was obtained from the Aramemnon database (Schwacke et al., 2003) and The Arabidopsis Information Resource Web site (Swarbreck et al., 2008). Alignments were created by ClustalX (Larkin et al., 2007) and phylogenetic trees calculated with the Phylip/Treeview software. Searches for related CDS sequences were conducted with the BLAST tool of the National Center for Biotechnology Information homepage. For prediction of transmembrane domains and protein secondary structure, ConPred II (Arai et al., 2004), a consensus approach combining the results of several proposed methods, was used. Predictions of subcellular localization were performed by employing the consensus prediction of the Aramemnon database and the algorithms of Multiloc (Hoeglund et al., 2006) and Mitopred (Guda et al., 2004).
Cultivation of Microorganisms
Escherichia coli TOP10 (Invitrogen) was used for cloning and propagation of Gateway (Invitrogen) compatible pENTR constructs. E. coli DB3.1 (Invitrogen) was used for propagating Gateway pDEST plasmids without insert and E. coli SURE (Stratagene) was used for cloning and propagation of destination vector constructs. All E. coli strains were cultivated in Luria-Bertani medium (Carl Roth) containing suitable antibiotics at 37°C. Agrobacterium tumefaciens GV3101::pMP90RK (Koncz and Schell, 1986) was used to transform plant material. The strain was cultivated in Luria-Bertani medium with suitable antibiotics at 28°C.
The Saccharomyces cerevisiae strain YBR029c was created by the Saccharomyces Genome Deletion project (Winzeler et al., 1999) and obtained from Euroscarf. Yeast cells were cultivated in minimal medium (1.7 g/L YNB [Bio101], 5 g/L ammonium sulfate [Applichem], and 0.66 g/L CSM [Bio101] without uracil and leucin) containing 2% Glc or Gal.
Construction of CDS Expression Vectors
The cDNA clones of CDS1/At1g62430 (GSLT-cDNA: BX813816), CDS2.1/At4g22340 (pda06248; RIKEN), CDS3/At4g26770 (GSLT-cDNA: BX826782), CDS4.1/At2g45150 (pda03915; RIKEN), and CDS5/At3g60620 (pda01154; RIKEN) were obtained from the RIKEN BioResource Center (Seki et al., 1998, 2002; Sakurai et al., 2005) and Genoscope/Life Technologies (INRA, The French National Resources Centre for Plant Genomics (http://cnrgv.toulouse.inra.fr; Castelli et al., 2004). cDNAs encoding CDS2.3, CDS4.2, and CDS4.3 were created by SOE-PCR (Warrens et al., 1997). The 5′ fragments of these cDNAs were amplified with pfu polymerase (Genecraft) using wild-type DNA of Arabidopsis as template and the primer pairs CDS2.3_F/CDS2.3_SOER, CDS4.1_F/CDS4.2_SOER, and CDS4.3_F/CDS4.3_SOER (Table I). Subsequently, the amplified 5′ fragments were fused to the corresponding fragments created with the primer pairs CDS2.3_SOEF/CDS2.1R, CDS4.2_SOEF/CDS4.1_R, and CDS4.3_SOEF/CDS4.1_R and the respective cDNA clones.
Table I. Names and sequences of the primers used for PCR amplification.
