Abstract
Soybean (Glycine max) RPG1-B (for resistance to Pseudomonas syringae pv glycinea) mediates species-specific resistance to P. syringae expressing the avirulence protein AvrB, similar to the nonorthologous RPM1 in Arabidopsis (Arabidopsis thaliana). RPM1-derived signaling is presumably induced upon AvrB-derived modification of the RPM1-interacting protein, RIN4 (for RPM1-interacting 4). We show that, similar to RPM1, RPG1-B does not directly interact with AvrB but associates with RIN4-like proteins from soybean. Unlike Arabidopsis, soybean contains at least four RIN4-like proteins (GmRIN4a to GmRIN4d). GmRIN4b, but not GmRIN4a, complements the Arabidopsis rin4 mutation. Both GmRIN4a and GmRIN4b bind AvrB, but only GmRIN4b binds RPG1-B. Silencing either GmRIN4a or GmRIN4b abrogates RPG1-B-derived resistance to P. syringae expressing AvrB. Binding studies show that GmRIN4b interacts with GmRIN4a as well as with two other AvrB/RPG1-B-interacting isoforms, GmRIN4c and GmRIN4d. The lack of functional redundancy among GmRIN4a and GmRIN4b and their abilities to interact with each other suggest that the two proteins might function as a heteromeric complex in mediating RPG1-B-derived resistance. Silencing GmRIN4a or GmRIN4b in rpg1-b plants enhances basal resistance to virulent strains of P. syringae and the oomycete Phytophthora sojae. Interestingly, GmRIN4a- or GmRIN4b-silenced rpg1-b plants respond differently to AvrB-expressing bacteria. Although both GmRIN4a and GmRIN4b function to monitor AvrB in the presence of RPG1-B, GmRIN4a, but not GmRIN4b, negatively regulates AvrB virulence activity in the absence of RPG1-B.
One of the myriad plant defense responses activated upon pathogen invasion is signaling induced via the activation of resistance (R) proteins. R gene-mediated resistance is generally activated in response to race-specific pathogen effectors, termed avirulence proteins (Avr), and often results in the development of a hypersensitive reaction at the site of pathogen entry (Dangl et al., 1996). The hypersensitive reaction is a form of programmed cell death that results in the formation of necrotic lesions around the site of pathogen entry and is thought to help prevent pathogen spread by confining it to the dead cells.
A majority of the known R proteins contain conserved structural domains, including N-terminal coiled coil (CC) or Toll-interleukin 1 receptor (TIR)-like domains, central nucleotide-binding site (NBS), and C-terminal Leu-rich repeat (LRR) domains (Martin et al., 2003). While some R proteins “perceive” pathogen presence via direct physical interactions with the cognate Avr proteins (Scofield et al., 1996; Jia et al., 2000; Leister and Katagiri, 2000; Deslandes et al., 2003), several others likely do so indirectly. This led to the suggestion that R proteins monitor the presence of Avr proteins by “guarding” other host proteins targeted by the pathogen effector (Van der Biezen and Jones, 1998; Innes, 2004; Jones and Dangl, 2006). Avr proteins enhance pathogen virulence in genetic backgrounds lacking cognate R proteins by targeting components of the host basal defense machinery, including “guardee” proteins (Chang et al., 2000; Guttman and Greenberg, 2001; Chen et al., 2004, Kim et al., 2005b; Ong and Innes, 2006; van Esse et al., 2007; Shan et al., 2008; Xiang et al., 2008). However, some Avr proteins were found to also target host proteins that do not contribute to the virulence function of the effector (Shang et al., 2006; Shabab et al., 2008; Zhou and Chai, 2008; Zipfel and Rathjen, 2008). This led to the proposition that plants express “decoy” proteins that mimic Avr-guardee recognition in the presence of the R protein. This decoy model suggests that, unlike guardees, decoy proteins do not directly contribute to host basal immunity, such that Avr-derived alterations of decoys do not enhance pathogen virulence in plants lacking the R protein (van der Hoorn and Kamoun, 2008).
A well-studied example of an indirect mode of effector recognition is that of the Arabidopsis (Arabidopsis thaliana) R protein, RPM1 (for resistance to Pseudomonas syringae pv maculicola 1). RPM1 mediates resistance against bacteria expressing two different Avr proteins, AvrRpm1 (AvrRpm1PmaM6) and AvrB (AvrB1Pgyrace4). Although RPM1 does not directly interact with either AvrRpm1 or AvrB, it does associate with RIN4 (for RPM1-interacting 4), which interacts with AvrRpm1 and AvrB. RIN4 is required for RPM1-induced resistance to AvrRpm1/AvrB-expressing P. syringae (Mackey et al., 2002). Both AvrRpm1 and AvrB induce the phosphorylation of RIN4, which is thought to induce RPM1-mediated resistance signaling. RIN4 also associates with a second Arabidopsis R protein, RPS2 (for resistance to P. syringae), which mediates resistance against P. syringae expressing AvrRpt2. RPS2-mediated signaling is activated when AvrRpt2 (AvrRpt2PtoJL1065), a Cys protease, cleaves RIN4 (Axtell and Staskawicz, 2003; Mackey et al., 2003; Kim et al., 2005a). The AvrRpt2-triggered loss of RIN4 compromises RPM1-mediated resistance, because RIN4 is not available for phosphorylation (Ritter and Dangl, 1996; Axtell and Staskawicz, 2003; Mackey et al., 2003).
The avirulence effector AvrB was first isolated from a P. syringae strain colonizing soybean (Glycine max) and used to identify the cognate resistance locus RPG1 in soybean (Staskawicz et al., 1987; Keen and Buzzell, 1991). This locus contains the RPG1-B (for resistance to P. syringae pv glycinea) gene, which encodes a CC-NBS-LRR protein conferring resistance to AvrB-expressing P. syringae in soybean (Bisgrove et al., 1994; Ashfield et al., 2004). Unlike RPM1, RPG1-B does not confer specificity to AvrRpm1 (Ashfield et al., 1995). However, as in Arabidopsis, the soybean RPG1-B-derived hypersensitive reaction to AvrB-expressing bacteria is inhibited by the presence of AvrRpt2-expressing bacteria (Axtell and Staskawicz, 2003, Mackey et al., 2003; Ashfield et al., 2004). This suggests that RPG1-B and RPM1 might utilize common signaling components even though they share very limited sequence identity. Therefore, we investigated the possible involvement of RIN4-like proteins in RPG1-B-mediated resistance signaling. In addition to Arabidopsis, RIN4-like proteins have also been identified in tomato (Solanum lycopersicum) and lettuce (Lactuca sativa; Jeuken et al., 2009; Luo et al., 2009). In tomato, the NBS-LRR protein, Prf (for Pseudomonas resistance and fenthion sensitivity), and its interacting protein kinase, Pto, mediate resistance to the AvrPto (AvrPto1PtoJL1065)-expressing strain of P. syringae (Scofield et al., 1996; Tang et al., 1996; Kim et al., 2002; Mucyn et al., 2006). AvrPto binds RIN4 proteins from both Arabidopsis (AtRIN4) and tomato (SlRIN4). Similar to AvrRpt2, AvrPto induces the proteolysis of RIN4, albeit only in the presence of Pto and Prf (Luo et al., 2009). However, in the case of AvrPto, degradation of RIN4 is the result of induced proteolytic activity in the plant, rather than that of AvrPto itself. In Lactuca (lettuce) species, the L. saligna RIN4 allele was recently shown to be essential for resistance to an avirulent strain of the downy mildew pathogen, Bremia lactucae (Jeuken et al., 2009).
Here, we report that two functionally nonredundant isoforms of soybean RIN4 (GmRIN4) function in RPG1-B-derived resistance as well as in the virulence activity of AvrB in the absence of RPG1-B.
