Abstract
Kiwifruit (Actinidia deliciosa) is a climacteric fruit sensitive to low concentrations of ethylene. To investigate the transcriptional mechanisms underlying kiwifruit ethylene response, transcription factors encoding four EIN3-Like (EILs) and 14 Ethylene Response Factors (ERFs) were cloned from kiwifruit. Expression of these transcription factors was examined during fruit development. The expression of transcripts of most AdERFs was higher during early fruit development, with the exception of AdERF3, which increased with maturity. Several AdERFs were apparently down-regulated by ethylene, as they were affected by the ethylene inhibitor 1-methylcyclopropene and by antisense suppression of ACO (for 1-aminocyclopropane-1-carboxylic acid oxidase) in the fruit. In contrast, AdEILs were constitutively expressed during fruit development and ripening. The transcription factors AdEIL2 and AdEIL3 activated transcription of the ripening-related genes AdACO1 and AdXET5 (xyloglucan endotransglycosylase gene) and, when overexpressed in Arabidopsis (Arabidopsis thaliana), stimulated ethylene production. The potential repressor AdERF9 suppressed this promoter activity. These results support a role for kiwifruit EILs and ERFs in transcriptional regulation of ripening-related genes and in the regulation of kiwifruit fruit-ripening processes.
Plant tissues, including fruit, respond to ethylene through an increasingly well-defined signal transduction pathway. The transcriptional response to ethylene includes the EIN3-Like (EIL) and Ethylene Response Factor (ERF) families of transcription factors (TFs). Members of both families act as positive controllers of the ethylene response and potentially are involved in cross talk with other signals (Guo and Ecker, 2004).
EILs encode a small TF gene family (Chao et al., 1997; Riechmann et al., 2000; Tieman et al., 2001; Rieu et al., 2003). Members of this family have been shown to directly bind to the primary ethylene response element (PERE) in the Arabidopsis (Arabidopsis thaliana) ERF1 promoter to regulate ERF1 expression. This was first verified by Solano et al. (1998), and subsequently, a consensus binding sequence (EIL conserved binding sequence [ECBS]) was also found for tobacco (Nicotiana benthamiana) TEIL, comprising A[A/C]G[A/T]A[A/C]CT (Kosugi and Ohashi, 2000). However, there is a need for more information on the interaction of EIN3/EILs and promoters of potential target genes.
Unlike EIN3/EILs, ERFs constitute one of the largest TF gene families in plants, with 122 members in Arabidopsis and 139 members in rice (Oryza sativa), containing a conserved DNA-binding domain (ERF domain; Nakano et al., 2006). ERFs were first identified from tobacco by interaction with a GCC box (core sequence is AGCCGCC; Ohem-Takagi and Shinshi, 1995). Besides the GCC box, a small group of ERFs (DREBs) also specifically bind to dehydration-responsive elements (DRE), with a core sequence of CCGAC (Hao et al., 2002; Sun et al., 2008).
These GCC and DRE motifs have been identified in many stress-related gene promoters associated with ethylene-mediated, and ethylene-independent, stress responses. Examples include CBF/DREB1 (for C-repeat-binding factor/dehydration responsive element-binding factor 1) genes in cold response (Novillo et al., 2004) and DREB2 in drought and heat stress response (Sakuma et al., 2006; Qin et al., 2008). Overexpression of sugarcane (Saccharum officinarum) SodERF3 has been shown to increase plant salt and drought tolerance (Trujillo et al., 2008), and wheat (Thinopyrum intermedium) TiERF1 is involved in disease resistance (Chen et al., 2008b). In addition to involvement in stress response, ERF genes have other distinct functions, such as involvement in lipid, cutin, and wax biosynthesis (Aharoni et al., 2004; Kannangara et al., 2007; Taketa et al., 2008).
EIN3/EIL genes show functional redundancy in modulating ethylene response (Tieman et al., 2001), despite the fact that transcripts have been found to be little affected by ethylene in some plants (Lee and Kim, 2003; Mao et al., 2006). Ethylene appears to modulate EIN3/EIL at the protein level; treatment with 1-aminocyclopropane-1-carboxylic acid (ACC) results in the accumulation of EIN3 protein in Arabidopsis (Yanagisawa et al., 2003). Recently, tobacco TEIL has been found to have some relationship with the basic PR gene, as basic PR5 was induced in TEIL-overexpressed plants; TEIL is also involved in flower development (Hibi et al., 2007). An important role for ethylene is in fruit ripening, and EIL genes have been isolated from fruit such as tomato (Solanum lycopersicum; Tieman et al., 2001; Yokotani et al., 2003) and kiwifruit (Actinidia deliciosa; Yin et al., 2008, 2009). In banana (Musa acuminata) fruit, MA-EIL2 is a ripening- and ethylene-inducible gene, unique within the EIL gene family (Mbéguié-A-Mbéguié et al., 2008). In kiwifruit, EIL genes have been shown to be induced by low temperature, suggesting an involvement in low temperature response (Yin et al., 2009). Tomato LeEIL genes show some functional redundancy and can modulate ethylene response and fruit development (Tieman et al., 2001). However, there has been little research on EIL function in transcriptional regulation in fruit.
Fruit ERF genes have been isolated from apple (Malus domestica) and plum (Prunus salicina; Wang et al., 2007; El-Sharkawy et al., 2009). In tomato, LeERF genes have GCC box DNA-binding activity and differential responses to ethylene and other stimuli; LeERF2 is induced by ethylene and suppressed in ripening-inhibited mutants (Wu et al., 2002; Tournier et al., 2003). Overexpression of LeERF1 shortens fruit postharvest life (Li et al., 2007), and LeERF2 regulates ethylene production in tomato and tobacco by modulating ethylene biosynthesis genes (Zhang et al., 2009b), transcriptional regulation being achieved by interaction of LeERF2 with promoters of NtACS3 and LeACO3. Autoregulation of ethylene biosynthesis is only one consequence of ethylene response in ripening fruit. Ethylene is involved to varying degrees in different fruit species in ripening-related processes such as softening (cell wall loosening), membrane breakdown, and aroma generation, but the involvement of the TFs of the EIL and ERF families in these processes is largely unknown.
Kiwifruit provides a good model for investigating ethylene response in fruit ripening, since it is very sensitive to ethylene (McDonald and Harman, 1982). Ripening-related genes have been isolated from kiwifruit, including those associated with ethylene biosynthesis (e.g. ACS and ACO; Whittaker et al., 1997; Xu et al., 1998), cell wall metabolism and fruit softening (e.g. polygalacturonase gene, PG; xyloglucan endotransglycosylase/hydrolase gene, XTH; expansin gene, EXP; Atkinson and Gardner, 1993; Yang et al., 2007; Atkinson et al., 2009), and membrane metabolism and aroma generation (e.g. lipoxygenase gene, LOX; Zhang et al., 2006, 2009a). We have also recently shown differential responses of ethylene receptors as well as CTR (for constitutive triple response) and EIL genes to ripening stimulation and low temperature in kiwifruit (Yin et al., 2008, 2009).
The Actinidia EST database (Crowhurst et al., 2008) provides an opportunity to investigate further the ethylene response during ripening of kiwifruit by selecting ERF genes and analyzing their expression. In this study, three AdEILs (data for a fourth, AdEIL1, have been published by Yin et al. [2008]) and 14 AdERF genes were isolated from kiwifruit ESTs. Tissue- and time-specific gene expression in ripening kiwifruit was analyzed, and the interaction of AdEIL and AdERF genes with target gene promoters was investigated.
