Abstract
Phosphatidylinositol phosphate kinase (PIPK) is an enzyme involved in the regulation of cellular levels of phosphoinositides involved in various physiological processes, such as cytoskeletal organization, ion channel activation, and vesicle trafficking. In animals, research has focused on the modes of activation and function of PIPKs, providing an understanding of the importance of plasma membrane localization. However, it still remains unclear how this issue is regulated in plant PIPKs. Here, we demonstrate that the carboxyl-terminal catalytic domain, which contains the activation loop, is sufficient for plasma membrane localization of PpPIPK1, a type I/II B PIPK from the moss Physcomitrella patens. The importance of the carboxyl-terminal catalytic domain for plasma membrane localization was confirmed with Arabidopsis (Arabidopsis thaliana) AtPIP5K1. Our findings, in which substitution of a conserved dibasic amino acid pair in the activation loop of PpPIPK1 completely prevented plasma membrane targeting and abolished enzymatic activity, demonstrate its critical role in these processes. Placing our results in the context of studies of eukaryotic PIPKs led us to conclude that the function of the dibasic amino acid pair in the activation loop in type I/II PIPKs is plant specific.
Phosphoinositides (PIs) are minor lipids found in membrane fractions but implicated in a wide variety of physiological regulations in eukaryotes (Di Paolo and De Camilli, 2006; Zonia and Munnik, 2006). Phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] is a major PI in animal plasma membranes, affecting the localization and activity of various kinds of proteins carrying phosphatidylinositol-binding domains, which in turn affect the regulation of cytoskeletal organization, vesicle trafficking, cell proliferation, and cell growth during development and stress responses (Doughman et al., 2003; Downes et al., 2005; Di Paolo and De Camilli, 2006; Zonia and Munnik, 2006; Heck et al., 2007). In addition, PtdIns(4,5)P2 is also a well-known substrate of phospholipase C, producing second messengers such as diacylglycerol, phosphatidic acid (PA), and inositol-1,4,5-trisphosphate, which are involved in the activation of intracellular signal transduction pathways (Zonia and Munnik, 2006). Transient accumulation of PtdIns(4,5)P2 has also been observed under various kinds of environmental stress (Pical et al., 1999; DeWald et al., 2001), suggesting an important role of this lipid in the regulation of stress signal transduction pathways also in plants. These findings indicate that PtdIns(4,5)P2 is multifunctional and involved in a variety of cellular processes. Therefore, elucidation of the mechanisms controlling the cellular levels of PtdIns(4,5)P2 is important in understanding the significance of PI signaling in eukaryotes.
PtdIns(4,5)P2 is synthesized by phosphatidylinositol phosphate kinases (PIPKs; Anderson et al., 1999; Doughman et al., 2003; Heck et al., 2007). Physiological roles of several plant PIPKs have been reported. In Arabidopsis (Arabidopsis thaliana), AtPIP5K3 is an essential regulator of tip growth of root hairs (Kusano et al., 2008; Stenzel et al., 2008), while AtPIPK4 and AtPIPK5 are essential for pollen germination and pollen tube elongation (Ischebeck et al., 2008; Sousa et al., 2008). In addition, AtPIP5K9 was shown to interact with the cytosolic invertase CINV1 to regulate sugar-mediated root cell elongation negatively (Lou et al., 2007). Rice (Oryza sativa) OsPIPK1 is proposed to be involved in shoot growth and floral initiation through the regulation of floral induction genes (Ma et al., 2004). In animals, membrane-associated type I PIPK mainly phosphorylates the D-5 hydroxyl group of PtdIns4P to produce PtdIns(4,5)P2 but also produces PtdIns(3,4)P2 and PtdIns(3,5)P2 from PtdIns3P with 5- and 4-kinase activity (Anderson et al., 1999; Heck et al., 2007), whereas type II PIPK prefers the D-4 position of PtdIns5P, producing PtdIns(4,5)P2 in the nucleus and at the endoplasmic reticulum (Clarke et al., 2007). Thus, in animals, type I and II PIPKs are involved in the generation of PtdIns(4,5)P2 via different pathways. Molecular biological analysis of plant PIPKs was initiated with AtPIP5K1 from Arabidopsis (Mikami et al., 1998), which phosphorylates PtdIns3P, PtdIns4P, and PtdIns(4,5)P2 to produce PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns (3,4,5)P3, respectively, with D-4- and D-5-kinase activity (Elge et al., 2001; Westergren et al., 2001; Im et al., 2007). Similar enzymatic activity was also reported for other PIPKs from Arabidopsis (Ischebeck et al., 2008; Kusano et al., 2008; Stenzel et al., 2008). In addition, a PIPK from the moss Physcomitrella patens (designated as PpPIPK1) preferred PtdIns4P, PtdIns3P, and PtdIns(3,4)P2 as substrates, but not PtdIns5P, producing PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,4,5)P3, respectively (Saavedra et al., 2009). These findings indicate that the substrate specificity of plant PIPKs is essentially the same as that of type I PIPKs. However, AtPIP5K1 has yet to be classified as either type I or type II based on sequence comparisons of the catalytic domain (CD; Mikami et al., 1998). This was confirmed by a genome-wide analysis of PIPK genes in Arabidopsis in which all 11 PIPKs were classified as type I/II based on sequence comparisons of the CDs, which were further subdivided into subtypes A and B (Mueller-Roeber and Pical, 2002). Therefore, it is suggested that typical type I and II PIPKs are absent in plants, although further confirmation is needed.
The conserved PIPK CD contains a short highly conserved region near its C-terminal end, designated the activation loop, which acts as the substrate-binding site and is responsible for the differences in substrate specificity and subcellular localization between animal type I and type II PIPKs (Kunz et al., 2000, 2002). Substrate specificities of animal type I and type II PIPKs, for example, are determined by a respective Glu and Ala at the corresponding positions in the activation loop. Moreover, it has been established that substitution of Glu to Ala results in a swap of substrate specificity and subcellular localization between the two types (Kunz et al., 2000, 2002). In contrast to animal PIPKs, a substitution in the activation loop of PpPIPK1 from Glu to Ala resulted in a nearly complete loss of type I/II activity; however, such a mutation did not fully convert the substrate specificity, although an enhancement of type II versus type I activity was observed (Saavedra et al., 2009). Since the corresponding amino acid residue is Glu in all plant PIPKs so far reported, it is suggested that there also is a plant-specific mode of substrate specificity regulation in plant type I/II PIPKs. However, enzymatic activity appears to be modified in similar ways between plant type I/II and animal type I PIPKs; that is, phosphorylation- and PA-dependent activation of PIPKs has been observed in both animals and plants (Moritz et al., 1992; Jenkins et al., 1994; Pical et al., 1999; Westergren et al., 2001; Perera et al., 2005; Saavedra et al., 2009).
