Abstract
SAG39 is a rice (Oryza sativa) gene that encodes a cysteine protease. SAG39 shares 55% homology with the Arabidopsis (Arabidopsis thaliana) senescence-associated protein SAG12. The promoter for SAG39 (PSAG39) was isolated, and SAG39 expression was determined to be relatively low in mature leaves, while not expressed in the endosperm. SAG39 mRNA levels increased as senescence progressed, with maximum accumulation of transcripts at late senescence stages. Gel retardation assays indicated that two cis-acting elements in PSAG39, HBOXCONSENSUSPVCHS and WRKY71OS, responded to leaf senescence. To test if PSAG39 could be useful for increasing rice yields by increasing cytokinin content and delaying senescence, homozygous transgenic plants were obtained by linking PSAG39 to the ipt gene and introducing it into Zhonghua 11. The chlorophyll level of the flag leaf was used to monitor senescence, confirming the stay-green phenotype in PSAG39:ipt transgenic rice versus wild-type plants. Changes in the cytokinin content led to early flowering and a greater number of emerged panicles 70 d after germination in the transgenic rice. Measurements of sugar and nitrogen contents in flag leaves demonstrated a transition in the source-sink relationship in transgenic plants triggered at the onset of leaf senescence, with the nitrogen content decreasing more slowly, while sugars were removed more rapidly than in wild-type plants. The importance of these changes to rice physiology, yield, and early maturation will be discussed.
Rice (Oryza sativa) is one of the most important food crops in the world. However, intensive cultivation of rice has severe consequences for the environment. Most critically, lowland rice production uses vast quantities of water, imparting an enormous strain on water resources, typically in countries that are already short on water supply (Mew et al., 2004). Yet, demand for rice continues to increase with estimates that world consumption is outstripping production (Matsumura et al., 2009). Therefore, researchers are actively pursuing methods to increase yield, without further environmental degradation.
One avenue for yield increase is to delay leaf senescence as a means of extending the available time for photosynthesis. Senescence is the regulated remobilization of nutrients and accompanying programmed cell death; however, it remains unclear how the variety of regulatory genes interact to control leaf senescence. Traditional methods for delaying senescence include directly spraying leaves with cytokinins (CKs) or the addition of nitrogen to the growth media at the seeding stage (Gan and Amasino, 1995). However, the development of biotechnology has allowed for more elegant molecular tools, utilizing CK to delay leaf senescence (Gan and Amasino, 1995). CKs control a variety of processes, including cell division and differentiation, plant growth, and nutrient mobilization, and CKs can act to inhibit the transcription of Cys proteases, delaying degradation of nucleic acids and proteins (Sakakibara et al., 2005; Werner et al., 2006). Isopentenyltransferase (ipt), an enzyme found in Agrobacterium tumefaciens, catalyzes a critical rate-limiting step in CK synthesis. This gene has been widely employed in stay-green engineering studies with varied results. Ipt expression under the control of a constitutive promoter (Li et al., 1992), heat shock promoter Phsp70 (Smigocki, 1991; Van Loven et al., 1993), or copper-induced promoter (McKenzie et al., 1998) resulted in abnormal phenotypes. Gan and Amasino (1995) generated an autoregulatory cycle for ipt expression using a senescence-specific promoter, PSAG12, from Arabidopsis (Arabidopsis thaliana). The PSAG12:ipt construct was introduced into tobacco (Nicotiana tabacum) plants, leading to a reduction in leaf and floral senescence, resulting in an increased number of flowers, seed yield, and biomass (80%, 50%, and 40%, respectively). In transgenic Arabidopsis (Huynh et al., 2005), petunia (Petunia hybrida; Chang et al., 2003), and alfalfa (Medicago sativa; Calderini et al., 2007) containing the PSAG12:ipt construct, the stay-green effects were achieved, along with a greater number of flowers, and an enhancement of drought resistance. The PSAG12:ipt construct promoted biomass production in rice (Lin et al., 2002a) and resulted in greater nitrate reductase activity in wheat (Triticum aestivum; Sýkorová et al., 2008), but no change in grain yield was found in either cereal. We speculate that it is crucial to find an appropriate promoter for ipt expression in order to gain yield improvement.
With the establishment of large international databases, unprecedented access has been given for the prediction and identification of promoters for novel innovations (Shahmuradov et al., 2003; Yamamoto and Obokata, 2008). The technology for promoter cloning by reporter genes and qualitative or quantitative analysis of promoters is well known, and a large number of specific promoters have been successfully verified (Tran et al., 2004). Through the use of several molecular tools, such as yeast one-hybrid, gel retardation assays, and DNase I footprint identification technologies, useful promoters can be identified and their expression analyzed (Hua et al., 2007; Saha et al., 2007; Cai et al., 2008).
In this study, a novel rice senescence-specific promoter, PSAG39, was isolated and identified from a Nipponbare (O. sativa sp. japonica) bacterial artificial chromosome clone. The expression pattern of the promoter was examined, including cis-element analysis. The promoter was also successfully linked to the ipt gene and introduced into the rice var Zhonghua 11. The stay-green phenotype was tested, along with modification to plant ontogeny and metabolism.
RESULTS
Identification and Characteristics of a Senescence-Specific Promoter PSAG39
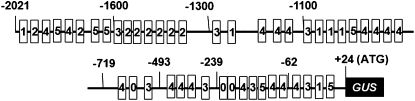
A 2.1-kb fragment harboring the −2,021 to +24 region (translation initiation site of SAG39 as +1) upstream of SAG39 was isolated from the rice var Nipponbare (Fig. 1). Sequence analysis showed that this fragment contained several basal regulatory elements including three TATA boxes (located at −220, −222, and −565, respectively) for RNA polymerase binding and 15 CAAT boxes (located at −92, −108, −229, −322, −457, −464, −501, −530, −573, −707, −1,047, −1,236, −1,735, −1,837, and −1,966, respectively) for transcription frequency regulation (Fig. 1). This fragment also harbored several known cis-elements that respond to hormone treatment and regulate stress-induced gene expression, including an ethylene and jasmonic acid response GCCCORE box (Brown et al., 2003); abscisic acid (ABA)-response boxes RYREPEATBNNAPA (Ezcurra et al., 1999), MYCATRD22 (Abe et al., 1997), and DPBFCOREDCDC3 (Kim et al., 1997); GA response box RYRIMIDINEBOXHVEPB1 (Cercos et al., 1999); OsWRKY71 binding site WRKY71OS that participates in GA signal repression (Zhang et al., 2004); drought tolerance response box MYBCORE (Urao et al., 1993); and a MYBST1 box (Baranowskij et al., 1994; Fig. 1).
Figure 1.