| Name | Primer Sequences |
| LBb1 (SALK) | 5′-GCGTGGACCGCTTGCTGCAACT-3′ |
| LB4 (FLAG) | 5′-CGTGTGCCAGGTGCCCACGGAATAGT-3′ |
| RP_115705 | 5′-AACCCATCCTCCAGCTAACAC-3′ |
| LP_115705 | 5′-TTCTCTTCTGCCTTATCGTCG-3′ |
| RP_568C12 | 5′-CGAATCTTCTTCAACATTTTGC-3′ |
| LP_568C12 | 5′-GTGCTCAAAAGTTGCTTGTCC-3′ |
| RTCDS4F | 5′-GCTGCTCCTGCGCTTAACACT-3′ |
| RTCDS4R | 5′-CTGTCGCAATTACACCGCTGAA-3′ |
| RTCDS4exF | 5′-GGCCGTTGCTCGAGCTGAATC-3′ |
| RTCDS4exR | 5′-ACCCATCCTCCAGCTAACACAAC-3′ |
| RTCDS5F | 5′-TCGTTGTCCTGGCCTCAATC-3′ |
| RTCDS5R | 5′-TGAGTGAGCCGGAGTCTTTGAC-3′ |
| RTCDS5exF | 5′-GTTGAAGTCTGTAGGTATAAGCCATT-3′ |
| RTCDS5exR | 5′-TGATTCAGCTCGAGCAACGGC-3′ |
| RTCDS2F | 5′-GACAGGCTGGCTCCTCTGTG-3′ |
| RTCDS2R | 5′-CACTGGCAAAGAAGCCACCAAA-3′ |
| CDS1_F | 5′-CACCATGGAGGAAGAGAATGTTACTAG-3′ |
| CDS1_R | 5′-CTATGAGAGCTTGTCCTTCAGCATT-3′ |
| CDS2.1_F | 5′-CACCATGCAGAAGGAAATTGCTGGTG-3′ |
| CDS2.1_R | 5′-CTAAGATCCAATAACCTTTTCCTGCAACATTTGG-3′ |
| CDS3_F | 5′-CACCATGGCAATGGAGAAAGATC-3′ |
| CDS3_R | 5′-TTAAAACCTCCCTTGCAACTTATGTT-3′ |
| CDS4.1_F | 5′-CACCATGGCGACTTTTGCTGAACTTG-3′ |
| CDS4.1_R | 5′-TCAAACTCCGTAAAGTTTTAGGG-3′ |
| CDS4.1_wo | 5′-AACTCCGTAAAGTTTTAGGGATG-3′ |
| CDS5_F | 5′-CACCATGGCGCCTTTTGTTGAAGTC-3′ |
| CDS5_R | 5′-TCAAACTCCATGAAGTCTTACGAACGAGTAG-3′ |
| CDS5_wo | 5′-AACTCCATGAAGTCTTACGAACGAGTAG-3′ |
| CDS2.2_F | 5’-CACCATGCTAGATGTACTCAAG-3′ |
| CDS2.3_F | 5′-CACCATGATAGGTGGTTTTGCTC-3′ |
| CDS2.3_SOEF | 5′-GTTTCGAACTCTTCCAACATGCAGAAGGAAATTGCTG-3′ |
| CDS2.3_SOER | 5′-CAGCAATTTCCTTCTGCATGTTGGAAGAGTTCGAAAC-3′ |
| CDS4.2_SOEF | 5′-GGTGATGATGACCACTCAAAGAATGTGGAA-3′ |
| CDS4.2_SOER | 5′-TTCCACATTCTTTGAGTGGTCATCATCACC-3′ |
| CDS4.3_F | 5′-CACCATGACAAATACGAACACAC-3′ |
| CDS4.3_SOEF | 5′-GGGGCATTTGGAGAGCAATGGCGACTTTTG-3′ |
| CDS4.3_SOER | 5′-CAAAAGTCGCCATTGCTCTCCAAATGCCCC-3′ |
| CDS4.4_R | 5′-TTATCTGTGAGGTATCATTCTTGGTGTTACCTTTCCACCG-3′ |
| 029c_Aconf | 5′-AAACGCCCATGTAAATAACTATCAA-3′ |
| 029c_Bconf | 5′-CCCTTTCTTAAACTACAAACGAACA-3′ |
| 029c_Cconf | 5′-CTTATACTTACTTGACGTGCCCTGT-3′ |
| 029c_Dconf | 5′-TGAGAAAATCATCCTAATTACTGCC-3′ |
| kanB | 5′-CTGCAGCGAGGAGCCGTAAT-3′ |
| kanC | 5′-TGATTTTGATGACGAGCGTAAT-3′ |
| pK7_F | 5′-GACAGTAGAAAAGGAAGGTGG-3′ |
| CDS4_intR | 5′-CGCAGGAGCAGCTAAAC-3′ |
| CDS5_intR | 5′-GCCAAACATCGTACTACTCA-3′ |
| M13F | 5′-GTAAAACGACGGCCAGT-3′ |
| M13R | 5′-GGAAACAGCTATGACCATG-3′ |
| T7c | 5′-TAATACGACTCACTATAGGG-3′ |
For expression studies, the coding sequences of the 10 different cDNA sequences were amplified by PCR employing pfu polymerase and the primer pairs CDS1_F/CDS1_R, CDS2.1_F/CDS2.1_R, CDS2.2_F/CDS2.1_R, CDS2.3_F/CDS2.1_R, CDS3_F/CDS3_R, CDS4.1_F/CDS4.1_R, CDS4.2_F/CDS4.1_R, CDS4.3_F/CDS4.1_R, CDS4.3_F/CDS4.4_R, and CDS5_F/CDS5_R given in Table I and inserted into the pENTR-SD-D-TOPO vector (Invitrogen) according to the manufacturer's instructions. To express CDS4 and CDS5 as N-terminal translation fusions with a fluorescing protein, the