RESULTS
RIN4 Orthologous Sequences in Soybean
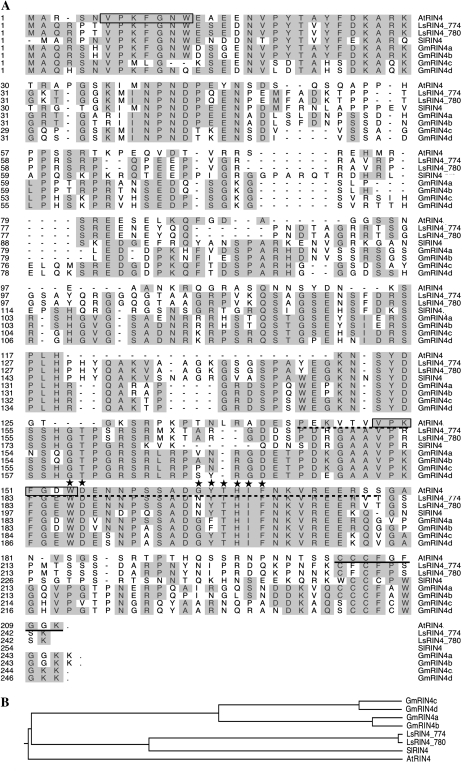
The soybean genome contains at least four genes that encode proteins with high similarities to Arabidopsis RIN4 (AtRIN4, At3g25070). These were designated GmRIN4a (Glyma03g19920), GmRIN4b (Glyma16g12160), GmRIN4c (Glyma18g36000), and GmRIN4d (Glyma08g46400). Amino acid alignments (Fig. 1A) showed that GmRIN4a and GmRIN4b were approximately 94% identical to each other, approximately 50% identical to AtRIN4, approximately 48% identical to two RIN4 isoforms from lettuce (LsRIN4_774/780-GQ497774/GQ497780; Jeuken et al., 2009), and approximately 45% identical to a tomato RIN4 (SlRIN4-TC174419; Luo et al., 2009). GmRIN4c and GmRIN4d were approximately 91% identical to each other, approximately 46%/48% identical to AtRIN4, respectively, approximately 49% identical to the two LsRIN4 proteins, and approximately 47% identical to SlRIN4. All four GmRIN4 proteins contained several conserved domains present in AtRIN4, including those involved in binding to AvrB, an amino acid sequence required for AvrRpt2-mediated cleavage, and a putative palmitoylation site for plasma membrane localization (Chisholm et al., 2005; Kim et al., 2005a; Takemoto and Jones, 2005, Desveaux et al., 2007; Fig. 1A). Phylogenetic analysis showed that all four GmRIN4 proteins clustered together and were more closely related to the Ls/SlRIN4 proteins than to AtRIN4. AtRIN4 was placed in a separate clade from all the other proteins analyzed (Fig. 1B).
Figure 1.
Sequence conservation and phylogenetic analysis of Arabidopsis (At; At3g25070), lettuce (Ls; GQ497774 and GQ497780), soybean (Gm), and tomato (Sl; TC174419) RIN4 proteins. A, Sequence conservation among the various RIN4 proteins. Numbers on the left denote amino acid positions. Identical residues are shaded in gray. The two AvrRPT2 cleavage sites (RCS1 and RCS2) are boxed. The AvrB-binding region is underlined by a dotted line. Residues that contact and/or form hydrogen bonds with AvrB are indicated by asterisks. A solid line underlines the palmitoylation site essential for plasma membrane localization. Sequence alignment was carried out using ClustalW in the Megalign program of the DNASTAR package. B, Phylogenetic analysis of the various RIN4 proteins. The tree was constructed using the program PAUP*, version 4b10.
GmRIN4 Proteins Interact with AvrB
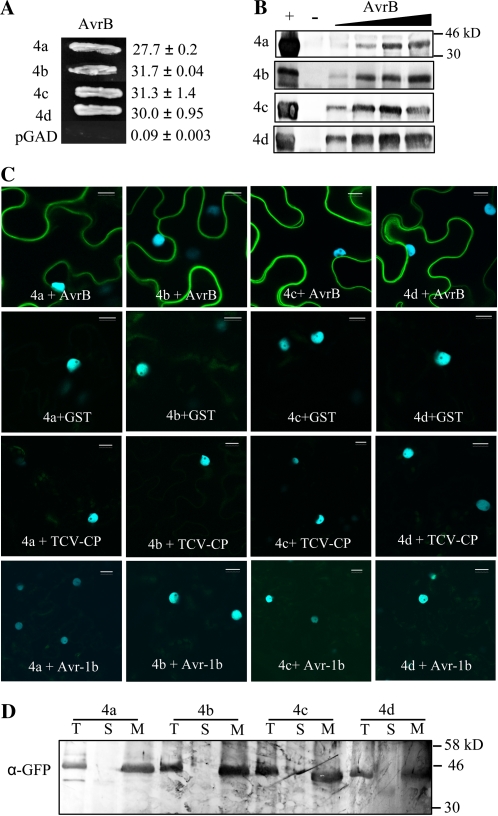
Since AtRIN4 associates with AvrB, we first tested binding between AvrB and the four GmRIN4 proteins using the yeast two-hybrid (Y2H) assay. AvrB was expressed as a GAL4 binding domain (BD) fusion protein, while GmRIN4a, GmRIN4b, GmRIN4c, and GmRIN4d were expressed as GAL4 activating domain (AD) fusion proteins. Yeast cells coexpressing AvrB-BD and GmRIN4a/b/c/d-AD were tested for His autotrophy or β-galactosidase activity (selectable markers for positive interactions). Y2H assay detected interactions between AvrB and all four GmRIN4 proteins but not the empty AD vector (Fig. 2A). GmRIN4a/b/c/d-AD also did not interact with the empty BD vector (data not shown). In vitro “pull-down” affinity assays confirmed these interactions. AvrB and GmRIN4a were expressed as His-tagged, GmRIN4b as Myc-His-tagged, and GmRIN4c and GmRIN4d as glutathione S-transferase (GST)-tagged proteins in bacteria. Affinity-purified GmRIN4a and GmRIN4b were immobilized on nickel-nitrilotriacetic acid agarose (Ni-NTA), while GmRIN4c and GmRIN4d were immobilized on glutathione Sepharose. The His tag on AvrB was cleaved, and increasing amounts (0.05–0.5 μg) of this AvrB were applied to the GmRIN4a, GmRIN4b, GmRIN4c, or GmRIN4d affinity columns. After extensive washing to remove nonspecifically bound protein, AvrB retained on the various affinity columns was visualized by immunoblot analysis using AvrB-specific antibodies. AvrB (0.5 μg) lacking the His tag was not retained on empty Ni-NTA lacking bait proteins or Ni-NTA preincubated with protein extracts from cells expressing the empty pET28A/pET30A vectors (Fig. 2B, – lanes; Supplemental Fig. S1A). AvrB was also not retained on empty glutathione Sepharose lacking bait proteins or glutathione Sepharose preincubated with protein extracts from cells expressing the empty pGEX-5X vector (Fig. 2B, – lanes; Supplemental Fig. S1A). In contrast, saturable binding was detected between AvrB and all four GmRIN4 proteins (Fig. 2B).
Figure 2.
GmRIN4 (GmRIN4a to GmRIN4d = 4a to 4d) proteins interact with AvrB. A, Y2H assay showing interactions between AvrB and 4a, 4b, 4c, or 4d. Growth on His−Trp−Leu− plates is shown. Numbers indicate β-galactosidase activity with sd (n = 3). B, Pull-down assays showing in vitro protein binding. Immobilized 4a/4b (Ni-NTA) and 4c/4d (glutathione) were incubated with increasing amounts (0.05–0.5 μg) of AvrB (His tag cleaved), and bound proteins were visualized using anti-AvrB antibodies. Purified AvrB was loaded as a positive control (+) or incubated with empty Ni-NTA/glutathione agarose as a negative control (−) on respective gels. C, BiFC assay showing in planta interactions. CFP and YFP overlay images (40× magnification) of micrographs at 48 h post infiltration from leaves coexpressing the indicated proteins are shown. Images are representative of three separate infiltrations from two independent experiments for each interaction. Bars = 10 μm. D, Western-blot analysis showing expression and localization of 4a/4b/4c/4d. Ten micrograms of protein each of the total (T), soluble (S), and membrane (M) fractions from CFP-H2B-expressing transgenic N. benthamiana leaves infiltrated with nEYFP-fused GmRIN4 proteins were subjected to western-blot analysis using anti-GFP antibodies.