The native SAG39 promoter with truncated promoter constructs and predicted regulatory elements. Numbers above the promoter represent the position at which the upstream region was deleted for the construct: −2,021 = entire promoter, P39; −1,600 = Pf16 construct; −1,300 = Pf13 construct; −1,100 = Pf10 construct; −719 = Pf7 construct; −493 = Pf5 construct; −239 = Pf2 construct; −62 = Pf06 construct. cis-Regulatory elements were predicted with the PLACE program (Prestridge, 1991; Higo et al., 1999; http://www.dna.affrc.go.jp/PLACE/signal-scan.html). 0, Predicted basal regulatory element TATA; 1, MYCCONSENSUSAT, predicted ABA response element; 2, GCCCORE, predicted jasmonic acid response element; 3, RYRIMIDINEBOXHVEPB1, predicted GA response element; 4, MYBST1, predicted Myb DNA binding domain; 5, WRKY71OS, predicted DNA binding WRKY domain.
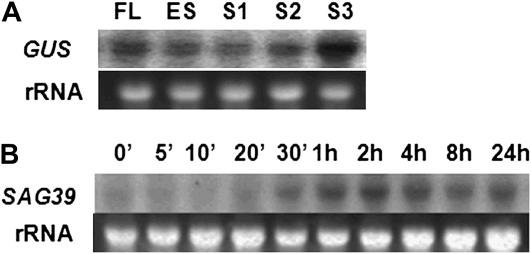
The SAG39 promoter (PSAG39) was linked to the GUS reporter gene. Using Southern-blot analysis in the T0 generation, single copy insert lines were selected for further study (Supplemental Figs. S1A and S2). The expression pattern of the PSAG39:GUS was examined under natural senescence conditions (Fig. 2A). Northern-blot analysis revealed that the greatest activity of the promoter was found under late senescing conditions. The promoter's activity was also investigated in wild-type plants following leaf-spraying with the senescence-inducing hormone ABA (Fig. 2B). SAG39 expression was strongly induced after 30 min, reaching a peak 2 h after application, indicating that the gene was involved in the ABA-induced senescence pathway. These results confirm that expression of the SAG39 promoter is senescence specific.
Figure 2.
Expression pattern of PSAG39. A, Northern-blot analysis of PSAG39:GUS under conditions of natural senescence. FL, Mature fully expanded leaves; ES, early senescence leaves with 90% chlorophyll; S1, senescing leaves with 70% chlorophyll; S2, senescing leaves with 60% chlorophyll; S3, senescing leaves with 40% chlorophyll. B, Northern-blot analysis of SAG39 following ABA treatment in wild-type rice plants.
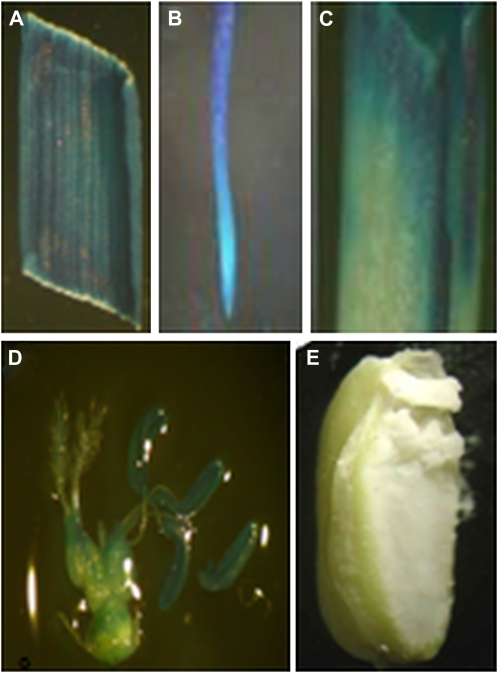
GUS staining analysis was used to determine the tissue-specific expression of PSAG39 (Fig. 3). Following chlorophyll destaining, deep-blue coloration was visible throughout the mature plant prior to seed harvesting, demonstrating PSAG39 expression (Fig. 3, A–D); however, no GUS staining was visible in the rice endosperm (Fig. 3E).
Figure 3.
GUS expression in mature transgenic plants with the P39 construct (entire promoter). A, Leaf; B, root; C, culm; D, flower; E, seed. Leaf, root, culm, and seed were sampled immediately before seed harvest.
Expression Pattern of 5′-Truncated Promoters
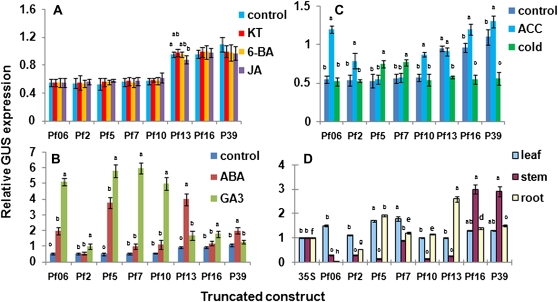
GUS expression levels in positive calli carrying a single copy of PSAG39:GUS or in seven truncated PSAG39:GUS constructs (Supplemental Table S1) were compared by quantitative analysis. Treatment with senescence-delaying kinetin and 6-benzylaminopurine or with senescence-inducing methyl jasmonate did not induce differences in GUS expression in the calli (Fig. 4A). The ethylene precursor 1-aminocyclopropane-1-carboxylic-acid (ACC) significantly increased GUS expression, compared to their respective controls, in most of the promoter constructs, except Pf5, Pf7, and Pf13 (Fig. 4B). Cold treatment increased GUS expression in constructs Pf5 and Pf7, while GUS expression was reduced in constructs Pf13, Pf16, and in the entire promoter, P39, relative to controls (Fig. 4B). Cold treatment had no effect on GUS expression linked to promoters Pf06, Pf2, and Pf10. ABA treatment had a pronounced affect, inducing a 2- to 4-fold increase in GUS expression in Pf06, Pf5, Pf13, and P39 compared to controls (Fig. 4C). GA3 significantly increased GUS expression in all constructs, except in the complete promoter, with as much as a 6-fold increase in GUS expression (Fig. 4C).
Figure 4.
GUS expression driven by a series of truncated promoters. A to C, Response to hormone and cold stress. See Figure 1 for description of truncated promoters. Control, Water treated; KT, kinetin; 6-BA, 6-benzylaminopurine; JA, methyl jasmonate. D, GUS activity in different organs of transgenic plants. Different letters refer to significantly different means within a construct (A–C) or between all constructs for a specific organ (D). P < 0.05, tested using ANOVA. Error bars refer to ±se of three replicates.
