Abstract
When attacked by insects, plants release mixtures of volatile compounds that are beneficial for direct or indirect defense. Natural variation of volatile emissions frequently occurs between and within plant species, but knowledge of the underlying molecular mechanisms is limited. We investigated intraspecific differences of volatile emissions induced from rosette leaves of 27 accessions of Arabidopsis (Arabidopsis thaliana) upon treatment with coronalon, a jasmonate mimic eliciting responses similar to those caused by insect feeding. Quantitative variation was found for the emission of the monoterpene (E)-β-ocimene, the sesquiterpene (E,E)-α-farnesene, the irregular homoterpene 4,8,12-trimethyltridecatetra-1,3,7,11-ene, and the benzenoid compound methyl salicylate. Differences in the relative emissions of (E)-β-ocimene and (E,E)-α-farnesene from accession Wassilewskija (Ws), a high-(E)-β-ocimene emitter, and accession Columbia (Col-0), a trace-(E)-β-ocimene emitter, were attributed to allelic variation of two closely related, tandem-duplicated terpene synthase genes, TPS02 and TPS03. The Ws genome contains a functional allele of TPS02 but not of TPS03, while the opposite is the case for Col-0. Recombinant proteins of the functional Ws TPS02 and Col-0 TPS03 genes both showed (E)-β-ocimene and (E,E)-α-farnesene synthase activities. However, differential subcellular compartmentalization of the two enzymes in plastids and the cytosol was found to be responsible for the ecotype-specific differences in (E)-β-ocimene/(E,E)-α-farnesene emission. Expression of the functional TPS02 and TPS03 alleles is induced in leaves by elicitor and insect treatment and occurs constitutively in floral tissues. Our studies show that both pseudogenization in the TPS family and subcellular segregation of functional TPS enzymes control the variation and plasticity of induced volatile emissions in wild plant species.
Plants emit a large variety of volatile organic compounds from their foliage and flowers. These volatiles serve a variety of functions ranging from the attraction of pollinating insects and fruit dispersers to direct and indirect defense against herbivores and microbial pathogens (Pichersky and Gershenzon, 2002; Dicke et al., 2005; Dudareva et al., 2006; Unsicker et al., 2009). Usually, plant volatiles are released as mixtures, which may be important in targeting a variety of different organisms. In this context, differences in the composition of volatile blends within and among species may be the result of adaptation to specific communities of organisms.
Terpenoids represent the largest and most diverse class of plant volatile metabolites. Low-Mr terpenes with a 10-carbon (monoterpenes) or 15-carbon (sesquiterpenes) skeleton are common constituents of floral and herbivore-induced leaf volatile blends (Dudareva et al., 2006). Monoterpenes and sesquiterpenes are produced from the central terpene precursors geranyl diphosphate (GPP) and farnesyl diphosphate (FPP), respectively, by the enzymatic activity of terpene synthases (TPSs; Tholl, 2006; Degenhardt et al., 2009). In the plant cell, terpene metabolism is compartmentalized, with GPP and monoterpene formation occurring primarily in plastids and FPP and sesquiterpenes being synthesized predominantly in the cytosol (Aharoni et al., 2005).
Studies of cultivated plants have given the first insight into the cellular, molecular genetic, and biochemical mechanisms controlling the variability of terpene volatile mixtures. For example, variation of monoterpene and sesquiterpene blends produced in the glandular trichomes of different basil (Ocimum basilicum) cultivars was attributed to the differential expression of TPS genes in these cultivars (Iijima et al., 2004). In maize (Zea mays), allelic variation of two TPSs was found to be responsible for the quantitative compositional differences of sesquiterpene volatile blends released from mature leaves and husks of different varieties (Köllner et al., 2004). Furthermore, a characterization of linalool/nerolidol synthases in snapdragon (Antirrhinum majus) flowers indicated that subcellular segregation of bifunctional TPSs in plastids and the cytosol can lead to a compartment-specific formation of monoterpenes and sesquiterpenes, respectively, thereby increasing the diversity of terpene volatile mixtures (Nagegowda et al., 2008).