The regulation of plasma membrane localization of mammalian type I PIPKs remains confusing. In addition to the involvement of a Glu residue as mentioned above, the substitution of two Lys residues in the activation loop to Asn residues changes the subcellular localization from the plasma membrane to the cytosol (Kunz et al., 2000, 2002). However, Arioka et al. (2004) also showed that the plasma membrane localization of type I PIPKs is regulated by another basic amino acid pair localized downstream of the activation loop in the CD, which is not found in type II PIPKs. Interestingly, the mechanism behind plasma membrane localization of plant PIPKs seems to differ significantly from the animal one. The obvious structural feature of plant PIPKs is the presence of a repetition of membrane occupation and recognition nexus (MORN) motifs at the N-terminal half, which is conserved across the B subfamily of plant type I/II PIPKs (Mueller-Roeber and Pical, 2002). The MORN motif was first identified in mammalian junctophilin, an endoplasmic reticulum-membrane-bound component of the junctional complex between the plasma membrane and the endoplasmic reticulum (Takeshima et al., 2000). Since MORN motifs are not found in PIPKs from nonplant organisms, a plant-specific mode of PIPK activation is speculated. Indeed, a regulatory role of the MORN domain was reported in the enzymatic activation of AtPIP5K1 (Im et al., 2007) and in root hair formation, but not in enzymatic activation, of AtPIP5K3 (Stenzel et al., 2008). Moreover, the MORN domain may play a role in the plasma membrane localization of OsPIPK1 from rice and AtPIP5K1 and AtPIP5K3 from Arabidopsis (Ma et al., 2006; Im et al., 2007; Kusano et al., 2008). However, stable transformation of tobacco (Nicotiana tabacum) cells to express an AtPIP5K1 MORN domain-GFP fusion did not allow visualization of the plasma membrane localization of this protein (Im et al., 2007). Thus, it is not clear if the MORN domain functions as a plasma membrane-targeting module.
Given the sequence conservation of the CD among eukaryotic PIPKs (Saavedra et al., 2009), we hypothesize that the CD is responsible for the plasma membrane localization of plant PIPKs. Thus, to gain further insight into the mechanisms regulating this issue, we dissected PpPIPK1 to determine the molecular determinants of plasma membrane localization. Here, we show that the MORN domain is not involved in the plasma membrane localization of PpPIPK1 and AtPIP5K1 in P. patens protoplasts and onion (Allium cepa) epidermal cells. We further demonstrate that two basic amino acids, but not Glu, conserved in the activation loop of the CD are required for plasma membrane localization. These findings demonstrate that the activation mode of type I/II PIPKs is plant specific and differs from that of the membrane-localized animal type I PIPKs.
RESULTS
Contribution of the CD in Plasma Membrane Localization of PpPIPK1
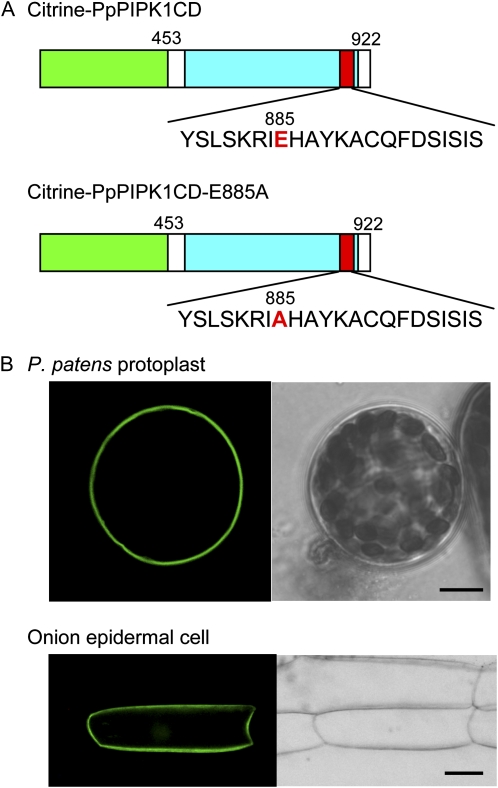
Subtype B plant PIPK consists of an N-terminal MORN domain (MD) and a C-terminal CD (Fig. 1), of which the MD is thought to be a plasma membrane-localizing module (Ma et al., 2006; Im et al., 2007; Kusano et al., 2008). Our demonstration that the two moss PIPKs, GFP-PpPIPK1 and GFP-PpPIPK2, localized to plasma membranes (Saavedra et al., 2009) raised the obvious question of whether the MD truly regulates plasma membrane localization of plant PIPKs. We addressed this issue by fluorescent microscopic analysis of P. patens protoplasts in which expression of Citrine-PpPIPK1 fusion proteins (Fig. 2A) was regulated under the direction of the cauliflower mosaic virus 35S promoter. Consistent with our previous results, plasma membrane localization of Citrine-tagged PpPIPK1 was confirmed (Fig. 2B, top), indicating that this system can be successfully used in our study.
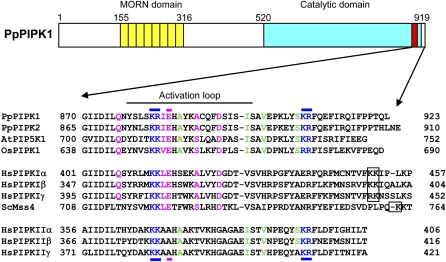
Figure 1.
Comparison of the activation loop in plant and animal PIPKs. Structural characteristics of PpPIPK1 are represented schematically to illustrate the presence of a repetition of eight MORN motifs (yellow boxes) and the conserved CD (blue box). The activation loop in the CD is shown by a red box. Numbers indicate amino acid positions. The bottom shows an amino acid sequence alignment of regions around the activation loop of PpPIPK1 (accession no. AB453863), PpPIPK2 (accession no. FJ024081), AtPIP5K1 (accession no. NP_173617), OsPIPK1 (accession no. CAD27794), Mss4 from Saccharomyces cerevisiae (accession no. NP_010494), and type I and type II isoforms of human PIPKs (type Iα, accession no. NP_001129110; type Iβ, accession no. NP_003549; type Iγ, accession no. NP_036530; type IIα, accession no. NP_005019; type IIβ, accession no. NP_003550; type IIγ, accession no. NP_079055). Dibasic amino acid pairs found in and downstream of the activation loop are shown in blue, and dibasic amino acid pairs farther downstream in type I PIPKs are indicated with a black frame. Positions of dibasic amino acid pairs (blue) and residues conferring substrate specificity (pink) are also indicated by bars above and below, respectively. Identical amino acid residues between plant and animal type I PIPKs are indicated in pink, while identical residues between plant and animal type II PIPKs are in green. The numbers to the left and right of the sequences indicate amino acid positions.
