Abstract
Artichoke (Cynara cardunculus subsp. scolymus) extracts have high antioxidant capacity, due primarily to flavonoids and phenolic acids, particularly chlorogenic acid (5-caffeoylquinic acid [CGA]), dicaffeoylquinic acids, and caffeic acid, which are abundant in flower bracts and bioavailable to humans in the diet. The synthesis of CGA can occur following different routes in plant species, and hydroxycinnamoyl-coenzyme A transferases are important enzymes in these pathways. Here, we report on the isolation and characterization of two novel genes both encoding hydroxycinnamoyl-coenzyme A quinate transferases (HQT) from artichoke. The recombinant proteins (HQT1 and HQT2) were assayed after expression in Escherichia coli, and both showed higher affinity for quinate over shikimate. Their preferences for acyl donors, caffeoyl-coenzyme A or p-coumaroyl-coenzyme A, were examined. Modeling and docking analyses were used to propose possible pockets and residues involved in determining substrate specificities in the HQT enzyme family. Quantitative real-time polymerase chain reaction analysis of gene expression indicated that HQT1 might be more directly associated with CGA content. Transient and stable expression of HQT1 in Nicotiana resulted in a higher production of CGA and cynarin (1,3-dicaffeoylquinic acid). These findings suggest that several isoforms of HQT contribute to the synthesis of CGA in artichoke according to physiological needs and possibly following various metabolic routes.
The incidence of many chronic disorders, such as cardiovascular diseases and certain cancers, could be reduced by means of improved nutrition, particularly through increased consumption of diets rich in fruit and vegetables. Most of the health-promoting properties of edible plants are related to the presence of secondary metabolites, known as phytonutrients (Segasothy and Phillips, 1999). Artichoke (Cynara cardunculus subsp. scolymus or Cynara cardunculus var scolymus), which originated in the Mediterranean basin (Sonnante et al., 2007), is an edible plant used in medicine since ancient times. Artichoke extracts possess many medicinal properties, including anticarcinogenic, anti-Human immunodeficiency virus, antioxidative, cholesterol-lowering, bile-expelling, hepatoprotective, and diuretic activities, as well as antifungal and antibacterial properties (Agarwal and Mukhtar, 1996; Gebhardt, 1997; Kraft, 1997; Brown and Rice-Evans, 1998; McDougall et al., 1998; Shimoda et al., 2003; Zhua et al., 2005). The high antioxidant capacity of artichoke is due primarily to flavonoids and phenolic acids, particularly 5-caffeoylquinic acid, also known as chlorogenic acid (CGA), 1,3-dicaffeoylquinic acid (cynarin), 1,5-dicaffeoylquinic acid, and caffeic acid, which are abundant in artichoke and bioavailable to humans through their diet (Azzini et al., 2007). CGA can be absorbed directly by the small intestine or hydrolyzed by the microflora of the large intestine to release caffeic acid (Plumb et al., 1999; Nardini et al., 2002; Stalmach et al., 2010), which shows the same antioxidant activity as CGA (Rice-Evans et al., 1997).
CGA is synthesized by many plants such as solanaceous species (e.g. tomato [Solanum lycopersicum], tobacco [Nicotiana tabacum], and eggplant [Solanum melongena]), apple (Malus domestica), pear (Pyrus communis), plum (Prunus domestica), coffee (Coffea arabica), and artichoke (Tamagnone et al., 1998a; Clifford, 1999; Wang et al., 2003; Schütz et al., 2004) and is believed to play important roles in free radical scavenging (Chen and Ho, 1997; Tamagnone et al., 1998b), enzymatic browning of fruits and vegetables (Walker, 1995), defense against fungal pathogens (Maher et al., 1994), and resistance to pathogenic insects (Dowd and Vega, 1996; Bushman et al., 2002). The biosynthetic routes leading to CGA in plants are not completely defined yet, and three possible paths have been proposed. In the first, hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase (HQT) catalyzes the formation of CGA from caffeoyl-CoA and quinic acid (route 1; Niggeweg et al., 2004). Caffeoyl-CoA is supplied by the combined activities of hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) and p-coumaroyl ester 3′ hydroxylase (C3′H) via a p-coumaroylshikimate intermediate (route 1b; Mahesh et al., 2007). The second proposed route is based on the synthesis of p-coumaroylquinate by HCT, followed by hydroxylation by C3′H (Ulbrich and Zenk, 1979, Hoffmann et al., 2003). The importance of this route has been questioned because HCT and C3′H are both active in Arabidopsis (Arabidopsis thaliana) and yet no accumulation of CGA occurs and because HCT does not show high affinity for quinate as an acyl acceptor (Hoffmann et al., 2003; Niggeweg et al., 2004). HCT could be supplemented by HQT in this pathway in some plants. In the third suggested route, caffeoyl glucoside serves as an activated intermediate (Villegas and Kojima, 1986).
Since CGA biosynthesis does not occur in all species, it is possible that different biosynthetic routes have evolved in diverged species, or indeed, that the different pathways have assumed various degrees of importance in distinct species. The cDNA encoding HQT has been isolated from tobacco and tomato; both transient expression and stable transformation established that HQT is the enzyme of greatest importance in the synthesis of chlorogenic acid in tomato (Niggeweg et al., 2004). In coffee, the expression of HQT is more closely correlated with CGA synthesis and accumulation than HCT (Lepelley et al., 2007).
In artichoke, a cDNA coding for HCT has been isolated (Comino et al., 2007). Biochemical characterization suggested that HCT could act either before or after the 3′-hydroxylation step and may contribute to CGA biosynthesis. Recently, after the initiation of this work, the isolation of a gene encoding HQT from artichoke was also reported (Comino et al., 2009).
A better understanding of CGA and cynarin production in artichoke is important to develop agronomic, genetic, or biotechnological tools for higher production of biologically active compounds. In this paper, we report on the isolation and characterization of two full-length cDNA sequences encoding HQT in artichoke, belonging to the BAHD transferase family (D'Auria, 2006). HQT1 and HQT2 have in vitro activities distinct from each other in terms of their specificities. Modeling and docking analyses based on the structures of two BAHD members from Dendranthema morifolium (2e1t; Unno et al., 2007) and Rauvolfia serpentina (2bgh; Ma et al., 2005) were undertaken to identify possible structural features defining catalytic specificities. Hqt transcript levels in different bract orders and the receptacle of the edible flower head, and in leaves in the vegetative and productive periods, were compared with CGA content in the same tissues. Transient and stable expression of hqt1 in Nicotiana induced a higher production of CGA and cynarin.
RESULTS
Isolation and Analysis of hqt1/hqt2 Genomic Sequences and Full-Length cDNAs
The hqt cDNA sequences from tobacco (AJ582651) and tomato (AJ582652) were used to search the lettuce (Lactuca sativa) and sunflower (Helianthus annuus) ESTs in The Gene Index database (http://compbio.dfci.harvard.edu/tgi/plant.html) because these species, like artichoke, are members of the Asteraceae family and were likely to have genes closely related structurally to HQT in artichoke, as has been reported for other genes (De Paolis et al., 2008). Moreover, no EST from artichoke was available when this work was started. The best hits obtained from the BLAST analysis grouped in two clusters and after alignment, TC22135, TC23302, TC25546, and TC10357 sequences were used to design the primers Hel-F3 and Hqt-R2 (Supplemental Table S1), which produced a 600-bp putative hqt sequence (named hqt1) from reverse transcription (RT)-PCR of artichoke leaf cDNA. Primers Hel_HQT_F6 and Hel_HQT_R4 were designed on the basis of the second group of sequences (TC16408, TC16209, and TC30763) and were used to isolate a 1,000-bp sequence named hqt2. Both fragments showed sequence similarity to tobacco and tomato hqt.
Partial sequences from artichoke were extended toward both the 3′ and 5′ ends by RACE-PCR. New sets of primers were designed around the start and stop codons for both hqt1 and hqt2. A 1,413-bp sequence composed of a 1,350-bp open reading frame encoding a 449-residue protein (estimated molecular mass of 49.59 kD) was obtained for hqt1, whereas the hqt2 sequence was 1,332 bp, encoding a 443-residue protein, with an estimated molecular mass of 48.89 kD.
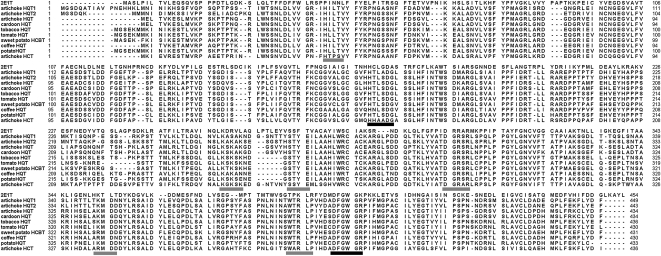
The artichoke hqt cDNA-encoded proteins were 80.4% identical and possessed the two conserved sequence motifs, HXXXDG and DFGWG (St-Pierre and De Luca, 2000; D'Auria, 2006), observed in other plant acyltransferases, for HQT1, HTLAD (170–174 amino acids) and DFGWG (396–400 amino acids), and for HQT2, HTLSD (162–166 amino acids) and DFGWG (390–394 amino acids), as shown in Figure 1. HQT1 showed the greatest levels of identity (71%) to the recently isolated HQT from artichoke and cardoon (Cynara cardunculus var altilis; Comino et al., 2009), to tobacco, tomato, coffee, and potato (Solanum tuberosum) HQTs (71%, 70%, 68%, and 68%, respectively), and to sweet potato (Ipomoea batatas) N-hydroxycinnamoyl benzoyltransferase (HCBT; 70%). HQT2 showed the greatest identity to HQT from artichoke (71%; Comino et al., 2009) and from cardoon (70%), to HCBT from sweet potato (70%), and to tobacco, tomato, and coffee HQTs (69%, 68%, and 66%, respectively). Both sequences displayed a lower level of identity to artichoke HCT (59% for HQT1 and 57% for HQT2).
Figure 1.
Protein sequence alignment of artichoke HQT1 and HQT2 with representative members of the HQT family, HCBT, artichoke HCT, and 2E1T. Accession numbers are as follows: artichoke HQT1, AM690438; artichoke HQT2, EU839580; artichoke HQT, ABK79689; cardoon HQT, ABK79690; tobacco HQT, Q70G33; tomato HQT, Q70G32; sweet potato HCBT, Q9SST8; coffee HQT, A4ZKM4; potato HQT, Q3HRZ5; artichoke HCT, AAZ80046. 2E1T refers to DmAT A4PHY4. Black bars indicate the conserved motifs for the acyl transferase family: HXXXDG and DFGWG. Gray bars identify conserved amino acid residues for HQT.