In planta binding between AvrB and GmRIN4 proteins was tested using the bimolecular fluorescence complementation (BiFC) assay (Fig. 2C). Proteins were expressed as fusions with the N- and C-terminal halves of enhanced yellow fluorescent protein (nEYFP and cEYFP) using the Gateway-compatible pSITE vectors (Martin et al., 2009). Agrobacterium tumefaciens cells expressing various combinations of the proteins fused to reciprocal halves of EYFP were coinfiltrated into leaves of transgenic Nicotiana benthamiana expressing the nuclear marker CFP-H2B (for cyan fluorescent protein fused to the histone 2B protein). Positive interactions between coinfiltrated proteins resulted in reconstitution of EYFP and thereby fluorescence detectable by confocal microscopy. A reduced inoculum of AvrB-expressing cells was used for infiltration to minimize autofluorescence from AvrB-induced cell death (Schechter et al., 2004; Supplemental Fig. S1B). BiFC assays confirmed interactions between AvrB and GmRIN4a, GmRIN4b, GmRIN4c, or GmRIN4d. AvrB did not interact with the membrane-localized TCV-CP [for coat protein (avirulence factor) of Turnip crinkle virus; Zhao et al., 2000; R.-D. Jeong and P. Kachroo, personal communication], even though both proteins were appropriately expressed in planta (Supplemental Fig. S1, C and D). No fluorescence was observed when the n/cEYFP-fused proteins were expressed individually or coexpressed with non/GST-fused halves of YFP (Fig. 2C). Furthermore, no fluorescence was observed when GmRIN4 proteins were coexpressed with two other membrane-localized proteins, TCV-CP and SYNV-G (for G protein of Sonchus yellow net virus; Martin et al., 2009; data not shown). The GmRIN4 proteins also did not interact with the Phytophthora sojae avirulence effector Avr1b, which is a secreted protein that can reenter cells due to an RXLR motif, thereby exhibiting cytoplasmic and nuclear localization in plant cells (Dou et al., 2008; Fig. 2C). Leaves coexpressing GmRIN4a, GmRIN4b, GmRIN4c, or GmRIN4d with GST, TCV-CP, or Avr1b contained detectable levels of the respective proteins, indicating that lack of fluorescence in BIFC assays was not due to insufficient protein expression (Supplemental Fig. S2, A–C). Consistent with the plasma membrane localization of AtRIN4, EYFP fluorescence in all BiFC assays expressing GmRIN4 proteins was confined to the periphery of leaf cells (Fig. 2C). Membrane localization of GmRIN4 proteins was further confirmed by immunoblot analysis. GmRIN4 proteins were only detected in the membranous fractions of N. benthamiana leaves expressing the corresponding nEYFP-fused proteins (Fig. 2D). Together, these data indicated that the membrane-localized GmRIN4a, GmRIN4b, GmRIN4c, and GmRIN4d specifically interact with AvrB.
GmRIN4 Proteins Interact with RPG1-B
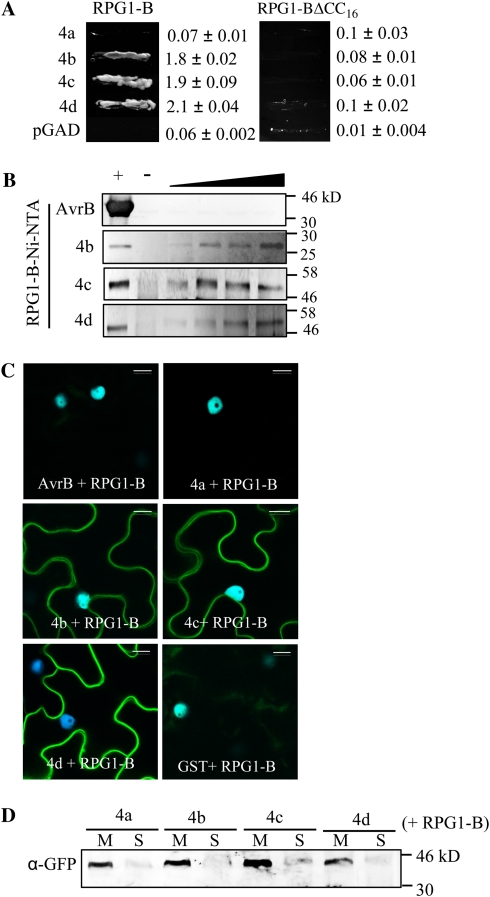
We next tested interactions between GmRIN4a, GmRIN4b, GmRIN4c, or GmRIN4d and RPG1-B. RPG1-B (CC domain, N-terminal 187 amino acids) was expressed as a GAL4 BD fusion protein, and its interaction with the GmRIN4a, GmRIN4b, GmRIN4c, or GmRIN4d AD protein was assayed in Y2H assays. Interestingly, RPG1-B interacted with GmRIN4b, GmRIN4c, and GmRIN4d but not GmRIN4a or AvrB (Fig. 3A; data not shown). Similar to RPM1, the CC domain of RPG1-B was sufficient for its interactions with GmRIN4b, GmRIN4c, or GmRIN4d. However, truncating 16 amino acid from the C terminus of the RPG1-B CC domain abolished the RPG1-B-GmRIN4b/c/d interactions (Fig. 3A). Likewise, in vitro pull-down assays detected binding between the CC domain of RPG1-B and GmRIN4b, GmRIN4c, or GmRIN4d but not AvrB. RPG1-B (CC domain) was expressed as a His-tagged protein in bacteria, immobilized on Ni-NTA, and incubated with increasing amounts of AvrB (0.05–0.5 μg) or GmRIN4b, GmRIN4c, or GmRIN4d (0.1–1μg). The His tag present on AvrB and GmRIN4b was removed prior to incubation with the RPG1-B affinity column. Bound proteins were visualized by immunoblot analysis using AvrB-specific, Myc (GmRIN4b)-specific, or GST (GmRIN4c or GmRIN4d)-specific antibodies. Saturable binding was detected between RPG1-B and GmRIN4b, GmRIN4c, or GmRIN4d (Fig. 3B). AvrB (0.5 μg) and GmRIN4b (1 μg) lacking the His tag or GST-tagged GmRIN4c (1 μg) and GmRIN4d (1 μg) were not retained on empty Ni-NTA or Ni-NTA preincubated with protein extracts from cells expressing the empty pET30A vector (used for expressing RPG1-B) when applied at maximum concentrations used in the assay (Fig. 3B, – lanes; Supplemental Fig. S3A).
Figure 3.
GmRIN4 (GmRIN4a to GmRIN4d = 4a to 4d) proteins interact with RPG1-B. A, Y2H assay between 4a, 4b, 4c, or 4d and RPG1-B (N-terminal 187 amino acids) or RPG1-B lacking 16 amino acids of the CC domain (N-terminal 171 amino acids; RPG1-BΔCC16). Growth on His−Trp−Leu− plates is shown. Numbers indicate β-galactosidase activity with sd (n = 3). B, Pull-down assays showing in vitro protein binding. RPG1-B was immobilized on Ni-NTA incubated with increasing amounts of AvrB (0.05–0.5 μg) or 4b, 4c, or 4d (0.1–1 μg), and bound proteins were visualized using anti-AvrB, anti-Myc (4b), or anti-GST (4c/4d) antibodies. Purified AvrB, 4b, 4c, or 4d was loaded as a positive control (+) or incubated with empty Ni-NTA as a negative control (−) on respective gels. C, BiFC assay showing in planta interactions. CFP and YFP overlay images (40× magnification) of micrographs at 48 h post infiltration from leaves coexpressing the indicated proteins are shown. Images are representative of three separate infiltrations from two independent experiments for each interaction. Bars = 10 μm. D, Western-blot analysis showing expression and localization of 4a/4b/4c/4d in 10 μg each of membrane (M) or soluble (S) fractions of N. benthamiana leaves coexpressing the four nEYFP-fused GmRIN4 isoforms and cEYFP-fused RPG1-B. Proteins were detected using anti-GFP antibodies that cross-react with nEYFP alone.
These interactions were further confirmed by in planta BiFC assays. Consistent with the Y2H and pull-down assays, florescence was detected only in N. benthamiana leaves coexpressing RPG1-B with GmRIN4b, GmRIN4c, or GmRIN4d but not AvrB, GST, or GmRIN4a (Fig. 3C). Western-blot analysis of total protein extracts from AvrB/GST/GmRIN4a and RPG1-B coexpressing leaves showed that RPG1-B was expressed at similar levels as in the GmRIN4b coexpressing leaves, indicating that lack of florescence was not due to inadequate protein expression (Supplemental Fig. S3B). These leaves also expressed detectable amounts of AvrB, GST, or GmRIN4a. Furthermore, GmRIN4a was detected at comparable levels as GmRIN4b, GmRIN4c, or GmRIN4d in the membrane fractions of N. benthamiana leaves coexpressing the respective proteins with RPG1-B (Fig. 3D). This indicated that the lack of interaction between GmRIN4a and RPG1-B was not due to the improper localization or insufficient expression of this protein in N. benthamiana. These data showed that three (GmRIN4b, GmRIN4c, and GmRIN4d) of the four GmRIN4 proteins directly interact with RPG1-B.