coding sequences of the CDS cDNAs were amplified with the same forward primers as indicated above, but the reverse primers CDS4.1_R and CDS5_R were substituted by CDS4.1_wo and CDS5_wo (Table I) and cloned into pENTR-SD-D-TOPO vector. After correctness of the pENTR constructs was verified by sequencing, they were used to set up an LR reaction (Invitrogen) with pYESDEST52 (Invitrogen) for yeast transformation or with pK7RWG2.0, pK7FWG2.0, or pH7WG2.0 (Karimi et al., 2002, 2007) for agrobacteria-mediated plant transformation.
Heterologous Expression of Arabidopsis CDS Genes in Yeast Mutant Cells
Cells of the yeast strain YBR029c were transformed with pYESDEST52 AtCDS constructs by electroporation (Bio-Rad electroporator; Bio-Rad Laboratories) and cultivated on minimal selection medium. Colonies containing the correct construct tested by PCR and restriction analysis were transferred to sporulation medium (1% potassium acetate and 0.005% zinc acetate) and cultivated at 28°C until 20% of the cells had sporulated (Winzeler et al., 1999). Eight to 10 ascospores of each culture harboring a different CDS construct were brought on water agar and dissected with a micromanipulator (Nikon Instruments). Dissected spores were transferred to minimal selection medium containing 2% Gal and cultivated at 28°C until spore germination and colony formation occurred. Genomic DNA was extracted from yeast cells (Harju et al., 2004), and the presence of the disrupted ScCDS1 gene was analyzed by PCR with the primers (029c_Aconf, 029c_Bconf, 029c_Cconf, 029c_Dconf, kanB, and kanC; Table I) deduced from the KanMX cassette and flanking regions (Winzeler et al., 1999).
Preparation of Subcellular Yeast Fractions
Microsomal and mitochondrial fractions were obtained according to Zinser and Daum (1995). In brief, the cells were harvested by centrifugation and washed and treated with Zymolyase-20T (Medac) for digestion of cell walls. The obtained spheroblasts were broken up in a Dounce homogenizer. After removing intact cells by low-speed centrifugation, crude mitochondria fraction was pelleted by centrifugation at 10,000g for 20 min, washed, resuspended in 10 mm Tris-HCl buffer, pH 7.4, frozen in liquid nitrogen, and stored at −80°C. The 10,000g supernatant fraction containing the microsomes was subjected to ultracentrifugation (120,000g, 60 min). Washed microsomal fractions were stored at −80°C. Protein concentration of the subcellular fractions was determined according to Bradford (1976).
Enzymatic Assays
CDS activities of subcellular fractions of yeast cells expressing an Arabidopsis CDS gene was determined at 30°C in a total volume of 50 μL. The reaction mixture was composed of 50 mm Bis-Tris-Propane-HCl, pH 7.5, 20 mm MgCl2, 1% (w/v) Triton X-100, 0.5 mm PA (from egg yolk; Sigma-Aldrich), 1.2 mm [5-3H]CTP (42 dpm/pmol; Perkin-Elmer), and up to 20 μg membrane protein.