The genomic sequences of the hqt1 and hqt2 genes established that both possess the conserved “Q intron” found in many members of the BAHD acyltransferase family (St-Pierre and De Luca, 2000), inserted between triplets encoding a Gln residue and a Val residue, in hqt1 at position 459 of the coding sequence and in hqt2 at position 438 of the coding sequence. The introns are 1,516 and 1,398 bp long for hqt1 and hqt2, respectively.
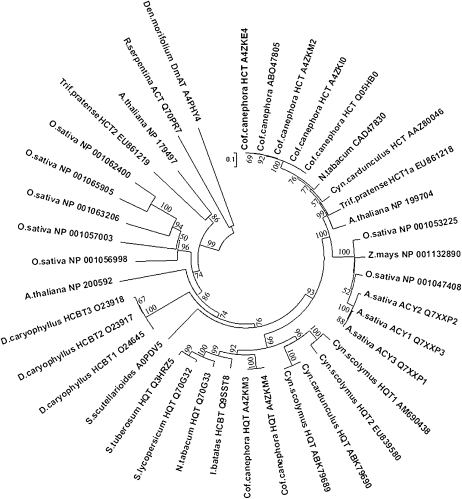
An alignment was obtained using acyltransferase sequences from different plant species, including the two artichoke HQTs and the amino acid sequences used for comparative modeling: D. morifolium acyltransferase DmAT (2e1t; Unno et al., 2007) and R. serpentina vinorine synthase (2bgh; Ma et al., 2005). In the deriving neighbor-joining phylogenetic tree (Fig. 2), the HQT1 and HQT2 proteins lie on a branch containing other HQT sequences and form a subcluster together with the HQT from artichoke and cardoon recently isolated by Comino et al. (2009). This HQT group is well separated from a cluster grouping HCT sequences from different species, including HCT from artichoke.
Figure 2.
Neighbor-joining phylogenetic tree of acyltransferase proteins from different plant species. Line lengths indicate the relative distances between nodes. Protein accession numbers are indicated. Den. morifolium DmAT and R. serpentina ACT refer to acyltransferase sequences for which the crystal structures are available (2e1t and 2bgh, respectively). ACY, Hydroxyanthranilate hydroxycinnamoyl transferase. Numbers on the nodes refer to bootstrap test values.
The promoters of hqt1 and hqt2 were isolated using restriction enzyme-based libraries. After screening several clones by sequencing, a 1,900-bp-long fragment containing the hqt1 promoter was identified in an EcoRV library, whereas a 1,400-bp-long fragment including the hqt2 promoter was found in a StuI library. We inspected 1,874 and 1,159 bp upstream the ATG codon of hqt1 and hqt2 respectively, by screening the PLACE database, and we found cis-regulatory elements present in other genes of the phenylpropanoid pathway, such as light-regulated elements (Narusaka et al., 2004) and pathogen elicitor and wounding-responsive elements (Soltani et al., 2006; De Paolis et al., 2008). Particularly, in both putative promoter regions, we detected different binding sites recognized by MYB transcription factors (P box, L box), light-regulated elements (I box, T box, GT1 consensus sequence, GATA box), dehydration-responsive elements (MYC recognition site, ACGT core sequence), drought-responsive elements (DRE core sequence), pathogen elicitor elements (ElRE, TGA box, E box), and DNA-binding sites for WRKY transcription factors (W box) and DOF proteins (DOF core site). The putative promoter region of hqt2 also contained a G box, a light-regulated element, and an ABRE-like sequence for abscisic acid responsiveness.
Expression of Artichoke HQT1 and HQT2 in Escherichia coli and Enzyme Assays
The hqt1 and hqt2 cDNAs were recombined into the expression vector pJAM1784 (Luo et al., 2007) to allow expression, in E. coli, of recombinant S-tag fusion proteins under the control of the T7 promoter. The S-tag was used to confirm the production of fusion proteins of approximately 50 kD (Supplemental Fig. S1) and to determine the concentration of target proteins in E. coli extracts by comparison of reconstituted activity with that of an S-peptide standard.
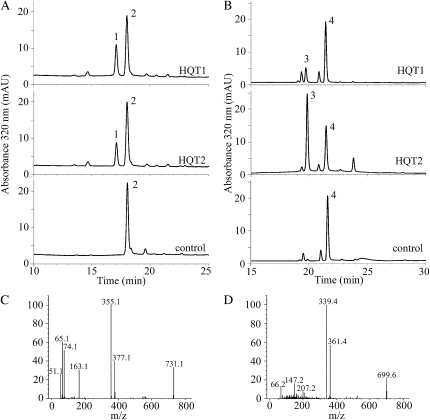
Enzymatic assays with the soluble fractions of the E. coli extracts showed that both HQT1 and HQT2 had acyltransferase activity, using caffeoyl-CoA or p-coumaroyl-CoA as acyl donor and quinate as the acyl acceptor. By means of HPLC and mass spectrometry analyses, reaction products were identified as CGA (Fig. 3, A and C) or p-coumaroylquinate (Fig. 3, B and D), depending on the acyl donor. The control reactions with the E. coli crude extracts transformed with the empty vector did not show any reaction product (Fig. 3, A and B).
Figure 3.
HPLC/LC/MS analysis of HQT1 and HQT2 acyltransferase assays. A and B, HPLC profiles of HQT1 and HQT2 reaction products without (control) or with recombinant protein obtained using caffeoyl-CoA and quinate (A) or p-coumaroyl-CoA and quinate (B). Peak 1, Chlorogenic acid (CGA); peak 2, caffeoyl-CoA; peak 3, p-coumaroylquinate; peak 4, p-coumaroyl-CoA. C, Mass spectrum of HPLC peak 1 obtained from HQT1 reaction; m/z (mass-to-charge ratio) = 355 ([M+H]+); M = CGA. D, Mass spectrum of HPLC peak 3 obtained from HQT1 reaction; m/z = 339 ([M+H]+); M = p-coumaroylquinate. mAU, Milliabsorbance units.
To investigate acyl acceptor specificity, shikimate was also tested. Both HQT1 and HQT2 were able to catalyze the reaction using caffeoyl-CoA and shikimate, whereas only HQT2 synthesized p-coumaroylshikimate using p-coumaroyl-CoA and shikimate as substrates. To further test HQT1 and HQT2 transferase activity, negative controls using sinapic and malic acids were performed. No activity was detected with either of these acyl acceptors. Generally, BAHD enzymes catalyze reversible reactions. To determine whether either HQT1 or HQT2 could catalyze the synthesis of cynarin, we assayed for their transferase activity on cynarin in the presence of CoA. Neither enzyme showed any detectable production of caffeoyl-CoA or CGA with these substrates under our experimental conditions.
Kinetic Parameters
The kinetic properties of the two recombinant artichoke HQTs were determined using caffeoyl-CoA or p-coumaroyl-CoA as the acyl-CoA donor and quinate and shikimate as acyl acceptors. HQT1 and HQT2 activities showed Michaelis-Menten kinetics, and the corresponding parameters are summarized in Table I.
Table I. Kinetic data of recombinant HQT1 and HQT2 from E. coli extracts.
The data represent means ± sd of at least three different experiments. 1 nkat = 1 nmol substrate s−1. NA, No activity.
| Varying Substrate | Saturating Substrate | HQT1 |
HQT2 |
||
| Km ± sd | Vmax ± sd | Km ± sd | Vmax ± sd | ||
| μm | nkat mg−1 | μm | nkat mg−1 | ||
| Quinate | Caffeoyl-CoA | 99 ± 41 | 197 ± 54 | 264 ± 66 | 145 ± 14 |
| Caffeoyl-CoA | Quinate | 1,391 ± 283 | 108 ± 28 | 6,196 ± 293 | 258 ± 83 |
| Quinate | p-Coumaroyl-CoA | 216 ± 76 | 329 ± 99 | 59 ± 4 | 430 ± 24 |
| p-Coumaroyl-CoA | Quinate | 1,034 ± 77 | 227 ± 65 | 1,065 ± 70 | 113 ± 7 |
| Shikimate | Caffeoyl-CoA | 5,088 ± 328 | 19 ± 4 | 6,120 ± 481 | 35 ± 8 |
| Shikimate | p-Coumaroyl-CoA | NA | NA | 15,690 ± 2,200 | 14 ± 1 |
| CGA | CoA | 130 ± 32 | 108 ± 5 | 251 ± 28 | 316 ± 79 |
| CoA | CGA | 72 ± 21 | 195 ± 18 | 29 ± 9 | 5,361 ± 375 |
Both HQT1 and HQT2 showed higher affinities for quinate than for shikimate using either acyl donor (Table I). The affinity of HQT1 for quinate in the presence of saturating caffeoyl-CoA was higher than that of HQT2 (Km = 99 ± 41 μm and 264 ± 66 μm, respectively), with in vitro efficiency in terms of Vmax/Km ratio of 1.99 and 0.55 nkat mg−1 μm−1, respectively. Conversely, HQT1 showed a lower affinity for quinate in the presence of saturating p-coumaroyl-CoA than HQT2 (Km = 216 ± 76 μm and 59 ± 4 μm, respectively), with Vmax/Km ratios of 1.52 and 7.29 nkat mg−1 μm−1, respectively. The affinity of HQT1 for caffeoyl-CoA in the presence of quinate was about 4.5-fold higher than for HQT2 (Km = 1,391 ± 283 μm and 6,196 ± 293 μm, respectively), with Vmax/Km ratios of 0.08 and 0.04 nkat mg−1 μm−1, respectively. On the other hand, HQT1 and HQT2 displayed similar affinities for p-coumaroyl-CoA in the presence of quinate (Km = 1,034 ± 77 μm and 1,065 ± 70 μm, respectively), with Vmax/Km ratios of 0.22 and 0.11 nkat mg−1 μm−1, respectively.
When shikimate was used as acyl acceptor, HQT2 had a higher affinity for shikimate at saturating caffeoyl-CoA levels than with saturating levels of p-coumaroyl-CoA (Km = 6,120 ± 481 μm and 15,690 ± 481 μm, respectively), although these Km values are very high and likely preclude HQT2 having any activity with shikimate in vivo. Therefore, neither HQT1 nor HQT2 was deemed likely to catalyze the synthesis of p-coumaroylshikimate or caffeoylshikimate in vivo.