RESULTS
Gene Isolation and Analysis
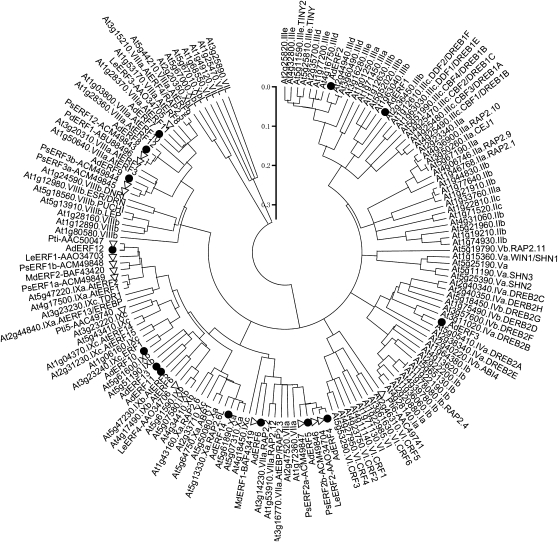
Fourteen ERF full-length cDNAs were isolated and designated as AdERF1 to AdERF14 (GenBank accession nos. GQ869852–GQ869865). Based on a phylogenetic tree, the 14 ERF genes are clustered into six subfamilies (Fig. 1). Within the three main subfamilies, AdERF4 to AdERF6 belong to subfamily VII, while AdERF7 to AdERF9 and AdERF10 to AdERF13 belong to subfamilies VIII and IX, respectively. Four genes were isolated from three other families: AdERF1 and AdERF2 (subfamily III), AdERF3 (subfamily IV), and AdERF14 (subfamily X; Fig. 1).
Figure 1.
Phylogenetic tree of ERFs. Fourteen kiwifruit AdERFs (black circles) were aligned with the Arabidopsis ERF family and other fruit ERFs (white triangles). The amino acid sequences were obtained from The Arabidopsis Information Resource or the National Center for Biotechnology Information database. The amino acid sequences were analyzed with Vector NTI (version 9.0.0; Invitrogen), and the phylogenetic tree was constructed with MEGA (version 3.1) using a bootstrap test of phylogeny with minimum evolution test and default parameters.
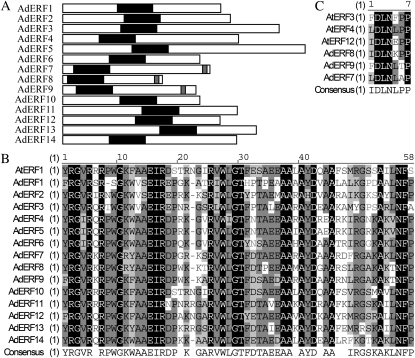
The sizes of the deduced amino acid sequences differed substantially (e.g. AdERF3 has 382 amino acids and AdERF8 has 157 amino acids). Alignments of full-length deduced proteins of AdERFs and AtERFs showed conserved motifs, including a DNA-binding domain called the ERF domain (amino acids 57–59; Fig. 2, A and B). The ERF domain is a defining character of the ERF transcriptional factor gene family (Fujimoto et al., 2000; Nakano et al., 2006). In subfamily VIII, AdERF8 and AdERF9 also had a repressor domain, the ERF-associated amphiphilic repression domain (EAR; Fig. 2, A and C). Unlike the 3′ end location of EAR repressor domains, the location of the ERF domain varied among genes, being near the 5′ end in AdERF8 and AdERF9 and near the middle of AdERF1 and AdERF2 sequences (Fig. 2A).
Figure 2.
Schematic analysis of AdERFs with ERF domains and EAR repressor domains. A, Location of ERF domains (black bars) and EAR repressor domains (gray bars). B, Comparison of ERF domains by deduced amino acids. C, Comparison of EAR repressor domains by deduced amino acids.
Although all AdERF genes have the conserved ERF domain, the similarities of the ERFs varied from 12% (AdERF5 and AdERF11) to 66% (AdERF8 and AdERF9). Genes in some subfamilies had higher similarity (e.g. AdERF7 had 61% and 57% similarity with AdERF8 and AdERF9, respectively), although, in contrast, those in subfamily VII were less similar (e.g. AdERF5 only had 36% and 26% similarity with AdERF4 and AdERF6; Supplemental Table S7).
Tissue Specificity of AdEIL and AdERF Gene Expression
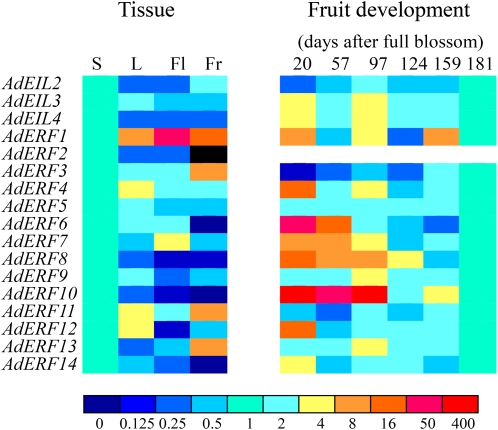
AdEIL and AdERF genes were all differentially expressed in various kiwifruit tissues (Fig. 3). Within the AdERF family, only AdERF2 was not detectable in mature fruit (Fig. 3). Among the different patterns, genes such as AdERF3 and AdERF5 were constantly expressed in all tissues, those such as AdERF4, AdERF11, and AdERF12 were higher in leaves, while AdERF1 was strongly expressed in flowers and mature fruit. In contrast, AdERF8 and AdERF10 were weakly expressed in reproductive tissues. In mature fruit, AdERF1, AdERF3, AdERF11, and AdERF12 were strongly expressed, while AdERF6, AdERF8, AdERF10, and AdERF14 were weakly expressed in mature fruit compared with other tissues (Fig. 3). Since AdERF2 was not detected in mature fruit, it was excluded in subsequent expression analysis of fruit during development and ripening.
Figure 3.
Tissue specificity and temporal expression pattern in fruit development. S, Stem; L, leaf; Fl, flower; Fr, mature fruit. Relative mRNA abundance was evaluated by real-time PCR from three biological repeats. Different colors indicate the average of relative mRNA abundance. Stem (S) and 181 DAFB were set as 1. The black area for AdERF2 in Fr means nondetectable. Data with se from three repeats are provided in Supplemental Figures S2 and S3.
Within the AdEIL gene family, AdEIL3 was constitutively expressed in all tissues, and AdEIL2 and AdEIL3 had similar expression patterns, with transcripts weak in leaves and flowers (Fig. 3).
Expression of AdEIL and AdERF Genes during Fruit Development
Six sampling times ranging from 20 d after full bloom (DAFB) to maturity (181 DAFB) covered the fruit development on the vine. Abundance of most AdERF transcripts was relatively high in fruit at the early stages of development (20 DAFB), including AdERF4, AdERF6, AdERF7, AdERF8, AdERF10, AdERF12, and AdERF14 (Fig. 3). However, expression patterns over time were slightly different, some decreasing through to 181 DAFB or peaking again at later stages; AdERF9 and AdERF13 had peaks in expression at 97 DAFB. In contrast, AdERF3, although at relatively low levels, was the only gene that increased in abundance during fruit development. AdERF5 was constantly expressed at low levels, while the abundance of AdERF1 varied during development, with three peaks (20, 97, and 159 DAFB; Fig. 3).
Within the AdEIL family, unlike the constitutively expressed AdEIL1 (Yin et al., 2008), AdEIL3 and AdEIL4 had two peaks at 20 and 97 DAFB, and AdEIL2 was weakly expressed at the earliest stage (Fig. 3).