Phosphatidyl ethanolamine methyl transferase was assayed according to Kuchler et al. (1986) at 30°C in a total volume of 100 μL for 2 h. PGP synthase assays were performed at 30°C in a total volume of 100 μL for 30 min as described before (Müller and Frentzen, 2001). Enzymic assays were stopped and lipids were extracted by addition of 500 μL choroform:methanol (1:1, v/v) and 250 μL 1 m KCl and 0.2 m H3PO4. The reaction products recovered in the organic phase were routinely quantified by scintillation counting (LS 5000 TD Beckman). To analyze the labeled reaction products of CDS assays, they were separated by thin-layer chromatography in chloroform:methanol:acetic acid (65:25:8,v/v), detected by a Bioimager (FLA 3000; Raytest), and identified by cochromatography with authentic CDP-DAG.
Intracellular Targeting of CDS Isoforms in Nicotiana tabacum BY2
The Gateway vectors that encode the CDS4 and CDS5 proteins as N-terminal translation fusions with RFP (pK7RWG2_CDS) or GFP (pK7FWG2_CDS) were introduced into A. tumefaciens GV3101::pMP90RK (Koncz and Schell, 1986) conferring gentamycin, kanamycin, and rifampicin resistance and an additional spectinomycin/streptomycin resistance in bacteria. To transiently transform tobacco BY2 suspension cultures (Nagata et al., 1992), they were mixed with transgenic agrobacteria in the presence of 200 μm acetosyringon (Sigma-Aldrich; Sack et al., 2007). After an incubation of 4 to 5 d at 24°C in the dark under constant shaking in MSBY medium (4.3 g MS salts [Duchefa], 100 mg myo-inositol [Duchefa], 1 mg thiamine hydrochloride [Sigma-Aldrich], 0.2 mg 2,4-dichlorophenoxyacetic acid [Duchefa], and 30 g Suc per liter), BY2 cells were examined and photographed with a fluorescence microscope (Nikon Eclipse 50i).
To obtain stably transformed BY2 cultures, they were treated with cefotaxim (500 μg/mL; Duchefa) to eliminate agrobacteria and plated on MSBY agar containing kanamycin (100 μg/mL; Duchefa). After an incubation period of 3 to 4 weeks in the dark at 24°C, the appearing calli were transferred to fresh agar plates. Two-week-old calli were brought into suspension in MSBY containing kanamycin (100 μg/mL; Sack et al., 2007).
Analysis and Transformation of T-DNA Insertional Mutant Lines
Seeds of the Arabidopsis T-DNA insertion line SALK_115705 (ecotype Columbia) carrying the insertion in the CDS4 gene were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000), and seeds of the FLAG_56C812 line (ecotype Wassilewskija) carrying the insertion in the CDS5 gene were from INRA, The French National Resources Centre for Plant Genomics (http://cnrgv.toulouse.inra.fr; Bechtold et al., 1993; Bouchez et al., 1993). Surface-sterilized seeds were germinated on 0.9% (w/v) agar-solidified MSG medium (Murashige and Skoog including Gamborg B5 vitamins; Duchefa) at 24°C and 60 μmol m−2 s−1 in a light/dark cycle of 8/16 h. After 8 to 14 d, seedlings were transferred to soil or MSG medium supplemented with 3% (w/v) Suc. To induce flowering, plants were cultivated in a 16/8-h photoperiod.
Mutant plants homozygous with regard to the T-DNA insertion were identified by PCR screening using the T-DNA-specific primers LBb1 (SALK line) and LB4 (FLAG line) and gene-specific LP and RP primers (LP/RP_115705, LP/RP_568C12) given in Table I. Insertion sites of the respective T-DNAs were ascertained by sequencing the PCR products.
To determine whether the T-DNA insertion caused a knockout of the target gene on the transcriptional level, total RNA was extracted from the mutant leaves (Chomczynski and Mackey, 1995) and used as template for cDNA synthesis with random primers and reverse transcriptase (MMLV; Promega) at 37°C for 1 h. PCR analysis of the cDNA was employed with the gene-specific primer pairs RTCDS4exF/RTCDS4exR and RTCDS5exF/RTCDS5exR flanking the T-DNA insertion site of cds4 and cds5, respectively, as well as with the primer pairs RTCDS4F/RTCDS4R and RTCDS5F/RTCDS5R binding downstream the insertion sites (Table I). The CDS2-specific primers RTCDS2F/RTCDS2R were used as a control (Table I).