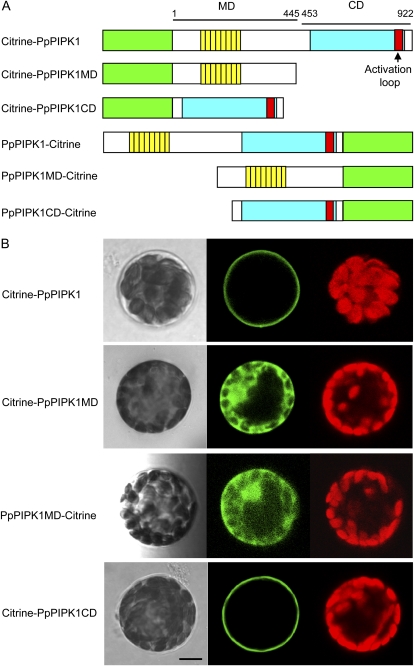
Figure 2.
Importance of the CD for plasma membrane localization of PpPIPK1. A, Recombinant Citrine fusion proteins whose intracellular localization was determined by transient expression in P. patens protoplasts. The green box indicates Citrine, and yellow, blue, and red boxes are the same as in Figure 1. The MD and CD of PpPIPK1 are indicated by bars with numbers of amino acid positions. B, Subcellular localization of Citrine fusion proteins. Confocal images of P. patens protoplasts transformed with expression plasmids for Citrine fusion proteins are shown. Left, center, and right panels show bright-field images, Citrine fluorescence, and autofluorescence of chloroplasts, respectively. Bar = 10 μm.
To further address the above-mentioned question, we first bisected PpPIPK1 and constructed plasmids for the expression of fusion proteins as shown in Figure 2A. The Citrine-tagged proteins were expressed in protoplasts of P. patens to compare their behavior. The living cell image analysis revealed that the CD of PpPIPK1 (amino acids 453–922) fused with Citrine appeared to predominantly localize to plasma membranes, whereas the MD of PpPIPK1 (amino acids 1–445) fused with Citrine remained diffusely spread within the cytoplasm (Fig. 2B), although fluorescent signals from PpPIPK1-Citrine and PpPIPK1CD-Citrine were not detected (data not shown), which is probably due to the instability of these fusion proteins. Plasma membrane localization of Citrine-PpPIPK1CD was further confirmed by colocalization with signals of FM4-64, a plasma membrane marker (Supplemental Fig. S1A, top). Since the behavior of Citrine-PpPIPK1CD was identical to that of Citrine-PpPIPK1, we concluded that the CD, but not the MD, is necessary to confer the plasma membrane localization of PpPIPK1.
AtPIP5K1 Is Localized to Plasma Membranes in a CD-Dependent Manner in Protoplasts of P. patens
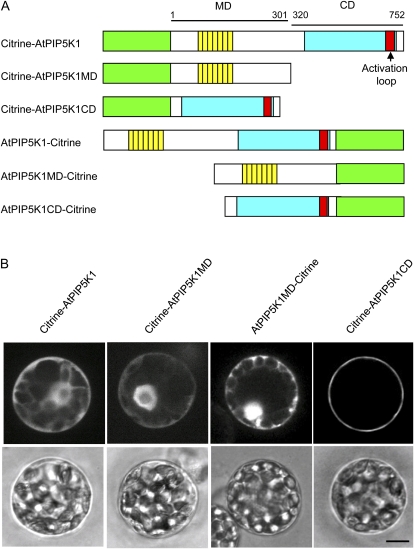
Despite the hypothesized involvement of the MD in the plasma membrane localization of OsPIPK1, AtPIP5K1, and AtPIP5K3 (Ma et al., 2006; Im et al., 2007; Kusano et al., 2008), plasma membrane localization of MD and CD could not be demonstrated using transgenic tobacco cells (Im et al., 2007). In order to determine whether the MD of AtPIP5K1 localizes to plasma membranes in protoplasts of P. patens, plasmids for the expression of Citrine fused with the full-length AtPIP5K1, MD (amino acids 1–301), and CD (amino acids 320–752) were created (Fig. 3A) and introduced into P. patens protoplasts. We could detect Citrine-AtPIP5K1CD localization to the plasma membrane, whereas Citrine-AtPIP5K1, Citrine-AtPIP5K1MD, and AtPIP5K1MD-Citrine signals were found in the cytoplasm and nucleus (Fig. 3B). In addition, Citrine-AtPIP5K1CD signals were colocalized with FM4-64 signals (Supplemental Fig. S1A, bottom). Fluorescent signals from AtPIP5K1-Citrine and AtPIP5K1CD-Citrine were not detected, as was the case for PpPIPK1 (data not shown). These results indicate that the CDs of both PpPIPK1 and AtPIP5K1 share the same function of plasma membrane targeting in P. patens protoplasts, although it is noteworthy that, in contrast to PpPIPK1, the full-length AtPIP5K1 did not localize to plasma membranes (Fig. 3B).
Figure 3.
Involvement of the CD in plasma membrane localization of AtPIP5K1. A, Recombinant Citrine fusion proteins whose intracellular localization was determined by transient expression in P. patens protoplasts. Green, yellow, blue, and red boxes are the same as in Figure 1. The MD and CD of AtPIP5K1 are also indicated by bars with numbers of amino acid positions. B, Subcellular localization of Citrine fusion proteins. Confocal images of P. patens protoplasts transformed with expression plasmids for Citrine fusion proteins are shown. Top and bottom panels show Citrine fluorescence and bright-field images, respectively. Bar = 10 μm.
Involvement of the CD in Plasma Membrane Localization of PpPIPK1 and AtPIP5K1 in Onion and Arabidopsis Cells
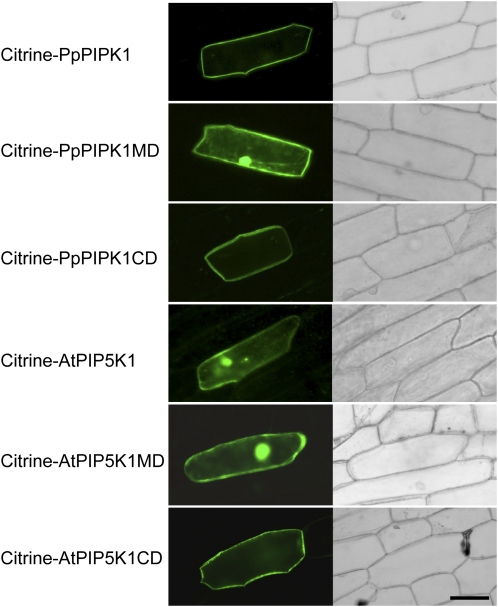
The above findings raise a further question of whether our observations were due to cell type specificity. To investigate the subcellular localization of plant PIPKs in other types of cells, we assessed the role of the CD in plasma membrane targeting by analyzing onion epidermal cells in which Citrine-tagged protein domains were expressed. As a result, the CDs from PpPIPK1 and AtPIP5K1 were shown to localize to plasma membranes, while the MDs did not (Fig. 4). Citrine-PpPIPK1CD and Citrine-AtPIP5K1CD were also localized to plasma membrane in plasmolyzed cells (Supplemental Fig. S1B). Moreover, the CD dependency of plasma membrane localization of AtPIP5K1 was also observed in root cortical cells of Arabidopsis (Supplemental Fig. S2). Since these results are fully consistent with those obtained with protoplasts of P. patens (Fig. 2B), the possibility of cell type dependency of CD function was excluded. In conclusion, the CD is crucial for targeting plant PIPKs to the plasma membrane.