Expression of AdEIL and AdERF Genes in Ripening Kiwifruit
Fruit physiology data for this experiment have been described in our previous publication (Yin et al., 2008). Ethylene treatment accelerates fruit ripening, promoting a climacteric rise beginning 6 d after harvest (DAH) and peaking at 9 DAH. Control fruit started the ethylene climacteric at 19 DAH and reached a peak at 21 DAH. In contrast, 1-methylcyclopropene (1-MCP), an ethylene receptor inhibitor, delayed fruit ripening, maintaining higher fruit firmness, with no ethylene being detected over the postharvest period.
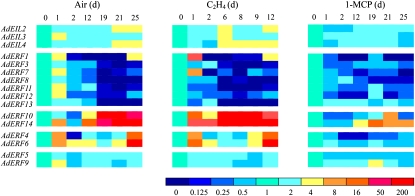
AdEIL2 and AdEIL4 showed some response to ethylene, with expression increasing slightly at the late climacteric stage in control fruit or at the time of the increase in ethylene production in ethylene-treated fruit (Fig. 4). AdEIL3 showed an early rise in expression in air and also increased at the time that ethylene increased in ethylene-treated fruit. 1-MCP treatment inhibited the ethylene-induced increases in expression (Fig. 4).
Figure 4.
Expression of the AdEILs and AdERFs in kiwifruit flesh during fruit ripening at 20°C. Gene transcript accumulation was evaluated by real-time PCR. Fruit were held at 20°C and treated with air (Air), 100 μL L−1 ethylene (C2H4), or 0.5 μL L−1 1-MCP (1-MCP) for 24 h. Relative mRNA abundance was evaluated by real-time PCR from three biological repeats. Different colors indicate the average of relative mRNA abundance. Harvest time (0 d) was set at 1. Data with se from three repeats are provided in Supplemental Figure S4.
Expression patterns of AdERF genes were more diverse and can be divided into four patterns. Most of the genes (seven members) followed a decreasing pattern during ripening (Fig. 4). AdERF1, AdERF7, AdERF11, and AdERF12 had slight increments at 1 DAH in control fruit, and ethylene treatment transiently increased AdERF1 and AdERF3 expression levels.
A second group (AdERF10 and AdERF14) showed very strong expression increases, with the increase in ethylene production at about 19 DAH in control fruit and 6 DAH in ethylene-treated fruit (Fig. 4). AdERF4 and AdERF6 expression was relatively high at the beginning of storage and again in the postclimacteric stage (25 DAH in control and 12 DAH in ethylene-treated fruit). Expression of AdERF5 and AdERF9 was constant and not affected by exogenous ethylene (Fig. 4).
1-MCP treatment was used to confirm the association of gene expression with ethylene production in the control fruit. 1-MCP treatment substantially inhibited the increases in AdERF4, AdERF6, AdERF10, and AdERF14 transcript levels toward the end of the ripening period and also inhibited the decrease in AdERF3 and AdERF13. 1-MCP also inhibited the 1-d increase in some transcript levels found in the control fruit, again confirming a likely association of the transcript increase and ethylene. 1-MCP treatment had little effect on the decrease in mRNA abundance of AdERF1, AdERF7, AdERF8, AdERF11, and AdERF12 in control fruit (Fig. 4; Supplemental Fig. S5).
We also took the opportunity to follow expression of the ERF genes in ACO knockout kiwifruit, which have inhibited ethylene production (R. Atkinson, unpublished data). Although these were of a related species (Actinidia chinensis), ethylene treatment of the fruit induced expression of AdERF1, AdERF7, and AdERF8 and suppressed that of AdERF3, AdERF11, AdERF12, and AdERF13 (Supplemental Fig. S6), confirming the above 1-MCP data, showing that decreases of AdERF3 and AdERF13 transcripts in ripening fruit were associated with ethylene, while decreases of AdERF1, AdERF7, AdERF8, AdERF11, and AdERF12 transcripts were independent of ethylene.
Interaction of AdEIL and AdERF Genes with Promoters of Ripening-Related Genes
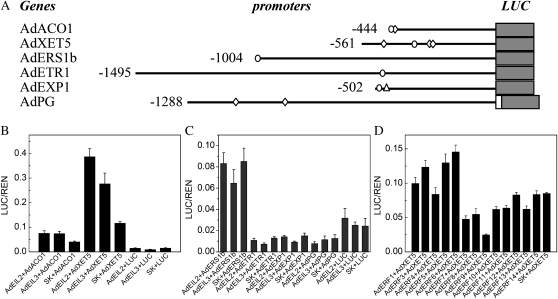
Promoters of six ripening-related genes were isolated from kiwifruit by genome walking with primers designed against existing sequences. By searching for cis-acting regulatory elements, some ethylene-related motifs were found in the promoters (Fig. 5A; Supplemental Fig. S1). However, a GCC box was only found in the promoter of AdEXP1 (Fig. 5A; Supplemental Fig. S1). An internal NcoI site in the promoter of AdPG meant that there was a 55-bp pGEM-T Easy vector sequence interval between the AdPG promoter and firefly luciferase (LUC; Fig. 5A).
Figure 5.
In vivo interaction of transcriptional factors and ripening-related gene promoters. A, Schematics of promoters are indicated with lines (promoter length), diamonds (PERE motif), circles (ECBS motif), and triangles (GCC box). B to D, In vivo associations of TFs and promoters were obtained from transient assays in tobacco leaves. Error bars indicate se from at least four replicates.
In vivo interactions between TFs and promoters were estimated by transient assays in tobacco leaves. AdEIL2 and AdEIL3 acted as activators inducing the AdACO1 and AdXET5 promoters, while no significant changes were found with other promoters (Fig. 5, B and C). Although the promoter of AdEXP1 had a GCC box, AdERF genes failed to affect promoter activity (data not shown). However, AdERF9 significantly suppressed AdXET5 promoter activity (Fig. 5D), and this was confirmed in a separate experiment described below. The other AdERFs had little effect on the AdXET5 promoter activity.
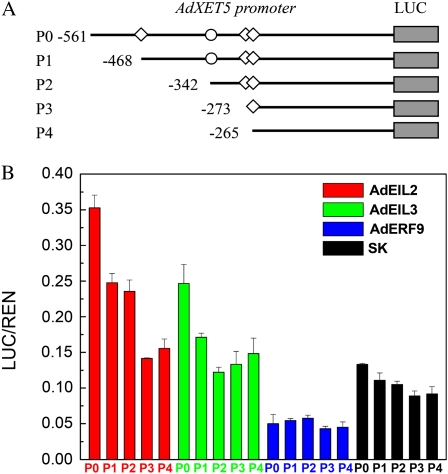
Promoter Deletions to Study Different Functions of PERE and ECBS Motifs and Possible Effective Regions for AdERF9
In order to study the functions of different motifs in the promoter of AdXET5, four deletions were made targeting four EIN3/EIL-related motifs (Fig. 6A). There was only a small effect of deletions on AdXET5 promoter activity, with 78.8% and 69.0% activity remaining for deletions P2 and P4. With respect to AdEIL2 and AdEIL3, the first deletion, P1, significantly reduced TF and promoter interaction, as there was only around 70% activity remaining for both AdEIL2 and AdEIL3 (Fig. 6B). The third deletion, P3, also significantly affected interaction of AdEIL2 and the AdXET5 promoter, resulting in only 40.1% and 60.1% of the LUC/Renilla luciferase (REN) ratio of P0 and P2, respectively (Fig. 6B). Nevertheless, except for P1, deletion P2 was also important for AdEIL3, with 49.6% and 71.4% compared with the LUC/REN ratio of P0 and P1 (Fig. 6B).
Figure 6.
