Abstract
After flower pollination, a programmed process called abscission occurs in which unwanted floral organs are actively shed from the main plant body. We found that a member of the DOF (for DNA binding with one finger) transcription factor family, Arabidopsis (Arabidopsis thaliana) DOF4.7, was expressed robustly in the abscission zone. The Arabidopsis 35S::AtDOF4.7 lines with constitutive expression of AtDOF4.7 exhibited an ethylene-independent floral organ abscission deficiency. In these lines, anatomical analyses showed that the formation of the abscission zone was normal. However, dissolution of the middle lamella failed to separate between the cell walls. AtDOF4.7 was identified as a nucleus-localized transcription factor. This protein had both in vitro and in vivo binding activity to typical DOF cis-elements in the promoter of an abscission-related polygalacturonase (PG) gene, PGAZAT. Overexpression of AtDOF4.7 resulted in down-regulation of PGAZAT. AtDOF4.7 interacted with another abscission-related transcription factor, Arabidopsis ZINC FINGER PROTEIN2. Taken together, our results suggest that AtDOF4.7 participates in the control of abscission as part of the transcription complex that directly regulates the expression of cell wall hydrolysis enzymes.
Organ shedding, or abscission, is a critical cell separation process that may occur throughout the life cycle of plants (Roberts et al., 2002; Lewis et al., 2006). Abscission can be an adaptation to environment stresses, such as water or nutrient deficiency, oxygen damage, and pathogen attack (Taylor and Whitelaw, 2001), or a developmentally controlled program that occurs after leaf senescence, flower pollination, and fruit maturation (van Doorn and Stead, 1997; Roberts et al., 2002). At the predetermined abscission position, an abscission zone (AZ) develops. The AZ is composed of a few layers of dense cytoplasmic cells (Sexton and Roberts, 1982; Patterson, 2001). The AZ cells recognize abscission signals that activate cell wall-loosening protein factors. Several wall-loosening proteins have been well documented for their AZ expression, including endoglucanases (del Campillo et al., 1990; Tucker et al., 1991; Tucker and Milligan, 1991; Taylor et al., 1994; del Campillo, 1999; Mishra et al., 2008), polygalacturonases (PGs; del Campillo et al., 1990; Taylor et al., 1991, 1993; Kalaitzis et al., 1995; del Campillo, 1999; Gonzalez-Carranza et al., 2002, 2007a; Jiang et al., 2008), and expansins (Cho and Cosgrove, 2000; Belfield et al., 2005). The middle lamellae of AZ cell walls are presumably loosened and dissolved by at least some of the listed wall proteins, resulting in organ detachment from the main body of the plant.
Floral organ abscission in Arabidopsis (Arabidopsis thaliana) has been used as a model system for studying the genetics underlying the abscission process (Bleecker and Patterson, 1997; Patterson, 2001). Certain genes involved in upstream signaling of abscission have been identified in this system. Ethylene is considered to be a fundamental regulator of the abscission rate (Bleecker and Patterson, 1997; Patterson, 2001; Taylor and Whitelaw, 2001; Patterson and Bleecker, 2004). Defects in the components of the ethylene perception and signaling pathways will delay abscission to various degrees. In ethylene-insensitive mutants, such as etr1 and ein2, both floral organ abscission and senescence are delayed (Patterson and Bleecker, 2004; Lewis et al., 2006).
Ethylene is not the only regulator of abscission. A ligand gene family, including INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and five IDA-LIKE (IDL) genes, participates in the control of abscission, and its function is not affected by exogenous ethylene. The loss-of-function mutant, ida, has a delayed abscission but normal senescence (Butenko et al., 2003), showing that abscission and senescence are not obligately coupled. Transgenic lines with IDA overexpression exhibit rapid shedding of floral organs and vestigial AZs of pedicels and cauline leaves (Stenvik et al., 2006, 2008). Two Leu-rich repeat receptor-like kinases (LRR-RLKs), HAESA (HAE) and HAESA-LIKE2 (HSL2), are activated by interactions with IDA or IDLs (Stenvik et al., 2008). The activity of HAE/HSL2 might be inhibited by another LRR-RLK, EVERSHED (EVR; Leslie et al., 2010). Moreover, an ARF-GAP protein, NEVERSHED, could regulate abscission by blocking the function of EVR (Liljegren et al., 2009; Leslie et al., 2010). A mitogen-activated protein kinase cascade is probably activated by HAE/HSL2 to participate in abscission signal transduction (Cho et al., 2008). In addition, some genes that relate leaf and floral organ patterning also influence the capacity for abscission. These genes include two well-known leaf-patterning factors, BOP1/BOP2 (McKim et al., 2008), a MADS box domain transcription factor, AGL15, and two chromatin regulators, ARP4 and ARP7 (Fernandez et al., 2000; Kandasamy et al., 2005a, 2005b). Moreover, the high-throughput transcriptome profiling revealed significantly regulated AZ genes that encoded multiple cell wall proteins and transcription factors. Of them, one zinc finger transcription factor, Arabidopsis ZINC FINGER PROTEIN2 (AtZFP2), was studied further with respect to its AZ-localized gene expression. Overexpression of AtZFP2 significantly delays floral organ abscission (Cai and Lashbrook, 2008).
Little is known about the regulation of enzymes that catalyze cell wall hydrolysis inside AZ. However, our study shows that AtDOF4.7, a member of the Arabidopsis DOF transcription factor family, may play a role in abscission regulation. Significant deficiencies in abscission were observed in the plants overexpressing the AtDOF4.7 gene. Based on the expression patterns and regulation of downstream genes, we propose that AtDOF4.7 participates in the transcriptional regulation of floral organ abscission via an effect on cell wall hydrolase gene expression.
RESULTS
The Expression Pattern of AtDOF4.7
To establish the tissue expression pattern of AtDOF4.7, total RNA was extracted from Arabidopsis seedlings, leaves, roots, stems, flowers, and young siliques. RNAs were subjected to semiquantitative reverse transcription (RT)-PCR analyses. ACTIN was used as the control gene. AtDOF4.7 transcripts from the equivalent cDNA from different organs were examined. As shown in Figure 1A, the AtDOF4.7 transcript was only detected in flowers and young siliques but not in other tissues after 30 cycles of PCR amplification. AtDOF4.7 tissue-specific expression was further examined using the GUS reporter gene under the control of the AtDOF4.7 promoter. GUS expression was detected strongly at the base of all organs of the flower, especially in the AZs of the petals, stamens, and sepals (Fig. 1B, a, d, and g). This tissue-specific distribution pattern of the AtDOF4.7 transcript suggested a potential contribution of AtDOF4.7 to abscission. Besides, low levels of the GUS stains were also detected in other sites, including the sepals (Fig. 1B, a), filaments (Fig. 1B, d), the stigmatic papillae and the tips of young elongation siliques (Fig. 1B, e), the base of pedicels (Fig. 1B, f), and the bases of leaf trichomes (Fig. 1B, c).
Figure 1.
Expression pattern of AtDOF4.7. A, RT-PCR analysis of AtDOF4.7 expression in root, stem, leaf, flower, and siliques from 4-week-old Arabidopsis plants and 1-week-old whole seedlings. ACTIN (At3g18780) was used as the internal control gene. B, Expression of PromoterAtDOF4.7::GUS. Reporter gene expression in plants harboring the PromoterAtDOF4.7::GUS fusion is as follows: GUS activity in transgenic inflorescences (a); GUS expression restricted to the base of the floral organ in the siliques at position 6 (b); GUS expression in leaf trichomes (c); GUS expression in flowers at position 2 (d); GUS expression in the style of position 2 flowers (e); GUS expression at the base of pedicels (f); and a closer view of siliques at position 6 showing GUS expression in floral organ AZ (g). Bars = 1.5 mm. C, Time course expression of PromoterAtDOF4.7::GUS. The time course of GUS expression from different developmental positions of flowers and siliques of 5-week-old Arabidopsis is shown. The flowers were counted along the inflorescence from position 1, assuming that the flower at position 1 had visible white petals at the top of the inflorescence.