Arabidopsis mutants homozygous for cds4 and heterozygous for cds5 (or vice versa) were transformed by the floral dip method (Clough and Bent, 1998) with A. tumefaciens GV3101::pMP90RK carrying either the constructs pK7FWG2_CDS4.1 or pH7WG2_CDS5. Transformants were selected by resistance to hygromycin, and presence of the transgenes was verified by PCR using the primer pairs pK7_F/CDS4intR and pK7_F/CDS5intR.
Arabidopsis mutant lines were crossed according to recommendations of the Nottingham Arabidopsis Stock Center (Scholl et al., 2000).
Microscopy Analysis
To determine the ultrastructure of chloroplasts, Arabidopsis leaves were fixed in 2% (w/v) glutaraldehyde (Sigma-Aldrich) and 2% (w/v) formaldehyde (freshly prepared from paraformaldehyde; Merck) in PBS (pH 7.2) for 48 h at 4°C. After rinsing the samples three times in 0.1 m sodium cacodylate (Sigma-Aldrich) containing 7% (w/v) Suc for 10 min on ice, they were rinsed twice in 0.1 m sodium cacodylate and then postfixed in 2% (w/v) OsO4 (Sigma-Aldrich) in 0.1 m sodium cacodylate for 2 h on ice. The samples were rinsed again in 0.1 m sodium cacodylate at room temperature and dehydrated in ascending concentrations of ethanol (30% and 40% for 15 min each, 50% for 60 min, 60%, 75%, and 90% for 30 min, 100% overnight, and 100% for 60 min). After dehydration, samples were equilibrated twice in propylene oxide (Serva) for 30 min, followed by 50% (w/v) propylene oxide and 50% (w/v) resign (Epon 812; Serva) overnight. The samples were incubated twice in 100% Epon for 2 h and then embedded in Epon 812. For light microscopy, semithin sections (1 μm) were stained with toluidine blue. For transmission electron microscopy, ultrathin sections were contrasted with aqueous solutions of 2% (w/v) uranyl acetate (Serva) for 20 min and 0.2% (w/v) lead citrate (Sigma-Aldrich) for 7 min and examined with a Zeiss EM10 electron microscope.
Lipid Analysis
Mutant and wild-type plants cultured for 4 weeks under sterile conditions on agar-solidified MSG medium containing Suc were used for in vivo labeling experiments with [33P]phosphate (Hartmann Analytik) and lipid analysis. In vivo labeling for 16 h was carried out essentially as described before (Babiychuk et al., 2003), but lipids were separated by thin-layer chromatography in chloroform:methanol:acetic acid (65:25:8, v/v). Unlabeled membrane lipids were separated by thin-layer chromatography in chloroform:methanol:acetic acid:water (91:30:4:4; v/v) and quantified via their methyl esters by gas-liquid chromatography (Babiychuk et al., 2003). Pigments were analyzed according to Lichtenthaler (1987).
The accession numbers for CDS1 to CDS5 are At1g62430 (BX813816), At4g22340 (pda06248), At4g26770 (BX826782), At2g45150 (pda03915), and At3g60620 (pda01154), respectively.
Acknowledgments
We thank Professor U. Klinner for advice on yeast spore separation and Dr. T. Rademacher for his help with BY2 cell transformation. We also thank Dr. R. Sadre for critically reading the manuscript. The cDNA clones pda06248, pda03915, and pda01154 were developed by the plant genome project of RIKEN Genomic Sciences Center and provided by the RIKEN BioResource Center through the National Bio-Resource Project of Ministry of Education, Culture, Sports, Science and Technology, Japan. The sequence region of pK7RWG2 encoding RFP was kindly provided by Dr. R.Y. Tsien (Howard Hughes Medical Institute, University of California, San Diego).