AdXET5 promoter deletions. A, Four deletions were designed to remove four EIN3/EIL-related motifs. B, The in vivo interactions between AdEIL2, AdEIL3, AdERF9, and AdXET5 promoters were tested with transient assays in tobacco leaves. SK represents empty vector. Error bars indicate se from at least four replicates.
As there was no GCC box in this promoter region, some unknown effective motif or region of the AdXET5 promoter might be involved in the interaction with AdERF9. AdERF9 constantly suppressed activity of the AdXET5 promoter, regardless of the different deletions (Fig. 6B), which suggests that unknown effective motifs or regions for AdERF9 exist at positions −265 to −1 of the AdXET5 promoter.
Overexpression of AdEIL2 and AdEIL3 in Relation to Ethylene Production in Arabidopsis
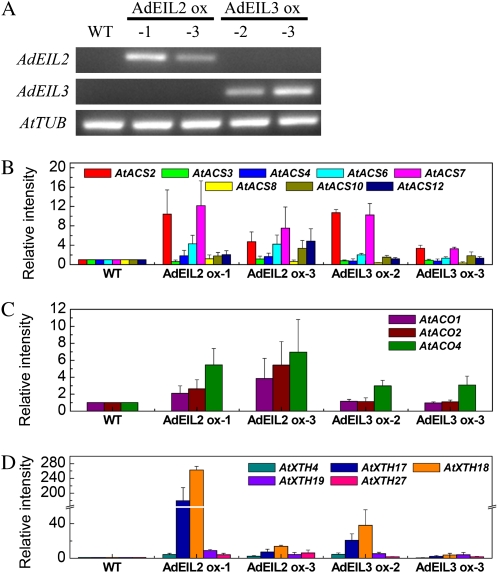
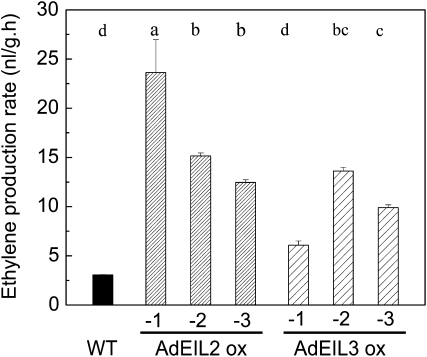
Since AdEIL2 and AdEIL3 can activate the promoter of AdACO1, the full-length sequences of AdEIL2 and AdEIL3 were introduced into Arabidopsis to study the relationship of ethylene production and AdEIL2 and AdEIL3. Transformed Arabidopsis plants were selected by kanamycin resistance and were verified by end-point PCR (Fig. 8). Long-term ethylene treatment was used to stimulate ethylene production of Arabidopsis 15-d-old seedlings. Overexpression of AdEIL2 and AdEIL3 significantly induced ethylene production in Arabidopsis. Generally, AdEIL2-overexpressed plants had higher ethylene production rates than equivalent AdEIL3 plants (Fig. 7). In order to analyze changes in ethylene biosynthesis genes in transformed Arabidopsis, AtACS and AtACO gene family members were chosen and transcript levels analyzed. The gene expression results show that both AtACS and AtACO genes are up-regulated in plants overexpressing AdEIL2 or AdEIL3 (Fig. 8, B and C). In addition, AtXTH genes were also up-regulated in transformed plants, with AtXTH17 and AtXTH18 most strongly affected by the transgene (Fig. 8D).
Figure 8.
Gene expression in transgenic Arabidopsis overexpressing (ox) AdEIL2 or AdEIL3. The Arabidopsis plants were germinated and grown in soil without any additional hormone or stress treatments. A, End-point PCR tests of AdEIL2 and AdEIL3 presence in transgenic Arabidopsis plants. B to D, Real-time PCR analysis changes of gene family members of AtACS (B), AtACO (C), and AtXTH (D) in transgenic EIL plants. Error bars indicate se from three biological replicates. WT, Wild type.
Figure 7.
Different ethylene production rates of wild-type (WT) and transformed (AdEIL) Arabidopsis. Three different independent overexpression (ox) lines were used. Fifteen-day-old plants on 1/2 MS medium were treated with around 80 μL L−1 ethylene for 24 h and then were exposed to ethylene-free air for 4 h. Before ethylene measurement, plants were sealed in air-tight jars for 18 h. Error bars indicate se from three replications. Different letters above the columns represent significant differences (P < 0.05) between different constructs and lines.
DISCUSSION
A model for ethylene signal transduction has been well established, with at least five gene families involved, including a receptor family, CTR1, EIN2, EIN3/EILs, and ERFs (Kendrick and Chang, 2008). While most of the research on this pathway has been done in Arabidopsis, analysis of the pathway has been extended to fruit species, particularly tomato (Barry and Giovannoni, 2007). Within the pathway, EIN3/EILs and ERFs are plant-specific nucleus-localized proteins, serving as TFs that bind conserved motifs in promoter regions of target genes, thus providing a route for activation of the ethylene signal at the level of target gene transcription.
Kiwifruit are generally accepted as being more physiologically sensitive to ethylene than other fruit, such as tomato, apple, or peach (Prunus persica; Wills et al., 2001). Unlike many other fruit, softening and ethylene production are temporally separated, as fruit initially soften, followed by a rise in ethylene production and rapid senescence. Because of this, the species provides a good model for investigating molecular responses to ethylene and fruit softening and to fruit ripening in general.
In previous work, we isolated members of three gene families, including five ethylene receptor genes, two CTR1-like genes, and four EIN3/EILs (Yin et al., 2008, 2009). Our analysis of the Actinidia EST database provided at least 36 nonredundant ERF-related sequences (R. Hellens, unpublished data). The 14 AdERFs cloned, characterized by the conserved ERF domain (Fujimoto et al., 2000; Nakano et al., 2006), were chosen because of their clustering into clades across six subfamilies. However, they were highly diverse in size, ranging from 157 to 382 amino acids, and therefore shared low sequence similarities, with the lowest at 12% and the highest at 66%. In subfamily VIII, AdERF7, AdERF8, and AdERF9 also had an EAR repressor domain, suggesting that these three members might act as transcription repressors (Ohta et al., 2001).
Spatial and Temporal Regulation of AdEIL and AdERF Expression
Among the three AdEILs and 14 AdERFs, only AdERF2 was undetectable in fruit tissue, while the others were widely expressed in various kiwifruit plant tissues. Three AdEILs were constitutively expressed in various tissues and during fruit development, with slightly higher abundance of AdEIL3 and AdEIL4 early in fruit development, as we found in a previous study of AdEIL1 (Yin et al., 2008).
As might be expected from the spread across subfamilies, the AdERFs were differentially expressed in various tissues, with, for example, AdERF1 and AdERF7 more highly expressed in flowers. Such differential expression patterns have also been found in other fruit species (Tournier et al., 2003; Wang et al., 2007) and match the multiple functions of ethylene in higher plants (Bleecker and Kende, 2000). The AdERFs were also differentially expressed during fruit development. Most of them, including AdERF4, AdERF6, AdERF7, AdERF8, AdERF10, and AdERF14, were expressed more strongly in early stages of development, as has been found with ERFs in plum (El-Sharkawy et al., 2009) and in tomato LeERF3b (Chen et al., 2008a), consistent with kiwifruit ethylene receptor and CTR1-like genes (Yin et al., 2008). This agrees with our knowledge of ethylene-dependent activity in the early stages of organ development. Generally, the first 10 weeks of kiwifruit development is a rapid growth stage, with about two-thirds of the fruit volume and weight gained in this period (Beever and Hopkirk, 1990). This was shown with the fruit used in our work, with kiwifruit having mean weights of 61.27 g at 97 DAFB and 87.16 g at 181 DAFB, indicating faster fruit growth before 97 DAFB. These genes appear to be associated with this rapid growth period.