Because the AZ morphology changes during the time course of floral organ abscission, a detailed analysis of the spatial and temporal GUS expression pattern along the inflorescence was performed. The time course of GUS accumulation revealed that the AtDOF4.7 expression could be detected at the site of floral organ abscission from the position 4 flower (we defined the position 1 flower as the first flower with visible white petals and then counted down along the inflorescence sequence). The expression of AtDOF4.7 continued until the silique at position 10 and disappeared at position 11, where the wild-type plants finished their flower abscission process (Fig. 1C).
Generation and Characterization of Transgenic Plants
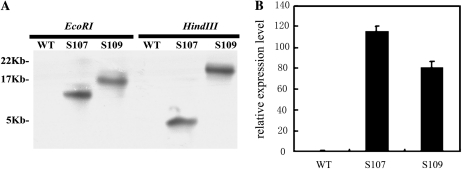
To investigate the potential function of the AtDOF4.7 gene, the AtDOF4.7 open reading frame sequence was cloned and fused with the cauliflower mosaic virus 35S promoter in a binary vector (pCAMBIA 1391). This construct was used to generate transgenic Arabidopsis plants, and over 30 independent hygromycin-resistant transformants (T1) were selected. Two homozygous single insertion T3 lines verified by Southern-blot analyses were selected and named S107 and S109 for further analysis (Fig. 2A). These two transgenic lines displayed AtDOF4.7 transcript levels that increased by 115.3-fold (S107) and 80.9-fold (S109) in comparison with wild-type plants in young siliques from positions 4 to 6 (Fig. 2B).
Figure 2.
Molecular identification of AtDOF4.7 overexpression plants. A, Southern hybridization of DNA from two AtDOF4.7 transgenic lines (S107 and S109) and the wild type (WT). Genomic DNA samples were digested with restriction enzymes HindIII and EcoRI and were then hybridized with a Hygromycin Phosphotransferase probe. B, Identification of AtDOF4.7 overexpression in 35S::AtDOF4.7 transgenic lines. The relative expression level of AtDOF4.7 was analyzed by quantitative real-time PCR from 4-week-old wild-type and transgenic plants. ACTIN was used as the internal control gene.
Delayed Abscission Induced by the Overexpression of AtDOF4.7
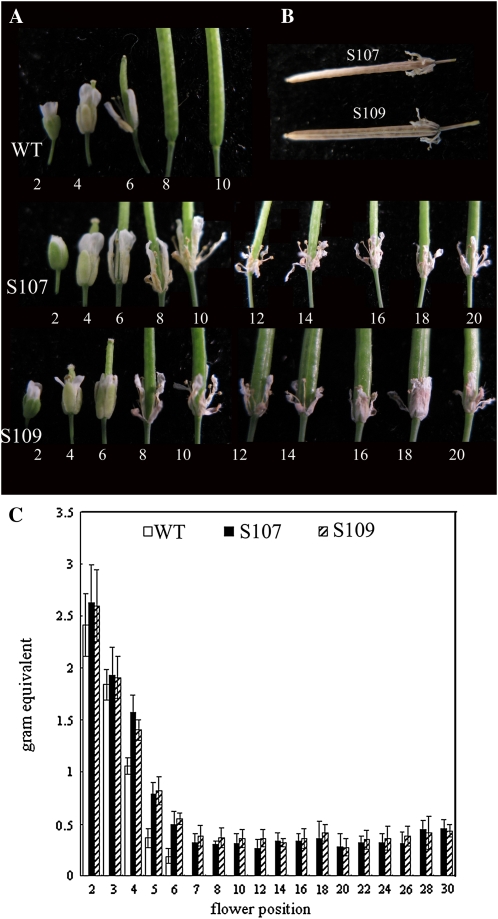
The failure of floral organ abscission is the most significant aberrant phenotype observed consistently in most transgenic lines, including single- and multiple-insertion lines, as well as in homozygous and hemizygous lines (Fig. 3A). After anthesis occurs in wild-type plants, the sepals, petals, and stamens normally abscise in a short period. As shown in Figure 3A, the floral organs were detached from the siliques at position 8, and statistical analysis showed that the flowers shed the sepals, petals, and stamens at position 8.23 ± 0.77 (Table I) in wild-type Arabidopsis plants. In contrast, in the AtDOF4.7 overexpression lines, the floral organs were attached closely at all positions, even on the mature and dry siliques past position 30 of the inflorescence (Fig. 3B).
Figure 3.
Floral organ abscission is delayed in plants with AtDOF4.7 overexpression. Flowers of 5-week-old plants were analyzed and compared. A, Flowers along the inflorescence in the wild type (WT) and AtDOF4.7 overexpression lines (S107 and S109). B, Flower organs were still attached to the dry siliques in S107 and S109. C, Petal breakstrength, which is the force required to peel the petals from the flowers, was measured from positions 2 to 30. Error bars indicate sd from measurements for 17 wild-type, S107, and S109 plants each, with a minimum of 15 measurements at each position. [See online article for color version of this figure.]
Table I. Characterization of floral organ abscission and senescence in wild-type and overexpression lines (n = 48).
| Line | Sepal Yellowing Position ± sd | Floral Organ Withering Position ± sd | Abscission Position ± sd |
| Wild type | 3.44 ± 0.67 | 6.60 ± 0.95 | 8.23 ± 0.77 |
| S107 | 3.81 ± 0.70 | 6.71 ± 0.68 | naa |
| S109 | 3.67 ± 0.62 | 6.71 ± 0.73 | naa |
Not observed even in the last position.
Senescence usually occurs synchronously with the abscission process in wild-type plants. Therefore, in some mutants with abscission-delayed genetic modifications, such as the etr1-1 mutant, the senescence process is also slowed down (Patterson and Bleecker, 2004). We examined the position of sepal yellowing and the position of floral organ withering in the wild type and the AtDOF4.7 overexpression lines. Unlike etr1-1, the floral organs of the AtDOF4.7 overexpression lines showed a normal progression of senescence similar to that of the wild type, which demonstrated yellowing sepals at positions 3 and 4 and withering sepals, petals, and stamens at positions 6 and 7 (Table I; Fig. 3A). These results suggest that ethylene has a significant effect on regulating senescence but is insufficient to induce abscission in AtDOF4.7 overexpression lines.
To measure the quantitative difference of abscission between wild-type and AtDOF4.7 overexpression plants, the force needed to remove the petals from the plants was assayed using a petal breakstrength meter (Lease et al., 2006). The breakstrength quantifications were highly consistent with the phenotype observations at different positions. In wild-type plants, the breakstrength force decreased rapidly from position 2 to position 6. After position 6, the breakstrength decreased to zero because the petal was already detached. However, in AtDOF4.7 overexpression plants, force was required to remove the petals from the plants at all tested positions, indicating that the decrease in the required breakstrength was significantly delayed (Fig. 3C).
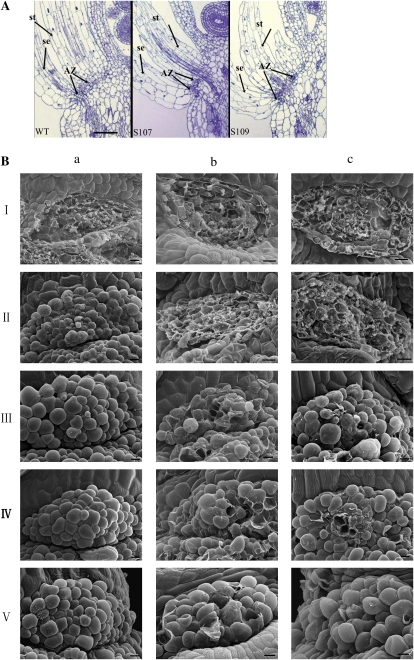
The programmed formation of the AZs is the foundation of normal abscission. Therefore, we characterized the AZs in flowers just before anthesis in two transgenic lines as well as in wild-type plants. Floral AZs were positioned at the junction between the bases of all floral organs and their receptacles. Among the wild-type and transgenic lines, the AZs were anatomically indistinguishable with respect to the size and organization of the AZ cells under light microscopy (Fig. 4A). These results suggest that the formation of AZs is normal in AtDOF4.7 overexpression lines. Therefore, the phenotype of delayed abscission may be caused by physiological events after AZ formation.