Figure 4.
Subcellular localization of Citrine fusion proteins in onion epidermal cells. Confocal images of onion epidermal cells transformed with expression plasmids for Citrine fusion proteins are shown. Left and right panels show Citrine fluorescence and bright-field images, respectively. Bar = 80 μm.
Identical to results using protoplasts of P. patens, AtPIP5K1, but not PpPIP5K1, localization to plasma membranes was inhibited when expressed in both onion epidermal and Arabidopsis root cortical cells (Fig. 4; Supplemental Fig. S2), suggesting an inhibitory effect of the MD, but not cell type specificity, on the plasma membrane localization of AtPIP5K1.
Enzymatic Activity Does Not Influence Plasma Membrane Localization of PpPIPK1
One remarkable feature of animal type I PIPKs is the reflection of substrate specificity to subcellular localization; substitution of the Glu residue conserved in the activation loop of type I PIPKs to the Ala found in type II PIPKs switched both substrate specificity and localization to those of type II PIPKs (Kunz et al., 2000, 2002). In contrast, our previous results demonstrated that Glu is also conserved in the activation loop of plant PIPKs, while its substitution at position 885 to Ala (PpPIPK1-E885A) resulted in drastically reduced activity toward PtdIns3P and PtdIns4P but with slight enhancement of activity toward PtdIns5P (Saavedra et al., 2009). These differences between animal and plant PIPKs led us to examine whether the Glu plays a role in the plasma membrane localization of PpPIPK1.
In this study, we isolated the CD from PpPIPK1-E885A (Saavedra et al., 2009) and then subsequently fused it with Citrine to create Citrine-PpPIPK1-E885A (Fig. 5A). When this fusion protein was expressed in both P. patens protoplasts and onion epidermal cells, it behaved like Citrine-PpPIPK1 and Citrine-PpPIPK1CD, localizing to plasma membranes (compare Figs. 2B and 5B). These results clearly indicate that the enzymatic activity resulting from the Glu conserved in the activation loop is dispensable in plasma membrane targeting of PpPIPK1, which differs from the function of the corresponding Glu in the activation loop of animal type I PIPKs (Kunz et al., 2000, 2002).
Figure 5.
Findings indicating that enzymatic activity is not required for plasma membrane localization of PpPIPK1. A, Recombinant Citrine fusion proteins whose intracellular localization was determined by transient expression in P. patens protoplasts. The Glu residue (E) at position 885, the target of a point mutation, and the Ala residue (A) resulting from mutation are highlighted in red. Green, yellow, blue, and red boxes are the same as in Figure 1. B, Subcellular localization of Citrine fusion proteins. Confocal images of P. patens protoplast and onion epidermal cells transformed with expression plasmids for Citrine-PpPIPK1-E885A are shown. Left and light panels show Citrine fluorescence and bright-field images. Bars = 10 and 40 μm for protoplasts and epidermal cells, respectively.
Identification of Determinants of PpPIPK1 Plasma Membrane Localization
Given that Glu at position 885 is not involved in the plasma membrane localization of plant PIPKs (Fig. 5B), we next attempted to identify the amino acid residues acting as determinants of the plasma membrane localization of PpPIPK1. In mammalian type I PIPKs, two dibasic amino acid pairs highly conserved in the activation loop (KK) and in the C-terminal flanking of the activation loop (KK; Fig. 1, boxed KKs) play important roles in plasma membrane localization, since substitutions of them to acidic amino acid pairs resulted in alteration of their localization to the cytoplasm (Kunz et al., 2000; Arioka et al., 2004). As shown in Figure 1, the amino acid sequences of the activation loops of plant PIPKs resemble those of type I PIPKs, whereas the C-terminal flanking of the activation loop of plant PIPKs is similar to that of type II PIPKs, indicating that the further downstream amino acid pair is absent in plant PIPKs (Fig. 1, boxed KKs). However, another dibasic amino acid pair, KR, exists just downstream of the activation loop in both plant and animal type II PIPKs (Fig. 1, indicated in blue). Thus, we examined the significance of these two dibasic amino acid pairs in plasma membrane targeting of PpPIPK1.
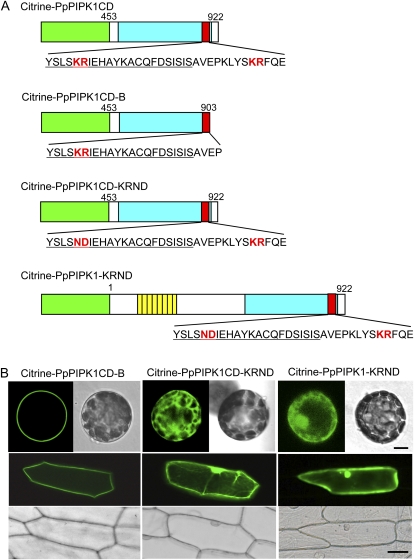
First, we made a deletion mutant of PpPIPK1CD lacking the C-terminal region containing the downstream KR pair at positions 908 and 909. We named the mutant PpPIPK1CD-B and subsequently constructed Citrine-PpPIPK1CD-B by fusing it to the C terminus of Citrine (Fig. 6A). When the fusion protein was expressed in both P. patens protoplasts and onion epidermal cells, we observed no effects of this mutation on plasma membrane localization (Fig. 6B), indicating that the downstream KR pair has no function in directing plasma membrane targeting of PpPIPK1.
Figure 6.
Identification of the molecular determinant of plasma membrane localization in PpPIPK1. A, Recombinant Citrine fusion proteins whose intracellular localization was determined by transient expression in P. patens protoplasts. The dibasic amino acid pairs used as targets of modification and the ND residues resulting from mutation are highlighted in red. Green, yellow, blue, and red boxes are the same as in Figure 1. The numbers indicate amino acid positions. The sequence of the activation loop is underlined. B, Subcellular localization of Citrine fusion proteins. Confocal images of P. patens protoplast and onion epidermal cells transformed with expression plasmids for Citrine-PpPIPK1CD-B and Citrine-PpPIPK1CD-KRND are shown by Citrine fluorescence and bright-field images. Bars = 10 and 40 μm for protoplasts and epidermal cells, respectively.