The reverse reaction, in which HQT1 and HQT2 catalyzed the breakdown of CGA to quinate and caffeoyl-CoA in the presence of CoA, was followed measuring caffeoyl-CoA synthesis. Km values for CGA were 130 ± 32 μm and 251 ± 28 μm for HQT1 and HQT2, respectively, and for CoA they were 72 ± 21 μm and 29 ± 9 μm, respectively.
Putative Comparative Homology Models of Artichoke HQT1, HQT2, and HCT, and a Proposed Common Binding Site
HQT1, HQT2, HCT, 2e1t, and 2bgh were added to other BAHD acyltransferase amino acid sequences sampled from different plant taxonomic groups. A multiple sequence alignment was generated using ClustalW (Persson, 2000), with the secondary structures of 2e1t and of 2bgh used to weigh the gap penalties to preferentially place insertions and deletions in loops. The PSIPRED server (McGuffin et al., 2000) provided the structural alignment between HQT1, HQT2, or HCT and the crystallized 2e1t and 2bgh proteins.
The alignment highlighted residues that were strongly conserved between the family members (Fig. 1; Supplemental Fig. S2), and these served as anchor points for the secondary structure alignment, which improved the quality of the pairwise alignment between HQT1, HQT2, and HCT on one side and 2e1t and 2bgh on the other. There was very little ambiguity in the alignment because of these highly conserved amino acid residues and motifs, such as [H/P][I/L/T][P/N/L/G]X[V/I/L], HXXXD[A/G], [K/R][A/S/Q/G][K/S/Q/A], S[S/T/R][Y/F][E/Q/T], [D/N][G/A/I]R[S/T/Q/A]R, [K/R][M/L]D[D/N], WXRX[P/S/T], and D[A/L]DFGWG[R/K] (Fig. 1; Supplemental Fig. S2).
D. morifolium acyltransferase 2e1t has been crystallized in a complex with malonyl-CoA (Unno et al., 2007); therefore, its malonyl-CoA binding pocket is known. Residues within 5 Å from malonyl-CoA in the crystallized structure of 2e1t were highlighted by means of PyMol, and the Pocket Finder tool was successfully used to establish whether it was able to predict the region involved in the binding of malonyl-CoA as a validation test. Then Pocket Finder was used to predict binding sites in HQT1, HQT2, and HCT, and it was found that they overlapped with the 2e1t malonyl-CoA binding region. Differences in amino acid composition of the predicted binding regions should be responsible for the experimentally observed different substrate specificities of HQT1, HQT2, and HCT (Supplemental Fig. S2).
For docking analyses, a list of potential substrates (caffeoylquinate, caffeoylshikimate, p-coumaroylquinate, p-coumaroylshikimate, sinapate, quinate, shikimate, and tetracaffeoylquinate) were docked in the proposed binding site using Autodock 1.5.2 (Goodsell et al., 1996) to investigate and support the identified substrate specificity found in the experimental tests. Autodock was chosen for its ability to carry out a virtual screening of chemical libraries and to output a list of the ligands ordered by docked energy from lowest to highest. Autodock suggested the preference of HQT1 for caffeoylquinate and of HQT2 for p-coumaroylquinate (shown experimentally) and proposed a preference of HCT for caffeoylshikimate.
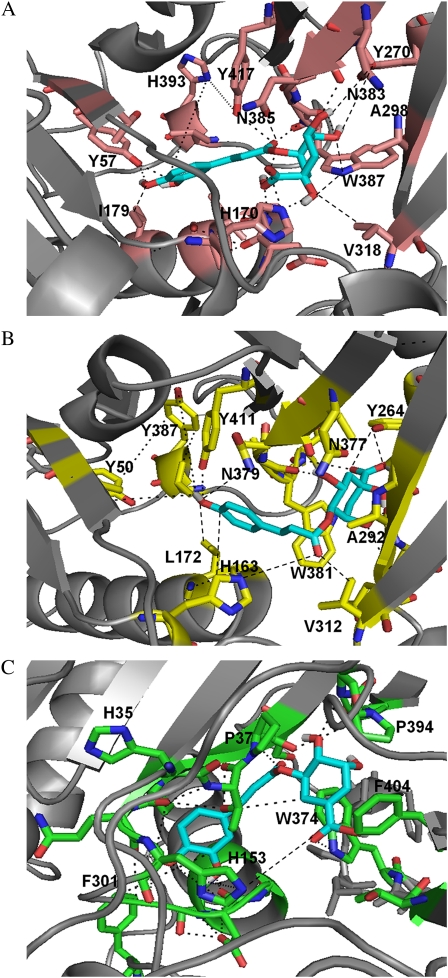
It was possible to highlight two distinct adjacent binding pockets in the proposed HQT1 and HQT2 binding regions that should be able to interact with the aromatic moiety of caffeoylquinate and p-coumaroylquinate, respectively. In HQT1, the first aromatic pocket is composed of residues His-393, Tyr-417, Tyr-57, Ile-179, and His-170, which together may bind the caffeoyl portion of caffeoylquinate, whereas the second binding pocket is composed of residues Tyr-270, Trp-387, Val-318, Asn-383, Ala-298, and Asn-385, which, together with His-170, should be able to bind the quinate portion (Fig. 4A). A set of similar residues surround p-coumaroylquinate in HQT2, where Tyr-387, Tyr-411, Tyr-50, Leu-172, Asn-379, and His-163 may bind the p-coumaroyl portion while Tyr-264, Ala-292, Val-312, Trp-381, Asn-377, and His-163 likely bind the quinate portion (Fig. 4B). Residues of the proposed binding sites are well conserved throughout the BAHD family members that accept hydroxycinnamoyl-CoAs as acyl donors and correspond or are very close to the sequence motifs highlighted in the multiple sequence alignment of Figure 1 (see also Supplemental Fig. S1): WXRXP (for HQT1387–391 and HQT2381–385), S[S/T]YE (for HQT1268–271 and HQT2262–265), DGRXR (for HQT1300–304 and HQT2294–298), and KX[K/S/A/Q]X[K/E/N/P/G] (for HQT1255–259 and HQT2250–254).
Figure 4.
Proposed binding sites for HQT1, HQT2, and HCT. Views of the comparative three-dimensional models of HQT1 (A), HQT2 (B), and HCT (C) highlighting the proposed binding sites are shown. α-Helices and β-sheets are shown in gray cartoon representation. The substrates caffeoylquinate (A), p-coumaroylquinate (B), and caffeoylshikimate (C) are shown in stick representation using the PyMOL color code: cyan, carbon atoms; red, oxygen atoms; white, hydrogen atoms; and blue, nitrogen atoms. Possible interactions between the substrates and residues of the proposed binding sites are indicated by black dashed lines. Residues of the binding sites are in pink cartoon and stick representation for HQT1, in yellow cartoon and stick representation for HQT2, and in green cartoon and stick representation for HCT.
Two equivalent binding pockets can be also identified in the proposed HCT binding region. In HCT, as might be expected, the binding pocket, which is predicted to prefer shikimate, is quite different from the quinate binding pockets in HQT1 and HQT2 and consists of residues Pro-394, Phe-404, and Trp-374 supported by Pro-37 and His-153 (Fig. 4C). Some differences also occur in the caffeoyl binding pocket of HCT that is composed of residues His-35, Phe-301, and His-153 compared with those of HQT1 and HQT2. However, these differences are less specific than those observed for the shikimate/quinate pockets between HCT and HQT1/2.
Expression of hqt1 and hqt2 Relative to CGA Content of Different Organs in Artichoke
A real-time PCR assay was developed using gene-specific primers that amplified exclusively hqt1 or hqt2 cDNAs. Very recently, another hqt sequence has been identified in artichoke (Comino et al., 2009), so we also confirmed the specificity of the hqt1 and hqt2 primers with respect to artichoke hct and hqt cDNAs. Real-time PCR was used to assay the expression of hqt1 and hqt2 in different bract orders (external, intermediate, and internal), the receptacle, and leaves at two developmental stages (vegetative and productive periods) in two artichoke genotypes. The cycle treshhold value used for gene expression analysis for each gene, tissue, and genotype was obtained by averaging the cycle treshhold values derived from all the biological and experimental replicates. The different expression levels of the target genes (hqt1 and hqt2) relative to the reference gene (elf1α) were calculated using Q-Gene software (Perikles, 2003). Normalization of the results against the expression of the constitutive elf1α and control of similar amplification efficiencies were performed to allow comparisons among tissues and between the genotypes.
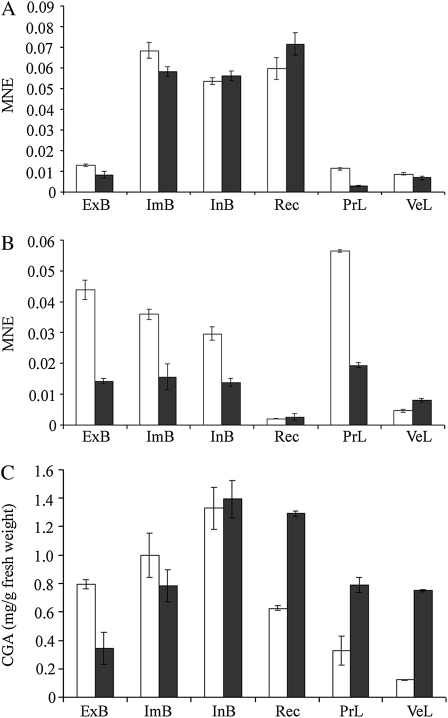
The expression profiles for hqt1 and hqt2 are shown in Figure 5, A and B. In both artichoke genotypes, the expression of hqt1 was higher in the receptacle and intermediate internal bracts of the flower head and lower in the external bracts and in the leaves. For one variety (Mola), the level of expression of hqt2 was quite similar to that of hqt1 in the different bract orders and productive leaves but lower in the vegetative leaves and in the receptacle. In the other variety, there was a similar trend, except for a higher level of expression in the productive leaves.
Figure 5.
Levels of expression of hqt1 and hqt2 and levels of chlorogenic acid in various artichoke tissues and two genotypes. A and B, Real-time PCR of hqt1 and hqt2 transcripts of external bracts (ExB), intermediate bracts (ImB), internal bracts (InB), receptacle (Rec), and leaves from the productive (PrL) or vegetative (VeL) period in two genotypes, S. Erasmo (white bars) and Mola (black bars). The data represent mean normalized expression (±sd) of biological and experimental replicates. C, Chlorogenic acid content as mg g−1 fresh weight of the same genotypes and tissues analyzed by real-time PCR. Values are means ± sd of three replicates.