GmRIN4b Complements the Arabidopsis rin4 Mutation
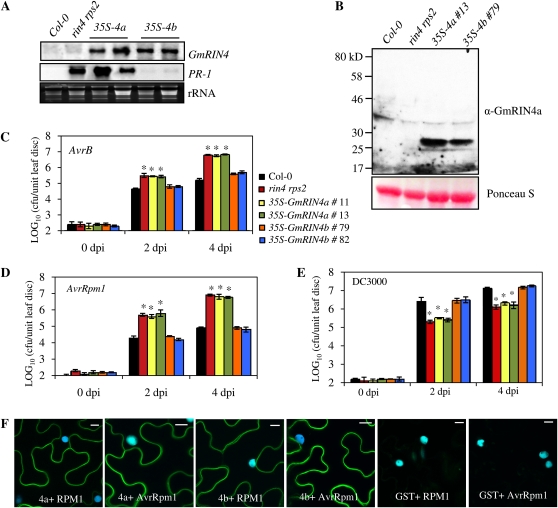
To determine if their differing affinities for RPG1-B reflected functional differences in the GmRIN4a and GmRIN4b proteins, we overexpressed the GmRIN4a and GmRIN4b isoforms in the Arabidopsis rin4 rps2 mutant. T2 plants containing GmRIN4a (35S-GmRIN4a) or GmRIN4b (35S-GmRIN4b) expressed the respective transgenes constitutively (Fig. 4A). Western-blot analysis of total protein extracts from the 35S-GmRIN4a and 35S-GmRIN4b lines showed that these plants also expressed the respective GmRIN4 proteins constitutively (Fig. 4B). The rin4 rps2 mutant exhibits constitutive PR-1 accumulation due to residual RPM1 activity (Belkhadir et al., 2004). Therefore, we assessed PR-1 expression in the transgenic plants as a measure of complementation of the rin4 mutation. Interestingly, increased PR-1 expression associated with the rin4 mutation was suppressed in 35S-GmRIN4b rin4 rps2 but not 35S-GmRIN4a rin4 rps2 transgenics (Fig. 4A). To assess RPM1-mediated defenses, we inoculated wild-type, mutant, and transgenic plants with AvrB- or AvrRpm1-expressing strains of P. syringae. The rin4 rps2 mutant and 35S-GmRIN4a rin4 rps2 transgenic plants accumulated approximately 40-fold more AvrB bacteria (P < 0.001) and approximately 100-fold more AvrRpm1 bacteria (P < 0.001) than the wild-type plants. In contrast, 35S-GmRIN4b rin4 rps2 plants contained wild-type-like levels of AvrB and AvrRpm1 bacteria (Fig. 4, C and D). We tested several (five to seven each) transgenic lines expressing the GmRIN4a or GmRIN4b transgene at various levels (high or low). For all lines tested, 35S-GmRIN4a lines always continued to show phenotypes similar to the rin4 rps2 mutant, while the 35S-GmRIN4b lines behaved like wild-type plants (data not shown). This suggested that complementation of the Arabidopsis rin4 mutant was not associated with levels of the GmRIN4b transcript.
Figure 4.
GmRIN4b rescues the rin4 mutation in Arabidopsis. A, Northern-blot analysis showing expression of GmRIN4a or GmRIN4b and PR-1 in wild-type (Col-0), rin4 rps2, 35S-GmRIN4a (35S-4a), and 35S-GmRIN4b (35S-4b) plants. Ethidium bromide staining of rRNA was used as a loading control. B, Western-blot analysis of protein extracts from the indicated genotypes. GmRIN4a and GmRIN4b proteins in the respective transgenic lines were visualized using GmRIN4a-specific antibodies. Ponceau-S staining of the membrane was used as a loading control. C to E, Response to AvrB (C), AvrRpm1 (D), or DC3000 (E) strains of P. syringae in the wild type (Col-0; black bars), a mutant (rin4 rps2; red bars), and two transgenic lines each expressing GmRIN4a (green and yellow bars) or GmRIN4b (orange and blue bars). Bacterial numbers at 0, 2, or 4 d post inoculation (dpi) are presented as log10 values of colony-forming units (cfu) per unit of leaf area. Error bars indicate sd (n = 5). Statistical significance was determined using Student's t test. Asterisks denote data significantly different from the wild type (Col-0), where P < 0.001 (n = 5). F, BiFC assay showing in planta interactions. CFP and YFP overlay images (40× magnification) of micrographs at 48 h post infiltration from leaves coexpressing the indicated proteins are shown. Images are representative of three separate infiltrations from two independent experiments for each interaction. Bars = 10 μm.
The rin4 rps2 mutant also exhibits enhanced resistance to virulent bacteria (Belkhadir et al., 2004; Kim et al., 2005b). Therefore, we next challenged the 35S-GmRIN4b rin4 rps2 and 35S-GmRIN4a rin4 rps2 transgenic plants with a virulent strain (DC3000) of P. syringae. Consistent with their response to avirulent P. syringae, the 35S-GmRIN4a rin4 rps2 plants behaved similar to rin4 rps2 mutant plants and exhibited enhanced resistance to virulent P. syringae. Both rin4 rps2 and 35S-GmRIN4a rin4 rps2 plants accumulated approximately 30-fold fewer bacteria than wild-type plants (P < 0.001). In comparison, the 35S-GmRIN4b rin4 rps2 transgenic plants showed a wild-type-like response (Fig. 4E). Together, these results suggest that GmRIN4b, but not GmRIN4a, was able to compensate for the loss of AtRIN4.
We next tested if the ability of GmRIN4b or the inability of GmRIN4a to complement the Arabidopsis rin4 mutation was associated with their respective abilities to bind the corresponding R protein, RPM1. In planta binding between GmRIN4a or GmRIN4b and full-length RPM1 as well as AvrRpm1 was tested using EYFP-fused proteins in BiFC assays. Fluorescence was detected when RPM1 was coexpressed with either GmRIN4a or GmRIN4b (Fig. 4E). This indicated that both GmRIN4a and GmRIN4b interacted with RPM1 in planta. Similarly, GmRIN4a and GmRIN4b also interacted with AvrRpm1 in planta (Fig. 4F). These data suggest that the inability of GmRIN4a to complement AtRIN4 function is not associated with its ability to interact with RPM1 or AvrRPM1.
GmRIN4a and GmRIN4b Are Required for RPG1-B-Mediated Resistance in Soybean
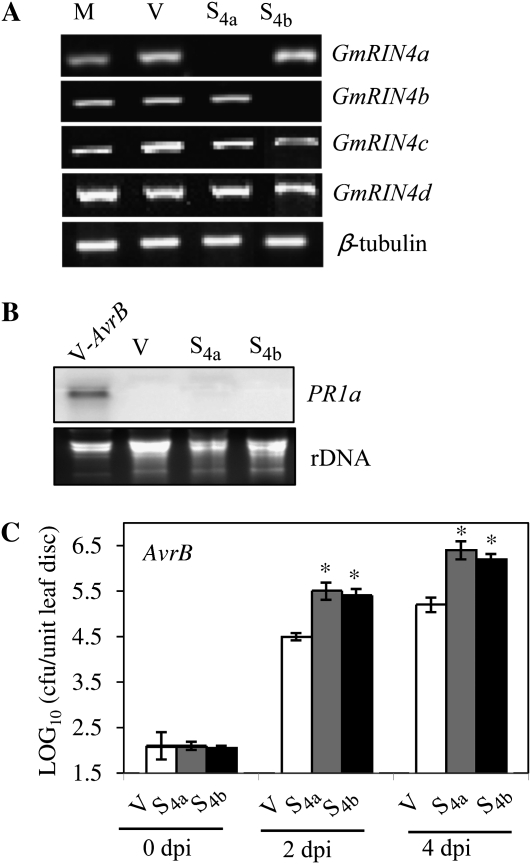
We next tested the functional requirements for GmRIN4a and GmRIN4b in RPG1-B-derived resistance signaling in soybean. Silencing GmRIN4a or GmRIN4b in plants (cv Harosoy) containing RPG1-B was achieved using the bean pod mottle virus (BPMV)-based vector (Zhang and Ghabrial, 2006; Kachroo et al., 2008; Fu et al., 2009). Plants were inoculated with recombinant vectors targeting GmRIN4a (S4a) or GmRIN4b (S4b). Similar expression of all four GmRIN4 isoforms observed in vector (V)- or mock (M)-inoculated plants suggested that BPMV infection did not alter GmRIN4 expression (Fig. 5A). Plants inoculated with S4a or S4b were silenced for the GmRIN4a or GmRIN4b isoform, respectively, but continued to express other GmRIN4 isoforms at basal levels (Fig. 5A). Unlike the Arabidopsis rin4 mutant, the S4a or S4b plants did not exhibit ectopic PR1a expression (Fig. 5B). Inoculation with an avirulent (AvrB) strain of P. syringae pv glycinea race 4 showed that S4a and S4b plants containing RPG1-B were significantly more susceptible than V plants, supporting 15- and 10-fold more growth (P < 0.001) of AvrB bacteria, respectively (Fig. 5C). Thus, RPG1-B-mediated resistance to AvrB-expressing P. syringae requires both GmRIN4a and GmRIN4b, even though RPG1-B does not interact with GmRIN4a.