Expression patterns of truncated PSAG39:GUS promoter constructs were tissue dependent (Fig. 4D). Pf5 and Pf7 had significantly higher expression than the control using Cauliflower mosaic virus 35S promoter (P35S) in roots and shoots but had significantly lower expression (Pf5 nearly 7 times lower) in the stem. All other promoters showed no differences in expression versus the control in the shoot. The entire P39 promoter and Pf16 showed a 1.5-fold greater root and nearly 3-fold higher stem expression than controls. Pf2, Pf10, and Pf13 had significantly lower stem expression. Pf13 had nearly three times more root expression than P35S, while Pf06 had nearly no root expression (Fig. 4D).
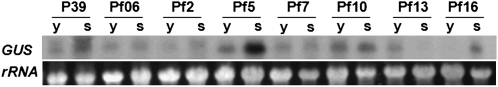
In naturally senescing leaves, transgenic plants carrying Pf5 and Pf16 showed a similar GUS expression pattern to the plants carrying the entire SAG39 promoter by northern-blot analysis (Fig. 5). The GUS level in the Pf5 transformants was the highest, while it was found to be lowest in the Pf16 transformants (Fig. 5). Interestingly, Pf13 showed an inverted response, with greater expression in young plants and no GUS expression as the plants aged. Apparently the upstream region of PSAG39-493:GUS contains negatively regulated cis-elements and the downstream contains positively regulated cis-elements for senescence.
Figure 5.
Northern-blot analysis of GUS expression driven by a series of truncated promoters during natural senescence. See Figure 1 for description of truncated promoters. Age of plants sampled denoted by either y, young plants (about 50 d after germination), or s, senescing plants (about 90 d after germination).
Identification of cis-Elements Controlling Senescence-Specific Expression
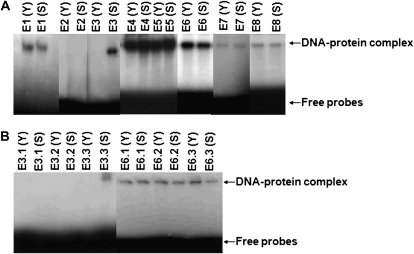
We compared cis-elements distributed upstream or downstream of −493 or −1,600, according to sequence analysis. Eight double-stranded DNA probes (Supplemental Table S2) covering the different regions of PSAG39 were used to study DNA-protein binding and to determine the senescence-specific sites of the promoter. Probes of 19 to 49 bp in length were used to interact with equal amounts of nuclear extracts from young (approximately 50 d after germination) or senescent (approximately 90 d after germination, with 40% chlorophyll) leaves. Gel mobility shift assays showed three key results (Fig. 6A): (1) probes E1, E2, E4, E5, E7, and E8 showed an interaction with the proteins from both young and senescent leaves; (2) probe E3 bound only proteins from senescent leaves; and (3) probe E6 bound intensely to the proteins from young leaves, but binding decreased with senescent leaves. This suggests that probes E3 and E6 contained cis-elements related to senescence.
Figure 6.
Gel mobility shift assays using nuclear proteins were from wild-type Zhonghua 11 plants. DNA probes were 19 to 49 bp in length. A, E1 and E2 are in the upstream of −493; E3 and E4 are in the downstream of −493; E5 and E6 are in the upstream of −1,600; E7 and E8 are in the downstream of −1,600. B, The truncated promoter region of E3 and E6 examined in greater detail. Age of plants sampled denoted by either y, young plants (about 50 d after germination), or s, senescing plants (about 90 d after germination).
Another group of six double-stranded short probes (Supplemental Table S2) were used to distinguish the senescence-specific elements in the promoter regions covered by probes E3 and E6 (Fig. 6B). Gel mobility shift assays showed that the binding capacity of E3.3 and E6.3 changed drastically when using proteins from senescent leaves as compared to proteins from young leaves.
Cosegregation Analysis of PSAG39:ipt Transgenic Plants
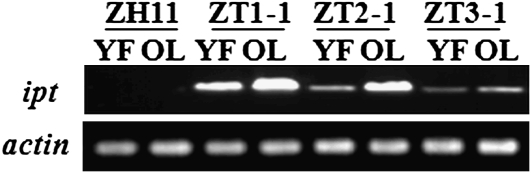
Transgenic plants carrying a single copy of PSAG39:ipt that were identified by Southern blots (Supplemental Fig. S2) and T2 homozygous lines were selected through rooting analysis. Ipt expression in young flag leaves or senescing fourth (from the top) leaves was detected just after rice plant heading by reverse transcription (RT)-PCR. Three lines named ZT1-1, ZT2-1, and ZT3-1 were ipt positive (Fig. 7). Their corresponding null lines from the same T0 transgenes were called ZT1-2, ZT2-2, and ZT3-2, respectively. As Figure 7 demonstrates, in the three selected lines, senescing leaves accumulated more ipt mRNA than young leaves, and in the three selected lines, ipt expression decreased gradually. These results show that ipt expression is under the control of the promoter PSAG39.
Figure 7.
RT-PCR analysis of ipt in wild-type (WT; Zhonghua 11 [ZH11]) or transgenic (ZT1-1, ZT2-1, and ZT3-1) plants. Age of plants sampled denoted by either YF, young flag leaves, or OL, senescing fourth (from the top) leaves.
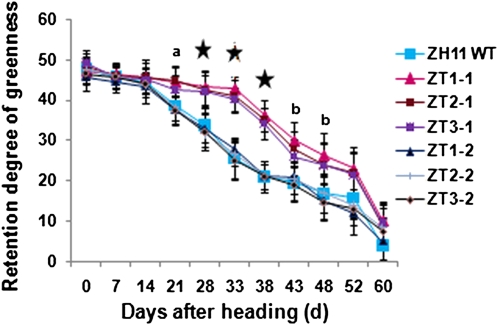
To corroborate the mRNA analysis, Soil Plant Analysis Development (SPAD) measurements were taken using the third leaf (from the top of the plant) on the primary tiller (Fig. 8). Twenty-eight days after heading, the three positive lines showed significantly higher SPAD values than either the wild-type or null lines. Only after 43 d following the heading date did the transgenic lines have SPAD values similar to either the wild-type or null lines.
Figure 8.
Comparison of SPAD values between field-grown wild-type (WT; Zhonghua 11 [ZH11]) and transgenic (ZT1-1, ZT2-1, and ZT3-1) rice plants using the third leaf. Null lines are shown for comparison (ZT1-2, ZT2-2, and ZT3-2). Different letters/asterisks refer to significantly different means: “a” denotes SPAD value for ZT2-1 was significantly greater than wild-type or null lines; “b” denotes SPAD value for ZT1-1 was significantly greater than wild-type or null lines; and asterisks denote SPAD values for all transgenic lines were significantly greater than wild-type or null lines. P < 0.05, tested using ANOVA. Error bars refer to ±se of three replicates, with each replicate being the average of eight to 10 different plants.