While the variation of herbivore-induced volatiles has been studied in some wild species (Halitschke et al., 2000; Gouinguene et al., 2001; Delphia et al., 2009), knowledge of the molecular mechanisms governing the natural diversity of volatile compounds is rather limited. We have been investigating the intraspecific and interspecific variation of volatile profiles in Arabidopsis species (Tholl et al., 2005; Abel et al., 2009) to explore the natural evolution of volatile mixtures in more detail. A previous survey of 37 accessions (ecotypes) revealed quantitative differences in floral sesquiterpene volatile compositions that are controlled by differences in TPS gene transcription and putative posttranslational modifications (Tholl et al., 2005). Here, we turn our attention to vegetative volatiles. Leaves of the Arabidopsis (Arabidopsis thaliana) accession Columbia (Col-0) release a simple three-compound blend consisting of the benzenoid compound methyl salicylate (MeSA), the C16-homoterpene 4,8,12-trimethyltridecatetra-1,3,7,11-ene (TMTT), and the sesquiterpene (E,E)-α-farnesene in response to treatment with the fungal peptide elicitor alamethicin and feeding by the crucifer specialist insects Pieris rapae and Plutella xylostella (Van Poecke et al., 2001; Herde et al., 2008). The induced volatile mixture is assumed to serve as an indirect defense signal by attracting parasitoids of P. rapae larvae. With the exception of Nossen (No-0; Fäldt et al., 2003), no other Arabidopsis accession has been investigated for elicitor- or insect-induced volatiles, and there is no information on the genetic and molecular mechanisms responsible for these differences.
Here, we report the analysis of induced volatile terpene emissions from rosette leaves of 27 Arabidopsis accessions. We show that several accessions such as Wassilewskija (Ws) emit the monoterpene (E)-β-ocimene and the sesquiterpene (E,E)-α-farnesene, while others such as Col-0 release (E,E)-α-farnesene without any or only traces of (E)-β-ocimene. We demonstrate that the difference in terpene volatile emission between Col-0 and Ws is caused by allelic variation leading to differential expression and subcellular targeting of two closely related bifunctional (E)-β-ocimene/(E,E)-α-farnesene synthases, TPS02 and TPS03. Our work provides evidence that natural diversity of herbivore-induced terpene volatiles evolves at multiple levels of TPS gene function and regulation, including the organelle-specific compartmentation of TPS enzymes.
RESULTS
Elicitor- and Insect-Induced Emission of the Terpene Volatiles (E)-β-Ocimene and (E,E)-α-Farnesene Vary among Different Arabidopsis Accessions, Including Col-0 and Ws
Since Arabidopsis accessions have been shown to differ substantially in their composition of secondary metabolites such as glucosinolates (Kliebenstein et al., 2001) and floral volatiles (Tholl et al., 2005), we investigated the ecotype-specific variation of terpene emissions from leaves of 27 accessions in response to treatment with coronalon, a synthetic mimic of jasmonic acid and other octadecanoid plant hormones. As previously demonstrated in accession Col-0, coronalon induces the emission of MeSA, (E,E)-α-farnesene, the homoterpene TMTT, and its precursor geranyllinalool, a response similar to that observed upon feeding damage by larvae of P. xylostella (Herde et al., 2008). By comparing emissions of volatile terpenes from leaves of intact plants treated with coronalon for 22 to 30 h, we found that 20 accessions released the monoterpene (E)-β-ocimene as the predominant volatile (approximately 75%–95% of the total amount of volatiles) at rates differing 80-fold between lowest and highest emitters (Table I). Ws was among the accessions with highest emission of (E)-β-ocimene. The remaining accessions, including Col-0, emitted no or only very small amounts of this monoterpene (Table I). The sesquiterpene (E,E)-α-farnesene was released from almost all accessions, including Col-0 and Ws, at emission rates approximately 10- to 100-fold lower than those of (E)-β-ocimene (Table I). In addition to (E)-β-ocimene and (E,E)-α-farnesene, TMTT and MeSA were detected in the induced volatile blends of all investigated ecotypes, although some accessions released these compounds only in trace amounts (Table I). No emission or only traces of terpene volatiles were found in untreated plants.
Table I. Emission of the four major volatile compounds from leaves of 27 Arabidopsis accessions in response to treatment with coronalon (coron).
Volatiles were collected for 8 h from single intact plants (see “Materials and Methods”). Emission was determined in ng g−1 fresh weight h−1. Mean values ± se are shown (n = 3). The order of accessions corresponds to increasing (E)-β-ocimene emission rates. 0.0 indicates values below 0.01 ng g−1 fresh weight h−1. n.d., Not detected.