Figure 5.
GmRIN4a and GmRIN4b are required for RPG1-B-mediated resistance signaling. A, RT-PCR analysis showing expression of the GmRIN4 isoforms in mock/vector (M/V)-inoculated and GmRIN4a (S4a)- or GmRIN4b (S4b)-silenced plants. β-Tubulin levels were used as an internal control for cDNA amounts. B, Northern-blot analysis showing expression of PR1a in V, S4a, or S4b plants or V plants inoculated with an avirulent (AvrB) strain of P. syringae pv glycinea (V-AvrB; 24 h). Ethidium bromide staining of rRNA was used as a loading control. C, Bacterial counts in RPG1-B (cv Harosoy) S4a (gray bars) or S4b (black bars) plants as compared with V (white bars) plants. Plants were infiltrated with AvrB bacteria. Log10 values of colony-forming units (cfu) per unit of leaf area from infected leaves at 0, 2, or 4 d post inoculation (dpi) are presented. Error bars indicate sd (n = 5). Statistical significance was determined using Student's t test. Asterisks denote data significantly different from control (V), where P < 0.001.
GmRIN4b Interacts with Itself as Well as with GmRIN4a, GmRIN4c, and GmRIN4d
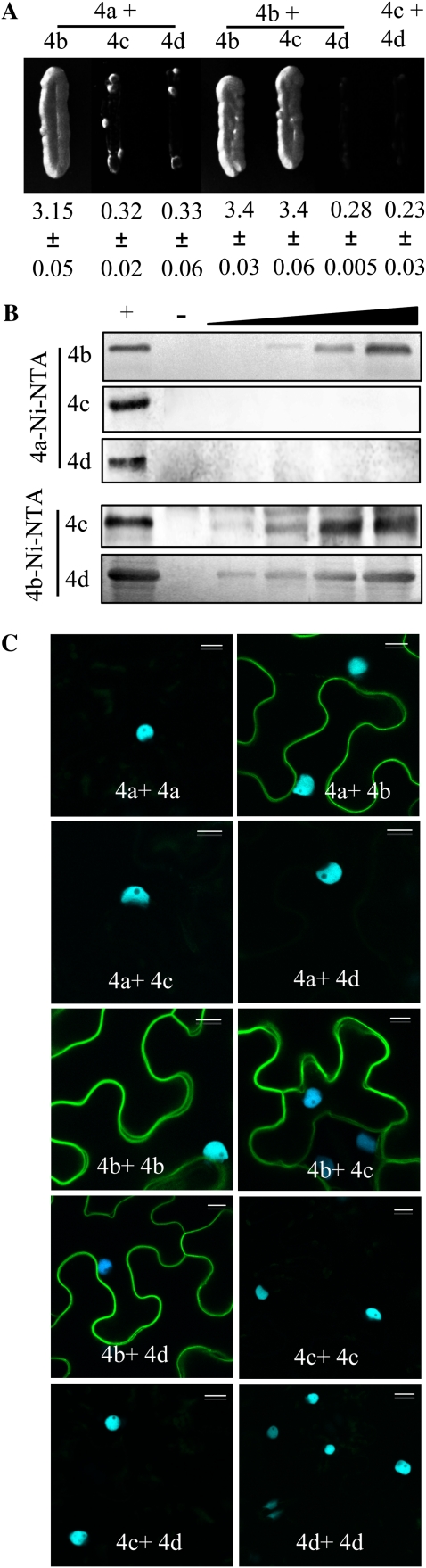
The fact that GmRIN4a was required for RPG1-B-mediated resistance to AvrB-expressing bacteria even though it did not interact with RPG1-B raised the possibility that the GmRIN4 proteins might function as a heteromeric complex. Indeed, Y2H analysis showed interactions between the GmRIN4a and GmRIN4b and between the GmRIN4b and GmRIN4c proteins (Fig. 6A). However, no interactions were detected for GmRIN4a with GmRIN4c or GmRIN4d or for GmRIN4b with GmRIN4d. Furthermore, only GmRIN4b was able to self-interact in Y2H assays (Fig. 6A). In vitro pull-down assays and in planta BiFC assays were used to confirm the Y2H interactions. For pull-down assays between GmRIN4a and GmRIN4b, GmRIN4c, or GmRIN4d, GmRIN4a-His was immobilized on Ni-NTA and incubated with GmRIN4b-Myc, GmRIN4c-GST, or GmRIN4d-GST. For pull-down assays between GmRIN4b and GmRIN4c or GmRIN4d, GmRIN4b-Myc-His was immobilized on Ni-NTA and incubated with the GST-tagged GmRIN4c or GmRIN4d. Bound proteins were visualized by immunoblot analysis using Myc- or GST-specific antibodies, as appropriate. GmRIN4b lacking a His tag, GmRIN4c-GST, or GmRIN4d-GST did not bind nonspecifically to Ni-NTA lacking bait proteins (Fig. 6B, – lanes) or Ni-NTA preincubated with protein extracts from cells expressing the empty pET28A (vector used for expressing GmRIN4a) or pET30A (vector used for expressing GmRIN4b) plasmids (Supplemental Fig. S3A). Pull-down assays showed that GmRIN4b interacted with itself and with GmRIN4c or GmRIN4d (Fig. 6B). Consistent with Y2H and pull-down assays, BiFC showed binding between GmRIN4b and GmRIN4a or GmRIN4c but not between GmRIN4a and GmRIN4c or GmRIN4d (Fig. 6C). In addition, except for GmRIN4b, other GmRIN4 isoforms did not self-interact even though all proteins were expressed at comparable levels (Fig. 6C; Supplemental Fig. S4, A and B). However, in contrast to Y2H assay, both pull-down and BiFC assays showed binding between GmRIN4b and GmRIN4d. As before, all four GmRIN4 proteins were localized to the membrane fraction (data not shown). Together, these data showed that the GmRIN4b protein interacts with itself and the other three GmRIN4 isoforms.
Figure 6.
GmRIN4 isoforms (GmRIN4a to GmRIN4d = 4a to 4d) interact with each other. A, Y2H assay showing interactions between the various GmRIN4 proteins. Growth on His−Trp−Leu− plates is shown. Numbers indicate β-galactosidase activity ± sd (n = 3). B, Pull-down assays showing in vitro binding between the GmRIN4 proteins. For pull-down assay between 4a and 4b, 4c, or 4d, 4a was immobilized on Ni-NTA and incubated with increasing amounts (0.1–1 μg) of 4b, 4c, or 4d. The His tag was cleaved from 4b before application to a 4a affinity column. For interactions between 4b and 4c or 4d, 4b was immobilized on Ni-NTA and incubated with increasing amounts (0.1–1 μg) of 4c or 4d. Purified 4b, 4c, or 4d was loaded as a positive control (+) and incubated with empty Ni-NTA/glutathione agarose as a negative control (−) on respective gels. Proteins were visualized by immunoblot analysis using anti-Myc (4b) or anti-GST (4c and 4d) antibodies. C, BiFC assay showing in planta interactions. CFP and YFP overlay images (40× magnification) of micrographs at 48 h post infiltration from leaves coexpressing the indicated proteins are shown. Images are representative of three separate infiltrations from two independent experiments for each interaction. Bars = 10 μm.