Kunz et al. (2000) examined the functional roles of two basic amino acids in the activation loop by substituting these to acidic amino acids. Thus, to test whether the first KR pair in the activation loop is involved in plasma membrane localization, amino acid substitutions from positively charged KR at positions 882 and 883 to negatively charged ND were performed by nucleotide mutagenesis in the activation loop of Citrine-PpPIPK1 and Citrine-PpPIPK1CD, producing Citrine-PpPIPK1-KRND and Citrine-PpPIPK1CD-KRND, respectively (Fig. 6A). Results indicate a drastic change in subcellular localization from the plasma membrane to the cytoplasm and nucleus upon expression of PpPIPK1-KRND and PpPIPK1CD-KRND in both P. patens protoplasts and onion epidermal cells (Fig. 6B). Thus, these findings suggest that the KR pair in the activation loop is essential for the plasma membrane localization of PpPIPK1.
The Dibasic Amino Acid Pair Is Also Involved in Enzymatic Activation of PpPIPK1
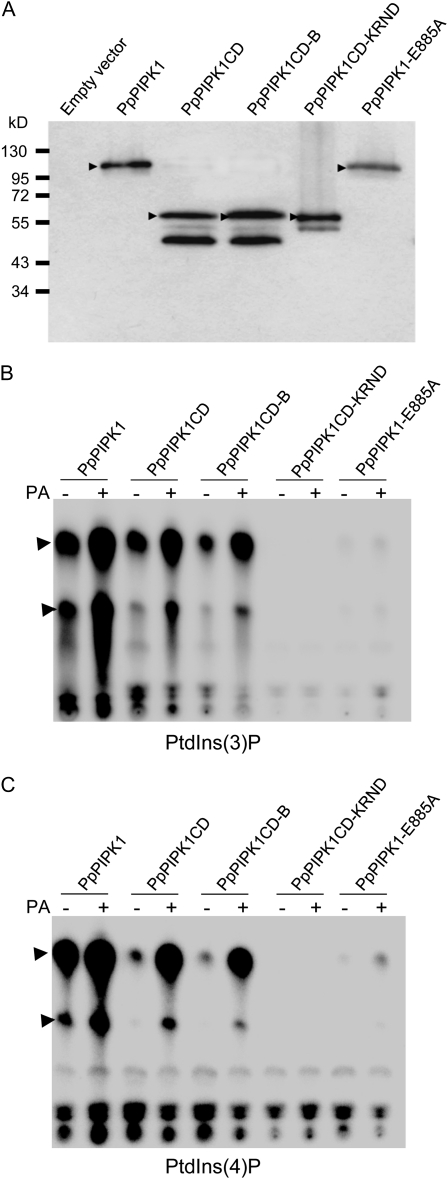
We further examined whether substitution of KR to ND had any effect on the enzymatic activity of PpPIPK1. When His-tagged PpPIPK1 mutants were purified after expression in Pichia pastoris (Fig. 7A), His-tagged PpPIPK1CD and PpPIPK1CD-B showed activity toward both PtdIns3P and PtdIns4P, although their activity was approximately 50% less for PtdIns3P and approximately 90% less for PtdIns4P than those of His-tagged PpPIPK1 (Fig. 7, B and C), indicating that the downstream KR pair is not essential for the enzyme activity. However, the missing MD may play a role in this respect. In contrast, enzymatic activity toward PtdIns3P and PtdIns4P was drastically reduced in His-tagged PpPIPK1CD-KRND and PpPIPK1-E885A. The CD-KRND mutation completely abolished enzyme activity and consequently PA was without effect, while the CD-E885A mutation still retained a low activity and PA slightly increased the activity (Fig. 7, B and C). These results indicate that although Glu is involved in the efficiency of enzymatic activity, as shown earlier (Saavedra et al., 2009), the KR pair in the activation loop is essential for enzymatic activity. The role of the MD is not clear; however, it may provide structural support for the CD, keeping the CD in an active conformation.
Figure 7.
Involvement of the dibasic amino acid pair in PA-dependent activation of PpPIPK1. A, Expression of recombinant proteins. Recombinant His-tagged proteins expressed in P. pastoris X-33 cells harboring the pPICZB-PpPIPK1, pPICZA-PpPIPK1CD, pPICZA-PpPIPK1CD-B, pPICZA-PpPIPK1CD-KRND, pPICZB-PpPIPK1-E885A, and pPICZB constructs were affinity purified and detected with anti-His antibodies. The arrowheads indicate bands corresponding to His-tagged proteins. Molecular mass standards are indicated in kD. The bands below PpPIPK1CD, PpPIPK1CD-B, and PpPIPK1CD-KRND are their degraded products produced during purification. B and C, In vitro lipid kinase activity and PA dependency of His-tagged proteins. The activities of 1 μg of recombinant proteins bound to nickel-nitrilotriacetic acid agarose beads were assayed with PtdIns3P (B) and PtdIns4P (C) as indicated. Activation of His-tagged proteins with 143 μm PA was also examined in lanes indicated by +. Top arrowheads represent reaction products PtdIns(3,4)P2 and PtdIns(4,5)P2, respectively, and bottom arrowheads represent reaction products lysoPtdIns(3,4)P2 and lysoPtdIns(4,5)P2, respectively. The reaction products PtdInsP2 and lysoPtdInsPIP2 were analyzed and their identities verified by HPLC (Saavedra et al., 2009). Products/spots close to the origin correspond, apart from residual [32P]ATP, to unidentified products.
DISCUSSION
Accumulating data indicate that the intracellular distribution of PIs and PI-producing enzymes is important in regulating development and environmental stress responses in plants (Zonia and Munnik, 2006). Despite the establishment of the roles of PIPKs in polarized tip growth of root hairs and pollen tubes via their localization at plasma membranes in Arabidopsis (Ischebeck et al., 2008; Kusano et al., 2008; Sousa et al., 2008; Stenzel et al., 2008), the regulatory mode of this plasma membrane localization remains elusive in plants. Here, we identified a dibasic amino acid pair, KR, in the activation loop of PpPIPK1, the biological relevance of which is 2-fold. First, substitution of these basic residues prevented plasma membrane localization (Fig. 6); furthermore, these mutations resulted in a complete loss of enzymatic activity toward PtdIns3P and PtdIns4P (Fig. 7). This dibasic amino acid pair in the activation loop of plant type I/II PIPKs is highly conserved (Saavedra et al., 2009); thus, it appears that the regulatory modes of translocation to plasma membranes and subsequent enzymatic activation of PIPKs are also conserved in different plants. It is noteworthy that the mode of membrane localization of PIPKs is different between plants and animals, since animal type I PIPKs require the conserved Glu residue in the activation loop and an additional dibasic amino acid pair for the plasma membrane localization (Kunz et al., 2002; Arioka et al., 2004).