Figure 4.
Morphological analysis of AZs on plants with AtDOF4.7 overexpression. A, Anatomical characteristics of the AZ in AtDOF4.7 overexpression plants are similar to those of the wild type (WT). A comparative anatomical analysis of the AZ showed the size and organization of cells in the AZs at the base of sepals (se) and stamens (st) in longitudinal sections of flowers at position 4. Bar = 600 μm. B, The morphology of stamen AZs. SEM of the fracture plane of AZ cells of petals in wild-type (a), S107 (b), and S109 (c) flowers at positions 2 (I), 6 (II), 8 (III), 15 (IV), and 20 (V) after forcible removal or natural abscission. Wild-type petals abscise naturally at position 8. Bars = 10 μm. [See online article for color version of this figure.]
Scanning electron microscopy (SEM) was used to examine the detailed morphological difference of AZs between the wild type and AtDOF4.7 overexpression lines by either manual removal or natural abscission of lateral floral organs (Fig. 4B). At position 2, cells of the stamen AZs in both the wild type and AtDOF4.7 overexpression lines were torn after the organ was removed forcibly from the flowers. The damage of the AZ cells reflected the strength of the cell wall connections between the cell layers. A difference appeared from position 6 downward. In wild-type lines, cells on both sides of the stamen AZ fracture plane were intact after abscission, suggesting that the middle lamella between the AZ cell walls was dissolved during the initiation of abscission. However, cells on the fracture plane were still broken at the same position in S107 and S109, which occurred at an earlier position in wild-type plants. These results correspond to a delayed abscission phenotype in the AtDOF4.7 overexpression lines. Compared with wild-type lines, the undissolved stamen AZ cells on the fracture plane margins were first observed at position 8 in overexpression lines and broken cells were observed in the AZs, especially in the middle region, until position 20. The number of torn AZ cells decreased with the progressive maturation of siliques (Fig. 4B). Taking these results together, we concluded that cells in the wild type were not disrupted after stamen abscission or removal but that the cells appeared to be torn apart when the stamens were removed from the AtDOF4.7 overexpression lines. These results suggest that the AtDOF4.7 overexpression plants have stamens with AZ cells that are still cemented together.
A deficiency in the rounding up process of the AZ cells could contribute to the delayed abscission (Patterson, 2001). However, this process was not interrupted by the overexpression of AtDOF4.7. We observed similar progression of cell rounding in both the wild type and the overexpression lines, beginning at position 8 (Fig. 4B, IV and V). The enlarged cells in the overexpression lines appeared to be more obscure than in the wild type, suggesting that the middle lamellae in AZ cells of AtDOF4.7 overexpression lines had not been completely dissolved (Fig. 4B, III–V).
Other Phenotypic Features of AtDOF4.7 Overexpression Plants
Other features of the AtDOF4.7 overexpression plants besides abscission were examined carefully. The development of flowers, siliques, and trichomes where AtDOF4.7 was expressed in situ was not significantly different between the wild type and the overexpression lines. However, the AtDOF4.7 ectopic expression lines exhibited an obvious defect in growth. The leaves of AtDOF4.7 overexpression plants at 20 d were substantially smaller than wild-type leaves under the same growth conditions, although the number and the morphology of leaves were not affected (Supplemental Fig. S1).
The AtDOF4.7 Overexpression Plants Are Sensitive to Ethylene
Abscission processes have been divided into ethylene-dependent and ethylene-independent types (Patterson and Bleecker, 2004). We tested the ethylene sensitivity of plants overexpressing AtDOF4.7 and determined whether delayed floral organ abscission in these plants could be reversed by exogenous ethylene application. First, a typical “triple-response” assay (Kieber et al., 1993) was used to determine whether the AtDOF4.7 overexpression plants have normal ethylene perception and response. Seeds germinated vertically in the dark on growth medium supplemented with 5 μm 1-aminocyclopropane-1-carboxylic acid (ACC), which is the natural precursor of ethylene. The AtDOF4.7 overexpression plants displayed similar triple-response morphological changes compared with the wild-type plants, including the inhibition of hypocotyl growth and root elongation, radical swelling of the hypocotyls, and exaggeration of the curvature of the apical hook (Fig. 5A). Therefore, the AtDOF4.7 overexpression seedlings did not show any deficiency in the perception of or response to ethylene.
Figure 5.
Ethylene responses of wild-type and AtDOF4.7 overexpression plants. A, The triple-response assay was conducted in growing seedlings of wild-type (WT) and AtDOF4.7 overexpression (S107 and S109) plants on vertical half-strength Murashige and Skoog (MS) agar plates containing 5 μm ACC for 3 d in the dark. B, Comparison of flowers of 4-week-old wild-type and AtDOF4.7 overexpression plants at positions 0 to 6 from left to right that were exposed to pure air or to air with 10 μL L−1 ethylene. [See online article for color version of this figure.]
Second, we exposed the inflorescences of wild-type and overexpression plants to air with 10 μL L−1 ethylene or pure air as the control. Incubation with ethylene significantly promoted the senescence of the floral organs as well as abscission in wild-type plants, as compared with incubation in pure air. As seen in wild-type plants, ethylene also accelerated the process of senescence in the plants overexpressing AtDOF4.7. However, ethylene incubation had no effect on delayed abscission in AtDOF4.7 overexpression plants. These observations suggest that exogenous ethylene application could not rescue the floral organ abscission deficiency in the AtDOF4.7 overexpression plants, even though these plants could perceive ethylene (Fig. 5B).
AtDOF4.7 Protein Is Located in the Nucleus
To gain insight into the function of the AtDOF4.7 gene, its coding region sequence was fused to GFP under the control of the cauliflower mosaic virus 35S promoter. The construct was expressed transiently in onion (Allium cepa) epidermal cells by gene gun transformation. The cells expressing the AtDOF4.7::GFP fusion proteins were examined with a confocal microscope (Fig. 6). For comparison, free GFP was used as a control for the localization of the protein in the nuclei and cytoplasm.
Figure 6.
Subcellular localization of AtDOF4.7. Transient expression in onion epidermal cells of free GFP and AtDOF4.7::GFP fusion proteins. A, Localization of free GFP fluorescence in onion epidermal cells. B, Localization of AtDOF4.7::GFP fluorescence in onion epidermal cells. Left to right: GFP fluorescence image (I), DAPI staining image (II), transmission image (III), and merged image (IV). Bar = 100 μm.
With GFP alone, fluorescence was detected strongly in the nucleoplasm, cytoplasm, and plasma membrane (Fig. 6A). In contrast, the AtDOF4.7::GFP fusion protein only colocalized with the nuclei-specific 4′,6-diamidino-2-phenylindole (DAPI) stain (Fig. 6B), indicating that AtDOF4.7 is a nucleus-localized protein and may function as a transcription factor.