GmRIN4a and GmRIN4b Mediate Basal Resistance in Soybean
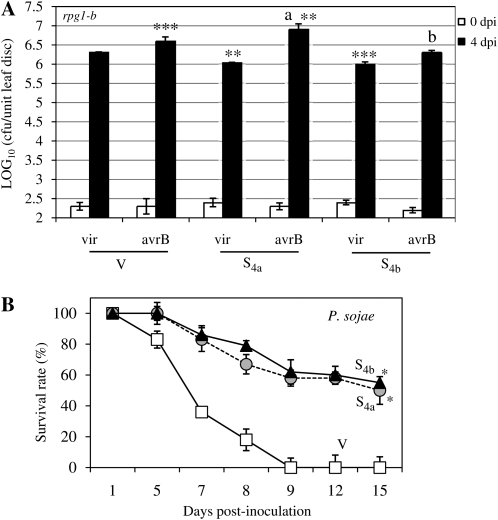
The RIN4 protein also mediates basal defense in Arabidopsis (Kim et al., 2005b). Therefore, we next tested basal resistance in S4a and S4b plants to virulent (vir) strains of P. syringae pv glycinea race 4 and the oomycete P. sojae. GmRIN4a (S4a) or GmRIN4b (S4b) was silenced in the Essex cultivar (lacking RPG1-B). Silencing of the respective genes was confirmed using reverse transcription (RT)-PCR analysis as described above (data not shown). All four GmRIN4 isoforms were expressed to similar levels in V and M plants. Thus, BPMV infection did not alter GmRIN4 expression in Essex plants, similar to that observed in Harosoy plants (data not shown). Essex (rpg1-b) V, rpg1-b S4a, and rpg1-b S4b plants were inoculated with vir bacteria. Both rpg1-b S4a and rpg1-b S4b plants consistently accumulated approximately 2-fold reduced levels of vir bacteria in comparison with V plants (Fig. 7A). Likewise, rpg1-b S4a and rpg1-b S4b plants also showed significantly enhanced basal resistance (P < 0.001) to virulent P. sojae than V plants; at 9 d, approximately 60% of the S4a or S4b plants survived infection versus none of the control plants (Fig. 7B). Together, these results suggest that GmRIN4a and GmRIN4b contribute to basal resistance to virulent pathogens in soybean.
Figure 7.
GmRIN4a and GmRIN4b mediate basal immunity in soybean. A, Bacterial counts in rpg1-b (cv Essex) plants silenced for GmRIN4a (S4a) or GmRIN4b (S4b) and those inoculated with the empty silencing vector (V). Plants were infiltrated with virulent (vir) or avirulent (avrB) strains of P. syringae pv glycinea. Log10 values of colony-forming units (cfu) per unit of leaf area from infected leaves at 0 d post inoculation (dpi; white bars) or 4 dpi (black bars) are presented. Error bars indicate sd (n = 5). Student's t test was used to determine statistical significance. Asterisks denote significant differences from V plants inoculated with vir bacteria as follows: ** P < 0.001, *** P < 0.0001. Significant difference between avrB-inoculated V and S4a plants is denoted by a, where P < 0.05. Significant difference between vir and avrB-inoculated S4b plants is denoted by b, where P < 0.0001. B, Percentage survival of V (white squares), S4a (gray circles), or S4b (black triangles) plants at 1, 5, 7, 8, 9, 12, and 15 d post inoculation with P. sojae. Error bars indicate sd (n = 3). Asterisks denote significant differences from V, where P < 0.001, as determined using Student's t test. Results are representative of three independent experiments.
Since AvrB enhances virulence on plants lacking RPG1-B (Ashfield et al., 1995; Ong and Innes, 2006), we next tested the response of the rpg1-b S4a and rpg1-b S4b plants to bacteria expressing AvrB. The AvrB-expressing strain of P. syringae was consistently 2-fold more virulent in rpg1-b V plants than the vir strain (P < 0.0001; Fig. 7A). AvrB bacteria also exhibited enhanced virulence in the rpg1-b S4b plants, consistently accumulating to 2-fold greater levels than the vir bacteria (P < 0.0001; Fig. 7A). In contrast, AvrB bacteria showed more pronounced virulence in the rpg1-b S4a plants, accumulating to levels approximately 4-fold higher than vir bacteria in rpg1-b V plants (P < 0.001). These levels were also 2-fold higher than the AvrB bacteria in rpg1-b V plants (P < 0.05). Together, these results suggest that GmRIN4a negatively regulates the virulence of AvrB bacteria on plants lacking RPG1-B.
DISCUSSION
We demonstrate that the soybean R protein RPG1-B recruits RIN4-like proteins for resistance signaling against AvrB-expressing P. syringae, similar to its nonorthologous counterpart, RPM1 in Arabidopsis. The Arabidopsis RIN4 protein is thought to serve as a guardee for RPM1, which presumably monitors the AvrB-mediated phosphorylation of RIN4 to induce resistance signaling. The lack of sequence conservation between RPM1 and RPG1-B indicates independent evolution of the two R proteins, in spite of their shared specificity for the same avirulence effector (Bisgrove et al., 1994; Grant et al., 1995; Ashfield et al., 2004). This is supported by the differences in their requirements for signaling components. For example, RPM1 but not RPG1-B requires HSP90, whereas RPG1-B but not RPM1 requires SGT1 for resistance signaling (Austin et al., 2002; Hubert et al., 2003; Fu et al., 2009). However, as we show here, RPM1 and RPG1-B do share their requirements for RIN4. Unlike in Arabidopsis, four AvrB-interacting GmRIN4 proteins are present in soybean, and three of these also interact with RPG1-B. Interestingly, in spite of their low sequence conservation, both RPM1 and RPG1-B interact with the respective Arabidopsis and soybean RIN4 proteins via their N-terminal CC domains (Mackey et al., 2002; this work). In soybean, more than one RIN4-like protein mediates effector-triggered immunity to AvrB-expressing P. syringae, since the absence of either GmRIN4a or GmRIN4b abrogates RPG1-B-derived resistance. The presence of multiple RIN4 proteins in soybean and the fact that more than one of them is required for RPG1-B-mediated resistance raises the possibility that multiple RIN4-like proteins may also be present in other genomes. Indeed, multiple RIN4-like proteins are reported in both tomato and lettuce species (Jeuken et al., 2009; Luo et al., 2009). Notably, the Arabidopsis genome also encodes several other as yet uninvestigated proteins with significant similarities to AtRIN4 (At3g25070), raising the possibility that some of these too might function in defense signaling.
Of particular interest is the fact that GmRIN4a is required for RPG1-B-mediated resistance even though this protein does not directly interact with RPG1-B. Moreover, GmRIN4a is unable to complement the Arabidopsis rin4 mutation even though it interacts with RPM1. Thus, the functioning of RIN4-like proteins in R-derived resistance does not rely on direct interactions with the R protein. However, GmRIN4a does interact with GmRIN4b, which in turn interacts with RPG1-B. Thus, the requirement for GmRIN4a in RPG1-B-derived resistance signaling could be because GmRIN4 proteins function as a heteromeric complex. GmRIN4b also interacts with two other RIN4 isoforms, GmRIN4c and GmRIN4d. It is likely that GmRIN4c and GmRIN4d also function in RPG1-B-derived resistance, since both interact with AvrB, RPG1-B, and GmRIN4b. However, silencing the GmRIN4c and GmRIN4d isoforms separately (without altering the expression of the other three isoforms) to test their individual functions in RPG1-B-mediated resistance has proven challenging.
A knockout mutation in RIN4 is lethal, while antisense reduction of RIN4 expression constitutively activates defense responses due to the ectopic induction of a second R protein, RPS2, in Arabidopsis (Mackey et al., 2002, 2003). Arabidopsis RIN4 also prevents the inappropriate activation of RPM1, because the absence of RIN4 enhances basal defense to virulent P. syringae (Belkhadir et al., 2004). This enhanced basal resistance and increased PR-1 expression in the rin4 rps2 double mutant plants were attributed to residual RPM1 activity, since both phenotypes are absent in the rin4 rps2 rpm1 triple mutant (Belkhadir et al., 2004). The soybean cv Harosoy (RPG1-B) did not exhibit constitutive induction of defense responses in response to silencing of GmRIN4a or GmRIN4b, suggesting that silencing GmRIN4a or GmRIN4b does not affect the activities of other R proteins in this cultivar. Furthermore, unlike RPM1 in Arabidopsis, silencing GmRIN4a or GmRIN4b does not inappropriately induce RPG1-B activity; the RPG1-B GmRIN4a- or GmRIN4b-silenced plants do not exhibit enhanced resistance to virulent bacteria (data not shown) and express basal levels of PR genes. However, silencing GmRIN4a or GmRIN4b in rpg1-b plants (cv Essex) does confer increased resistance to virulent bacteria and P. sojae. Strikingly, the GmRIN4a- or GmRIN4b-silenced rpg1-b plants showed much better resistance to virulent P. sojae than to P. syringae. The nominal increase in resistance to virulent P. syringae might result from derepression of basal immunity, albeit only in cv Essex. Alternatively, the various GmRIN4 proteins might function additively in basal defense to P. syringae, such that absence of all or multiple isoforms is required for the induction of a more robust response. Analyzing basal defense in plants simultaneously silenced for multiple GmRIN4 isoforms would help address this. The more pronounced resistance to P. sojae might be associated with the ectopic activation of other unidentified R/R-like protein(s) in cv Essex that are specifically active against P. sojae.