To evaluate the relationship between the expression of the hqt1 and hqt2 genes and CGA content, caffeoylquinic acid was measured in the same tissues used for real-time expression analysis. Within the S. Erasmo variety, the CGA content of various tissues generally reflected the expression of the hqt1 gene, except for the outer bracts, which showed a relatively higher content of CGA (Fig. 5C). In the Mola variety, CGA content was most closely related to hqt1 expression. CGA content was relatively high in internal bracts, receptacle, and intermediate bracts compared with hqt1 transcript levels. CGA content was not clearly associated with the level of hqt2 expression in different organs in either variety.
In Vivo Expression of hqt1 for Functional Analysis
In order to assess the function of artichoke hqt in planta, both transient and stable expression systems were set up. For transformation experiments, hqt1 was chosen since its transcript levels appeared to be more closely related to CGA content in artichoke tissues compared with hqt2. Both Nicotiana benthamiana and N. tabacum are reported to synthesize CGA (Tamagnone et al., 1998a; Niggeweg et al., 2004).
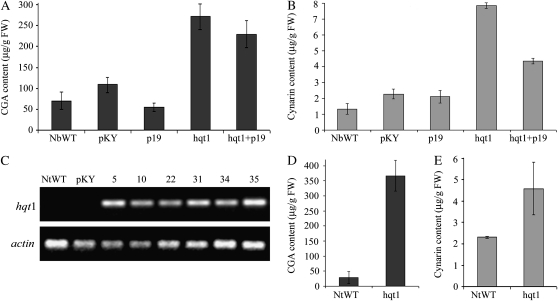
For transient hqt1 transformation, N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens carrying constructs of either hqt1 or the silencing-suppressing p19 gene (Voinnet et al., 2003) or both. Alongside, control plants were agroinfiltrated with the empty vector pKYLX71:35S2 (Schardl et al., 1987). Four days after infiltration, untransformed and transformed leaves were assayed for CGA and cynarin content by HPLC analysis (Fig. 6). N. benthamiana leaves transiently expressing hqt1 accumulated CGA to levels that were about three times higher than the wild type (NbWT), rising from 70 ± 20 μg g−1 fresh weight to 270 ± 30 μg g−1 fresh weight. Leaves infiltrated with the empty vector or with the p19-harboring construct showed similar CGA levels to the NbWT (Fig. 6A). Infiltration of the silencing-inhibiting p19 gene construct in addition to hqt1 did not increase accumulation of CGA compared with hqt1-overexpressing leaves (Fig. 6A), indicating that inhibition of A. tumefaciens-mediated hqt1 gene expression by posttranscriptional gene silencing was not a major factor under our experimental conditions. Interestingly, N. benthamiana leaves infiltrated with hqt1 also showed increased levels of cynarin, even though we have found no earlier reports of the synthesis of cynarin in the literature, probably due to the very low content of this compound in Nicotiana leaves. Cynarin accumulated to an average of 7.85 ± 0.18 μg g−1 fresh weight in the hqt1-expressing leaves, while NbWT leaves had a content of 1.33 ± 0.33 μg g−1 fresh weight (Fig. 6B).
Figure 6.
Transient and stable overexpression of artichoke hqt1 in N. benthamiana and N. tabacum, respectively, molecular characterization of stable transgenic lines, and determination of CGA and cynarin contents. A and B, Contents of CGA (A) and cynarin (B) as μg g−1 fresh weight (FW) in N. benthamiana wild-type plants (NbWT) and in transiently transformed leaves. Transformants either carried the empty vector (pKY) or expressed the silencing inhibitor protein p19 (p19), HQT1 (hqt1), or both HQT1 and p19 proteins (hqt1+p19). C, RT-PCR analysis of hqt1 (top) and the tobacco actin gene used as control (bottom) in a tobacco wild-type plant (NtWT), in a tobacco line transformed with the empty vector (pKY), and in hqt1-overexpressing lines (lines 5, 10, 22, 31, 34, and 35). D and E, Contents of CGA (D) and cynarin (E) as μg g−1 fresh weight in N. tabacum wild-type plants (NtWT) and in stably HQT1-transformed leaves (hqt1). The data represent means ± sd of eight transiently transformed leaves and leaves from three independent, stably transformed lines (lines 5, 31, and 35).
Stable transformation of N. tabacum cv Samsun NN was achieved using the same A. tumefaciens recombinant strains carrying the binary plasmid pKYhqt1 or the empty vector used for transient expression studies. Several regenerants were obtained for both constructs, which were checked for T-DNA insertion by PCR amplification with primer pairs for the kanamycin resistance gene nptII and, only for hqt1 transformants, for the region including the hqt1 gene and 57 nucleotides of the downstream T-DNA sequence (data not shown). In vivo-grown plants of six randomly selected independent transformants were checked for expression of the hqt1 transgene by semiquantitative RT-PCR using specific primers for artichoke hqt1 (Fig. 6C). Wild-type plants (NtWT) and plants transformed with the empty vector were also analyzed by RT-PCR with the same primer pairs. Expression of the transgene was demonstrated in all hqt1 transformants, while no amplification was detected in NtWT or pKYLX71:35S2-transformed plants (Fig. 6C).
Three independent transformants (lines 5, 31, and 35; Fig. 6C) as well as NtWT cv Samsun NN were used to measure the amount of CGA and cynarin. Similar to the results observed in transiently expressing leaves, transgenic plants stably expressing hqt1 accumulated high levels of CGA (Fig. 6D) compared with the NtWT, which increased from 30 ± 20 μg g−1 fresh weight in the control to 370 ± 50 μg g−1 fresh weight, averaged over the three independent transformants. Interestingly, as in the case of transient expression, stable transformation of hqt1 also resulted in the accumulation of cynarin, from 2.31 ± 0.05 μg g−1 fresh weight in the control plants to an average of 4.59 ± 1.22 μg g−1 fresh weight in the transformants, most notably in the lines with the highest CGA levels (Fig. 6E).
DISCUSSION
The accumulation of CGA and cynarin is thought to underpin many of the health-promoting properties of artichoke, effects that result from the consumption of immature inflorescences or extracts of leaves (Bundy et al., 2008).
HQT is considered one of the key enzymes for the synthesis of CGA in plants (Niggeweg et al., 2004). The predicted proteins encoded by hqt1 and hqt2 showed a high level of similarity to other hqt sequences available in the databases, and the encoded proteins group with other HQTs in phylogenetic analysis, whereas the HCT acyltransferase from artichoke (Comino et al., 2007) belongs to a distinct branch. Recently, another hqt cDNA has been reported from artichoke (Comino et al., 2009), although it encodes a protein distinct from HQT1 and HQT2 described here, with just 71% identity. Therefore, in artichoke, HQT enzymes seem to be encoded by a gene family composed of at least three members. A similar condition is likely to occur also in other genera of the Asteraceae, given that we found two groups of EST sequences leading to artichoke hqt1 and hqt2, both in sunflower and in lettuce.
Both HQT and HCT enzymes belong to the superfamily of BAHD acyltransferases, a recently described family of enzymes that utilize CoA thioesters and catalyze the formation of a diverse group of plant metabolites (St-Pierre and De Luca, 2000). The acyl donors and acyl acceptors used as substrates by BAHD acyltransferases are diverse, and different BAHD family members show a range of substrate specificities. Similar substrate specificity may have evolved independently in different species for some BAHD acyltransferases; therefore, experimental validation tests are needed to definitively identify acyltransferase function (D'Auria, 2006; Luo et al., 2007). Enzymes with HQT activity and showing a preference for quinate over shikimate as acyl acceptor cluster together in phylogenetic trees and are clearly distinct from HCT enzymes, which generally prefer shikimate to quinate as an acyl acceptor and are thought to work principally in lignin biosynthesis (Hoffmann et al., 2003, 2004; Sullivan, 2009). This clear conservation of catalytic specificity within phylogenetically distinct clades of the BAHD family is surprising given that not all plant species accumulate caffeoylquinate; it might have been predicted that HQT activity would have evolved de novo in species that develop the ability to synthesize CGA, as reported for acylation of anthocyanins (Suzuki et al., 2004). Therefore, rather than being derived by convergence, the ability to synthesize caffeoyl or p-coumaroylquinate would appear to be an ancient function conserved in the HQT clade of proteins, which has been lost in some species, such as Arabidopsis (Niggeweg et al., 2004).
Both HQT1 and HQT2 from artichoke are able to synthesize CGA, p-coumaroylquinate, and caffeoylshikimate in vitro, but only HQT2 can catalyze the reaction to produce p-coumaroylshikimate. In general, the calculated Km and Vmax for HQT1 and HQT2 are in the range of HQT enzymes from other plants (Niggeweg et al., 2004). Both HQT1 and HQT2 have the highest affinity for quinate, and their strong preference for quinate over shikimate aligns both artichoke HQT1 and HQT2 most closely with HQT activities from other plants (Hoffmann et al., 2003; Niggeweg et al., 2004). Our kinetic results and in silico homology modeling docking analyses suggest that the two enzymes might be involved in different steps in the synthesis of CGA: HQT1 likely catalyzes the step leading directly to CGA from caffeoyl-CoA and quinic acid preferentially, whereas HQT2 is more likely involved in the synthesis of p-coumaroylquinate. This reaction product could then be hydroxylated by a CYP98A type of C3′H for the production of CGA (Fig. 7). To a lesser extent, artichoke HQT1 and HQT2 could also catalyze the synthesis of caffeoylshikimate from caffeoyl-CoA and shikimic acid in vitro. However, the much higher affinity of HQT1 and HQT2 for quinate makes it improbable that they use shikimate as an acyl acceptor in vivo, as has also been suggested for tobacco and tomato (Niggeweg et al., 2004). The HQT2 reaction leading to p-coumaroylshikimate is even less probable, given the substrate specificity shown by this enzyme in vitro.
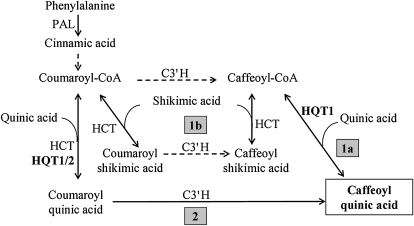
Figure 7.
Proposed pathways for the biosynthesis of chlorogenic acid in artichoke (redrawn from Mahesh et al., 2007). PAL, Phe ammonia-lyase.