References
- Andrews J, Mudd JB. (1984) Characterization of CDP-DG and PG synthesis in pea chloroplasts. Siegenthaler PA, Eichenberger W, , Structure, Function and Metabolism of Plant Lipids. Elsevier North Holland Biomedical Press, Amsterdam, pp 131–134 [Google Scholar]
- Andrews J, Mudd JB. (1985) Phosphatidylglycerol synthesis in pea chloroplasts. Plant Physiol 79: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. (2004) ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res 32: W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Müller F, Eubel H, Braun HP, Frentzen M, Kushnir S. (2003) Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J 33: 899–909 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III 316: 1194–1199 [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al. (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M. (1993) A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C R Acad Sci III 316: 1188–1193 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Castelli V, Aury JM, Jaillon O, Wincker P, Clepet C, Menard M, Cruaud C, Quétier F, Scarpelli C, Schächter V, et al. (2004) Whole genome sequence comparisons and “full-length” cDNA sequences: a combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res 14: 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Mackey K. (1995) Modification of the TRI Reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19: 942–945 [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Daum G, Vance JE. (1997) Import of lipids into mitochondria. Prog Lipid Res 36: 103–130 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Hebb DM, Richards KD, Lin JM, Ryan PR, Gardner RC. (1999) Cloning and expression of a wheat (Triticum aestivum L.) phosphatidylserine synthase cDNA. J Biol Chem 274: 7082–7088 [DOI] [PubMed] [Google Scholar]
- Dowhan W. (1997) CDP-diacylglycerol synthase of microorganisms. Biochim Biophys Acta 1348: 157–165 [DOI] [PubMed] [Google Scholar]
- Frentzen M. (2004) Phosphatidylglycerol and sulfoquinovosyldiacylglycerol: anionic membrane lipids and phosphate regulation. Curr Opin Plant Biol 7: 270–276 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS. (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2: 413–425 [DOI] [PubMed] [Google Scholar]
- Guda C, Guda P, Fahy E, Subramanian S. (2004) MITOPRED: a web server for the prediction of mitochondrial proteins. Nucleic Acids Res 32: 372–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagio M, Gombos Z, Várkonyi Z, Masamoto K, Sato N, Tsuzuki M, Wada H. (2000) Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol 124: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagio M, Sakurai I, Sato S, Kato T, Tabata S, Wada H. (2002) Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol 43: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Hanenberg A, Heim S, Wissing JB, Wagner K. (1993) Characterization of cytidine triphosphate:phosphatidate cytidyltransferase from suspension cultured Catharanthus roseus cells. Plant Sci 88: 13–18 [Google Scholar]
- Harju S, Fedosyuk H, Peterson KR. (2004) Rapid isolation of yeast genomic DNA: Bust n′Grab. BMC Biotechnol 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH. (2005) Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol 139: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeglund A, Doennes P, Blum T, Adolph HW, Kohlbacher O. (2006) MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22: 1158–1165 [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Turkenburg M, Poll-The BT, Karall D, Pérez-Cerdá C, Morrone A, Malvagia S, Wanders RJ, Kulik W, Vaz FM. (2009) The enigmatic role of tafazzin in cardiolipin metabolism. Biochim Biophys Acta 1788: 2003–2014 [DOI] [PubMed] [Google Scholar]
- Icho T, Sparrow CP, Raetz CRH. (1985) Molecular cloning and sequencing of the gene for CDP-diglyceride synthetase of Escherichia coli. J Biol Chem 260: 12078–12083 [PubMed] [Google Scholar]
- Inglis-Broadgate SL, Ocaka L, Banerjee R, Gaasenbeek M, Chapple JP, Cheetham ME, Clark BJ, Hunt DM, Halford S. (2005) Isolation and characterization of murine Cds (CDP-diacylglycerol synthase) 1 and 2. Gene 356: 19–31 [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P. (2007) Recombinational cloning with plant Gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kelley MJ, Carman GM. (1987) Purification and characterization of CDP-diacylglycerol synthase from Saccharomyces cerevisiae. J Biol Chem 262: 14563–14570 [PubMed] [Google Scholar]
- Kinney AJ. (1993) Phospholipid head groups. Moore TS, , Lipid Metabolism in Plants. CRC Press, Boca Raton, FL, pp 259–284 [Google Scholar]