Conflicting suggestions are at hand regarding the prediction of a plasma membrane-targeting module in plant PIPKs. In contrast to our data (Figs. 2–4), for example, it has been reported that the MD, but not the activation loop containing the dibasic amino acid pair, regulates the plasma membrane localization of plant PIPKs (Ma et al., 2006; Im et al., 2007; Kusano et al., 2008). However, our findings closely resemble those of Kunz et al. (2000) on the membrane-targeting machinery of mammalian type I PIPKs, in which the dibasic amino acid pair in the activation loop also acts as a determinant of plasma membrane localization. They showed that the substitution of the dilysine motif KK, corresponding to KR in PpPIPK1, to two Asn residues also eliminated both plasma membrane localization and phosphorylation of PtdIns3P, although activity toward PtdIns4P was unchanged. Since substitutions of the two basic amino acids eliminated activity toward both PtdIns3P and PtdIns4P in PpPIPK1 (Fig. 7), it appears that the mode of membrane localization and substrate specificity is specific for plant type I/II PIPKs.
A role of basic amino acid residues in plasma membrane targeting has also been found in the C2 domains of phospholipase A2 and protein kinase Cα, for which it was proposed that electric interactions by cationic residues initially bring proteins to anionic membrane surfaces (Stahelin and Cho, 2001; Stahelin et al., 2003). On the basis of these findings, it is postulated that the molecular basis of plasma membrane localization via basic amino acid residues is widely conserved among membrane-targeting proteins in eukaryotes. Therefore, it is necessary to determine what the target of these dibasic amino acid pairs is at plasma membranes in eukaryotic PIPKs.
Our results also indicate the involvement of the MD in the enhancement of enzymatic activity but not in PA-dependent activation (Fig. 7). It is inconsistent with AtPIP5K1, the MD of which represses enzymatic activity (Im et al., 2007). Since deletion of the MD from AtPIP5K3 had no effect on its enzymatic activity (Stenzel et al., 2008), it is possible that its contribution to the regulation of PIPK catalytic activity varies among enzymes.
We found that localization of the intact AtPIP5K1 is not restricted to plasma membranes, which is distinct from the behavior of intact PpPIPK1 at plasma membranes (Figs. 3 and 4). These findings suggest that PpPIPK1 and AtPIP5K1 are conformationally distinct, possibly influencing localization in both P. patens protoplasts and onion epidermal cells. The determining factor of cytoplasmic AtPIP5K1 conformation is thought to be the MD, since the CD from both AtPIP5K1 and PpPIPK1 targeted plasma membranes equally well (Figs. 2–4; Supplemental Fig. S1). Thus, the MD of AtPIP5K1 appears to play a role in modulating the subcellular localization by preventing plasma membrane localization. We propose that this is one reason why Im et al. (2007) did not observe plasma membrane localization of the MD of AtPIP5K1. However, it remains unclear why the CD of AtPIP5K1 and AtPIP5K3 did not target plasma membranes (Im et al., 2007; Kusano et al., 2008).
Although PA stimulation of the animal type I and plant type I/II PIPKs has been shown (Moritz et al., 1992; Jenkins et al., 1994; Ishihara et al., 1998; Perera et al., 2005; Im et al., 2007; Saavedra et al., 2009), the mechanism by which PA stimulates PIPK activity has yet to be studied in detail. However, Stace et al. (2008) recently demonstrated that basic amino acids upstream of the activation loop in the catalytic region are involved in PA-dependent activation of human PIP5K-1β. In our experiments, the mutation of a dibasic amino acid pair resulted in the complete inhibition of enzymatic activity of PpPIPK1 (Fig. 7), suggesting that the loss of PA dependency is a secondary effect of the total inhibition of PpPIPK1 activity. Moreover, PA-dependent activation was still observed in the PpPIPK1-E885A mutant (Fig. 7). Therefore, amino acids other than the dibasic amino acid pair and the Glu in the activation loop are critical for PA dependency. More detailed analysis of the PA-dependent activation in PpPIPK1 is necessary to resolve this question.
Another example of a membrane-localizing protein that is affected by PA is Raf1, a mitogen-activated protein kinase kinase kinase involved in the development of zebrafish embryos (Ghosh et al., 2003). PA binds to the short tetrapeptide motif RKxR of Raf-1 and enhances translocation of this kinase to plasma membranes (Ghosh et al., 1996; Rizzo et al., 2000). Thus, electric charge is thought to be important for electrostatic interaction between the RKxR motif and PA. Since PA regulates the affinity of PIPK to substrates (Jarquin-Pardo et al., 2007), it is predicted that PA first binds to the basic amino acids in the catalytic region and then recruits substrates to enhance enzymatic activity in plant PIPKs. It is noteworthy that PA is produced by diacylglycerol kinase via phosphorylation of diacylglycerol, a product of phospholipase C action on PtdIns(4,5)P2, and also by phospholipase D action on phosphatidylcholine (Zonia and Munnik, 2006). In addition, PtdIns(4,5)P2 positively regulates phospholipase D activity (Potocký et al., 2003; Ode Weernink et al., 2007). Thus, it is necessary to investigate the function of PIPKs in relation to the positive feedback machinery consisting of signaling cascades involving PIPK-phospholipase C-diacylglycerol kinase and phospholipase D.
Animal type I PIPKs direct various physiological regulations such as actin organization, vesicle trafficking, and ion channel activity at plasma membranes, whereas animal type II PIPKs, which are found only in metazoans, are localized in the cytosol, nucleus, endoplasmic reticulum, and actin cytoskeleton (Clarke et al., 2007; Heck et al., 2007). Thus, members of animal type I and type II PIPKs are thought to be functionally nonredundant, localizing to different cellular compartments. In contrast, our previous studies (Mikami et al., 1998; Saavedra et al., 2009) and our results here clearly indicate that plants have no typical type I and type II PIPKs but rather have a type I/II class of PIPKs. However, in terms of enzymatic activity, plant type I/II PIPKs resemble animal type I PIPKs, with a lack of activity toward the PtdIns5P found in animal type II PIPKs. In contrast to the structural and biochemical characteristics, little is known about the function of plant type I/II PIPKs in physiological processes; AtPIP5K3 regulates tip growth of root hairs (Kusano et al., 2008; Stenzel et al., 2008), whereas AtPIP5K4 and AtPIP5K5 are involved in apical growth of pollen tubes (Ischebeck et al., 2008; Sousa et al., 2008). In P. patens PIPKs, although the functions of PpPIPKs are not clear, it is likely that PpPIPK1 and PpPIPK2 are functionally distinct because of the unique enzymatic activity of PpPIPK2 as a PtdIns kinase (Saavedra et al., 2009). In addition, osmostress-inducible expression of the PpPIPK1 gene suggests the involvement of PpPIPK1, but not PpPIPK2, in stress responses (Saavedra et al., 2009). Thus, generation of gene-targeting disruption mutants for the PpPIPK genes is now under way to investigate the functions of PpPIPKs during the development of P. patens by comparison of phenotypes of gene-disruption mutants of PpPIPK1 and PpPIPK2 genes.