The overall sequence similarity between the HQT and HCT members of the BAHD acyltransferase family allowed comparative models of the structures of the enzymes to be built based on the structures of DmAT (2e1t), complexed with malonyl-CoA (Unno et al., 2007), and vinorine synthase 2bgh from R. serpentina (Ma et al., 2005). A critical determinant in the accuracy of comparative modeling is the quality of the pairwise alignment between the target proteins and the crystallized templates. As the similarity between HQT1, HQT2, or HCT and the crystallized templates is lower than normally considered appropriate for comparative modeling (24%–26%), a multiple sequence alignment approach was used to improve pairwise alignment.
A binding site is speculated for HQT1 and HQT2, using Pocket Finder predictions and multiple sequence alignment comparisons. Docking analysis suggested that the best substrate for the proposed binding site of HQT1 is caffeoylquinate (CGA), while the best one for HQT2 is p-coumaroylquinate. Within each enzyme-binding site, two aromatic pockets have been detected that should be able to bind the two aromatic moieties of CGA and/or p-coumaroylquinate.
In the putative first aromatic pocket, HQT1 has His-393 and Ile-179, which are substituted by Tyr-387 and Leu-172 in HQT2. These differences may be not sufficient to explain the differing substrate specificities of the two enzymes. However, in the alignment, some other hydrophobic/hydrophilic or basic/hydrophilic differences were observed close to the first aromatic pocket: the Ala of peptide 170HTLA173 in HQT1 is substituted with a Ser in the HQT2 peptide 163HTLS166, and an important hydrophilic/basic difference is observed in a cavity that leads to the binding site where the Ser of the HQT1 peptide 255KASAK259 is substituted with a Lys in the HQT2 peptide 250KAKAK254. These residues, together with neighboring residues, as well as the observed amino acid variability in these portions could be responsible for substrate specificity and for redirecting the substrate toward the binding pockets and so might influence substrate specificity and enzyme efficiency. Unfortunately, at present, it is not possible to transform artichoke; therefore, experiments using transgenic lines to provide further evidence on substrate specificities are not yet feasible.
The amino acid composition of the putative second aromatic pocket is identical in HQT1 and HQT2, and it is predicted that this pocket is able to bind the quinate portion of CGA with analogous efficiency. The preference of both HQT1 and HQT2 for quinate over shikimate was also evident from the docking analysis. Very recently, another HQT coding sequence has been isolated from artichoke (DQ915589; Comino et al., 2009). This represents a fourth gene coding for a hydroxycinnamoyl transferase in artichoke: its cDNA and the encoded protein are different from those of HQT1 and HQT2. For alignment purposes, we used the sequence DQ915589 retrieved from the National Center for Biotechnology Information (NCBI) database, even though the authors show a 10-amino acid longer sequence in their paper. Comino et al. (2009) reported that their recombinant HQT could use either caffeoyl-CoA or p-coumaroyl-CoA as acyl donor in the presence of quinate, whereas it was inactive with shikimate. However, our results cannot be compared directly with artichoke HQT identified by Comino et al. (2009), since the affinity of their HQT for caffeoyl-CoA or p-coumaroyl-CoA was not investigated. A cDNA encoding HCT has also been isolated from artichoke. Enzyme assays of recombinant HCT suggested it to be more similar to HQT from tobacco and tomato than to HCT from tobacco or Arabidopsis, despite it aligning in the HCT clade. It was claimed that HCT showed a preference for quinate over shikimate (Comino et al., 2007), but this claim was based on Km values for p-coumaroyl-CoA at saturating levels of quinate or shikimate. Km values for HCT for quinate and shikimate need to be established to verify this claim. Our modeling and docking analyses suggest a preference of artichoke HCT for shikimate, in agreement with the predicted binding site pocket for HCT, which is clearly different from that of HQT.
To clarify the respective roles of hqt1 and hqt2 in CGA accumulation in artichoke, transcript levels of the two genes were analyzed in flower heads and leaves. hqt1 and hqt2 showed different patterns of expression, the former being more highly expressed in younger bracts and receptacle, while the latter was expressed almost equally in the different bract orders and productive leaves.
To gain further insight into the relationship between expression of hqt1 and CGA synthesis, in vivo functional analysis was carried out through transient and stable overexpression in N. benthamiana and N. tabacum, respectively, since artichoke is recalcitrant to genetic transformation. The results demonstrated that HQT1 is directly involved in the synthesis of CGA, given that the synthesis of this compound increased substantially in both transiently and stably transformed leaf tissues. In addition, increased accumulation of the health-promoting compound cynarin indicated that it is likely synthesized from CGA. Indeed, synthesis of 3,5-dicaffeoylquinic acid has been proposed to proceed from CGA in sweet potato, although the catalyzing enzyme was able to synthesize only 3,5-dicaffeoylquinic acid out of the three possible dicaffeoylquinic acid isomers (Kojima and Kondo, 1985).
The synthesis of CGA can theoretically occur through different routes operating in a metabolic grid (Fig. 7; Mahesh et al., 2007). Our data suggest that the route of CGA synthesis in artichoke is likely modulated according to substrate availability, and probably in vivo, CGA synthesis occurs preferentially via route 1, involving HCT, C3′H, and HQT (Fig. 7). This is also suggested by the evidence that a recently isolated C3′H enzyme from artichoke proved to be more efficient, within route 1b, in the conversion of p-coumaroylshikimate into caffeoylshikimate (Moglia et al., 2009). However, other possible isoforms of C3′H could be involved in route 2, as in coffee (Mahesh et al., 2007).
The route taken for CGA biosynthesis in any particular cell will depend on the substrate specificities of the acyltransferases and the C3′H and on the availability of cytoplasmic pools of the alternative acyl donors and acceptors. In artichoke, we have identified two new HQT enzymes with different specificities, which suggest that they may operate in different parts of the grid (Fig. 7). HCT is likely most important in route 1, section 1b (Fig. 7), and its relative contribution to CGA synthesis in different organs will depend on the cells in which it is active. If, like HCT in tobacco and Arabidopsis, it is expressed predominantly in vascular tissues in artichoke, then its contribution to CGA synthesis may be restricted to these tissues. The specificity of HQT1 suggests that it may operate in route 1, specifically section 1a, although it may also function in route 2 (Fig. 7). The substrate specificity of HQT2 suggests that it probably operates in route 2. The specificity of different C3′H enzymes operating in different cells may also influence the primary route for CGA synthesis, as reported for developing coffee beans (Mahesh et al., 2007). In addition, the role of the newly reported HQT enzyme (Comino et al., 2009) can be better clarified only after experimental determination of its catalytic specificities.
In conclusion, we have demonstrated that HQT1 and HQT2 are both involved in the synthesis of CGA in artichoke. The recent report of an additional HQT from artichoke shows that several HQT isoforms exist in this plant and that they likely operate in a metabolic grid to ensure high levels of CGA synthesis, probably by multiple metabolic routes. Indeed, artichoke may have evolved multiple HQT isoforms with differing specificities to enhance CGA accumulation by optimizing flux down the different biosynthetic routes. Putative homology modeling and docking analyses have allowed us to predict, to our knowledge for the first time, the binding sites of HQT and HCT enzymes, which could be of general interest for further studies of these enzymes in plants.
The exciting observation that elevated expression of hqt1 in leaves of Nicotiana species not only increases the levels of CGA but also those of cynarin lends support to previous suggestions that cynarin is synthesized from CGA and also offers a means of increasing production of this important plant phytonutrient.
MATERIALS AND METHODS
Plant Materials
Plants of artichoke (Cynara cardunculus subsp. scolymus ‘Mola’ and ‘S. Erasmo’) were grown in the experimental field of the Institute of Plant Genetics, Policoro, Italy. For each variety, material was collected from three independent plants; young leaves were collected during the vegetative (mid October) and productive (mid March) phases; flower heads were harvested at the commercial stage and separated into external, intermediate, and internal bracts and receptacle (De Paolis et al., 2008).
Nicotiana benthamiana and tobacco (Nicotiana tabacum ‘Samsun NN’) plants were grown in soil in a growth chamber at 23°C and 16/8-h photoperiod. Transgenic N. tabacum plants were grown in vitro on Murashige and Skoog solid medium supplemented with 3% (w/v) Suc and 100 mg L−1 kanamycin at 23°C and 16/8-h light/dark photoperiod for rooting. In vitro-grown plants were then transferred to soil and acclimated to in vivo conditions in a growth chamber with the same temperature and photoperiod.
Isolation and Cloning of Artichoke hqt cDNAs
The hqt nucleotide sequences from tobacco (AJ582651) and tomato (Solanum lycopersicum; AJ582652) were used to search the lettuce (Lactuca sativa) and sunflower (Helianthus annuus) The Gene Index databases (http://compbio.dfci.harvard.edu/tgi/plant.html). After alignment of the best tentative consensus sequences, primers Hel-F3 and Hqt-R2 for hqt1 and HEL_HQT_F6 and HEL_HQT_R4 for hqt2 (Supplemental Table S1) were designed to amplify expected fragments of about 600 and 1,000 bp, respectively. Amplification reactions contained 1 μL of cDNA from leaves, 1× PCR buffer, 0.4 mm deoxyribonucleotide triphosphates (dNTPs), 400 nm each primer, and 1 unit of JumpStart (Sigma-Aldrich) Taq DNA polymerase, in a final volume of 25 μL. The thermal cycling program included a step at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. PCR fragments were cloned using the GENEJet PCR Cloning kit (Fermentas) and subsequently sequenced. Sequences were subjected to a BLAST search against the GenBank plant database (http://www.ebi.ac.uk/blastall/index.html).
Full-Length cDNA and Genomic Sequences, and Phylogenetic Analysis
New primers were synthesized for amplification of the 5′ and 3′ ends of hqt1 and hqt2 transcripts using 5′ and 3′ RACE (Invitrogen), respectively. To identify the 5′ ends, 5RACE_HQT1 primer for hqt1 or 5RACE_HQTa1 for hqt2 (Supplemental Table S1) and total RNA from cv Mola leaf were used for cDNA synthesis. After treatment with RNAse H and cDNA column purification, a homopolymeric tail was added to the cDNA 3′ end. A first round of PCR was performed using the nested antisense primer 5RACE_HQT2 for hqt1 and 5RACE_HQTa2 for hqt2 (Supplemental Table S1) along with the RACE-specific primer furnished by the manufacturer. Amplification reactions contained the tailed cDNA, 1× Tuning PCR buffer, 0.2 mm dNTPs, 400 nm each primer, and 2.5 units of PCR Extender Polymerase (5 PRIME) in a final volume of 50 μL. The thermal cycling program included a step at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 60°C (hqt1) or 50°C (hqt2) for 30 s, and 72°C for 2 min, and a final extension step at 72°C for 10 min. A second round of PCR was performed, under the same conditions except for the annealing temperature (60°C for hqt1 or 56°C for hqt2), using a 1:1,000 dilution of the first amplificate and the nested primer 5RACE_HQT3 for hqt1 or 5RACE_HQTa3 for hqt2 (Supplemental Table S1) along with the universal RACE-specific primer. Amplified cDNA fragments were cloned and sequenced.