Differential Expression of AdEILs and AdERFs during Fruit Ripening
The ERF gene family is the third biggest TF family in Arabidopsis (Riechmann et al., 2000). Subfamilies III and IV have been associated with the stress response (Novillo et al., 2004; Sakuma et al., 2006; Qin et al., 2008), while subfamilies VII, VIII, and IX contain members that are ethylene responsive (Gu et al., 2000; Tournier et al., 2003; Yang et al., 2005; Oñate-Sánchez et al., 2007; Champion et al., 2009). Subfamily VII genes have been particularly associated with fruit ripening; for example, the tomato LeERF2 (Tournier et al., 2003), apple MdERF1 (Wang et al., 2007), and plum PsERF2a and PsERF2b (El-Sharkawy et al., 2009) genes accumulate during fruit ripening. This supports the hypothesis that there might be a group-specific expression pattern in subfamily VII. However, we found no clearly consistent pattern of expression in the kiwifruit ERFs across the three subfamilies (VII, VIII, and IX). For example, in subfamily VII, AdERF4 and AdERF5 transcripts decreased while AdERF6 accumulated at the late ripening stage. However, as with other fruit, there were four genes, AdERF4, AdERF6, AdERF10, and AdERF14, that generally increased during fruit ripening. AdERF10 and AdERF14 showed a strong association with the late rise in ethylene production, which was confirmed by a reduction in expression with the 1-MCP treatment and the positive response to external ethylene. AdERF4 and AdERF6 also increased during fruit ripening, but not concomitantly with the rise in ethylene, and were markedly stimulated at the later senescence stage. Thus, AdERF10 and AdERF14 might be ripening related, while AdERF4 and AdERF6 are more likely associated with senescence or other fruit processes.
One of the most interesting of our observations was the decrease in expression of some ERF genes with ripening. Most reported fruit ERFs have been shown to be induced with ripening or ethylene treatment, including seven from plum, apple MdERF1, and tomato LeERF1, LeERF2, LeERF4, JERF3, and Pti4 (Thara et al., 1999; Tournier et al., 2003; Huang et al., 2004; Wang et al., 2007; El-Sharkawy et al., 2009). However, AdERF1, AdERF7, AdERF11, and AdERF12 transcripts were slightly higher directly after harvest in control fruit and then declined over ripening. An apparent stimulation of AdERF1 and AdERF7 at 1 DAH in control fruit could be related to trace ethylene accumulation over 24 h in the controls for the ethylene and 1-MCP treatments. The decreasing expression patterns of these genes might not depend on ethylene, confirmed by 1-MCP treatment and ACO knockout fruit. 1-MCP treatment inhibited the reduction in AdERF3 and AdERF13 expression, while the other five were less affected by 1-MCP. Similarly, in ACO knockout fruit, only AdERF3 and AdERF13 were suppressed by long-term ethylene treatment and maintained mRNA levels without ethylene (external and internal ethylene). Therefore, the decline in AdERF3 and AdERF13 is likely to be associated with ethylene, while the decline in AdERF1, AdERF7, AdERF8, AdERF11, and AdERF12, while ripening specific, is independent of ethylene. There is only one other report of such a decline in ERF expression, where tomato LeERF3b transcripts decreased during ripening of wild-type fruit, and in Neverripe mutant and ACO1 cosuppression lines (Chen et al., 2008a). Expression patterns that paralleled the ethylene rise, such as AdERF10 and AdERF14, might only be ethylene associated. These genes might not necessarily be involved in controlling ripening but perhaps in other fruit processes. In contrast, the five ripening-specific ERF genes that decreased could be involved in specific ripening-related processes. A decline in any repressor activity will result in increased gene activation of the TF targets. However, the roles of these five ERF genes need to be further investigated.
AdEIL2 and AdEIL3 Act as Transcriptional Activators by Interaction with Ripening-Related Genes
AdEIL genes showed less response to ethylene and 1-MCP and were constitutively expressed during fruit development and ripening, similar to findings with EIN3/EILs in other plants (Shinshi, 2008). However, the EIN3/EIL gene family is considered to have a role in transcriptional regulation of target genes, such as Arabidopsis ERF1 (Solano et al., 1998) and tobacco basic PR (Hibi et al., 2007). In order to explore the relation between AdEIL genes and kiwifruit ripening-related genes, six such genes were selected for promoter isolation: AdACO1 (ACC oxidase, similar to the gene used in the ACO knockout kiwifruit; Xu et al., 1998), AdXET5 (xyloglucan endotransglycosolase; Schröder et al., 1998; Atkinson et al., 2009), AdEXP1 (Yang et al., 2007), AdPG (the promoter of AdPG was confirmed in tomato; Atkinson and Gardner, 1993), and AdERS1b and AdETR1 (ethylene receptor genes; Yin et al., 2008). All these genes are associated with fruit ripening and softening. As described previously, the PERE and ECBS motifs are two different EIN3-interactive motifs that have been identified in Arabidopsis and tobacco, respectively (Solano et al., 1998; Kosugi and Ohashi, 2000). Despite the constant expression levels of the kiwifruit AdEIL (Fig. 4), these genes were confirmed as TFs, especially as transcriptional activators, inducing activities of the promoters of AdACO1 and AdXET5. A recent publication on melon (Cucumis melo) fruit shows that CmEIL1 and CmEIL2 can activate the CmACO1 promoter in leaf discs (Huang et al., 2010). Such activation of EIL genes from both kiwifruit and melon fruit confirms that this gene family is active in regulating genes associated with ripening, such as those involved in softening and ethylene synthesis.
AdEIL2 and AdEIL3 Triggered Ethylene Production in Overexpressed Arabidopsis
Since both AdEIL2 and AdEIL3 were shown to activate the promoter of AdACO1, the effect of both genes on ethylene biosynthesis was tested. As kiwifruit transformation, through to fruit production, is a long process, AdEIL2 and AdEIL3 were overexpressed in Arabidopsis. Because of low basal levels of ethylene production in Arabidopsis, external ethylene was used to stimulate synthesis, following reports showing that ACC treatment can accelerate Arabidopsis ethylene production rates (Pieterse et al., 1998) and inhibit EIN3 protein degradation (Potuschak et al., 2003). The external ethylene treatment increased plant ethylene production. Moreover, gene expression results using transformed Arabidopsis showed that AdEIL2 and AdEIL3 were not only involved in the regulation of ACO gene expression but also up-regulated ACS genes. The increased ethylene production in transgenic Arabidopsis and transcriptional regulation of ethylene biosynthesis genes confirms the suggestion that AdEIL genes function within the signal transduction pathway connecting ethylene signaling and ripening processes.
Nonredundant Roles of Different Motifs in TF and Promoter Interaction
Although AdEIL2 and AdEIL3 modulated AdACO1 and AdXET5 promoters and ethylene production in Arabidopsis, there were also four promoters that did not respond. Furthermore, another 15 promoters, selected from a resource created by Dare et al. (2008), containing PERE and/or ECBS motifs, were also tested using the transient assay, but none could be induced by AdEIL2 or AdEIL3 (data not shown). These results suggest that the motifs are not equal in the interaction between TFs and promoters, both between and within promoters. AdXET5 promoter deletions indicated different functions in relation to the AdEILs; for AdEIL2, the first and third PERE motifs were important, while for AdEIL3, the first PERE and ECBS motifs were more active (Fig. 6).