MATERIALS AND METHODS
Construction of Expression Plasmids for Subcellular Localization Experiments
Expression plasmids for Citrine fusion proteins were constructed using pUGW0 for C-terminal fusion and pUGW2 for N-terminal fusion (Nakagawa et al., 2007) through Gateway technology (Invitrogen). PpPIPK1CD-KRND was made by introducing point mutations in codons corresponding to KR residues using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) with primers 5′-CAAAATTACAGTCTCAGCAACGACATAGAGCACGCATACAAG-3′ and 5′-CTTGTATGCGTGCTCTATGTCGTTGCTGAGACTGTAATTTTG-3′ (nucleotides responsible for point mutations are underlined). To make entry plasmids, appropriate portions of open reading frames corresponding to the full length, MD, and CD of PpPIPK1 and AtPIP5K1 were amplified with the sets of primers listed in Supplemental Table S1; primer sets used for amplification of point-mutated and deleted CDs, such as PpPIPK1CD-E885A, PpPIPK1CD-B, and PpPIPK1CD-KRND, are also indicated. Each amplified DNA was directly subcloned into the pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning Kit (Invitrogen). Recombination reactions were performed to make Citrine fusions with entry plasmids and pUGW0 or pUGW2 according to the manufacturer's instructions. Resultant constructs are schematically represented in Figures 2A, 3A, 5A, and 6A and Supplemental Figure S1.
Transient Transformation
Polyethylene glycol-mediated transformation of protoplasts from the wild-type strain of Physcomitrella patens subspecies patens (Ashton and Cove, 1977) was performed as described previously (Nishiyama et al., 2000) except that 20 μg of circular plasmids was used for transient transformation followed by the culture of protoplasts for 1 or 2 d under polarized white light. Transient transformation of onion (Allium cepa) epidermal cells by particle bombardment was performed as described previously for gametophytic cells of the red alga Porphyra yezoensis (Mikami et al., 2009). For root cortical cells of Arabidopsis (Arabidopsis thaliana), gold particles (1.6 μm in diameter) coated with plasmids were bombarded into leaves of 3-week-old plants by a PDS-1000/HE Biolistic Delivery System (Bio-Rad Laboratories) as described by Charon et al. (1997) except for the helium pressure of 1,550 p.s.i., the target distance of 90 mm, and the vacuum of 28 mm of mercury per inch. After the bombardment, the leaves were allowed to recover at 22°C in the dark for 16 h.
Light and Fluorescence Microscopy
Multiwavelength confocal microscopy images of Citrine fluorescence derived from the Citrine-PIPK fusions and chloroplast autofluorescence were obtained with a Leica TCS-SP2 inverted confocal laser microscope. Acquisition was along one focal line using 488-nm (laser power, 16%) and 546-nm (laser power, 100%) excitation with collection of the 500- to 535-nm emission spectrum for Citrine and the 640- to 685-nm emission spectrum for autofluorescence of chloroplasts, respectively, as mentioned by Hiwatashi et al. (2008). For the Citrine fusion proteins of AtPIP5K and its derivatives, observation was performed as described previously (Saavedra et al., 2009) using a UPLSAPO 60XW lens mounted on an inverted IX70 microscope (Olympus) equipped with a CSU21 confocal system (Yokogawa; a 488-nm excitation laser). Images were captured with a Cool SNAP HQ camera (Nihon Roper) running on Meta Morph version 7.1 (Nihon Molecular Devices). Simultaneous observations of Citrine and FM4-64 (Molecular Probes) signals in P. patens protoplasts were performed as described by Oda et al. (2009) except for treatment of cells with 50 μm FM4-64 for 3 min. In addition, observations of Citrine fluorescent signals in onion epidermal cells with and without plasmolysis were performed as described previously for gametophytic cells of P. yezoensis (Mikami et al., 2009), except for the use of a fluorescent microscope (DM5000B; Leica) equipped with a fluorescence L5 filter to yield GFP signals. Images were captured with a Leica DFC 300FX camera running on the Leica Application Suite as described previously (Mikami et al., 2009).
In Vitro Analysis of PIPK Enzymatic Activity
For the expression of PpPIPK1 and PpPIPK1-E885A in Pichia pastoris X-33 cells, pPICZB-PpPIPK1 and pPICZB-PpPIPK1-E885A (Saavedra et al., 2009) were used. For the expression of PpPIPK1CD, PpPIPK1CD-B, and PpPIPK1CD-KRND in yeast cells, we employed Gateway technology (Invitrogen). To produce the destination vector, pPICZA (Invitrogen) was digested with NotI in the multicloning site, filled with Klenow enzyme, and ligated with a reading frame cassette amplified using primers Rfc-U (5′-ATCAAACAAGTTTGTACAAAAAAGCTG-3′) and Rfb-L (5′-ATCAACCACTTTGTACAAGAAAGC-3′). The resultant plasmid was designated pPICZA-DES. Recombination reactions were then performed with the entry plasmids mentioned above and pPICZA-DES according to the manufacturer's instructions, producing plasmids pPICZA-PpPIPK1CD, pPICZA-PpPIPK1CD-B, and pPICZA-PpPIPK1CD-KRND.
Transformation of P. pastoris X-33 cells with the above expression plasmids, colony PCR of transformants, and, following expression, purification and western-blot analysis of His-tagged recombinant proteins were performed as described previously (Saavedra et al., 2009). The PIPK activity assay using 1 μg of purified His-tagged proteins and liquid substrates PtdIns3P and PtdIns4P (Echelon) was carried out as described by Cooke et al. (1998) and Dove and Michell (1998) with the modifications indicated by Saavedra et al. (2009). When PA (Echelon) was added to the assay, the final concentration was 143 μm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Visual confirmation of plasma membrane localization of the CDs from P. patens and Arabidopsis PIPKs.
Supplemental Figure S2. Importance of the CD for plasma membrane localization of AtPIPK1 in Arabidopsis root cortical cells.
Supplemental Table S1. List of oligonucleotide primers.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tsuyoshi Nakagawa of Shimane University for kindly providing pUGW0 and pUGW2, Dr. Naotsune Saga of Hokkaido University for valuable discussion, and Daiske Tanaka of Hokkaido University for technical assistance.