Unlike their increased resistance to DC3000, rin4 rps2 plants do not exhibit enhanced resistance to P. syringae expressing AvrB or AvrRPM1 (Belkhadir et al., 2004). Likewise, rin4 rps2 rpm1 triple mutant plants accumulate similar levels of bacteria or induce chlorosis like the rps2 rpm1 double mutant plants in response to AvrRPM1- or AvrB-expressing strains of the weak pathogen P. syringae pv maculicola M6CΔE, respectively (Nimchuk et al., 2000; Rohmer et al., 2003; Belkhadir et al., 2004). Thus, although RIN4 is a negative regulator of immunity, it is not required for the virulence activities of either AvrRPM1 or AvrB in Arabidopsis (Nimchuk et al., 2000; Belkhadir et al., 2004). This is also true for the two GmRIN4 proteins tested here; both GmRIN4a- and GmRIN4b-silenced rpg1-b plants support more growth of AvrB-expressing P. syringae than the virulent strain. Interestingly, however, GmRIN4a-silenced rpg1-b plants consistently accumulate higher levels of AvrB bacteria than the control plants, suggesting that GmRIN4a negatively regulates the virulence function of AvrB. Thus, the virulence activity of AvrB is enhanced in plants lacking both the R protein and GmRIN4a. A likely possibility is that GmRIN4a serves as a decoy to mimic AvrB interaction with its target and thereby limit pathogen fitness. This scenario is in agreement with the proposed function for decoy proteins that might serve to both perceive and compete for binding with the effector (van der Hoorn and Kamoun, 2008). A second possibility is that the Essex background contains another R protein with specificity for AvrB and that GmRIN4a but not GmRIN4b is required for the function of this unknown R protein. Indeed, besides RPM1, the Arabidopsis TIR-NBS-LRR protein TAO1 also exhibits AvrB specificity but does not require RIN4 for its function (Eitas et al., 2008). Comparing pathogen fitness in RPG1-b and rpg1-b plants silenced for either GmRIN4a or GmRIN4b versus those silenced for both GmRIN4a and b simultaneously could help resolve the functions of the encoded proteins and their relative contributions to the perception of AvrB and the virulence activity of AvrB.
MATERIALS AND METHODS
Plant Growth Conditions
Soybean (Glycine max ‘Harosoy’ [RPG1-B] and ‘Essex’ [rpg1-b]) were grown in the greenhouse with day and night temperatures of 25°C and 20°C, respectively. For silencing experiments, inoculation of recombinant BPMV vectors and confirmation of silencing were carried out as described before (Kachroo et al., 2008). Arabidopsis (Arabidopsis thaliana) plants (wild type or rin4 rps2 mutant in the Columbia [Col-0] ecotype) were grown in MTPS 144 Conviron walk-in chambers at 22°C, 65% relative humidity, and 14-h photoperiod.
Y2H Assays
Y2H assays were carried out using standard protocols for the GAL4 system (Clontech). Interactions were tested in the yeast strain Y190, with AvrB or RPG1-B (N-terminal 187 amino acids) expressed from the GAL4 bait (pGADT7) plasmid and GmRIN4 proteins expressed from the prey (pGBKT7) plasmid.
In Vitro Binding Assays
The AvrB, GmRIN4a, GmRIN4b, and RPG1-B (CC domain, N-terminal 187 amino acids) proteins were expressed as His-tagged fusions in Escherichia coli (AvrB-pET28A, GmRIN4a-, and RPG1-B-pET30). For GmRIN4b, a Myc tag was also added at the C terminus (pBAD). GmRIN4c and GmRIN4d were expressed as GST fusion proteins in E. coli (pGEX-5X). Proteins were purified using Ni-NTA (His) and glutathione Sepharose (GST) affinity chromatography. For binding between AvrB and GmRIN4a, -b, -c, -d, or RPG1-B, the His tag was removed from AvrB using thrombin. To ensure complete cleavage of the His tag, 0.5 μg of the cleaved AvrB was applied to empty Ni-NTA. Lack of nonspecific retention was assessed by immunoblot analysis using AvrB-specific antibodies. One microgram each of GmRIN4a-His, GmRIN4b-His, and RPG1-B-His was immobilized on Ni-NTA and GmRIN4c-GST or GmRIN4d-GST immobilized on glutathione agarose. The immobilized proteins were incubated with increasing amounts (50, 100, 250, and 500 ng) of AvrB. For pull down between GmRIN4b, GmRIN4c, or GmRIN4d and RPG1-B, the His tag was cleaved from GmRIN4b using thrombin, and nonretention of the cleaved protein on empty Ni-NTA or Ni-NTA bound to protein extracts from cells expressing the empty pET28A/pET30A vectors was assessed using Myc-specific antibodies. One microgram of RPG1-B-His was immobilized on Ni-NTA and incubated with 100, 250, and 500 ng and 1 μg of GmRIN4b-Myc (His tag removed), GmRIN4c-GST, or GmRIN4d-GST. For pull-down assay between GmRIN4a and GmRIN4b, GmRIN4c, or GmRIN4d, GmRIN4a-His was immobilized on Ni-NTA incubated with increasing amounts (0.1–1 μg) of GmRIN4b-Myc (His tag removed), GmRIN4c-GST, or GmRIN4d-GST. For pull-down between GmRIN4b and GmRIN4c or GmRIN4d, GmRIN4b-Myc-His was immobilized on Ni-NTA and incubated with increasing amounts (0.1–1 μg) of GmRIN4c-GST or GmRIN4d-GST. Purified proteins were loaded as positive controls (+) and incubated with empty Ni-NTA/glutathione Sepharose as negative controls (−) on the respective gels. Purified proteins were also incubated with Ni-NTA preincubated with extracts from cells expressing the empty pET28A/pET30A vectors or with glutathione Sepharose (GE Healthcare) preincubated with extract from cells expressing the empty pGEX-5X vector. Affinity columns were washed extensively with sodium phosphate buffer (0.01 m, pH 7.0, 150 mm NaCl, 10 mm imidazole) to remove unbound proteins, and immunoblot analysis of bound proteins was carried out using anti-AvrB, anti-Myc (GmRIN4b), and anti-GST (GmRIN4c and GmRIN4d) antibodies. Results presented are representative of two to three independent binding assays using proteins from two separate preparations. Approximate protein molecular masses are as follows: AvrB (36 kD), RPG1-BCC (20.68 kD), GmRIN4a (27.12 kD), GmRIN4b (27.08 kD), GmRIN4c-GST (52.66 kD), and GmRIN4d-GST (53.51 kD).
BiFC Assays
BiFC assays were carried out as described before (Martin et al., 2009). Briefly, the various proteins were fused to the N/C-terminal halves of EYFP (nEYFP/cEYFP) using the pSITE-BiFC-C1cec/nec (EYPF) vectors (Martin et al., 2009) and introduced in Agrobacterium tumefaciens strain C58C1. A. tumefaciens strains were then infiltrated into CFP-H2B-tagged Nicotiana benthamiana plants (transgenic plants expressing nucleus-localized CFP). At 48 h later, water-mounted sections of leaf tissue were examined by confocal microscopy using a water-immersion PLAPO60XWLSM (numerical aperture, 1.0) objective on a FV1000 point-scanning/point-detection laser scanning confocal microscope (Olympus) equipped with lasers spanning the spectral range of 405 to 633 nm. CFP and YFP overlay images (40× magnification) were acquired at a scan rate of 10 ms pixel−1. Olympus FLUOVIEW 1.5 was used to control the microscope, image acquisition, and the export of TIFF files. All proteins were expressed as both nEYFP and cEYFP fusions, and all interactions were tested using both combinations of n/cEYFP-fused reciprocal proteins. Results presented are representative of three separate infiltrations for every combination tested. All interactions (positive and negative) were tested at least twice.
Plant Protein Extraction and Immunoblot Analysis
Proteins were extracted in buffer containing 50 mm Tris-HCl, pH 7.5, 10% glycerol, 150 mm NaCl, 10 mm MgCl2, 5 mm EDTA, 5 mm dithiothreitol (DTT), and 1× protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was measured by the Bio-Rad protein assay. For Ponceau-S staining, polyvinylidene difluoride membranes were incubated in Ponceau-S solution (40% methanol [v/v], 15% acetic acid [v/v], and 0.25% Ponceau-S). The membranes were destained using deionized water. Membrane proteins were extracted using buffer containing 50 mm Tris-MES, pH 8.0, 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, 10 mm EGTA, 10 mm ascorbic acid, 5 mm DTT, and 1× protease inhibitor cocktail. Total protein extracts were centrifuged at 10,000g followed by a second centrifugation at 125,000g. The microsomal fractions were suspended in a buffer containing 5 mm potassium phosphate, pH 7.8, 2 mm DTT, 1× protease inhibitor cocktail, and 1% Triton X-100. Proteins (30–50 μg) were fractionated on a 7% to 10% SDS-PAGE gel and subjected to immunoblot analysis using anti-TCV-CP or anti-GFP antibody. Approximate protein molecular masses were as follows: AvrB-nEYFP (53 kD), RPG1-BCC-nEYFP (37.67 kD), GmRIN4a-nEYFP (44.12 kD), GmRIN4b-nEYFP (44.08 kD), GmRIN4c-nEYFP (43.66 kD), GmRIN4d-nEYFP (44.5 kD), GST-nEYFP (43 kD), Avr1b-nEYFP (32.7 kD), GST-cEYFP (33 kD), and TCV-CP-cEYFP (45 kD).