For the completion of the 3′ ends of each cDNA, total RNA and the adapter primer (AP) from the kit were used for cDNA synthesis. A first PCR was performed using primer HQT-F2 for hqt1 or 3RACE_HQTa1 for hqt2 (Supplemental Table S1) along with the anchored RACE primer. Reactions contained 2 μL of cDNA, 1× Tuning PCR buffer, 0.2 mm dNTPs, 400 nm each primer, and 2 units of PCR Extender Polymerase (5 PRIME) in a final volume of 50 μL. The amplification program started with a step at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 3 min, and a final extension at 72°C for 10 min. A second round of PCR was performed, under the same conditions, with the exception of the annealing temperature (60°C for hqt1 or 54°C for hqt2), using a 1:500 dilution of the first amplification and the nested primers 3RACE_HQT1 for hqt1 or 3RACE_HQTa2 for hqt2 (Supplemental Table S1). Amplified cDNA fragments were cloned and sequenced.
Full-length cDNA clones were generated using end-specific primers for hqt1 and hqt2 (HQT_ATG_F/HQT_STOP_R and Hqta_ATG_F/Hqta_STOP_R, respectively; Supplemental Table S1). Amplification reactions contained 2 μL of cDNA from leaves, 1× PCR buffer, 0.4 mm dNTPs, 400 nm each primer, and 2.5 units of recombinant Taq DNA polymerase (Invitrogen) in a final volume of 50 μL. Amplification conditions were 94°C for 3 min and 30 cycles at 94°C for 45 s, 55°C (hqt1) or 62°C (hqt2) for 30 s, and 72°C for 90 s; the reaction mixture was then incubated at 72°C for 10 min. Fragments were cloned and five clones were sequenced.
The same primers were used to amplify artichoke genomic DNA by prolonging the extension step to 4 min. Genomic DNA fragments were cloned and sequenced.
Full-length cDNA and genomic sequences of hqt1 and hqt2 were aligned and separately subjected to BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/). The hqt cDNA sequences were translated in silico using the program TRANSEQ (http://bioweb.pasteur.fr/seqanal/interfaces/transeq.html), and the deduced protein sequences were aligned to protein sequences from other species. Phylogenetic analysis was performed using the program MEGA4 (Tamura et al., 2007). The tree was constructed using the neighbor-joining method of Saitou and Nei (1987), with 10,000 bootstrap replicates.
For the isolation of hqt1 and hqt2 promoters, the Genome Walker Universal kit (Clontech) was used, following the manufacturer's instructions. After artichoke genomic DNA digestion with DraI, EcoRV, PvuII, and StuI and ligation to adaptors as indicated in the manufacturer's instructions, a first round of PCR was performed for each digested-ligated DNA library, using the AP1 primer from the kit and the primer specific for hqt1 or hqt2 (GWhqtGSP1 or GWhqt2GSP1, respectively; Supplemental Table S1). Amplification reactions contained 1 μL of ligated DNA, 1× PCR buffer, 0.5 mm dNTPs, 400 nm each primer, and 2 units of PCR Extender Polymerase (5 PRIME) in a final volume of 50 μL. The thermal cycling program included seven cycles of 94°C for 25 s and 69°C (hqt1) or 68°C (hqt2) for 3 min, followed by 37 cycles of 94°C for 25 s and 64°C for 3 min. After the 37 cycles, the reaction mixture was incubated at 67°C for 7 min. A 1:50 dilution of the amplificates was used as a template for a nested PCR, using primers GWhqtGSP2 and GWhqt2GSP2 (Supplemental Table S1), and the following thermal cycling program: five cycles of 94°C for 25 s and 72°C (hqt1) or 66°C (hqt2) for 3 min, followed by 20 cycles of 94°C for 25 s and 67°C (hqt1) or 62°C (hqt2) for 3 min, followed by an incubation step at 67°C for 7 min. Amplicons were resolved on a low-melting-point agarose gel, and fragments longer than 1 kb were excised from the gel, purified, cloned, and sequenced.
The analysis of promoter sequences for the two genes was performed using the database PLACE (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999).
Heterologous Expression in Escherichia coli and Immunoblot Analysis
Full-length cDNAs of hqt1 and hqt2 were introduced into the Gateway system (Invitrogen) as follows. The cDNAs were amplified using the primer pairs A-HQT-GW_F/A-HQT-GW_R and A_HQTa_GW_F/A_HQTa_GW_R, respectively (Supplemental Table S1). The entry clones (pDONR207-hqt1 and pDONR207-hqt2) were obtained through recombination of the PCR products with pDONR207 (Invitrogen). Error-free clones were then introduced into the Gateway-compatible S-tag vector pJAM1784 (Luo et al., 2007) to produce the expression vectors pJAM1784-hqt1 and pJAM1784-hqt2. Recombinant proteins with an N-terminal S-tag were expressed in E. coli BL-21 Rosetta cells. Cells were grown at 37°C with shaking at 250 rpm until the optical density at 600 nm was 0.6. Isopropyl β-d-1-thiogalactopyranoside was added to 0.4 mm. Cells were cultured at 20°C with shaking at 250 rpm for 16 h. Cells were harvested, resuspended in 100 mm potassium phosphate, pH 7.0, with 1 mm EDTA, broken using a French press, and centrifuged at 14,000 rpm for 30 min at 4°C. Total protein was assayed from crude extracts. Samples containing 10 μg of proteins were separated by SDS-PAGE, and proteins were electrophoretically transferred to nitrocellulose. S-tag fusion peptides were immunodetected with 1/5,000 AP-conjugated S-Protein (Novagen). Negative controls used comparable extracts of E. coli harboring an empty vector.
Transient and Stable Heterologous Expression of hqt1 in Nicotiana
Full-length cDNA of hqt1 was amplified with the primers f-HQT-HindIII and r-HQT-XbaI (Supplemental Table S1) to introduce HindIII and XbaI restriction sites and cloned into the HindIII-XbaI polylinker sites of the binary vector pKYLX71:35S2 under the transcriptional control of the cauliflower mosaic virus 35S promoter with double enhancer (Schardl et al., 1987). Correct insertion was verified by sequencing. The resulting recombinant pKYhqt1 plasmid was introduced into the Agrobacterium tumefaciens strain LBA4404 by the freeze-thaw method (Hofgen and Willmitzer, 1988).
For transient transformation, A. tumefaciens LBA4404 cultures carrying the plasmid pKYhqt1, or the 35S:p19 plasmid harboring the gene encoding the silencing inhibitor protein p19 (Voinnet et al., 2003), or the empty vector pKYLX71:35S2 were grown overnight at 28°C with vigorous shaking. Pelleted cells were resuspended in 10 mm MgCl2 and 150 μg mL−1 acetosyringone to an optical density at 600 nm of 1.0 and incubated at room temperature for 3 h (Voinnet et al., 2003). For agroinfiltration, equal amounts of the cultures containing pKYhqt1, or 35S:p19, or pKYhqt1 plus 35S:p19, or the empty pKYLX71:35S2 vector were used. Each construct or construct combination was infiltrated into the abaxial air spaces of four leaves of each of two N. benthamiana plants. After 4 d, the infiltrated leaf material was collected.
The recombinant A. tumefaciens LBA4404 strain harboring either the pKYhqt1 plasmid or the empty vector was also used to stably transform N. tabacum cv Samsun NN plants according to standard methods (Horsh et al., 1987). Kanamycin-resistant regenerated shoots were excised and transferred to kanamycin-supplemented solid medium for rooting and then transferred to soil and acclimated to in vivo conditions in a growth chamber. Three-week-old transgenic and control plants were used for molecular characterization and HPLC analysis (see below).
Stable insertion of the exogenous expression cassette was checked in kanamycin-resistant plants by PCR amplification with primers for the nptII gene (TransF and TransR; Supplemental Table S1). The presence of the hqt1 transgene was also assessed by amplifying DNA of kanamycin-resistant plants with specific primers for hqt1 (f-HQT-Hind III) and for pKYLX71:35S2 polylinker (pKYLX_R; Supplemental Table S1).
Enzyme Assays and HPLC/Liquid Chromatography/Mass Spectrometry Analyses
S-tag fusion peptides for HQT1 and HQT2 were measured using the S-tag rapid assay kit (Novagen) by comparison with the S-tag peptide standard.
For CGA enzyme assays, the standard reaction mixture (100 μL) consisted of 50 mm potassium phosphate buffer, pH 7.0, 60 μm caffeoyl-CoA or p-coumaroyl-CoA, 120 μm quinate or shikimate, 1 mm EDTA, and enzyme as 1 to 5 μL of the crude E. coli extract. For cynarin assay, 1 mm cynarin and 500 μm CoA were used. Each reaction was started by addition of the acyl acceptor. After incubation at 30°C for 10 min, the reaction was terminated by adding 100 μL of ice-cold 0.5% (v/v) trifluoroacetic acid.
The reaction mixture was analyzed by liquid chromatography/mass spectrometry (LC/MS) using an Agilent 1100 LC system equipped with diode array UV light and single quadrupole 1100 MSD detectors. Ten microliters of the reaction was analyzed on a 100- × 2-mm 3μ Luna C18(2) column (Phenomenex) using a gradient of methanol (B) versus 0.1% formic acid (A) in water run at 25°C and 0.25 mL min−1: 0 min, 98% A, 2% B; 30 min, 30% A, 70% B; 30.5 min, 98% A, 2% B; 38 min, 98% A, 2% B. Spray chamber conditions were 10 L min−1 drying gas at 350°C, 25-p.s.i. nebulizer pressure, and 4,000-V (positive mode) or 3,500-V (negative mode) spray voltage. Masses were scanned from 50 to 1,500 atomic mass units, and the fragmentor voltage was 75 V.