Impact of PSAG39:ipt on the Panicle
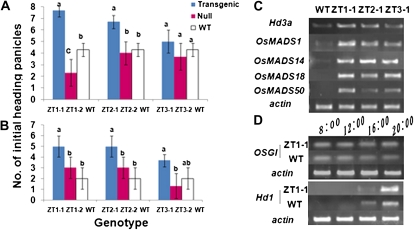
Transgenic rice expressing the PSAG39:ipt construct showed an earlier heading date in either well-watered or poorly watered field conditions. Figure 9 demonstrates that the number of initial heading panicles, 70 d after germination, was significantly higher in ZT1-1 and ZT2-1 than in control plants under both watering conditions. While ZT3-1 did not show significant differences from wild-type plants, it followed the trend of having more initial heading panicles, similar to ZT1-1 and ZT2-1.
Figure 9.
Comparison of heading date and flowering related genes between field-grown wild-type (WT) and transgenic rice plants. A, Number of initial heading panicles under well-watered field conditions 70 d after germination. B, Number of initial heading panicles under water-limited field conditions 70 d after germination. C, RT-PCR analysis of several flowering-associated genes in wild-type and transgenic plants. D, RT-PCR analysis of the flowering-associated genes OSGI and Hd1 in wild-type and transgenic plants. Hd3a (heading date gene); OsMADS1 (MADS box floral gene); OsMADS14 (MADS box floral gene); OsMADS18 (MADS box floral gene); OsMADS50 (MADS box flowering regulator); OsGI (O. sativa GIGANTEA); Hd1 (floral regulator gene). Different letters in A and B refer to significantly different means determined by comparing the wild-type plants to the transgenic and null lines for each construct. P < 0.05, tested using ANOVA. Error bars refer to ±se of three replicates, with each replicate being the average of eight to 10 different plants.
To understand why the initial heading panicle number was greater in the transgenic plants, expression levels of flowering-regulated genes and circadian-regulated genes were investigated (Fig. 9). There was an accumulation of RNA transcripts for Hd3a, OsMADS1, OsMADS14, OsMADS18, and OsMADS50 during flowering in the positive transgenic lines ZT1-1, ZT2-1, and ZT3-1, although the degree of accumulation differed between lines (Fig. 9C). OsGI and Hd1 showed no difference in the amount of RNA that accumulated (Fig. 9D).
Sugar and Nitrogen Metabolic Analysis of PSAG39:ipt Transgenic Plants
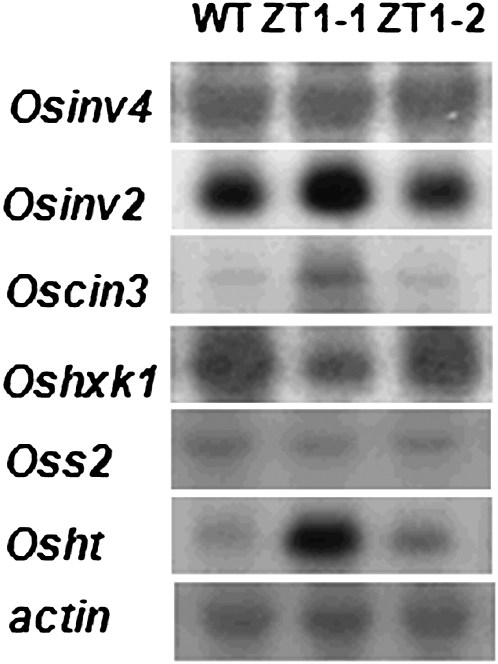
Since ZT1-1 showed the strongest stay-green phenotype among the three transgenic lines, the sugar contents of the flag leaf, second leaf, and third leaf (from the top) were determined at various stages of development in order to test the hypothesis that carbon metabolism had been augmented in these transgenic plants (Supplemental Fig. S3). The sugar content in the flag leaf was similar in the transgenic and wild-type plants until the heading stage, after which the flag leaf sugar content was lower in the transgenic plants versus the wild type. The sugar content of the third leaf reached a peak at the milky stage (seeding 1, approximately 90 d after germination) in the transgenic plants, while the sugar content of the wild-type plants peaked at the heading stage. These changes in sugar content likely demonstrate an alteration in the carbon source and recycling of sugar in the plants. The expression patterns of several genes involved in sugar biosynthesis and transport were examined by RNA-blot analysis (from the third leaf at the heading stage; Fig. 10). Northern-blot results showed that expression of OsINV4 (apoplastic invertase) and OsS2 (Suc synthase2) showed no differences between transgenic and control plants. Hexokinase I (OsHXK1) expression decreased in transgenic plants, while OsINV2 (vacuolar acid invertase), OsCIN3 (cell wall invertase), and OsHT (hexose transporter) expression increased in transgenic plants with respect to the wild type. This expression pattern, along with the sugar analysis, strongly indicates a shift in carbon flow, toward maintaining metabolic activity in the lower leaves of the transgenic plants, while the flag leaf exports carbon to the primary sink, the developing grain.
Figure 10.
Northern-blot analysis of sugar-associated genes in wild-type (WT) and transgenic plants. Null line (ZT1-2) shown for comparison. Osinv4 (apoplastic invertase); Osinv2 (vacuolar acid invertase); Oscin3 (cell wall invertase); Oshxk1 (hexokinase I); Oss2 (Suc synthase2); Osht (hexose transporter).
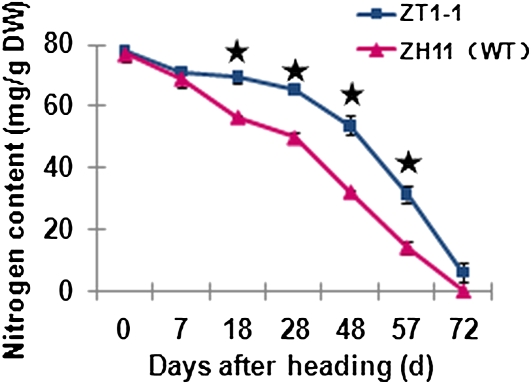
The nitrogen content of ZT1-1 was also measured under the control field conditions in fully expanded flag leaves (starting about 63 d after germination; Fig. 11). The nitrogen content of the wild-type plants was significantly lower than in the transgenic rice from the heading to wax ripeness stages (approximately 30–57 d after heading). This was likely in large part due to a lower rate of chlorophyll degradation and reduced nitrogen remobilization in the transgenic plants.
Figure 11.