References
- Anderson RA, Boronenkov IV, Doughman SD, Kunz J, Loijens JC. (1999) Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J Biol Chem 274: 9907–9910 [DOI] [PubMed] [Google Scholar]
- Arioka M, Nakashima S, Shibasaki Y, Kitamoto K. (2004) Dibasic amino acid residues at the carboxyl-terminal end of kinase homology domain participate in the plasma membrane localization and function of phosphatidylinositol 5-kinase gamma. Biochem Biophys Res Commun 319: 456–463 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ. (1977) The isolation and preliminary characterization of auxotrophic and analogue resistant mutants in the moss Physcomitrella patens. Mol Gen Genet 154: 87–95 [Google Scholar]
- Charon C, Johansson C, Kondorosi E, Kondorosi A, Crespi M. (1997) enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc Natl Acad Sci USA 94: 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JH, Richardson JP, Hinchliffe KA, Irvine RF. (2007) Type II PtdInsP kinases: location, regulation and function. Biochem Soc Symp 74: 149–159 [DOI] [PubMed] [Google Scholar]
- Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. (1998) The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol 8: 1219–1222 [DOI] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H. (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Doughman RL, Firestone AJ, Anderson RA. (2003) Phosphatidylinositol phosphate kinase put PI4,5P(2) in its place. J Membr Biol 194: 77–89 [DOI] [PubMed] [Google Scholar]
- Dove SK, Michell RH. (1998) Analysis of polyphosphorylated inositol lipids of S. cerevisiae. Milligan G, , Signal Transduction: A Practical Approach. Oxford University Press, Oxford, pp 255–281 [Google Scholar]
- Downes CP, Gray A, Lucocq JM. (2005) Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol 15: 259–268 [DOI] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Mueller-Roeber B. (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 26: 561–571 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Moore S, Bell RM, Dush M. (2003) Functional analysis of a phosphatidic acid binding domain in human Raf-1 kinase: mutations in the phosphatidate binding domain lead to tail and trunk abnormalities in developing zebrafish embryos. J Biol Chem 278: 45690–45696 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Strum JC, Sciorra VA, Daniel L, Bell RM. (1996) Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid. J Biol Chem 271: 8472–8480 [DOI] [PubMed] [Google Scholar]
- Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, Anderson RA. (2007) A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol 42: 15–39 [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y, Obara M, Sato Y, Fujita T, Murata T, Hasebe M. (2008) Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 20: 3094–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Davis AJ, Perera IY, Johannes E, Allen NS, Boss WF. (2007) The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J Biol Chem 282: 5443–5452 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Stenzel I, Heilmann I. (2008) Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell 20: 3312–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. (1998) Type I phosphatidylinositol-4-phosphate 5-kinases: cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J Biol Chem 273: 8741–8748 [DOI] [PubMed] [Google Scholar]
- Jarquin-Pardo M, Fitzpatrick A, Galiano FJ, First EA, Davis JN. (2007) Phosphatidic acid regulates the affinity of the murine phosphatidylinositol 4-phosphate 5-kinase-Ibeta for phosphatidylinositol-4-phosphate. J Cell Biochem 100: 112–128 [DOI] [PubMed] [Google Scholar]
- Jenkins GH, Fisette PL, Anderson RA. (1994) Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem 269: 11547–11554 [PubMed] [Google Scholar]
- Kunz J, Fuelling A, Kolbe L, Anderson RA. (2002) Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J Biol Chem 277: 5611–5619 [DOI] [PubMed] [Google Scholar]
- Kunz J, Wilson MP, Kisseleva M, Hurley JH, Majerus PW, Anderson RA. (2000) The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol Cell 5: 1–11 [DOI] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JE, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T. (2008) The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW. (2007) PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19: 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Lou Y, Lin WH, Xue HW. (2006) MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res 16: 466–478 [DOI] [PubMed] [Google Scholar]
- Ma H, Xu SP, Luo D, Xu ZH, Xue HW. (2004) OsPIPK 1, a rice phosphatidylinositol monophosphate kinase, regulates rice heading by modifying the expression of floral induction genes. Plant Mol Biol 54: 295–310 [DOI] [PubMed] [Google Scholar]
- Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K. (1998) A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J 15: 563–568 [DOI] [PubMed] [Google Scholar]
- Mikami K, Uji T, Li L, Takahashi M, Yasui H, Saga N. (2009) Visualization of phosphoinositides via the development of the transient expression system of a cyan fluorescent protein in the red alga Porphyra yezoensis. Mar Biotechnol 11: 563–569 [DOI] [PubMed] [Google Scholar]
- Moritz A, De Graan PN, Gispen WH, Wirtz KW. (1992) Phosphatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase. J Biol Chem 267: 7207–7210 [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C. (2002) Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M. (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Oda Y, Hirata A, Sano T, Fujita T, Hiwatashi Y, Sato Y, Kadota A, Hasebe M, Hasezawa S. (2009) Microtubules regulate dynamic organization of vacuoles in Physcomitrella patens. Plant Cell Physiol 50: 855–868 [DOI] [PubMed] [Google Scholar]
- Ode Weernink PA, López de Jesús M, Schmidt M. (2007) Phospholipase D signaling: orchestration by PIP2 and small GTPases. Naunyn Schmiedebergs Arch Pharmacol 374: 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Davis AJ, Galanopoulou D, Im YJ, Boss WF. (2005) Characterization and comparative analysis of Arabidopsis phosphatidylinositol phosphate 5-kinase 10 reveals differences in Arabidopsis and human phosphatidylinositol phosphate kinases. FEBS Lett 579: 3427–3432 [DOI] [PubMed] [Google Scholar]
- Pical C, Westergren T, Dove SK, Larsson C, Sommarin M. (1999) Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J Biol Chem 274: 38232–38240 [DOI] [PubMed] [Google Scholar]
- Potocký M, Eliás M, Profotová B, Novotná Z, Valentová O, Zárský V. (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130 [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Watkins SC, Romero G. (2000) The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem 275: 23911–23918 [DOI] [PubMed] [Google Scholar]
- Saavedra L, Balbi V, Dove SK, Hiwatashi Y, Mikami K, Sommarin M. (2009) Characterization of phosphatidylinositol kinases from the moss Physcomitrella patens: PpPIPK1 and PpPIPK2. Plant Cell Physiol 50: 595–609 [DOI] [PubMed] [Google Scholar]
- Sousa E, Kost B, Malhó R. (2008) Arabidopsis phosphatidylinositol-4-monophosphate 5-kinase 4 regulates pollen tube growth and polarity by modulating membrane recycling. Plant Cell 20: 3050–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace C, Manifava M, Delon C, Coadwell J, Cockcroft S, Ktistakis NT. (2008) PA binding of phosphatidylinositol 4-phosphate 5-kinase. Adv Enzyme Regul 48: 55–72 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Cho W. (2001) Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions: a surface plasmon resonance study on phospholipases A2. Biochemistry 40: 4672–4678 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Rafter JD, Das S, Cho W. (2003) The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J Biol Chem 278: 12452–12460 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Ischebeck T, König S, Hołubowska A, Sporysz M, Hause B, Heilmann I. (2008) The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20: 124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. (2000) Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell 6: 11–22 [DOI] [PubMed] [Google Scholar]
- Westergren T, Dove SK, Sommarin M, Pical C. (2001) AtPIP5K1, an Arabidopsis thaliana phosphatidylinositol phosphate kinase, synthesizes PtdIns(3,4)P(2) and PtdIns(4,5)P(2) in vitro and is inhibited by phosphorylation. Biochem J 359: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T. (2006) Cracking the green paradigm: functional coding of phosphoinositide signals in plant stress responses. Subcell Biochem 39: 207–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.