Determination of Kinetic Parameters
To evaluate the substrate specificity of HQT1 and HQT2, caffeoyl-CoA and p-coumaroyl-CoA were tested as acyl donors, while quinate, shikimate, sinapate, and malate were used as acyl acceptors. The standard reaction mixture (100 μL) consisted of 50 mm potassium phosphate buffer, pH 7.0, 1 μm to 24 mm of the different substrates, and 0.6 to 1.4 μg of S-tag fusion proteins from crude extracts of E. coli. Enzyme activity was measured as the decrease in time of acyl donor A340 or A360 for p-coumaroyl-CoA or caffeoyl-CoA reaction, respectively, using a FlexStation3 Microplate Reader (Molecular Devices). These wavelengths were chosen because they correspond to the maxima of the difference spectra between the thioesters and the corresponding quinic or shikimic esters (Lotfy et al., 1992; Niggeweg et al., 2004). The following concentrations of enzymes and substrates were used. Km for quinate: 57 and 25 ng μL−1 for HQT1 and HQT2, respectively, 2.5 mm caffeoyl-CoA, 0.05 to 1 mm quinate or 2.5 mm p-coumaroyl-CoA, and 0.001 to 0.6 mm quinate; Km for shikimate: 140 ng μL−1 for HQT1 or HQT2, 2.5 mm caffeoyl-CoA/p-coumaroyl-CoA, and 1 to 24 mm shikimate; Km for caffeoyl-CoA: 140 ng μL−1 for HQT1 or HQT2, 10 mm quinate, and 0.1 to 4.5 mm caffeoyl-CoA; Km for p-coumaroyl-CoA: 71 ng μL−1 for HQT1 or HQT2, 10 mm quinate, and 0.1 to 3 mm p-coumaroyl-CoA. For reverse reactions, the Km for CoA was determined using 10 ng μL−1 HQT1 or 6 ng μL−1 HQT2, 0.5 mm CGA, and 0.01 to 0.3 mm CoA. The Km for CGA was determined using 10 ng μL−1 HQT1 or 6 ng μL−1 HQT2, 0.4 mm CoA, and 0.1 to 0.6 mm CGA.
No activity was detected with sinapic acid or malic acid as acyl acceptor, using 140 ng μL−1 HQT1 or HQT2, and 10 mm sinapic acid or malic acid. Km and Vmax values were calculated from Lineweaver-Burk double reciprocal plots. CGA, CoA, quinate, shikimate, sinapate, and malate were from Sigma-Aldrich. Caffeoyl-CoA and p-coumaroyl-CoA were purchased from TransMIT.
Comparative Modeling and Docking Investigations
Protein sequences of plant BAHD acyltransferases were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/). The homologs of the artichoke proteins HQT1 and HQT2 were aligned using ClustalW with the secondary structure of the anthocyanin acyltransferase from Dendranthema morifolium (DmAT), complexed with malonyl-CoA, and vinorine synthase from Rauvolfia serpentina. These crystallized structures were used to weigh gap penalties. The three-dimensional structures of the two cited proteins are available at the Protein Data Bank (Berman et al., 2002) under the code names 2e1t (Unno et al., 2007) and 2bgh (Ma et al., 2005), respectively. The pairwise alignment of each artichoke protein (HQT1 and HQT2) with DmAT was taken from the multiple sequence alignment and improved by comparison with the PSIPRED secondary structure alignment prediction output. Then, Modeller (Sánchez and Sali, 2000) was used to calculate structural models of artichoke HQT1 and HQT2 proteins. For comparative purposes, the artichoke HCT protein (Comino et al., 2007) was also analyzed. Five hundred steps of energy minimization were repeated to generate 100 optimized HQT1, HQT2, and HCT structures. The structural properties of the HQT1, HQT2, and HCT models with the best energy function were evaluated using the biochemical/computational tools of the WHAT IF Web server (Vriend, 1990).
Final models were examined in PyMOL (http://www.pymol.org/) and SwissPDBViewer (http://spdbv.vital-it.ch/), and where side chain packing led to clashes or gaps in the local secondary structure, alternative side chain rotamers were evaluated.
Pocket Finder was used to predict the potential binding sites of the HQT1, HQT2, and HCT best models. For docking analysis, potential substrates were docked in the proposed binding site using Autodock 1.5.2 (Goodsell et al., 1996), which allows selection of a three-dimensional region within which the software will search for the best interactions between all the possible substrate conformations and the selected three-dimensional region proposed as the binding site.
Expression Analysis in Artichoke and in Transgenic N. tabacum
Levels of hqt1 and hqt2 transcripts in different artichoke tissues and physiological stages were analyzed using real-time PCR in a Rotor-Gene 6000 (Corbett Research). Artichoke elongation factor 1α (Elf1α; GenBank accession no. EU442190) was used as a housekeeping gene. Each reaction contained 1.2 μL of a 1:6 cDNA dilution, 5 μL of Power SybrGreen mix (Applied Biosystems), with 300 nm gene-specific primers RT-HQT-F2 and RT-HQT-R2 for hqt1, RT-HQT2-F and RT-HQT2-R for hqt2, or EL-RT-F and EL-RT-R for Elf1α (Supplemental Table S1), in a total volume of 10 μL. To reduce pipetting errors, a CAS-1200 liquid-handling system (Corbett) was used. In all experiments, appropriate negative controls containing no template were subjected to the same procedure. Analysis was performed on three independent plants for each genotype, on three replicates for each sample, and reactions were repeated twice to verify reproducibility. Real-time PCR cycles were 95°C for 10 min, followed by 40 cycles of three-step reactions (95°C for 15 s, 60°C for 30 s, and 72°C for 30 s). The specificity of products was verified by melting curve analysis (60°C–95°C). Standard curves for the genes of interest and the housekeeping gene were generated on the basis of a six-point cDNA dilution series by plotting the threshold cycle versus relative template concentration. Mean normalized expression of hqt1 and hqt2 was calculated using Q-Gene software (Perikles, 2003) by averaging the normalized expression values derived from biological and experimental replicates.
For the semiquantitative RT-PCR analysis of expression of the hqt1 transgene in stably transformed tobacco plants, cDNAs were synthesized from 1.0 μg of total RNA from leaves of six hqt1 transformants, one plant transformed with the empty pKYLX71:35S2 vector and one wild-type plant, using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Semiquantitative RT-PCR amplifications were performed using specific primers for artichoke hqt1 (RT-HQT-F2 and RT-HQT-R2; Supplemental Table S1) and tobacco actin as a housekeeping gene (f-ACT and r-ACT; Supplemental Table S1). Reactions contained 1× PCR buffer, 0.2 mm dNTPs, 1.5 mm MgCl2, 1.5 units of Taq DNA polymerase (Invitrogen), 500 nm each primer, and 1 μL of cDNA template in a final volume of 50 μL. The amplification program started with a step at 95°C for 5 min, followed by 30 cycles of three steps (95°C for 15 s, 60°C for 30 s, and 72°C for 10 s), followed by an elongation step at 72°C for 5 min. Amplified products were separated and visualized on a 2% agarose gel.
Determination of CGA Content in Artichoke and Nicotiana Tissues
The same artichoke plant parts used for real-time PCR experiments were analyzed for their CGA content, following Fratianni et al. (2007), with some modifications. Frozen plant tissues were weighed, ground, and incubated at 4°C for 2 h with 5% (w/v) of an extraction mixture composed of acetone:methanol:ethanol (70:15:15). The supernatant was collected and stored at 4°C, while the pellet was reextracted in ethyl acetate. The two supernatants were mixed and dried in a Rotavapor at 25°C, resuspended in methanol, and 10 μL was injected for HPLC analysis using a Luna C18 Phenomenon column in a Gold System chromatograph equipped with a UV light detector (Beckman). The mobile phase included HPLC-grade water (containing 0.01% TCA; solvent A) and 95% acetonitrile (containing 0.01% TCA; solvent B) in the following gradient system: initial, 0% B; at 2 min, 36% B for 33 min; at 35 min, 36% B for 10 min; at 45 min, 10% B for 2 min. The flow rate was 1 mL min−1, and the detection wavelength was set at 310 nm.
Soluble phenolics were extracted from transiently transformed and wild-type N. benthamiana leaves and from N. tabacum leaves of three independent stable transformants. Freshly harvested tissues were homogenized on ice in 80% aqueous methanol solution (1:3, w/v) and then sonicated three times for 1 min. Soluble phenolics were separated by centrifuging at 13,000 rpm for 5 min and stored at −20°C until use. HPLC analysis was performed as follows. Twenty microliters of each extract was analyzed by HPLC (LC 10; Shimadzu) with a diode array detector and a Prodigy column (5 μm ODS3 100A, 250 × 4.60 mm; Phenomenex) at a flow rate of 0.8 mL min−1. The mobile phase was a mixture of water:formic acid (99.8:0.2, v/v; A) and methanol:acetonitrile (40:60, v/v; B). Phenolic compound elution was achieved using the following linear gradient: starting condition, 85% A, 15% B; 6 min, 75% A, 25% B; 16 min, 70% A, 30% B; 20 min, 60% A, 40% B; 24 min, 50% A, 50% B; 32 min, 40% A, 60% B; 35 min, 20% A, 80% B; 38 min, 75% A, 15% B. Chromatograms were recorded at 365 nm for phenolic acids.
Standard for cynarin (1,3-dicaffeoylquinic acid) was from Carl Roth, and CGA was from Sigma-Aldrich.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AM690438 (hqt1 cDNA), EU839580 (hqt2 cDNA), EU697935 (hqt1 genomic DNA), FM244907 (hqt2 genomic DNA), and EU442190 (Elf1α).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of recombinant artichoke HQT1 and HQT2 proteins in E. coli.
Supplemental Figure S2. Multiple sequence alignment of several plant acyltransferases.
Supplemental Table S1. List of primer names and sequences.
Supplementary Material
Acknowledgments
We thank A. De Lisi, D. Nigro, and A. Visconti for HPLC analyses of CGA and cynarin content; A. Morgese for DNA sequencing; and L. Hill for LC/MS analyses.