Analysis of nitrogen content in the flag leaf of wild-type (WT; Zhonghua 11 [ZH11]) and transgenic plants starting 63 d after germination. For each sampling date, the nitrogen content was compared between transgenic and wild-type plants. Asterisks refer to significantly different means of nitrogen content. P < 0.05, tested using Student's t test. Error bars refer to ±se of three replicates, with each replicate being the average of eight to 10 different plants. [See online article for color version of this figure.]
Agronomic Traits of PSAG39:ipt Transgenic Plants
Panicle number, yield, and shoot dry mass production varied significantly among the three transgenic lines and the wild type at the end of the field experiment (Table I). Two of the transgenic lines (ZT2-1 and ZT3-1) and one of the null lines (ZT2-2) had significantly more panicles than all other genotypes tested. These three lines also were found to have the greatest amount of shoot dry matter production and seed yield per plant. ZT1-1 performed in many cases similar to the wild type, although ZT1-1 had a more modest yield and greater 1,000-grain weight than the wild type.
Table I. Agronomic traits for field-grown transgenic and wild-type rice, grown under well-watered conditions.
Each value represents the mean ± se of 20 to 24 replicates. Different superscripted letters refer to significantly different means within an agronomic trait examined. P < 0.05, tested using ANOVA.
| Genotype | Panicle No. | Grain Yield per Plant | Shoot Dry Mass per Plant | 1,000-Grain Weight |
| g | g | |||
| Zhonghua 11 (wild type) | 12.25 ± 0.78b | 28.72 ± 1.53ab | 24.02 ± 1.41d | 23.40 ± 0.26bc |
| ZT1-1 (transgenic) | 11.81 ± 0.76b | 24.07 ± 1.49c | 24.50 ± 1.38d | 25.28 ± 0.26a |
| ZT1-2 (null) | 11.65 ± 0.73b | 23.82 ± 1.42c | 26.74 ± 1.31bcd | 23.65 ± 0.24bc |
| ZT2-1 (transgenic) | 16.68 ± 0.74a | 30.61 ± 1.46ab | 28.85 ± 1.34abc | 24.61 ± 0.25a |
| ZT2-2 (null) | 16.55 ± 0.78a | 31.02 ± 1.53a | 32.46 ± 1.41a | 23.84 ± 0.26b |
| ZT3-1 (transgenic) | 14.71 ± 0.71a | 29.31 ± 1.39ab | 29.03 ± 1.29ab | 23.92 ± 0.24b |
| ZT3-2 (null) | 12.52 ± 0.73b | 26.63 ± 1.42bc | 25.18 ± 1.31cd | 23.03 ± 0.24c |
DISCUSSION
SAG39 is a senescence-associated gene in rice that encodes a Cys protease involved in protein degradation. The promoter for SAG39, PSAG39, was determined to be active during senescence. Northern-blot analysis of SAG39, along with GUS expression analysis of PSAG39, determined that the gene increased in activity with natural leaf senescence, peaking in the late phases of senescence (Fig. 2). While the promoter was active in most plant tissues, it was not detected in the rice endosperm (Fig. 3). Combining the SAG39 promoter with the IPT gene resulted in transgenic plants that demonstrated significantly earlier heading dates under both well-watered and water-limited field conditions (Fig. 9).
SAG39 Is Regulated by Positive and Negative Responsive cis-Elements
From a series of expression models of truncated promoters, Pf5 and Pf16 were found to act as senescence-specific promoters, similar to the entire PSAG39 promoter. Other truncated constructs showed either constitutive expression or peak expression during younger phases of plant growth, with reduced expression as senescence progressed (Fig. 5). Although there was great variation in gene expression, all the constructs shared the commonality of not expressing in the endosperm, implying that this tissue localization signal was not present in the regions affected by the truncated promoter. Moreover, the truncated Pf5 and Pf16 promoter constructs demonstrated the appropriate stay-green phenotype, making them more likely candidates as models for plant breeding projects (see below).
Comparative analysis of the functions of native and truncated promoters indicated that PSAG39 contained cis-acting elements with responses that were tissue specific, senescence induced, or regulated by hormone application. While GUS staining in leaves was generally consistent among all the tested promoters, the GUS activity in stems was drastically reduced by deleting the region between −1,300 and −1,600 (Fig. 4D). In roots, regions −1,100 to −1,300, −239 to −493, and −62 to −239 appeared critical for enhancing expression (Fig. 4D). Addition of ABA and GA3 generated the strongest GUS responses (Fig. 4B). Interestingly ABA and GA have opposing effects on senescence, yet SAG39 appears to respond to both hormones. Deletions encompassing the regions between −239 and −439, and −1,100 and −1,300 affected GUS expression for both ABA and GA3. Gubler and Jacobsen (1992) found a similar ABA/GA cis-acting element in barley (Hordeum vulgare), which may imply that this dual response to hormones is a common mechanism for plant promoters. Application of the ethylene precursor ACC and cold stress resulted in significant changes in GUS expression; however, they were considerably less dramatic than ABA or GA3 (Fig. 4C). While the SAG39 promoter may be activated by ethylene or cold alone, it is more likely that the corresponding response elements in the promoter enhance gene activity initiated by other factors, such as water limitation or senescence. Such additive gene effects have already been found during heat and drought stress (Rizhsky et al., 2004) and likely form the basis of plant adaptation strategies.
Senescence is a complex process and can be induced by stress (pathogen, salt, or extreme temperature) or hormone treatment. In this sense, cis-acting elements can possibly regulate senescence-specific expression in response to stress or hormone treatment, demonstrated by the gel mobility shift assays. As Figure 6 showed, probe E3.3, located in the −438 to −430 region, specifically bound only proteins from senescent leaves, but probe E6.3, located in the −1,899 to −1,892 region, showed a decreased binding activity to nuclear proteins in the same leaves. According to PLACE analysis, HBOXCONSENSUSPVCHS was embodied in probe E3.3 and WRKY71OS (Zhang et al., 2004) was embodied in probe E6.3. H-box is essential for both light regulation and elicitor induction. Previous results in tobacco suffering from wounding or abiotic stress have shown that a special transcription factor KAP-2 binds to the H-box and can induce gene transcription (Loake et al., 1992; Mhiri et al., 1997; Lindsay et al., 2002). WRKY71OS is the binding site of rice WRKY71, a transcriptional repressor of the GA signaling pathway (Zhang et al., 2004; Cai et al., 2007). In young leaves, WRKY71OS bound to the protein strongly, but in senescent leaves, the binding was repressed and gene expression was stimulated. These results suggested that H-box may function as a positive regulatory element and WRKY71OS may function as a negative regulatory element in the coordination of senescence-specific expression.