- Kleppinger-Sparace KF, Moore TS. (1985) Biosynthesis of cytidine 5′-diphosphate-diacylglycerol in endoplasmic reticulum and mitochondria of castor bean endosperm. Plant Physiol 77: 12–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kopka J, Ludewig M, Müller-Röber B. (1997) Complementary DNAs encoding eukaryotic-type cytidine-5′-diphosphate-diacylglycerol synthases of two plant species. Plant Physiol 113: 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Daum G, Paltauf F. (1986) Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J Bacteriol 165: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C. (1988) Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc Natl Acad Sci USA 85: 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lykidis A, Jackowski S. (2001) Regulation of mammalian cell membrane biosynthesis. Prog Nucleic Acid Res Mol Biol 65: 361–393 [DOI] [PubMed] [Google Scholar]
- Mackenzie SA. (2005) Plant organellar protein targeting: a traffic plan still under construction. Trends Cell Biol 15: 548–554 [DOI] [PubMed] [Google Scholar]
- Martin D, Gannoun-Zaki L, Bonnefoy S, Eldin P, Wengelnik K, Vial H. (2000) Characterization of Plasmodium falciparum CDP-diacylglycerol synthase, a proteolytically cleaved enzyme. Mol Biochem Parasitol 110: 93–105 [DOI] [PubMed] [Google Scholar]
- Moore TS. (1982) Phospholipid biosynthesis. Annu Rev Plant Physiol 33: 235–259 [Google Scholar]
- Müller F, Frentzen M. (2001) Phosphatidylglycerophosphate synthases from Arabidopsis thaliana. FEBS Lett 509: 298–302 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. (1992) Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Sack M, Paetz A, Kunert R, Bomble M, Hesse F, Stiegler G, Fischer R, Katinger H, Stoeger E, Rademacher T. (2007) Functional analysis of the broadly neutralizing human anti-HIV-1 antibody 2F5 produced in transgenic BY-2 suspension cultures. FASEB J 21: 1655–1664 [DOI] [PubMed] [Google Scholar]
- Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K. (2005) RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res 33: D647–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Hagio M, Wada H, Tsuzuki M. (2000) Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc Natl Acad Sci USA 97: 10655–10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge U-I, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K. (1998) High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J 15: 707–720 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Shen H, Heacock PN, Clancey CJ, Dowhan W. (1996) The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem 271: 789–795 [DOI] [PubMed] [Google Scholar]
- Sparrow CP, Raetz CRH. (1985) Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. J Biol Chem 260: 12084–12091 [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta M, Bulfone A, Gattuso C, Rossi E, Mariani M, Consalez GG, Zuffardi O, Ballabio A, Banfi S, Franco B. (1999) Identification and characterization of CDS2, a mammalian homolog of the Drosophila CDP-diacylglycerol synthase gene. Genomics 55: 68–77 [DOI] [PubMed] [Google Scholar]
- Wada H, Murata N. (2007) The essential role of phosphatidylglycerol in photosynthesis. Photosynth Res 92: 205–215 [DOI] [PubMed] [Google Scholar]
- Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WB, Mackenzie SA. (2009) Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrens AN, Jones MD, Lechler RI. (1997) Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186: 29–35 [DOI] [PubMed] [Google Scholar]
- Weeks R, Dowhan W, Shen H, Balantac N, Meengs B, Nudelman E, Leung DW. (1997) Isolation and expression of an isoform of human CDP-diacylglycerol synthase cDNA. DNA Cell Biol 16: 281–289 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Bruno A, Bangham R, Benito R, Boeke JD, Bussey H, et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wu F, Yang Z, Kuang T. (2006) Impaired photosynthesis in phosphatidylglycerol-deficient mutant of cyanobacterium Anabaena sp. PCC7120 with a disrupted gene encoding a putative phosphatidylglycerophosphatase. Plant Physiol 141: 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Niemeyer B, Colley N, Socolich M, Zuker CS. (1995) Regulation of PLC-mediated signalling in vivo by CDP-diacylglycerol synthase. Nature 373: 216–222 [DOI] [PubMed] [Google Scholar]
- Xu C, Härtel H, Wada H, Hagio M, Yu B, Eakin C, Benning C. (2002) The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol 129: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yu B, Cornish AJ, Froehlich JE, Benning C. (2006) Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3- phosphate acyltransferase. Plant J 47: 296–309 [DOI] [PubMed] [Google Scholar]
- Xue HW, Chen X, Mei Y. (2009) Function and regulation of phospholipid signalling in plants. Biochem J 421: 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W. (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10: 407–409 [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11: 493–536 [DOI] [PubMed] [Google Scholar]