Abscission-Related Gene Expression in AtDOF4.7 Overexpression Plants
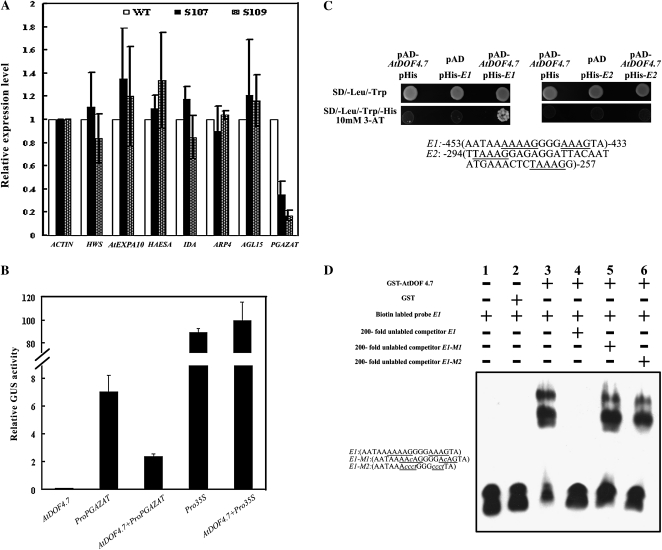
Because AtDOF4.7 belongs to a family of transcription factors, we were interested in determining whether it regulates the expression of genes in abscission. Several components with putative signaling roles or functional enzymatic activities in abscission were selected to analyze the regulation of their expression by AtDOF4.7 overexpression. Their transcript levels in positions 4 to 6 young siliques were compared between the wild type and the AtDOF4.7 overexpression lines by quantitative RT-PCR (Fig. 7A). For most of the selected genes, including HWS, HAESA, IDA, AGL15, AtEXP10, and ARP4, no significant changes (P > 0.05) were observed, except for a PG gene, PGAZAT. The relative PGAZAT expression ratios between the wild type and the overexpression lines were 0.35 ± 0.12 in the S107 line and 0.17 ± 0.04 in the S109 line. These results suggested that PGAZAT expression might be suppressed significantly by overexpression of AtDOF4.7.
Figure 7.
PGAZAT is regulated by AtDOF4.7. A, Abscission-related gene expression in AtDOF4.7 overexpression lines. Real-time RT-PCR analysis was performed to quantify the transcript abundance of abscission-related genes in wild-type (WT) and AtDOF4.7 overexpression (S107 and S109) plants. Relative expression levels were calculated and normalized with respect to the corresponding gene transcript levels in the wild type. ACTIN was used as the internal control gene. The relative transcript levels were averaged over three biological replicates and are shown with the sd (error bars). B, Regulation of AtDOF4.7 on the PGAZAT promoter. The promoter activities are shown as relative GUS activity in transient cotransfection assays with the 35S::GUS control as a reference. C, The AtDOF4.7 protein binds to cis-DNA elements in the promoter region of PGAZAT. The underlined sequences indicate the core sequence of the DOF-binding elements. The reporter pHIS2 vector carrying the corresponding fragment and the effector pAD-AtDOF4.7 vector were cotransfected into yeast Y187 cells. Growth of the transfected yeast cells on 3-AT medium indicates that the AtDOF4.7 protein can bind to the PGAZAT promoter in a sequence-specific manner. D, Gel-shift analysis of the AtDOF4.7 protein. Electrophoretic mobility shift assays of the recombinant AtDOF4.7 protein fused to GST and the PGAZAT promoter fragment including the AAAG motif are shown. E1, A copy of the PGAZAT promoter fragment; E2, single-base mutation of the AAAG motif in E1; E3, four-base mutation of the AAAG motif in E1.
Expression of PGAZAT Is Suppressed by AtDOF4.7
To confirm that PGAZAT expression is down-regulated by AtDOF4.7, a transcription factor-promoter interaction was analyzed by transient cotransformation in cultured Arabidopsis cells. A 1,476-bp fragment of the PGAZAT promoter, which was identified previously as the AZ-expressed promoter (Gonzalez-Carranza et al., 2002), was fused to drive the GUS reporter gene. The constitutively expressed 35S::GUS construct was used as the control in the assay. As shown in Figure 7B, the background expression level of the 35S-driven AtDOF4.7 effector construct was clear. However, cells cotransformed with 35S::AtDOF4.7 and PromoterPGAZAT::GUS had significantly lower GUS activity in comparison with those transformed with the PromoterPGAZAT::GUS reporter alone (P < 0.05). By contrast, the activity of 35S::GUS was not affected significantly by cotransforming with 35S::AtDOF4.7 (Fig. 7B).
To examine whether the AtDOF4.7 protein regulates PGAZAT expression by directly binding to the promoter region, the PGAZAT promoter sequence was investigated. Two regions containing the DOF protein-binding cis-element AAAG (Yanagisawa, 2002, 2004) were selected and subsequently synthesized as probes for evaluation by yeast one-hybrid assays. Each probe (probe E1, the sequence from positions −453 to −433 of the PGAZAT promoter; probe E2, the sequence corresponding to positions −294 to −257) was inserted upstream of the reporter gene HIS3 in the reporter plasmid pHIS2 and cotransfected into the yeast cells with the pAD-AtDOF4.7 effector plasmid. The binding of AtDOF4.7 and the corresponding DNA sequence was indicated by the growth of transfected yeast cells on nutrient-deficient medium (synthetic dextrose/-Trp-Leu-His) plus 3-amino-1,2,4-triazole (3-AT). The yeast cells harboring the effector AtDOF4.7 plasmid and reporter plasmid containing probe E1 grew well on 3-AT medium, whereas the yeast cells harboring AtDOF4.7 and probe E2 did not grow. These results indicated that AtDOF4.7 could bind strongly to the PGAZAT promoter sequence at positions −453 to −433 (probe E1) but not to the sequence at positions −294 to −257 (probe E2; Fig. 7C).
To further examine the binding of AtDOF4.7 to the PGAZAT promoter probe E1 sequence, the glutathione S-transferase (GST)-fused AtDOF4.7 recombinant protein was used to test in vitro binding activity by a gel-shift assay. As shown in Figure 7D, GST-AtDOF4.7 but not the GST control protein significantly reduced the mobility of the probe E1 fragment (Fig. 7D, lanes 2 and 3), indicating that the AtDOF4.7 protein could bind to the PGAZAT promoter. Competition experiments with unlabeled probe versus labeled probe further supported this result (Fig. 7D, lane 4). Replacement of either a single base (Fig. 7D, lane 5) or four bases (Fig. 7D, lane 6) of the AAAG cis-element completely abolished or severely reduced the competition ability of the unlabeled probes in the competition analysis, indicating that the binding activity of AtDOF4.7 acts in a specific sequence-dependent manner.
AtDOF4.7 Interacts with AtZFP2
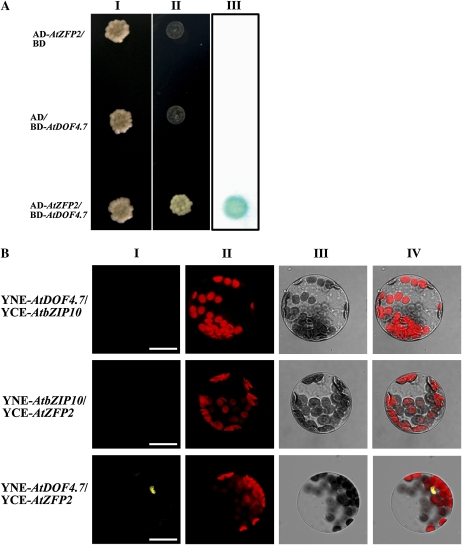
DOF proteins often associate with other transcription factors to form a transcription complex that regulates the expression of downstream genes. Therefore, we performed a yeast two-hybrid screen with AtDOF4.7 as the bait to identify potential interaction proteins. The full-length protein and several fragments corresponding to an abscission-related putative transcription factor gene, AtZFP2, were identified in multiple interacting clones. AtZFP2 is a TFIIIA-type zinc finger protein that is expressed in AZs. A previous study has shown that AtZFP2 overexpression can delay the abscission process (Cai and Lashbrook, 2008). To verify this result, we tested the interaction between AtZFP2 and AtDOF4.7 by directed yeast two-hybrid and bimolecular fluorescence complementation (BiFc) assays (Walter et al., 2004). The directed yeast two-hybrid assay confirmed the AtDOF4.7-AtZFP2 interaction found in the initial yeast two-hybrid screen (Fig. 8A). To verify the interaction between AtDOF4.7 and AtZFP2 independently, we fused two proteins with the N- or C-terminal portion of yellow fluorescent protein (YFP) and cotransformed these plasmids into Arabidopsis protoplasts. Another transcription factor, AtbZIP10 (for Arabidopsis basic Leu zipper 10), was fused to the N or C terminus of YFP as a control (Kaminaka et al., 2006). Restored YFP fluorescence localized in the nuclei was observed when YNE-AtDOF4.7 and YCE-AtZFP2 were cotransformed. However, the YFP signal was not detected with the combination of AtDOF4.7 and AtbZIP10 or AtZFP2 and AtbZIP10 (Fig. 8B). These results suggest that AtDOF4.7 and AtZFP2 form a transcript complex in planta.