Despite sequence similarity to many previously identified cis-elements, there were only a few regions that acted accordingly. Clearly, rigorous testing of cis-elements is necessary in different organisms to assign functional regions to promoters. As demonstrated here, such work is complicated by regions that may share functions or that operate by enhancing the activity of nearby elements that have bound trans-acting factors.
Potential Use of PSAG39 in Rice Breeding
In our study, OsMADS50 was induced in the PSAG39:ipt transgenic plants (Fig. 9C). OsMADS50 is an upstream regulator of many floral development genes, including OsMADS1, OsMADS14, OsMADS18, and Hd3a (Lee et al., 2004). Enhanced expression of these genes led to the first demonstration of earlier flowering by a SAG:ipt construct, under both well-watered and water-limited conditions (Fig. 9, A and B). In addition, the similarity in gene expression of OsGI (development related gene) and Hd1 (negative regulator of heading date) in control and transgenic plants indicates that neither gene is likely to be part of the regulatory pathway involving CKs. Advancing the heading date of cereals can prove to be beneficial under some environmental conditions (Haro and Allan, 1997). Under drought conditions, it has been found that a delay in heading date is negatively correlated with yield (Zou et al., 2007). Clearly, additional research is necessary to unravel the regulatory elements in the promoter, along with the pathways that led to an earlier flowering date with PSAG39:ipt, as opposed to the commonly observed delayed flowering response with other SAG:ipt constructs (Nguyen et al., 2008; Sýkorová et al., 2008, and refs. therein)
Interestingly, the transgenic lines did not outperform all null and wild-type genotypes at the end of the field experiment (Table I). While the transgenic lines ZT2-1 and ZT3-1 had significantly more panicles, aboveground shoot mass, and overall grain yield, the null line ZT2-2 performed equally as well. The performance of the third transgenic line, ZT1-1, was similar to the null line, ZT1-2, and the wild-type plants. Yet, the yield performance is misleading, since all the plants were harvested 135 d following germination. Carbon and nitrogen analysis, along with RNA-blot analysis from the ZT1-1 genotype, offers strong evidence that the lack of difference in agronomic performance is a result of a more rapidly maturing transgenic plant than either null or wild-type lines.
Changes in carbon metabolism were detected in ZT1-1 plants following anthesis in both the flag leaf and the third leaf (Supplemental Fig. S3). These changes included a delayed remobilization of sugars in the third leaf, caused by CK stimulation of extracellular invertase, vacuolar invertase, and a putative hexose transporter (Fig. 10). Such a coordinated response by sugar-related genes to CK has been previously demonstrated in other plant systems (Roitsch and González, 2004). The continued metabolic activity in the third leaf of the transgenic plants was in contrast to the flag leaf that rapidly exported sugars to the developing grain following anthesis. Wild-type plants maintained normal metabolic activity in the third and flag leaves, showing carbon export first in the lower leaf and subsequently in the flag leaf.
Finally, delayed chlorophyll degradation and mobilization of nitrogen likely provided the necessary energy for increased carbon flow throughout the plant under the earlier maturation of the transgenic plants (Figs. 8 and 11). While movement of nitrogen was postponed, with a significant difference visible 57 d after heading, by 72 d following anthesis, the nitrogen content was similar in transgenic and wild-type plants. This indicates that nitrogen was ultimately fully remobilized in the transgenic plants. Given these results, it is possible that the expression of ipt altered the photosynthetic apparatus of the transgenic rice, similar to the mechanism described by Rivero et al. (2009) in tobacco, providing a modified mechanism for photosynthesis. While photosynthesis was not prolonged in the transgenic rice, metabolism was apparently modified.
Delaying leaf senescence in order to increase the time for producing and transferring nutrients to the grain has been a source for a variety of investigations since it was determined that CKs can produce a stay-green phenotype (Ray et al., 1983; Gan and Amasino, 1995; Sýkorová et al., 2008). However, under current conditions of climate change, it is also important to understand how to adjust crop lifecycles in order to cope with variable growing season lengths. Current estimations for increasing temperatures, combined with the possibility of shortened growing seasons, could potentially severely damage rice yields, particularly in Southeast Asia (Wassmann et al., 2009). The transgenic PSAG39:ipt plants were found to have similar yields as wild-type plants under field conditions. The combination of an earlier flowering date, along with accelerated export of carbon from the flag leaf, while other leaves remained green, appears to have allowed these plants to mature more rapidly without a yield penalty. While continued work is necessary to understand the complex regulatory networks associated with senescence, these results provide a highly promising platform for mitigating the effects of a changing climate by reducing the growing season without compromising rice yields.
MATERIALS AND METHODS
Promoter Analysis
The protein with highest similarity to Arabidopsis (Arabidopsis thaliana) SAG12 was found in the rice (Oryza sativa) genome, obtained through the online software BLASTP (http://www.ncbi.nlm.nih.gov/). This protein was encoded by the SAG39 gene. SAG39 is contained in a bacterial artificial chromosome clone, OSJNBa0052O21, from the rice cv Nipponbare (O. sativa sp. japonica). The promoter region of this gene was predicted using the software TSSP, provided at the Softberry Web site (http://linux1.softberry.com/berry.phtml?topic%810%867=case_study_plants). The regulatory elements found in this promoter region were predicted using the online software PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html).
Isolation of the Whole and 5′-Truncated Promoters of SAG39 Gene and Construction of PSAG39:GUS and PSAG39:ipt
The promoter of the SAG39 gene had a size of 2.1 kb, and it was named PSAG39. The whole promoter (PSAG39) and seven truncated fragments from the 5′-end terminal region were obtained by PCR amplification of OSJNBa0052O21 using the primers shown in Supplemental Table S1. Their respective products were named PSAG39-62, PSAG39-239, PSAG39-493, PSAG39-719, PSAG39-1100, PSAG39-1300, and PSAG39-1600 in turn, according to their start positions at the promoter (Fig. 1). All forward primers contained an EcoRI site at the 5′ end, and the reverse primers contained a BglII site at the 5′ end. The PCR program included initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 60 s, 58°C for 60 s, and 72°C for 120 s, and then a final extension at 72°C for 7 min. The PCR products were digested with BglII and EcoRI and ligated to vector pCAMBIA1301 to drive the expression of reporter gene GUS instead of Cauliflower mosaic virus 35S promoter. The fused genes were designated PSAG39:GUS, PSAG39-62:GUS, PSAG39-239:GUS, PSAG39-493:GUS, PSAG39-719:GUS, PSAG39-1100:GUS, PSAG39-1300:GUS, and PSAG39-1600:GUS (Fig. 1).