AdERF9 Acts as a Transcriptional Repressor, and the Possible Non-GCC Motif Is Located Near the Start Code Region
In the above six kiwifruit promoters, only AdEXP1 had a GCC box, which is one of the targets for ERF genes. However, there was no interaction between AdERFs and AdEXP1. Surprisingly, AdERF9 significantly suppressed activity of the AdXET5 promoter, which does not have a GCC or DRE box. This result might be explained by recognizing that AdERF9 is a repressor TF. In tomato, Pti4, an ERF-like TF, could regulate defense-related gene expression via GCC and non-GCC boxes (Chakravarthy et al., 2003), suggesting that there are potential unknown motifs in relation to ERFs. In order to identify unknown motifs in the AdXET5 promoter, the four deletions, based on PERE and ECBS motifs, were also applied to AdERF9. The transient assay results confirmed the suppression of AdERF9 on the AdXET5 promoter. The possible effective motif for AdERF9 is probably in the −265 to −1 region of the AdXET5 promoter, but the specific region needs to be further investigated. Alternatively, GCG boxes may exist farther upstream from the transcriptional start site in the AdXET5 promoter.
CONCLUSION
Four EIN3-like genes and 14 ERF genes isolated from kiwifruit encoded two separate levels of the ethylene signaling transduction pathway and have been shown to function as TFs; AdEIL2, AdEIL3, and AdERF9 could interact with promoters of several ripening-related genes. Expression studies of these genes revealed different behavior patterns in various tissues and during fruit development and ripening. In particular, five AdERF genes had reduced expression with the progression of fruit ripening, confirmed by 1-MCP treatment. These may be more important in fruit ripening than other ethylene-associated AdERFs. Transient assays of AdEIL2 and AdEIL3 verified that these genes were transcriptional activators in vivo, while AdERF9 acted as a transcriptional repressor. Although AdEIL genes were constitutively expressed during fruit ripening, we could show that they up-regulated promoters of AdACO1 and AdXET5 and also up-regulated AtACS and AtACO gene expression, thus potentially triggering ethylene production in AdEIL2- and AdEIL3-overexpressed Arabidopsis. In contrast, AdERF9 suppressed the AdXET5 promoter, although no GCC box was identified in this promoter. AdERF9 constantly suppressed the activity of the AdXET5 promoter, with the region from −256 to −1 being important for this interaction. Future research will focus on cis-elements for these TFs and the broader function of ERFs in kiwifruit responses. This research has taken us a step farther in understanding the nature of transcriptional regulation involved in the later stages of the ethylene signal transduction pathway in ripening fruit.
MATERIALS AND METHODS
Plant Material
To allow analysis of tissue-specific gene expression, different kiwifruit (Actinidia deliciosa var deliciosa ‘Hayward’) plant tissues, including stem, leaf, and whole flowers, were collected from the Plant and Food Research Orchard, Te Puke, New Zealand, over the 2006 to 2007 growing season.
For a fruit development series, Hayward kiwifruit at different developmental stages were picked from the same site. Full blossom was on November 15, 2006, and there were six subsequent sampling times: 20, 57, 97, 124, 159, and 181 DAFB (Yin et al., 2008).
To test postharvest treatments, kiwifruit with a mean soluble solids concentration (SSC) of 6.7% (commercial maturity) were harvested from a commercial orchard in Patumahoe, South Auckland. Three postharvest treatments, ethylene treatment (100 μL L−1, 24 h), 1-MCP treatment (0.5 μL L−1, 24 h; SmartFresh), and a control treatment (air), were performed as described previously (Yin et al., 2008). After treatment, fruit were held at 20°C until reaching a firmness of less than 2 n and development of the ethylene climacteric, although the latter did no occur in 1-MCP-treated fruit. At each sampling time, ethylene production, fruit firmness, and SSC were recorded, as described by Yin et al. (2008). For ethylene production, eight replicates (two fruit for each replicate) were performed, and 10 replicates (one fruit for each replicate) were performed for fruit firmness and SSC. After measurement, fruit flesh (without skin, seeds, or core) was bulked for the 10 fruit, frozen in liquid nitrogen, and stored at −80°C for further use.
RNA Extraction and cDNA Synthesis
Total RNA was isolated from the frozen tissues following the protocol described by Chang et al. (1993b). Approximately 1.8 or 0.5 g of tissue was used for fruit flesh or vegetative and flower tissues, respectively. Total RNA was treated with TURBO DNase (Ambion) to remove contaminating DNA. After DNase treatment, 2.5 μg of RNA was used for cDNA synthesis with the SuperScript III First Strand Synthesis Kit (Invitrogen) following the manufacturer's protocol. In order to reduce the between-fruit or between-tissue variability, for each time point, three batches of RNA were isolated as three biological repeats for separate cDNA synthesis.
Gene Isolation
Fourteen ERF genes were isolated from the Plant and Food Research EST database of Actinidia species using the BLAST algorithm (Crowhurst et al., 2008). Five of the 14 ERF genes were full-length EST sequences in the database, with complete start and stop codes, while full-length sequences of the other nine ERF genes were obtained by RACE with the SMART RACE cDNA Amplification Kit (Clontech). The details of the primers used for RACE are described in Supplemental Table S1. Full-length proofreadings were performed by the primers described in Supplemental Table S2 and by PCR product resequencing confirmation. Four EIN3-like genes, AdEIL1 to AdEIL4, were also isolated from the EST database, and these have been described in our previous publications (Yin et al., 2008, 2009). Alignments were carried out on Vector NTI (version 9.0.0 [Invitrogen], using ClustalW), and a phylogenetic tree was generated with MEGA version 3.1 (Kumar et al., 2004).
Real-Time PCR
For real-time PCR, oligonucleotide primers were designed according to each gene's untranslated region with Primer3 (version 0.4.0; http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The sizes of all PCR products ranged from 118 to 196 bp. All primers were tested with melting peaks and dissociation curves to confirm that there was only one product for each pair of primers. In order to verify that primers were specific amplifications of target genes, all PCR products were purified and resequenced. The sequences of all primers used for real-time PCR analysis are described in Supplemental Table S3.
The PCR mixture (10-μL total volume) comprised 5 μL of 2× LightCycler 480 SYBR Green I Master Mix (Roche), 1 μL of each primer (5 μm), and 3 μL of diluted cDNA. PCR was performed on a LightCycler 480 instrument (Roche), initiated by 5 min at 95°C, then followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s, and completed with a melting curve analysis program. No-template controls and melting curve analyses were included in every reaction. Kiwifruit actin was used as the housekeeping gene to quantify cDNA abundance.
Mature fruit sampled on the harvest day for the ripening fruit series and fruit sampled 181 DAFB for the fruit development series were set as calibrators (set to 1) for calculating relative expression levels. For the tissue-specific experiment, expression in the stem tissue was used as the calibrator. Three separate biological replicates were performed for relative intensities. All relative intensities were calculated from standard curves for each gene.
Genome Walking
Genomic DNA was extracted from kiwifruit leaves using the DNeasy Plant Mini Kit (Qiagen). Genomic DNA, 2 μg for each, was digested with eight restriction enzymes (DraI, Ecl136II, EcoRV, HpaI, MscI, StuI, SspI, and ScaI) separately. After enzyme digestion, all products were cleaned up using a DNA Clean and Concentration Kit (ZYMO Research) and then were ligated to an adaptor (Clontech).
Six known genes were chosen for promoter work: two ethylene receptor genes (AdETR1 and AdERS1b; Yin et al., 2008), one expansin-like gene (AdEXP1; Yang et al., 2007), one xyloglucan endotransglycosylase gene (AdXET5; Schröder et al., 1998), one polygalacturonase gene (AdPG; Atkinson and Gardner, 1993), and one ACC oxidase gene (AdACO1; Xu et al., 1998). Among these six genes, the AdPG promoter was cloned by Atkinson and Gardner (1993) and the other five 5′ upstream genomic sequences were generated using a GenomeWalker kit (Clontech) with nested PCR. The nested PCR analysis was performed with two sets of primers, including two adaptor primers that were obtained from the kit and two gene-specific primers that were designed by Primer3, as described in Supplemental Table S4.