Figure 8.
AtDOF4.7 interacts with AtZFP2 in both yeast and Arabidopsis cells. A, AtDOF4.7 interacts with AtZFP2 in yeast. Yeast cells were cotransformed with pBD-GAL4 cam and pAD-GAL4-AtZFP2, pBD-GAL4-AtDOF4.7 and pAD-GAL4-2.1, and pBD-GAL4-AtDOF4.7 and pAD-GAL4-AtZFP2. Transformants were cultured and then dipped at the same concentration on a plate containing synthetic dropout selection medium that lacked either Trp and Leu (line I) or Trp, Leu, and His but supplemented with 5 mm 3-AT (line II). Line III shows the β-galactosidase activity assay of the line I yeast. B, BiFc visualization of AtDOF4.7 interaction with AtZFP2 in the nuclei of Arabidopsis mesophyll protoplasts. Line I, YFP fluorescence image of Arabidopsis protoplasts transfected or cotransfected with pUC-SPYNE-AtDOF4.7 and pUC-SPYCE-AtZFP2 or their corresponding YNE/YCE-AtbZIP10 controls; line II, background chlorophyll fluorescence of the protoplast; line III, bright-field image of the protoplast; line IV, merged images of three channels. Bars = 20 μm.
DISCUSSION
The working model of Arabidopsis floral abscission can be divided into four steps: AZ differentiation, abscission signal response, abscission activation, and postabscission transdifferentiation (Patterson, 2001). The activation of cell wall modification proteins is essential for abscission activation, and these proteins are key components for determining shedding actions. Many cell wall modification proteins have been reported to be involved in abscission (Roberts et al., 2002), and the regulation of these proteins is important for elucidation of the molecular mechanisms underlying abscission. Some abscission-related PGs, such as PGAZAT and QUARTET2 from Arabidopsis, CAW471 from Brassica napus, and TAPG1, TAPG2, and TAPG4 from tomato (Solanum lycopersicum), accumulate after ethylene treatment (Kalaitzis et al., 1995, 1997; Gonzalez-Carranza et al., 2002; Ogawa et al., 2009). The expression of some endoglucanases that are related to abscission is also regulated by abscission signals, such as ethylene and hydrogen peroxide (del Campillo et al., 1990; Mishra et al., 2008; Sakamoto et al., 2008). However, the regulatory mechanisms of these proteins in abscission have not yet been established in either the ethylene-dependent or the ethylene-independent pathway. Therefore, there is a gap in our understanding of the molecular mechanisms between signal transduction and cell wall dissolution in abscission. Our results using the AtDOF4.7 gene provide novel insight into the regulation of abscission enzyme activation and demonstrate a role for the AtDOF4.7 protein in the control of abscission.
Overexpression of AtDOF4.7 Induces an Ethylene-Independent Floral Organ Abscission Deficiency
Distinct from several other abscission-delayed mutants, such as etr1-1 and ein2, we observed that the floral organs remain attached to siliques even after the plants finish their life cycles in more than 30 independent 35S::AtDOF4.7 transgenic lines (data not shown). Thus, overexpression of AtDOF4.7 leads to the failure of floral organ abscission rather than a simple delay, clearly demonstrating that AtDOF4.7 is involved in the control of floral abscission. Both the differentiation of AZs and cell separation activation could determine organ shedding, but these processes are regulated by independent control mechanisms (Stenvik et al., 2006; McKim et al., 2008). The bop1/bop2 double mutants exhibit complete absence of abscission because of the absence of AZs (McKim et al., 2008). However, in AtDOF4.7 overexpression lines, we did not observe any disruption of organ patterning. Furthermore, the AZs in these lines appeared anatomically normal, indicating that AZ differentiation was not the reason for the shedding failure in AtDOF4.7 overexpression lines. Therefore, we can rule out the possibility that AtDOF4.7 controls abscission by regulating the AZ differentiation process.
The SEM data revealed more detailed information about how cell separations were interrupted in the AtDOF4.7 overexpression lines. In the wild type, the AZ cells in the fracture plane were not torn from position 4. By contrast, some AZ cells in the overexpression lines, especially the cells in middle of the AZs, were still broken until position 20 with forcibly removed petals (Fig. 4B). These results indicate that transgenic AZ cells retain normal levels of adhesion between neighboring cells while wild-type AZs exhibit reduced adhesion. Furthermore, compared with the smooth surface of AZs in wild-type plants, the surfaces of AZs in AtDOF4.7 overexpression lines appeared to be rough or filled with fragments of cell wall material in a similar manner to ida mutants (Butenko et al., 2003). In addition, a significantly larger force was required to peel petals in the AtDOF4.7 overexpression lines. These results suggest that incomplete dissolution of the middle lamellae in transgenic plants is one of the principal reasons for the failure of floral organ abscission.
It is well known that the plant hormone ethylene participates in the regulation of the abscission rate. In Arabidopsis, the blockage of ethylene signal transduction causes a delay in floral abscission (Bleecker et al., 1988; Wilkinson et al., 1997; Patterson and Bleecker, 2004; Lewis et al., 2006). However, the AtDOF4.7 overexpression lines did not appear to be insensitive to ethylene, because their seedlings showed the same triple response as the wild-type seedlings. In addition, the senescence process was also accelerated by ethylene in the AtDOF4.7 overexpression lines, similar to that in the wild type. These results suggest that the response to ethylene is not interrupted in the floral organs of transgenic plants. However, in contrast to wild-type plants, exogenous ethylene does not accelerate any part of the floral organ abscission process in the AtDOF4.7 overexpression lines (Fig. 5B). Therefore, we can conclude that overexpression of the AtDOF4.7 gene leads to abscission failure in an ethylene-independent manner.
The Spatial and Temporal Expression Pattern of AtDOF4.7 Is Consistent with Its Involvement in the Process of Abscission
In a gene family with a high sequence similarity between its members, spatial and temporal regulation of gene expression is a common strategy to avoid functional redundancy. As a large and conserved plant-specific gene family, the expression patterns of transcription factor members of the DOF family are often related closely to their functions. The DOF protein CDF1, which is a photoperiodic flowering controller in Arabidopsis, has an evident light/dark expression cycle (Imaizumi et al., 2005). The restricted expression of the barley (Hordeum vulgare) DOF family members BPBF and SAD is coincident with their roles in the regulation of GA-induced endosperm development (Mena et al., 1998; Isabel-LaMoneda et al., 2003). Consistent with a role for the AtDOF4.7 gene in abscission, the GUS reporter was detected at the base of the floral organ, with an especially high abundance at the AZ sites in the AtDOF4.7 promoter::GUS transgenic plants (Fig. 1B, b). GUS expression was also found in other tissues, including the petals of young floral buds and the bases of the pedicel and leaf trichome (Fig. 1B, c and f). However, phenotypic changes were not observed in these tissues in the plants that overexpressed AtDOF4.7. The expression of AtDOF4.7 in these tissues other than AZs was also similar to the expression patterns of other genes proposed to function in cell separation processes, such as AtEXPA10, PGAZAT, HWS, AtZFP2, and IDLs (Cho and Cosgrove, 2000; Gonzalez-Carranza et al., 2007a, 2007b; Cai and Lashbrook, 2008; Stenvik et al., 2008). Notably, some abscission-related genes, such as PGAZAT and AtZFP2 (Gonzalez-Carranza et al., 2007a; Cai and Lashbrook, 2008), which were shown to be related to AtDOF4.7 in this study, have the common feature of expression at the bases of the trichome and pedicel. These results suggest the possibility that there may be the same regulatory mechanism with floral organ abscission for the evolutionary loss of certain abscission processes at these sites. AtEXPA10, which is only expressed at the bases of the pedicel and trichome but not at the floral organ AZs, may represent the vestigial remnant of abscission that was lost during evolution (Cho and Cosgrove, 2000). However, AtEXP10 expression is not regulated by AtDOF4.7 (Fig. 7A). Therefore, elucidation of another expansin downstream of AtDOF4.7 might help us to understand the potential evolutionary mechanisms involved in ancient cell separation processes.