Ipt was amplified from the plasmid psag516 (Lin et al., 2002a) using the specific primers p39IPT-F (5′-CGGAATTCAGATCTATGGATCTGCGTCTAATTTTCGG-3′) and p39IPT-R (5′-AGGTAACCCTAATACATTCCGAATGGATGAC-3′) containing a BglII site and a BstEII site, respectively, at the 5′ end (underlined). The PCR program included initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 60 s, 58°C for 60 s, and 72°C for 60 s, and then a final extension at 72°C for 7 min. The PCR products were digested with BglII and BstEII and were fused to the same digested PSAG39:GUS. Ipt gene expression was therefore under the control of the PSAG39 promoter (Supplemental Fig. S1).
Plant Transformation
The various constructs described above were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation. The promoter:GUS constructs and the PSAG39:ipt construct were then transferred into rice var Zhonghua 11 (O. sativa sp. japonica) by Agrobacterium-mediated transformation (Lin et al., 2002b). The positive transformants were selected by PCR using the hygromycin phosphotransferase gene-specific primers hpt-F (5′-ATTTGTGTACGCCCGACAGT-3′) and hpt-R (5′-GGATATGTCCTGCGGGTAAA-3′).
The copy number of PSAG39:ipt in transgenic plants was determined by DNA gel-blot analysis as described by Lin and Zhang (2005). The probe was amplified with primers hpt-F and hpt-R. Selection of homozygous ipt transgenic lines with single copy insertion and plot test was conducted as described previously by Lin et al. (2002b).
Hormone Treatment
Seeds of Zhonghua 11 were grown in a greenhouse with a 14-h-light/10-h-dark cycle for hormone treatment. The leaves of 1-month-old seedlings were sprayed with 100 μm ABA solutions. Samples were harvested after ABA treatment at 0 min, 5 min, 10 min, 20 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 24 h.
Expression Pattern of 5′-Truncated Promoters
The transient expression of GUS in rice calli was determined as follows: The calli generated from the seeds of Zhonghua 11 were cocultured for 16 h with the Agrobacterium strain EHA105, which carried the constructs of promoter:GUS fusion. The calli were treated in 1-mL solutions of ABA (100 μm), jasmonate (100 μm), ethylene (ACC, 100 μm), GA3 (100 μm), kinetin (1μm), 6-benzylaminopurine (1 μm), or water (control) for 1 h. Low temperature treatment (cold) was for 24 h. GUS activity in the calli was measured immediately after treatments and compared to the GUS activity of calli containing P35S treated with water, generating a relative expression value for each treatment. Each GUS activity was measured using three samples, with each sample containing five pieces of calli of approximately 0.5 g. Histochemical staining and quantitative analyses of GUS activity were conducted as described previously by Cai et al. (2007).
Identification of cis-Elements by Gel Mobility Shift Assay
Young or senescent leaves from transgenic plants were harvested at the seedling stage and nuclear extracts were isolated using the method reported by Qiu et al. (2007). The gel mobility shift assay was applied essentially following Cai et al. (2007). For that, eight long DNA probes and six truncated probes were synthesized commercially (Sangon Biotechnology, Shanghai, China) and their sequences are as shown in Supplemental Table S2.
Cosegregation Analysis of PSAG39:ipt Transgenic Plants
Aliquots (20 μg) of total RNA were used for RNA gel-blot analysis (Qiu et al., 2007). The probe for GUS was amplified with the primers gus-F and gus-R (see Supplemental Table S3 for primer sequences), while for SAG39 the primers used were sag39i-F and sag39i-R (see Supplemental Table S3 for primer sequences). First-strand cDNA was generated from 5 μg of total RNA using 100 units of M-MLV reverse transcriptase (Promega). For the amplification and the analysis of the ipt expression, iptF and iptR primers were used (see Supplemental Table S3 for primer sequences). Actin was used as internal control for the normalization of the ipt expression changes. The PCR program included initial denaturation at 94°C for 3 min, followed by 28 cycles of 94°C for 45 s, 55°C for 60 s, and 72°C for 60 s, and then a final extension at 72°C for 5 min.
For stay-green phenotype determination, chlorophyll concentration of flag leaves was measured nondestructively using a chlorophyll meter (SPAD-502; Minolta) after heading.
Sugar and Nitrogen Contents Determination
Flag leaves, second leaves, and third leaves were harvested and immediately grinded in a chilled mortar and pestle. The total leaf nitrogen content was determined by the Kjeldahl method (Makino et al., 1994) using a continuous flow analyzer (Futura 2000; Alliance Instruments). The sugar content was determined as described previously by Cowan et al. (2005).
Field Experimental Setup
Rice seedlings (wild-type Zhonghua 11, three homozygous transgenic lines, and the accompanying three null lines) were planted in the experimental farm of Huazhong Agricultural University, Wuhan, China, in May, 2006. Seedlings were arranged in a randomized complete block design, with three blocks. Plants were kept approximately 13 cm apart and in 5 to 10 cm of water throughout the growth period. The field was dried following seed formation. A total of 252 kg per acre of compound fertilizer (ammonium phosphate, potassium chloride, and urea) was applied upon transplanting seedlings into the field. An additional 108 kg per acre was applied at the onset of tillering, and a final 100 kg per acre was applied at booting. All plants were harvested at the same time, 135 d after germination, in October, 2006.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The native promoter constructs of PSAG39.
Supplemental Figure S2. The copy number of T0 transformants determined by Southern-blot analysis.
Supplemental Figure S3. Analysis of sugar content in wild-type and transgenic plants.
Supplemental Table S1. Primers for PSAG39 and truncated promoters.
Supplemental Table S2. The sequence of probes for electrophoretic mobility shift assay.
Supplemental Table S3. The sequence of primers for RT-PCR and probes for northern blotting.