Conserved cis-element motifs of promoters were predicted using PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and Plant-CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) databases.
Promoter Modification
In order to test the interaction between AdEILs and motifs, promoter deletions were designed. Four deletions of the AdXET5 promoter covered four EIN3/EIL-related motifs, including three PERE motifs and one ECBS motif. Promoter deletions were conducted with PCR using the primers described in Supplemental Table S5.
Transient Assay
A transient assay, also known as a dual luciferase assay, was performed with tobacco (Nicotiana benthamiana) to study the interaction of TFs and promoters, which has been widely used in our laboratory (Hellens et al., 2005; Espley et al., 2007; Palapol et al., 2009). Two pGreenII vectors were used for the transient assay, pGreenII 0800-LUC vector and pGreenII 0029 62-SK vector, and all related information on the vectors can be found in Hellens et al. (2005). For promoters, primers (Supplemental Table S4) were designed according to the sequences obtained by genome walking, and a NcoI site was introduced at the 3′ end of each promoter. PCR products were cloned into pGEM-T Easy vector (Promega) and resequenced to confirm the sequence. Promoters were then cut from the pGEM-T Easy vector and inserted into a pGreenII 0800-LUC vector at the 5′ end of a LUC gene. As the AdPG promoter region also contains a NcoI site, ApaI and SpeI were used to digest the vector, so there existed a 55-bp pGEM-T Easy vector sequence between the AdPG promoter and LUC. The interaction between TFs and promoters was indicated by luminescence activity. pGreenII 0800-LUC vector carried a REN gene under the control of the 35S promoter as a positive control. The ratios of LUC and REN were expressed as activation or repression of the promoters by the TFs. Full-length cDNAs of the TFs (AdEILs and AdERFs) were generated by primers (described in Supplemental Table S2) and were cloned into pGreenII 0029 62-SK.
All constructs were electroporated into Agrobacterium tumefaciens GV3101(MP90). The bacterial strains were stored as glycerol stock at −80°C. Agrobacterium was streaked on a Luria-Bertani plate for 2 d from glycerol stock and then was restreaked on a new Luria-Bertani plate for 1 d. Agrobacterium cultures were prepared with infiltration buffer (10 mm MgCl2 and 0.5 μm acetosyringone) to an optical density at 600 nm from 0.7 to 1.0. Agrobacterium culture mixtures of TFs (1 mL) and promoters (100 μL) were infiltrated into tobacco leaves by needleless syringes. Tobacco plants were grown in a glasshouse, under natural light with daylight extension to 16 h. Three days after infiltration, LUC and REN were assayed using the dual luciferase assay reagents (Promega). The analysis was carried out using an Orion Microplate Luminometer (Berthold Detection System) according to the method described by Palapol et al. (2009), with at least triplicate transient assay measurements for each plant.
Transformation of Arabidopsis
The constructs with full-length genes and pGreenII 0029 62-SK, as used in the transient assay experiment, were used for transformation in Arabidopsis (Arabidopsis thaliana; overexpression). All constructs were electroporated into Agrobacterium GV3101 (MP90) and then introduced into Arabidopsis (cv Columbia) by floral dip (T1).
T1 seeds were germinated on half-strength Murashige and Skoog (1/2 MS) medium and were selected with kanamycin resistance. Then, 2- to 3-week-old kanamycin-resistant plants were transferred to soil and grown in a greenhouse under general conditions (T2). Transgene expression was verified by end-point PCR, and the cycles were limited to 35. Changes of transcript abundance of select target genes were studied with real-time PCR using a Bio-Rad C1000 thermal cycler. The methods for end-point PCR and real-time PCR were as described by Yin et al. (2008). The sequences of all primers used for Arabidopsis analysis are described in Supplemental Table S6.
Ethylene Production of Transformed Arabidopsis
Fifteen-day-old plants (wild type, AdEIL2 overexpressed, and AdEIL3 overexpressed; T3, generated from T2 seeds) on 1/2 MS medium were treated with 80 μL L−1 ethylene for 24 h and then were exposed to ethylene-free air for 4 h. Before ethylene measurement, plants were sealed in air-tight jars (0.5 L) for 18 h at 23°C. Head-space gas (1 mL) was removed from each jar using a syringe, and ethylene was measured by gas chromatography using a gas chromatograph (Hewlett-Packard 5890 series II) fitted with a flame ionization detector (Philips PU4500; Unicam) and an alumina F1 column (1.5 m × 6 mm). The injector, detector, and oven temperatures were 160°C, 200°C, and 130°C, respectively.
The amount of plant material was determined by the weight of aboveground parts (stems and leaves) without roots. Three technical replicates were performed for ethylene production measurement. The statistical analysis was performed with DPS software (version 3.11).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ869852 to GQ869865.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Motif analysis of ripening-related gene promoters.
Supplemental Figure S2. Tissue specificity expression patterns during fruit development.
Supplemental Figure S3. Temporal expression patterns during fruit development.
Supplemental Figure S4. Expression of AdEIL and AdERF genes in kiwifruit flesh during fruit ripening at 20°C.
Supplemental Figure S5. Decreasing expression of AdERF1, AdERF3, AdERF7, AdERF8, AdERF11, AdERF12, and AdERF13 during kiwifruit ripening.
Supplemental Figure S6. Study of the relation between decreasing expression of AdERFs and ethylene by ACO knockout kiwifruit.
Supplemental Table S1. Primers for RACE.
Supplemental Table S2. Primers for full-length amplification.
Supplemental Table S3. Primers for real-time PCR.
Supplemental Table S4. Primer sequences (5′–3′) for promoter isolation.
Supplemental Table S5. Primers for promoter modification.
Supplemental Table S6. Primers for transformed Arabidopsis examination.
Supplemental Table S7. Similarity of AdERFs based on deduced amino acids.