The temporal regulation of AtDOF4.7 expression in wild-type plants is also consistent with its role in abscission. The activity of the AtDOF4.7 promoter was first detected at the AZs in position 4 flowers and was maintained at a high level throughout the abscission process. As lateral floral organs were shed from the siliques between positions 8 and 10, the AtDOF4.7 promoter activity also decreased and ultimately became undetectable. This highly synchronal spatial and temporal expression pattern strongly supports the hypothesis that AtDOF4.7 regulates AZ cell separation in a negative manner at the later stages of abscission.
AtDOF4.7 Functions as an Inhibitor of Transcription in the Regulation of Abscission
The DOF domain protein family is a family of transcription factors that is unique to plants (Yanagisawa, 2004). They are involved in various plant-specific physiological processes and regulate the expression of downstream genes. Consistent with a recent report of the subcellular location of AtDOF4.7 in Arabidopsis protoplasts (Krebs et al., 2010), we observed that the AtDOF4.7 protein is located in the nuclei by transient expression in onion epidermal cells, suggesting that AtDOF4.7 is potentially involved in the regulation of gene transcription. Considering the specific influence of AtDOF4.7 in the abscission process, the target genes were screened in our known abscission-related genes. We determined that a PG gene with AZ expression, PGAZAT, is down-regulated in AtDOF4.7 overexpression plants. In addition, two fragments of the PGAZAT promoter containing abundant AAAG elements were tested for their ability to bind AtDOF4.7. Notably, AtDOF4.7 bound to the E1 probe but not to the E2 probe sequence (Fig. 7C). Although there are more AAAG cis-element copies in the E2 probe sequence than in the E1 probe sequence, the specific binding activity of AtDOF4.7 may also depend on the flanking sequences of the core cis-element. Further analysis showed that AtDOF4.7 could bind to the PGAZAT promoter in vitro in a sequence-specific manner to inhibit its activation. Knockout plants of PGAZAT displayed a delay in abscission but still carried out the shedding process. One possible explanation is that PGAZAT is a functional abscission PG protein, but it may not be the only one (Gonzalez-Carranza et al., 2007a). An alternative explanation is that PGAZAT, a pectolytic protein, needs to be coordinately expressed with nonpectolytic proteins to dismantle the AZ (Lashbrook and Cai, 2008). In either case, the inhibition of PGAZAT expression could be an example of how AtDOF4.7 regulates cell wall modification genes during abscission. To support this hypothesis, several cell wall modification enzymes that are expressed in AZs during abscission (Cai and Lashbrook, 2008) were chosen to examine their expression profiles in AtDOF4.7 overexpression plants. Compared with wild-type plants, the expression levels of another PG gene (At4g23820) and three endotransglucosylase/hydrolase (At4g37800, At5g57530, At1g14720) genes are all down-regulated in the same manner as PGAZAT in AtDOF4.7 overexpression plants (Supplemental Fig. S2). A large number of AAAG cis-elements that could be specifically recognized by DOF family members were also detected in the promoter regions of these genes (data not shown). Furthermore, the smaller plant size observed in the lines that overexpressed AtDOF4.7 in our study might result from the inhibition of the cell wall-loosening capability induced by the ectopic expression of AtDOF4.7. Taken together, our results show a logical relationship between decreased activities of cell wall hydrolysis enzymes and a deficiency in primary cell wall processes, including the dissolution of the middle lamellae, in AtDOF4.7 overexpression plants. Therefore, AtDOF4.7 plays a direct role in the regulatory network of abscission as a transcriptional inhibitor of cell wall modification enzymes.
The Regulation of AtDOF4.7 Functions
Because AtDOF4.7 directly regulates cell wall-loosening proteins, it should function downstream of abscission early signal transduction. The ethylene-independent abscission process in AtDOF4.7 overexpression plants suggests that AtDOF4.7 either may be involved in the ethylene-independent abscission signal transduction pathway or may act at a unique downstream point in the ethylene-dependent abscission signal transduction pathway. IDA/IDL-HAE/HSL2 activation is recognized as an early event in the signal transduction within the ethylene-independent abscission process (Cho et al., 2008; Stenvik et al., 2008). However, failed abscission via overexpression of AtDOF4.7 seems mechanistically different from the abscission failure in the ida or hae hsl2 mutant. In the latter, the petal breakstrength decreases gradually from position 2 to 10 of the siliques but then increases again after position 12 (Butenko et al., 2003; Cho et al., 2008; Stenvik et al., 2008). However, in AtDOF4.7 overexpression lines, the petal breakstrength profile exhibited a continuous decreasing curve, similar to the prolonged profile obtained from the wild type (Fig. 3C). The same petal breakstrength profile has also been observed in MKK4-MKK5 RNA interference plants (Cho et al., 2008). Compared with the irregular or delayed AZ cell-rounding process exhibited in the failure of shedding in the ida mutant (Butenko et al., 2003; Patterson and Bleecker, 2004), the rounding of AZ cells in AtDOF4.7 overexpression lines started at approximately position 8, which was the same as that observed in wild-type plants. These results suggest that AtDOF4.7 may not be the direct target of early ethylene-independent signal transduction. One step of ethylene-independent abscission signal transduction, MPK3/MPK6 activation, has also been shown to be involved in the ethylene signal transduction pathway (Cho et al., 2008; Yoo et al., 2008). It would be interesting to examine the phosphorylation of AtDOF4.7 by MPK3/MPK6 in the future, because it may serve as a downstream signaling event common in both the ethylene-dependent and ethylene-independent abscission processes. Further analysis of AtDOF4.7 promoter::GUS expression in the ethylene-independent abscission deficiency mutant ida and the ethylene-insensitive mutant etr1-1 will provide some clues concerning the role of AtDOF4.7 downstream of early abscission signal transductions.
In addition to its DNA-binding activity, the DOF domain is also recognized for its role in protein-protein interactions. Several DOF proteins, such as SAD2, BPBF, and OsDOF3, have been reported to take part in the transcriptional regulation of downstream genes by forming a complex with other transcription factors (Diaz et al., 2002; Isabel-LaMoneda et al., 2003; Washio, 2003; Isabel et al., 2005; Kandasamy et al., 2005a). Because the interaction with other transcription factors may also affect the regulation activity of AtDOF4.7, the binding partners of AtDOF4.7 were also investigated. Our results revealed that a zinc finger transcription factor, AtZFP2, bound to AtDOF4.7, and the AtZFP2 gene had a similar expression pattern as AtDOF4.7. Overexpression of AtZFP2 also induces the failure of floral organ abscission (Cai and Lashbrook, 2008). However, the coexpression of AtZFP2 with AtDOF4.7 did not show an increased inhibitory effect on the activity of the PGAZAT promoter compared with the overexpression of AtDOF4.7 alone (data not shown). These results suggest that the AtDOF4.7-AtZFP2 complex might regulate the expression of cell wall-loosening proteins other than PGAZAT and/or that there may be other unidentified regulatory proteins that participate in a transcription complex with AtDOF4.7 and AtZFP2.
Preventing precocious shedding in crops is the primary practical goal of fundamental studies of abscission control. Research on Arabidopsis has identified several important positive regulators in abscission control, but not all of them are suitable to help directly with crop improvement because of differences in genome structure between crops and the model plant. Other than AtDOF4.7, two negative regulators have been reported in Arabidopsis. However, both of them lead to morphological changes in other parts of their overexpressing plants (Fang and Fernandez, 2002; Cai and Lashbrook, 2008). Because of its more abscission-specific phenotypes, it will be of great interest to test the utility of modifying AtDOF4.7 gene expression for improvement of abscission behavior with less negative side effects on crop growth and development.