Supplementary Material
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L. (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. (2003) A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132: 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S. (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31: 86–96 [DOI] [PubMed] [Google Scholar]
- Cai M, Wei J, Li X, Xu C, Wang S. (2007) A rice promoter containing both novel positive and negative cis-elements for regulation of green tissue-specific gene expression in transgenic plants. Plant Biotechnol J 5: 664–674 [DOI] [PubMed] [Google Scholar]
- Calderini O, Bovone T, Scotti C, Pupilli F, Piano E, Arcioni S. (2007) Delay of leaf senescence in Medicago sativa transformed with the ipt gene controlled by the senescence-specific promoter SAG12. Plant Cell Rep 26: 611–615 [DOI] [PubMed] [Google Scholar]
- Cercos M, Gomez-Cadenas A, Ho THD. (1999) Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J 19: 107–118 [DOI] [PubMed] [Google Scholar]
- Chang H, Jones ML, Banowetz GM, Clark DG. (2003) Overproduction of cytokinins in petunia flowers transformed with PSAG12-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol 132: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan AK, Freeman M, Bjorkman PO, Nicander B, Sitbon F, Tillberg E. (2005) Effects of senescence-induced alteration in cytokinin metabolism on source-sink relationships and ontogenic and stress-induced transitions in tobacco. Planta 221: 801–814 [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Ellerstrom M, Wycliffe P, Stalberg K, Rask L. (1999) Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol Biol 40: 699–709 [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM. (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV. (1992) Gibberellin-responsive elements in the promoter of a barley high-pl α-amylase gene. Plant Cell 4: 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro ES, Allan RE. (1997) Effects of heading date on agronomic performance of winter wheat isolines. Crop Sci 37: 346–351 [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Lu Q, Cai M, Xu C, Zhou DX, Li X, Zhang Q. (2007) Analysis of rice genes induced by striped stemborer (Chilo suppressalis) attack identified a promoter fragment highly specifically responsive to insect feeding. Plant Mol Biol 65: 519–530 [DOI] [PubMed] [Google Scholar]
- Huynh LN, VanToai T, Streeter J, Banowetz G. (2005) Regulation of flooding tolerance of SAG12: ipt Arabidopsis plants by cytokinin. J Exp Bot 56: 1397–1407 [DOI] [PubMed] [Google Scholar]
- Kim SY, Chung HJ, Thomas TL. (1997) Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J 11: 1237–1251 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G. (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ. (1992) Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev Biol 153: 386–395 [DOI] [PubMed] [Google Scholar]
- Lin YJ, Cao ML, Xu CG, Chen H, Wei J, Zhang QF. (2002a) Cultivating rice with delaying leaf- senescence by PSAG12-ipt gene transformation. Acta Bot Sin 44: 1333–1338 [Google Scholar]
- Lin YJ, Chen H, Cao YL, Wu CY, Wen J, Li YF, Hua HX. (2002b) Establishment of high-efficiency Agrobacterium-mediated genetic transformation system of Mudanjiang 8. Acta Agron Sin 28: 294–300 [Google Scholar]
- Lin YJ, Zhang Q. (2005) Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23: 540–547 [DOI] [PubMed] [Google Scholar]
- Lindsay WP, McAlister FM, Zhu Q, He XZ, Droge-Laser W, Hedrick S, Doerner P, Lamb C, Dixon RA. (2002) KAP-2, a protein that binds to the H-box in a bean chalcone synthase promoter, is a novel plant transcription factor with sequence identity to the large subunit of human Ku autoantigen. Plant Mol Biol 49: 503–514 [DOI] [PubMed] [Google Scholar]
- Loake GJ, Faktor O, Lamb CJ, Dixon RA. (1992) Combination of H-box [CCTACC (N) 7CT] and G-box (CACGTG) cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proc Natl Acad Sci USA 89: 9230–9234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Nakano H, Mae T. (1994) Responses of ribulose-1, 5-bisphosphate carboxylase, cytochrome f, and sucrose synthesis enzymes in rice leaves to leaf nitrogen and their relationships to photosynthesis. Plant Physiol 105: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura KI, Hijmans RJ, Chemin Y, Elvidge CD, Sugimoto K, Wu W, Lee YW, Shibasaki R. (2009) Mapping the global supply and demand structure of rice. Sustain Sci 4: 301–313 [Google Scholar]
- McKenzie MJ, Mett V, Reynolds PHS, Jameson PE. (1998) Controlled cytokinin production in transgenic tobacco using a copper-inducible promoter. Plant Physiol 116: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mew TW, Leung H, Savary S, Vera Cruz CM, Leach JE. (2004) Looking ahead in rice disease research and management. Crit Rev Plant Sci 23: 103–127 [Google Scholar]
- Mhiri C, Morel JB, Vernhettes S, Casacuberta JM, Lucas H, Grandbastien MA. (1997) The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol Biol 33: 257–266 [DOI] [PubMed] [Google Scholar]
- Nguyen KH, Jordi W, van Dun K, Schepers F, Davelaar E, Stoopen G, Dix PJ, Kane EJ. (2008) Delayed senescence in cauliflower transformed with an autoregulated isopentenyl transferase gene. Int J Plant Sci 169: 339–347 [Google Scholar]
- Prestridge DS. (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Ray S, Mondal WA, Choudhuri MA. (1983) Regulation of leaf senescence, grain-filling and yield of rice by kinetin and abscisic acid. Physiol Plant 59: 343–346 [Google Scholar]
- Rivero RM, Shulaev V, Blumwald E. (2009) Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol 150: 1530–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, González MC. (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9: 606–613 [DOI] [PubMed] [Google Scholar]
- Saha P, Chakraborti D, Sarkar A, Dutta I, Basu D, Das S. (2007) Characterization of vascular-specific RSs1 and rolC promoters for their utilization in engineering plants to develop resistance against hemipteran insect pests. Planta 226: 429–442 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T. (2005) Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 102: 9972–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV. (2003) PlantProm: a database of plant promoter sequences. Nucleic Acids Res 31: 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigocki AC. (1991) Cytokinin content and tissue distribution in plants transformed by a reconstructed isopentenyl transferase gene. Plant Mol Biol 16: 105–115 [DOI] [PubMed] [Google Scholar]
- Sýkorová B, Kurešová G, Daskalova S, Trčková M, Hoyerová K, Raimanová I, Motyka V, Trávníčková A, Elliott MC, Kamínek M. (2008) Senescence-induced ectopic expression of the A. tumefaciens ipt gene in wheat delays leaf senescence, increases cytokinins content, nitrate influx, and nitrate reductase activity, but does not affect grain yield. J Exp Bot 59: 377–387 [DOI] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loven K, Beinsberger SEI, Valcke RLM, Van Onckelen HA, Clijsters HMM. (1993) Morphometric analysis of the growth of P hsp70-ipt transgenic tobacco plants. J Exp Bot 44: 1671–1678 [Google Scholar]
- Wassmann R, Jagadish SVK, Sumfleth K, Pathak H, Howell G, Ismail A, Serraj R, Redona E, Singh RK, Heuer S. (2009) Regional vulnerability of climate change impacts on Asian rice production and scope for adaptation. Adv Agron 102: 91–133 [Google Scholar]
- Werner T, Kollmer I, Bartrina I, Holst K, Schmulling T. (2006) New insights into the biology of cytokinin degradation. Plant Biol 8: 371–381 [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Obokata J. (2008) ppdb: a plant promoter database. Nucleic Acids Res 36: D977–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Xie Z, Zou X, Casaretto J, David Ho T, Shen QJ. (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134: 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GH, Liu HY, Mei HW, Liu GL, Yu XQ, Li MS, Wu JH, Chen L, Luo LJ. (2007) Screening for drought resistance of rice recombinant inbred populations in the field. J Integr Plant Biol 49: 1508–1516 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.