Supplementary Material
Acknowledgments
We thank Dr. N. Lallu and Dr. J. Burdon (Plant and Food Research) for advice on postharvest experiments; Dr. R.J. Schaffer and Dr. R.G. Atkinson (Plant and Food Research) for supplying ACO knockout kiwifruit cDNA for real-time PCR; S. Dejnoprat, K. Lin-Wang, R.M. Wu, and A. Dare (Plant and Food Research) for technical support with the tobacco transient experiments and Arabidopsis transformation; and Dr. B. Zhang and P. Wang (Zhejiang University) for assistance with gene RACE. We also acknowledge the laboratory and greenhouse staff at Plant and Food Research for their help and guidance. The China Scholar Council supported X.-r.Y. with living expenses for his overseas study.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Gardner RC. (1993) A polygalacturonase gene from kiwifruit (Actinidia deliciosa). Plant Physiol 103: 669–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Johnston SL, Yauk YK, Sharma NN, Schröder R. (2009) Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol Technol 51: 149–157 [Google Scholar]
- Barry CS, Giovannoni JJ. (2007) Ethylene and fruit ripening. J Plant Growth Regul 26: 143–159 [Google Scholar]
- Beever DJ, Hopkirk G. (1990) Fruit development and fruit physiology. Warrington IJ, Weston GC, , Kiwifruit: Science and Management. New Zealand Society for Horticultural Science, Auckland, pp 97–126 [Google Scholar]
- Bleecker AB, Kende H. (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D'Ascenzo MD, Fobert PR, Després C, Martin GB. (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15: 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion A, Hebrard E, Parra B, Bournaud C, Marmey P, Tranchant C, Nicole M. (2009) Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas. Mol Plant Pathol 10: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993b) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chao QM, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen GP, Hu ZL, Grierson D. (2008a) Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J Plant Physiol 165: 662–670 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang ZY, Liang HX, Liu HX, Du LP, Xu HJ, Xin ZY. (2008b) Overexpression of TiERF1 enhances resistance to sharp eyespot in transgenic wheat. J Exp Bot 59: 4195–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, Ampomah-Dwamena C, Atkinson RG, Beuning LL, Bulley SM, Chagne D, Marsh KB, Matich AJ, et al. (2008) Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics 9: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dare AP, Schaffer RJ, Lin-Wang K, Allan AC, Hellens RP. (2008) Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I, Sherif S, Mila I, Bouzayen M, Jayasankar S. (2009) Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J Exp Bot 60: 907–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. (2007) Red coloration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. (2000) Arabidopsis ethylene-response element binding factors act as transcriptional activator or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang CM, Thara VK, Zhou JM, Martin GB. (2000) Pti4 is induced by ethylene and salicylic acid, and its production is phosphorylated by the Pto kinase. Plant Cell 12: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HW, Ecker JR. (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Hao DY, Yamasaki K, Sarai A, Ohme-Takagi M. (2002) Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41: 4202–4208 [DOI] [PubMed] [Google Scholar]
- Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, Karunairetnam S, Gleave A, Laing W. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi T, Kosugi S, Iwai T, Kawata M, Seo S, Mitsuhara I, Ohashi Y. (2007) Involvement of EIN3 homologues in basic PR gene expression and flower development in tobacco plants. J Exp Bot 58: 3671–3678 [DOI] [PubMed] [Google Scholar]
- Huang SZ, Sawaki T, Takahashi A, Mizuno S, Takezawa K, Matsumura A, Yokotsuka M, Hirasawa Y, Sonoda M, Nakagawa H, et al. (2010) Melon EIN3-like transcription factors (CmEIL1 and CmEIL2) are positive regulators of an ethylene- and ripening-induced 1-aminocyclopropane-1-carboxylic acid oxidase gene (CM-ACO1). Plant Sci 178: 251–257 [Google Scholar]
- Huang ZJ, Zhang ZJ, Zhang XL, Zhang HB, Huang DF, Huang RF. (2004) Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett 573: 110–116 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P. (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick MD, Chang C. (2008) Ethylene signalling: new levels of complexity and regulation. Curr Opin Plant Biol 11: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res 28: 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim WT. (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132: 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Zhu BZ, Xu WT, Zhu HL, Chen AJ, Xie YH, Shao Y, Luo YB. (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Mao CZ, Wang SM, Jia QJ, Wu P. (2006) OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Mol Biol 61: 141–152 [DOI] [PubMed] [Google Scholar]
- Mbéguié-A-Mbéguié D, Hubert O, Fils-Lycaon B, Chillet M, Baurens FC. (2008) EIN3-like gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Grande Naine). Physiol Plant 133: 435–448 [DOI] [PubMed] [Google Scholar]
- McDonald B, Harman JE. (1982) Controlled-atmosphere storage of kiwifruit. I. Effect on fruit firmness and storage life. Sci Hortic (Amsterdam) 17: 113–123 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohem-Takagi M, Shinshi H. (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Anderson JP, Young J, Singh KB. (2007) AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol 143: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palapol Y, Ketsa S, Lin-Wang K, Ferguson IB, Allan AC. (2009) A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen (Garcinia mangostana L.) fruit during ripening. Planta 229: 1323–1334 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LSP, Maruyma K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono KI, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligase and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rieu I, Mariani C, Weterings K. (2003) Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J Exp Bot 54: 2239–2244 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder R, Atkinson RG, Langenkämper G, Redgwell RJ. (1998) Biochemical and molecular characterization of xyloglucan endotransglycosylase from ripe kiwifruit. Planta 204: 242–251 [DOI] [PubMed] [Google Scholar]
- Shinshi H. (2008) Ethylene-regulated transcription and crosstalk with jasmonic acid. Plant Sci 175: 18–23 [Google Scholar]
- Solano R, Stepanova A, Chao QM, Ecker JR. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Yu JP, Chen F, Zhao TJ, Fang XH, Li YQ, Sui SF. (2008) TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connection the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J Biol Chem 283: 6261–6271 [DOI] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, et al. (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105: 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thara VK, Tang XY, Gu YQ, Martin GB, Zhou JM. (1999) Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J 20: 475–483 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ. (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26: 47–58 [DOI] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech JC, Bouzayen M. (2003) New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett 550: 149–154 [DOI] [PubMed] [Google Scholar]
- Trujillo LE, Sotolongo M, Menéndez C, Ochogavía ME, Coll Y, Hernández I, Borrás-Hidalgo O, Thomma BPHJ, Vera P, Hernández L. (2008) SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol 49: 512–525 [DOI] [PubMed] [Google Scholar]
- Wang A, Tan DM, Takahashi A, Li TZ, Harada T. (2007) MdERFs, two ethylene-response factors involved in apple fruit ripening. J Exp Bot 58: 3743–3748 [DOI] [PubMed] [Google Scholar]
- Whittaker DJ, Smith GS, Gardner RC. (1997) Expression of ethylene biosynthetic genes in Actinidia chinensis fruit. Plant Mol Biol 34: 45–55 [DOI] [PubMed] [Google Scholar]
- Wills RBH, Warton MA, Mussa DMDN, Chew LP. (2001) Ripening of climacteric fruits initiated at low ethylene levels. Aust J Exp Agric 41: 89–92 [Google Scholar]
- Wu KQ, Tian LN, Hollingworth J, Brown DCW, Miki B. (2002) Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol 128: 30–37 [PMC free article] [PubMed] [Google Scholar]
- Xu ZC, Ikoma Y, Yano M, Ogawa K, Hyodo H. (1998) Varietal differences in the potential to produce ethylene and gene expression of ACC synthase and ACC oxidase between ‘Kui mi’ and ‘Hong xin’ of Chinese kiwifruit. J Jpn Soc Hortic Sci 67: 204–209 [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J. (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yang SL, Xu CJ, Zhang B, Li X, Chen KS. (2007) Involvement of both subgroups A and B of expansin genes in kiwifruit ripening. HortScience 42: 315–319 [Google Scholar]
- Yang Z, Tian LN, Latoszek-Green M, Brown D, Wu KQ. (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Zhang B, Wu RM, Burdon J, Wang P, Ferguson IB, Chen KS. (2009) Ethylene-related genes show a differential response to low temperature during ‘Hayward’ kiwifruit ripening. Postharvest Biol Technol 52: 9–15 [Google Scholar]
- Yin XR, Chen KS, Allan AC, Wu RM, Zhang B, Lallu N, Ferguson IB. (2008) Ethylene-induced modulation of genes associated with the ethylene signaling pathway in ripening kiwifruit. J Exp Bot 59: 2097–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Tamura S, Nakano R, Inaba A, Kubo Y. (2003) Characterization of a novel tomato EIN3-like gene (LeEIL4). J Exp Bot 54: 2775–2776 [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen K, Bowen J, Allan A, Espley R, Karunairetnam S, Ferguson I. (2006) Differential expression within the LOX gene family in ripening kiwifruit. J Exp Bot 57: 3825–3836 [DOI] [PubMed] [Google Scholar]
- Zhang B, Yin XR, Li X, Yang SL, Ferguson IB, Chen KS. (2009a) Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production. J Agric Food Chem 57: 2875–2881 [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Zhang HW, Quan RD, Wang XC, Huang RF. (2009b) Transcriptional regulation of ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol 150: 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.