MATERIALS AND METHODS
Plasmid Construction and Plant Transformation
A detailed description of the construction of the vectors can be found in Supplemental Materials and Methods S1. The sequences of all primers used in this report are listed in Supplemental Table S1.
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used for gene transformation using the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Hygromycin B (30 μg mL−1) was used to screen the transgenic plants. Independent primary transformants were randomly chosen for a test of hygromycin-resistant segregation in the progeny. The lines displaying a 3:1 segregation (hygromycin resistant:hygromycin sensitive) were allowed to set seeds, and the homozygous T3 lines were selected for further analysis.
Light Microscopy and SEM
Morphology analysis of AZ was performed according to a previous report (Kandasamy et al., 2005b). Flowers from position 2 were fixed sequentially with 3% glutaraldehyde and 1% osmium tetroxide and then were embedded in Spurr's resin. Tissue sections with a thickness of 2.5 μm were generated by a Leica EM UC6 ultramicrotome and stained with 0.05% toluidine blue. The morphology of the AZs was analyzed using an Olympus BX41 light microscopy system.
GUS expression was analyzed by staining different tissues in GUS reaction solution, which contained 100 mm sodium phosphate, 10 mm EDTA, 0.1% (v/v) Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Sigma) buffer, pH 7.0, and then clearing them in 75% ethanol. The histochemical GUS stain was observed with a Nikon SMZ-645 microscope.
For SEM analysis, different flowers or siliques were fixed in 4% (w/v) glutaraldehyde in 0.5 m potassium phosphate buffer at pH 7.5 before being dehydrated in a graded ethanol series. After the exchange of ethanol to isoamyl acetate by a graded ethanol-isoamyl acetate series, the samples were critical-point dried in liquid CO2 and then mounted on steel plates that were covered with double-sided adhesive tape and coated with palladium. The samples were viewed at an accelerating voltage of 10,000 on a Hitachi S-4800 scanning electron microscope. For the morphology analyses, the samples were collected from different individual plants at least three times.
Southern Blotting and Quantitative Real-Time PCR Analysis
For the Southern-blot assay, genomic DNA of two transgenic lines and the wild type was collected from at least four individual plants and isolated and purified by a DNeasy Plant Mini Kit (Qiagen). The DNA was then digested with EcoRI or HindIII. The 665-bp fragment corresponding to the coding sequence of the Hygromycin Phosphotransferase gene was amplified by PCR (Supplemental Table S2) and used as the hybridization probe. DNA probe labeling, hybridization, washing, and chemiluminescence were performed using a North2South Complete Biotin Random Prime Labeling and Detection Kit (ThermoFisher Scientific).
For quantitative RT-PCR analysis, total RNAs were isolated from siliques of positions 4 to 6 and other plant parts with an RNeasy Plant Mini Kit (Qiagen) and reverse transcribed to cDNAs using a ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs). Quantitative RT-PCR was run on a MyIQ thermal cycler using real-time PCR reagents (Bio-Rad). After normalization to an internal control (ACTIN), the relative levels of gene expression were calculated with the Delta-Delta Ct method (Livak and Schmittgen, 2001). Gene-specific primers used for the quantitative RT-PCR are listed in Supplemental Table S3. Each relative expression assay was repeated at least three times.
Subcellular Location Analysis
For the transient gene expression assay in onion (Allium cepa) epidermal cells, the pBI221-AtDOF4.7-GFP plasmid and empty 35S-GFP were introduced into onion epidermal cells on solid plates by the particle bombardment method as described previously with slight modifications (Varagona et al., 1992). The bombarded onion cells were incubated for 18 h at 25°C in the dark. Nuclei were stained with DAPI (1 mg mL−1 in water) before observation.
The fluorescence signal was analyzed and images were obtained using a Zeiss Meta 510 confocal system with the following parameters: excitation at 488 nm and emission at 505 to 530 nm for GFP and GFP fusion proteins, and excitation at 364 nm and emission at 460 nm for DAPI staining. Each transient expression experiment was repeated at least two times.
Petal Breakstrength
The force required for peeling a petal from a flower was measured by a petal breakstrength meter (Lease et al., 2006). The values for the petal breakstrength from each indicated floral position were analyzed from 17 plants each from the wild type (Columbia) and the overexpression lines (S107 and S109). The maximum and minimum values were considered as outliers.
Ethylene Response
Ethylene treatment was performed as described previously (Butenko et al., 2003). For the triple response, 5 μm of the ethylene precursor ACC was added to Murashige and Skoog medium for germination of wild-type or transgenic seeds in the dark for 3 d. For an assessment of the ethylene response of flowers, mature plants were incubated with pure air or 10 μL L−1 ethylene in an air-tight growth chamber for 3 d. Each treatment was repeated at least three times.
Yeast Two-Hybrid Screen and BiFc
A yeast two-hybrid screen was performed according to the protocol of the HybriZAP-2.1 Two-Hybrid system (Stratagene). The BD-AtDOF4.7 fusion protein was used as the bait to screen for interaction candidates in an Arabidopsis flower cDNA library (Horwitz and Ma Two-Hybrid cDNA Library, Arabidopsis Biological Resource Center Stock Center). After the transformed library was induced by polyethylene glycol, cells were plated on a selection medium that lacked Trp, Leu, and His but was supplemented with 5 mm 3-AT to reduce any artificial interaction. The selected clones were sequenced by a T7 primer. The full-length sequences of candidates were cloned from Arabidopsis cDNA and used to form pAD-GAL4-2.1 constructs. Cotransformed cells with bait and prey constructs were dropped onto 5 mm 3-AT selection medium lacking Trp, Leu, and His. β-Galactosidase activities were assayed as described previously (Gindullis et al., 1999).
The BiFc assay was performed as described previously (Walter et al., 2004). Cotransformations of pUC-SPYNE-AtDOF4.7 and pUC-SPYCE-AtZFP2 or their corresponding controls and the sequential examination of YFP fluorescence were performed as described previously (Miao et al., 2006). The polyethylene glycol-induced method was used to transform Arabidopsis mesophyll protoplasts according to Asai et al. (2002). All transformations were repeated at least three times using Arabidopsis mesophyll protoplasts that were isolated from different batches of plants.
Transient Transcription Factor-Promoter Interaction Assay
The transient interaction assay was performed as described previously (Berger et al., 2007). The reporter vector, PromoterPGAZAT::GUS, and the effector vector, 35S::AtDOF4.7, were constructed as described previously. Together with the controls (35S::GUS, empty GUS vector, and anti-silence p19 vector; Voinnet et al., 2003), these plasmids were transformed into Agrobacterium tumefaciens LBA4404.pBBR1MCS virGN54D by electroporation (van der Fits et al., 2000). Colonies were selected on agar that contained 20 μg mL−1 rifampicin, 75 μg mL−1 chloramphenicol, and 50 μg mL−1 kanamycin at 28°C and were verified by PCR with gene-specific primers. Positive colonies were grown in 5 mL of AB medium with the appropriate antibiotics for 48 h at 28°C to an optical density at 600 nm of 0.5.
After 7 d, suspended Arabidopsis Columbia cells were harvested by a 45-μm cell strainer and were then diluted by 5-fold in culture medium. The reporter and effector strains were collected and washed with suspension cell culture medium and finally resuspended to 25% of their original volume. Each fresh Arabidopsis cell aliquot was added to the corresponding p19 strains. After 4 d of coculture, the cells were collected and the total proteins were isolated to assay GUS activity as described (Jefferson et al., 1987). All transformations were repeated at least three times.
Yeast One-Hybrid Assay and Electrophoretic Mobility Shift Assays
The effector pAD-AtDOF4.7 construct and the reporter pHIS2 containing the cis-DNA element were cotransfected into Y187 yeast strains by the LiAc yeast transformation method. As a negative control, the pAD empty vector or the empty pHIS2 plasmid was substituted into pAD-AtDOF4.7 or pHIS2, respectively. These constructs contained the cis-DNA elements that were cotransfected into yeast. The transfected yeast was selected from the synthetic dextrose/-Trp-Leu medium and then dropped onto synthetic dextrose/-Trp-Leu-His medium with the optimal concentration of 3-AT for 4 d.