Construction of Viral Vectors, in Vitro Transcription, and Plant Inoculation
Generation of silencing vectors, in vitro transcription, and rub inoculation of soybean leaves was carried out as before (Kachroo et al., 2008). A 159-bp fragment (Gly-184 to Gln-236) of GmRIN4a and a 243-bp fragment (Thr-59 to Pro-139) of GmRIN4b were used to generate vectors targeting GmRIN4a and GmRIN4b, respectively.
RNA Extraction, Northern Blot, and RT-PCR Analysis
RNA from leaf tissues of soybean plants at the V1/V2 growth stage was extracted using the Trizol reagent (Invitrogen) per the manufacturer's instructions. Northern-blot analysis and synthesis of randomly primed probes were as described before (Kachroo et al., 2001). RT and first-strand cDNA synthesis were carried out using SuperScript II (Invitrogen). Two to three independent RNA preparations were analyzed at least twice by RT-PCR using a reduced number of cycles (15–20) for evaluating relative differences in transcript levels.
Trypan Blue Staining and Pathogen Inoculations
Trypan blue staining of N. benthamiana leaves was performed as described (Chandra-Shekara et al., 2006). Pseudomonas syringae inoculation of soybean and monitoring of bacterial proliferation were carried out as described before (Fu et al., 2009). Mock inoculations were carried out with 10 mm MgCl2 in 0.04% Silwett L-77. Experiments were repeated three to four times. For Arabidopsis inoculations, bacterial suspensions in 10 mm MgCl2 were infiltrated to the abaxial surface of leaves using a 1-mL needleless syringe. After inoculations, plants were transferred to a Conviron PGV36 walk-in chamber maintained at 22°C with 65% relative humidity and a 10-h photoperiod. For soybean Phytophthtora sojae inoculations, race 1, strain P6497 was grown on V8 agar at 25°C in the dark. P. sojae infections were carried out as described before (Kachroo et al., 2008). Experiments were repeated three times with 15 to 20 plants tested per wild-type or silenced line per experiment.
Plant Transformation
Full-length cDNAs corresponding to GmRIN4a or GmRIN4b under the control of a double 35S promoter and the 35S terminator were cloned into the binary vector pBAR1. Constructs were introduced into A. tumefaciens strain MP90 by electroporation and transformed into Arabidopsis using vacuum infiltration. Transgenic seeds were selected on soil sprayed with the herbicide BASTA.
The sequences described have been submitted to the GenBank database (accession nos. GU132851–GU132854).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AvrB-induced cell death and controls for AvrB pull-down and BiFC assays.
Supplemental Figure S2. GmRIN4 proteins do not interact with GST, TCV-CP, or Avr1b in BiFC assays.
Supplemental Figure S3. Controls for GmRIN4 pull-down and BiFC assays.
Supplemental Figure S4. GmRIN4 isoforms interact with each other.
Supplementary Material
Acknowledgments
We thank Raghuprakash Kastoori for help with yeast transformations, Ludmila Lapchyk for help with plant transformations, Rae-Dong Jeong for the TCV-CP construct, Michael Goodin for the pSITE vectors, the SYNV-G construct, and CFP-H2B-expressing transgenic N. benthamiana, Alan Collmer for AvrB antibodies, Jeff Dangl for the avrB P. syringae pv tomato strain, David Mackey for rin4 mutant lines, Jack Morris for TCV-CP antibodies, Roger Innes for useful discussions and sharing the GmRIN4c and GmRIN4d sequences, Adam Bogdanove for the P. syringae pv glycinea strains, Pradeep Kachroo and Said Ghabrial for critical review of the manuscript, and Amy Crume for managing the plant growth facility.
References
- Ashfield T, Keen NT, Buzzell RI, Innes RW. (1995) Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. (2004) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. (1994) A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Gupte M, Navarre D, Raina S, Raina R, Klessig D, Kachroo P. (2006) Light-dependent hypersensitive response and resistance signaling against Turnip crinkle virus in Arabidopsis. Plant J 45: 320–334 [DOI] [PubMed] [Google Scholar]
- Chang JH, Rathjen JP, Bernal AJ, Staskawicz BJ, Michelmore RW. (2000) avrPto enhances growth and necrosis caused by Pseudomonas syringae pv. tomato in tomato lines lacking either Pto or Prf. Mol Plant Microbe Interact 13: 568–571 [DOI] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN. (2004) The Pseudomonas syringae type III effector AvrRpt2 functions downstream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J 37: 494–504 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. (2005) Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci USA 102: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. (1996) Death don't have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. (2007) Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog 3: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Jiang RH, Bruce NA, Arredondo FD, Zhang X, Tyler BM. (2008) RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20: 1930–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Nimchuk ZL, Dangl JL. (2008) Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA 105: 6475–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DQ, Ghabrial S, Kachroo A. (2009) GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean. Mol Plant Microbe Interact 22: 86–95 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Guttman DS, Greenberg JT. (2001) Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol Plant Microbe Interact 14: 145–155 [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes R. (2004) Guarding the goods: new insights into the central alarm system of plants. Plant Physiol 135: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken MJW, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RGF, Niks RE. (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21: 3368–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Fu DQ, Havens W, Navarre D, Kachroo P, Ghabrial SA. (2008) An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol Plant Microbe Interact 21: 564–575 [DOI] [PubMed] [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Buzzell RI. (1991) New resistance genes in soybean against Pseudomonas syringae pv. glycinea: evidence that one of them interacts with a bacterial elicitor. Theor Appl Genet 81: 133–138 [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. (2005a) The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA 102: 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. (2005b) Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Leister RT, Katagiri F. (2000) A resistance gene product of the nucleotide binding site-leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J 22: 345–354 [DOI] [PubMed] [Google Scholar]
- Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. (2009) Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell 21: 2458–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alfonso JM, Ecker JR, Dangl JL. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, III, Wiig A, Dangl JL. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. (2003) Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54: 23–61 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. (2006) The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18: 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363 [DOI] [PubMed] [Google Scholar]
- Ong LE, Innes RW. (2006) AvrB mutants lose both virulence and avirulence activities on soybean and Arabidopsis. Mol Microbiol 60: 951–962 [DOI] [PubMed] [Google Scholar]
- Ritter C, Dangl JL. (1996) Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L, Kjemtrup S, Marchesini P, Dangl JL. (2003) Nucleotide sequence, functional characterization, and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol 47: R22–R24 [DOI] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol 186: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. (1996) Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274: 2063–2065 [DOI] [PubMed] [Google Scholar]
- Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, Chintha R, Harzen A, Colby T, Kamoun S, van der Hoorn RA. (2008) Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell 20: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Li X, Cui H, He P, Thilmony R, Chintamanani S, Zwiesler-Vollick J, Gopalan S, Tang X, Zhou JM. (2006) RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA 103: 19200–19205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B, Dahlbeck D, Keen N, Napoli C. (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol 169: 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Jones DA. (2005) Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol Plant Microbe Interact 18: 1258–1268 [DOI] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JD. (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Kamoun S. (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse HP, Bolton MD, Stergiopoulos I, de Wit PJ, Thomma BP. (2007) The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol Plant Microbe Interact 20: 1092–1101 [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Zhang C, Ghabrial SA. (2006) Development of bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344: 401–411 [DOI] [PubMed] [Google Scholar]
- Zhao Y, DelGrosso L, Yigit E, Dempsey DA, Klessig DF, Wobbe KK. (2000) The amino terminus of the coat protein of Turnip crinkle virus is the AVR factor recognized by resistant Arabidopsis. Mol Plant Microbe Interact 13: 1015–1018 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Chai J. (2008) Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol 11: 179–185 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Rathjen JP. (2008) Plant immunity: AvrPto targets the frontline. Curr Biol 18: R218–R220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.