References
- Agarwal R, Mukhtar H. (1996) Cancer chemoprevention by polyphenols in green tea and artichoke. Adv Exp Med Biol 401: 35–50 [DOI] [PubMed] [Google Scholar]
- Azzini E, Bugianesi R, Romano F, Di Venere D, Miccadei S, Durazzo A, Foddai MS, Catasta G, Linsalata V, Maiani G. (2007) Absorption and metabolism of bioactive molecules after oral consumption of cooked edible heads of Cynara scolymus L. (cultivar Violetto di Provenza) in human subjects: a pilot study. Br J Nutr 97: 963–969 [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. (2002) The Protein Data Bank. Acta Crystallogr D Biol Crystallogr 58: 899–907 [DOI] [PubMed] [Google Scholar]
- Brown JE, Rice-Evans CA. (1998) Luteolin-rich artichoke extract protects low density lipoprotein from oxidation in vitro. Free Radic Res 29: 247–255 [DOI] [PubMed] [Google Scholar]
- Bundy R, Walker AF, Middleton RW, Wallis C, Simpson HC. (2008) Artichoke leaf extract (Cynara scolymus) reduces plasma cholesterol in otherwise healthy hypercholesterolemic adults: a randomized, double blind placebo controlled trial. Phytomedicine 15: 668–675 [DOI] [PubMed] [Google Scholar]
- Bushman BS, Snook ME, Gerke JP, Szalma SJ, Berhow MA, Houchins KE, McMullen MD. (2002) Two loci exert major effects on chlorogenic acid synthesis in maize silks. Crop Sci 42: 1669–1678 [Google Scholar]
- Chen JH, Ho CT. (1997) Antioxidant activities of caffeic acid and its related hydroxycinnamic acid compounds. J Agric Food Chem 45: 2374–2378 [Google Scholar]
- Clifford MN. (1999) Chlorogenic acids and other cinnamates: nature, occurrence, and dietary burden. J Sci Food Agric 79: 362–372 [Google Scholar]
- Comino C, Hehn A, Moglia A, Menin B, Bourgaud F, Lanteri S, Portis E. (2009) The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol 9: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comino C, Lanteri S, Portis E, Acquadro A, Romani A, Hehn A, Larbat R, Bourgaud F. (2007) Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- De Paolis A, Pignone D, Morgese A, Sonnante G. (2008) Characterization and differential expression analysis of artichoke phenylalanine ammonia-lyase coding sequences. Physiol Plant 132: 33–43 [DOI] [PubMed] [Google Scholar]
- Dowd PF, Vega FE. (1996) Enzymatic oxidation products of allelochemicals as a basis for resistance against insects: effects on the corn leafhopper Dalbulus maidis. Nat Toxins 4: 85–91 [DOI] [PubMed] [Google Scholar]
- Fratianni F, Tucci M, De Palma M, Pepe R, Nazzaro F. (2007) Polyphenolic composition in different parts of some cultivars of globe artichoke: Cynara cardunculus L. var. scolymus (L.) Fiori. Food Chem 104: 1282–1286 [Google Scholar]
- Gebhardt R. (1997) Antioxidative and protective properties of extracts from leaves of artichoke (Cynara scolymus L.) against hydroperoxide-induced oxidative stress in cultured rat hepatocytes. Toxicol Appl Pharmacol 144: 279–286 [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Morris GM, Olson AJ. (1996) Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 9: 1–5 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxyl cinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M. (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L. (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsh RB, Fry JE, Hoffmann NL, Eicholtz D, Rogers SH, Fraley RT. (1987) A simple and general method for transferring genes in plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Kojima M, Kondo T. (1985) An enzyme in sweet potato root which catalyzes the conversion of chlorogenic acid, 3-caffeoylquinic acid, to isochlorogenic acid, 3,5-dicaffeoylquinic acid. Agric Biol Chem 49: 2467–2469 [Google Scholar]
- Kraft K. (1997) Artichoke leaf extract: recent findings reflecting effects on lipid metabolism, liver and gastrointestinal tracts. Phytomedicine 4: 369–378 [DOI] [PubMed] [Google Scholar]
- Lepelley M, Cheminade G, Tremillon N, Simkin A, Caillet V, McCarthy J. (2007) Chlorogenic acid synthesis in coffee: an analysis of CGA content and real-time RT-PCR expression of HCT, HQT, C3H1, and CCoAOMT1 genes during grain development in C. canephora. Plant Sci 172: 978–996 [Google Scholar]
- Lotfy S, Fleuriet A, Macheix JJ. (1992) Partial purification and characterization of hydroxycinnamoyl-CoA: transferases from apple and date fruits. Phytochemistry 31: 767–772 [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al. (2007) Convergent evolution in the BAHD family of acyltransferases: identification and characterization of anthocyanin acyltransferases from Arabidopsis thaliana. Plant J 50: 678–695 [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J. (2005) Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem 280: 13576–13583 [DOI] [PubMed] [Google Scholar]
- Maher EA, Bate NJ, Ni W, Elkind Y, Dixon RA, Lamb CJ. (1994) Increased disease susceptibility of transgenic tobacco plants with suppressed levels of preformed phenylpropanoid products. Proc Natl Acad Sci USA 91: 7802–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesh V, Million-Rousseau R, Ullmann P, Chabrillange N, Bustamante J, Mondolot L, Morant M, Noirot M, Hamon S, de Kochko A, et al. (2007) Functional characterization of two p-coumaroyl ester 3′-hydroxylase genes from coffee tree: evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol Biol 64: 145–159 [DOI] [PubMed] [Google Scholar]
- McDougall B, King PJ, Wu BW, Hostomsky Z, Manfred G, Robinson WE., Jr (1998) Dicaffeoylquinic acid and dicaffeoyltartaric acid are selective inhibitors of human immunodeficiency virus type 1 integrase. Antimicrob Agents Chemother 42: 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Moglia A, Comino C, Portis E, Acquadro A, De Vos RC, Beekwilder J, Lanteri S. (2009) Isolation and mapping of a C3'H gene (CYP98A49) from globe artichoke, and its expression upon UV-C stress. Plant Cell Rep 28: 963–974 [DOI] [PubMed] [Google Scholar]
- Nardini M, Cirillo E, Natella F, Scaccini F. (2002) Absorption of phenolic acids in humans after coffee consumption. J Agric Food Chem 50: 5735–5741 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M, Enju A, Shinozaki K. (2004) Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 55: 327–342 [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C. (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 2: 746–754 [DOI] [PubMed] [Google Scholar]
- Perikles S. (2003) Q-Gene processing quantitative real-time RT-PCR data. Bioinformatics 19: 1439–1440 [DOI] [PubMed] [Google Scholar]
- Persson B. (2000) Bioinformatics in protein analysis. EXS 88: 215–231 [DOI] [PubMed] [Google Scholar]
- Plumb GW, Garcia-Conesa MT, Kroon PA, Rhodes M, Ridley S, Williamson G. (1999) Metabolism of chlorogenic acid by human plasma, liver, intestine and gut microflora. J Sci Food Agric 79: 390–392 [Google Scholar]
- Rice-Evans CA, Miller NJ, Paganga G. (1997) Antioxidant properties of phenolic compounds. Trends Plant Sci 2: 152–159 [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sánchez R, Sali A. (2000) Comparative protein structure modeling: introduction and practical examples with Modeller. Methods Mol Biol 143: 97–129 [DOI] [PubMed] [Google Scholar]
- Schardl C, Byrd AD, Benzion GB, Altschuler MA, Hildebrand DF, Hunt AG. (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–11 [DOI] [PubMed] [Google Scholar]
- Schütz K, Kammerer D, Carle R, Schieber A. (2004) Identification and quantification of caffeoylquinic acids and flavonoids from artichoke (Cynara scolymus L.) heads, juice, and pomace by HPLC-DAD-ESI/MS. J Agric Food Chem 52: 4090–4096 [DOI] [PubMed] [Google Scholar]
- Segasothy M, Phillips PA. (1999) Vegetarian diet: panacea for modern lifestyle diseases? Q J Med 92: 531–544 [DOI] [PubMed] [Google Scholar]
- Shimoda H, Ninomiya K, Nishida N, Yoshino T, Morikawa T, Matsuda H, Yoshikawa M. (2003) Anti-hyperlipidemic sesquiterpenes and new sesquiterpene glycosides from the leaves of artichoke (Cynara scolymus L.): structure requirement and mode of action. Bioorg Med Chem Lett 13: 223–228 [DOI] [PubMed] [Google Scholar]
- Soltani BM, Ehlting J, Hamberger B, Douglas CJ. (2006) Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 224: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Sonnante G, Pignone D, Hammer K. (2007) The domestication of artichoke and cardoon: from Roman times to the genomic age. Ann Bot (Lond) 100: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalmach A, Steiling H, Williamson G, Crozier A. (2010) Bioavailability of chlorogenic acids following acute ingestion of coffee by humans with an ileostomy. Arch Biochem Biophys (in press) [DOI] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V. (2000) Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv Phytochem 34: 285–315 [Google Scholar]
- Sullivan ML. (2009) A novel red clover hydroxycinnamoyl transferase has enzymatic activities consistent with a role in phaselic acid biosynthesis. Plant Physiol 150: 1866–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Sawada S, Watanabe K, Nagae S, Yamaguchi MA, Nakayama T, Nishino T. (2004) Identification and characterisation of a novel anthocyanin malonyltransferase from scarlet sage (Salvia splendens) flowers: an enzyme that is phylogenetically separated from other anthocyanin acyltransferases. Plant J 38: 994–1003 [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. (1998a) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Stacey N, Plaskitt K, Parr A, Chang C, Lynn D, Dow JM, Roberts K, Martin C. (1998b) Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell 10: 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Ulbrich B, Zenk MH. (1979) Partial purification and properties of hydroxycinnamoyl-CoA:quinate hydroxycinnamoyl transferase from higher plants. Phytochemistry 18: 929–933 [Google Scholar]
- Unno H, Ichimaida F, Suzuki H, Takahashi S, Tanaka Y, Saito A, Nishino T, Kusunoki M, Nakayama T. (2007) Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J Biol Chem 282: 15812–15822 [DOI] [PubMed] [Google Scholar]
- Villegas RJA, Kojima M. (1986) Purification and characterization of hydroxycinnamoyl D-glucose: quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. J Biol Chem 261: 8729–8733 [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8: 52–56 [DOI] [PubMed] [Google Scholar]
- Walker JRL. (1995) Enzymatic browning in fruits: its biochemistry and control. Lee CY, Whitaker JR, , Enzymatic Browning and Its Prevention: ACS Symposium Series 600. American Chemical Society, Washington, DC, pp 8–22 [Google Scholar]
- Wang M, Simon JE, Aviles IF, He K, Zheng QY, Tadmor Y. (2003) Analysis of antioxidative phenolic compounds in artichoke (Cynara scolymus L.). J Agric Food Chem 51: 601–608 [DOI] [PubMed] [Google Scholar]
- Zhua XF, Zhanga HX, Lob R. (2005) Antifungal activity of Cynara scolymus L. extracts. Fitoterapia 76: 108–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.