For the gel-shift assay, the AtDOF4.7 protein was expressed in Escherichia coli [Rosetta 2 (DE3) pLysS strain]. Recombinant GST-AtDOF4.7 and GST proteins were induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside for 12 h at 18°C with supplementation by zinc chloride to a final concentration of 50 μm. Protein purification of the GST fusion protein from bacterial extracts was achieved by affinity chromatography with Glutathione Sepharose 4B resin (GE Healthcare) following the instructions of the manufacturer. Cells carrying the pGEM-4T-1 empty vector were processed as negative controls in an identical manner. Complementary single-stranded oligonucleotides of two copies of the E1 probe and its mutants were synthesized, labeled with biotin, and annealed to make probes. The electrophoretic mobility shift assays were performed with a LightShift Chemiluminescent EMSA Kit (ThermoFisher Scientific) according to the manufacturer's instructions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The growth of AtDOF4.7 overexpression plant is different from the wild type.
Supplemental Figure S2. Expression of potential abscission-related genes in AtDOF4.7 overexpression lines.
Supplemental Table S1. The primers used for vector construction.
Supplemental Table S2. The primers used for Southern blots.
Supplemental Table S3. The primers used for quantitative real-time PCR.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Dr. Jun-Jun Liu (Canadian Forest Service) and Dr. Xian-Ting Wu (University of California, Berkeley-U.S. Department of Agriculture Agricultural Research Service) for critical reviews on the manuscript. We thank David Baulcombe (Sainsbury Laboratory, John Innes Centre) for providing the p19 vector, Johan Memelink (Leiden University) for providing A. tumefaciens strain LBA4404.pBBR1MCS virGN54D, and Michael Walter (Universität Tübingen) for providing the BiFc vectors.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S. (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56: 817–823 [DOI] [PubMed] [Google Scholar]
- Berger B, Stracke R, Yatusevich R, Weisshaar B, Flugge UI, Gigolashvili T. (2007) A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J 50: 911–916 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE. (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB. (2003) INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lashbrook CC. (2008) Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol 146: 1305–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Campillo E. (1999) Multiple endo-1,4-beta-D-glucanase (cellulase) genes in Arabidopsis. Curr Top Dev Biol 46: 39–61 [DOI] [PubMed] [Google Scholar]
- del Campillo E, Reid PD, Sexton R, Lewis LN. (1990) Occurrence and localization of 9.5 cellulase in abscising and nonabscising tissues. Plant Cell 2: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P. (2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 29: 453–464 [DOI] [PubMed] [Google Scholar]
- Fang SC, Fernandez DE. (2002) Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol 130: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC. (2000) The embryo MADS domain factor AGL15 acts postembryonically: inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12: 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis F, Peffer NJ, Meier I. (1999) MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Carranza ZH, Elliott KA, Roberts JA. (2007a) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58: 3719–3730 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA. (2007b) HAWAIIAN SKIRT: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol 144: 1370–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Carranza ZH, Whitelaw CA, Swarup R, Roberts JA. (2002) Temporal and spatial expression of a polygalacturonase during leaf and flower abscission in oilseed rape and Arabidopsis. Plant Physiol 128: 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Isabel D, Manuel M, Ines IL, Ignacio RS, Pilar C. (2005) The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 42: 652–662 [DOI] [PubMed] [Google Scholar]
- Isabel-LaMoneda I, Diaz I, Martinez M, Mena M, Carbonero P. (2003) SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J 33: 329–340 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CZ, Lu F, Imsabai W, Meir S, Reid MS. (2008) Silencing polygalacturonase expression inhibits tomato petiole abscission. J Exp Bot 59: 973–979 [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Koehler SM, Tucker ML. (1995) Cloning of a tomato polygalacturonase expressed in abscission. Plant Mol Biol 28: 647–656 [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Solomos T, Tucker ML. (1997) Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol 113: 1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaka H, Nake C, Epple P, Dittgen J, Schutze K, Chaban C, Holt BF, Merkle T, Schafer E, Harter K, et al. (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25: 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB. (2005a) Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J 41: 845–858 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Deal RB, Meagher RB. (2005b) Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol 138: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Krebs J, Mueller-Roeber B, Ruzicic S. (2010) A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. J Plant Physiol 167: 583–586 [DOI] [PubMed] [Google Scholar]
- Lashbrook CC, Cai S. (2008) Cell wall remodeling in Arabidopsis stamen abscission zones: temporal aspects of control inferred from transcriptional profiling. Plant Signal Behav 3: 733–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease K, Cho SK, Walker J. (2006) A petal breakstrength meter for Arabidopsis abscission studies. Plant Methods 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie ME, Lewis MW, Youn JY, Daniels MJ, Liljegren SJ. (2010) The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 137: 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Liljegren SJ. (2006) Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol 9: 59–65 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Leslie ME, Darnielle L, Lewis MW, Taylor SM, Luo R, Geldner N, Chory J, Randazzo PA, Yanofsky MF, et al. (2009) Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 136: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- McKim SM, Stenvik GE, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, Aalen RB, Haughn GW. (2008) The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Khare S, Trivedi PK, Nath P. (2008) Ethylene induced cotton leaf abscission is associated with higher expression of cellulase (GhCel1) and increased activities of ethylene biosynthesis enzymes in abscission zone. Plant Physiol Biochem 46: 54–63 [DOI] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. (1998) An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J 16: 53–62 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM. (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE. (2001) Cutting loose: abscission and dehiscence in Arabidopsis. Plant Physiol 126: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE, Bleecker AB. (2004) Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134: 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH. (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158 [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Munemura I, Tomita R, Kobayashi K. (2008) Involvement of hydrogen peroxide in leaf abscission signaling, revealed by analysis with an in vitro abscission system in Capsicum plants. Plant J 56: 13–27 [DOI] [PubMed] [Google Scholar]
- Sexton R, Roberts JA. (1982) Cell biology of abscission. Annu Rev Plant Physiol 33: 133–162 [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JKC, Aalen RB. (2006) Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Coupe SA, Picton S, Roberts JA. (1994) Characterization and accumulation pattern of an mRNA encoding an abscission-related β-1,4-glucanase from leaflets of Sambucus nigra. Plant Mol Biol 24: 961–964 [DOI] [PubMed] [Google Scholar]
- Taylor JE, Tucker GA, Lasslett Y, Smith CJS, Arnold CM, Watson CF, Schuch W, Grierson D, Roberts JA. (1991) Polygalacturonase expression during leaf abscission of normal and transgenic tomato plants. Planta 183: 133–138 [DOI] [PubMed] [Google Scholar]
- Taylor JE, Webb STJ, Coupe SA, Tucker GA, Roberts JA. (1993) Changes in polygalacturonase activity and solubility of polyuronides during ethylene-stimulated leaf abscission in Sambucus nigra. J Exp Bot 44: 93–98 [Google Scholar]
- Taylor JE, Whitelaw CA. (2001) Signals in abscission. New Phytol 151: 323–340 [Google Scholar]
- Tucker ML, Baird SL, Sexton R. (1991) Bean leaf abscission: tissue-specific accumulation of a cellulase mRNA. Planta 186: 52–57 [DOI] [PubMed] [Google Scholar]
- Tucker ML, Milligan SB. (1991) Sequence analysis and comparison of avocado fruit and bean abscission cellulases. Plant Physiol 95: 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Deakin EA, Hoge JHC, Memelink J. (2000) The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol Biol 43: 495–502 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Stead AD. (1997) Abscission of flowers and floral parts. J Exp Bot 48: 821–837 [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4: 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Susana R, Pere M, David B. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walter M, Christina C, Katia S, Oliver B, Katrin W, Christian N, Dragica B, Christopher G, Karin S, Claudia O, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Washio K. (2003) Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133: 850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol 15: 444–447 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2002) The Dof family of plant transcription factors. Trends Plant Sci 7: 555–560 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45: 386–391 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.