| Ecotype | Compound |
Total Coron | ||||||||
| (E)-β-Ocimene |
(E,E)-α-Farnesene |
TMTT |
MeSA |
|||||||
| Coron | Control | Coron | Control | Coron | Control | Coron | Control | |||
| 1 | Bl-1 | n.d. | n.d. | 0.1 ± 0.0 | 0.0 ± 0.0 | 1.8 ± 0.2 | 0.0 ± 0.0 | 0.7 ± 0.1 | 0.0 ± 0.0 | 2.5 ± 0.4 |
| 2 | Lip-0 | n.d. | n.d. | 1.8 ± 0.6 | n.d. | 3.1 ± 1.0 | 0.2 ± 0.0 | 4.1 ± 1.8 | n.d. | 9.0 ± 3.4 |
| 3 | Pi-0 | n.d. | n.d. | 0.8 ± 0.3 | 0.1 ± 0.1 | 1.1 ± 0.2 | 0.0 ± 0.0 | 1.5 ± 0.2 | 0.0 ± 0.0 | 3.4 ± 0.6 |
| 4 | Tsu-1 | n.d. | n.d. | n.d. | 0.0 ± 0.0 | 0.4 ± 0.3 | 0.0 ± 0.0 | 1.3 ± 1.0 | 0.0 ± 0.0 | 1.8 ± 1.4 |
| 5 | Can-0 | 0.2 ± 0.1 | n.d. | n.d. | n.d. | 6.5 ± 2.5 | 0.0 ± 0.0 | 3.1 ± 1.0 | 0.0 ± 0.0 | 9.7 ± 3.6 |
| 6 | Bla-10 | 0.2 ± 0.0 | n.d. | n.d. | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | n.d. | 0.3 ± 0.1 |
| 7 | Col-0 | 0.7 ± 0.7 | n.d. | 2.4 ± 0.6 | n.d. | 8.8 ± 2.0 | 2.4 ± 0.7 | 18 ± 5.2 | 0.3 ± 0.1 | 29.9 ± 8.5 |
| 8 | Di-g | 2.6 ± 0.8 | n.d. | 0.2 ± 0.1 | n.d. | 0.4 ± 0.1 | 0.0 ± 0.0 | 1.8 ± 1.0 | n.d. | 5.0 ± 2.0 |
| 9 | Ri-0 | 9.4 ± 4.4 | n.d. | 0.7 ± 0.4 | n.d. | 0.6 ± 0.2 | 0.0 ± 0.0 | 1.7 ± 1.2 | 0.0 ± 0.0 | 12.3 ± 6.2 |
| 10 | Stw-0 | 9.7 ± 1.3 | 0.3 ± 0.6 | 0.2 ± 0.1 | n.d. | 1.2 ± 0.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | n.d. | 11.1 ± 1.5 |
| 11 | Lu-1 | 11.1 ± 2.8 | 0.3 ± 0.3 | 0.4 ± 0.2 | n.d. | 1.9 ± 0.7 | 0.1 ± 0.0 | 1.2 ± 0.5 | 0.0 ± 0.0 | 14.6 ± 4.1 |
| 12 | Est-0 | 18.6 ± 8.8 | n.d. | 1.3 ± 0.6 | n.d. | 0.6 ± 0.1 | n.d. | 2.6 ± 1.0 | n.d. | 23.2 ± 10.5 |
| 13 | Sei-0 | 19.9 ± 5.8 | 0.1 ± 0.0 | 0.3 ± 0.1 | n.d. | 0.2 ± 0.0 | n.d. | 0.2 ± 0.1 | n.d. | 20.5 ± 6.0 |
| 14 | Chi-0 | 20.4 ± 9.1 | 0.3 ± 0.1 | 0.4 ± 0.2 | n.d. | 0.5 ± 0.2 | 0.0 ± 0.0 | 0.6 ± 0.2 | n.d. | 21.9 ± 9.7 |
| 15 | Hodja | 20.8 ± 6.7 | 0.2 ± 0.2 | 0.6 ± 0.2 | 0.0 ± 0.0 | 0.6 ± 0.2 | 0.0 ± 0.0 | 1.5 ± 0.7 | n.d. | 23.5 ± 7.8 |
| 16 | Bla-1 | 29.9 ± 4.9 | 0.1 ± 0.1 | 1.2 ± 0.3 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 | 31.3 ± 5.3 |
| 17 | Pog-0 | 36.2 ± 10.9 | n.d. | 2.9 ± 0.9 | 0.3 ± 0.2 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 0.2 | 0.2 ± 0.1 | 39.8 ± 12.0 |
| 18 | An-1 | 37.5 ± 4.5 | 0.3 ± 0.2 | 1.2 ± 0.0 | 0.1 ± 0.1 | 0.4 ± 0.1 | 0.0 ± 0.0 | 1.2 ± 0.2 | 0.1 ± 0.1 | 40.3 ± 4.8 |
| 19 | Tul-0 | 39.6 ± 11.6 | n.d. | 3.9 ± 1.1 | n.d. | 0.0 ± 0.0 | n.d. | 3.4 ± 1.1 | n.d. | 46.8 ± 13.9 |
| 20 | Mt-0 | 45.9 ± 19.5 | 0.1 ± 0.0 | 5.9 ± 2.5 | 0.1 ± 0.1 | 2.2 ± 0.1 | 0.2 ± 0.1 | 1.4 ± 0.5 | 0.0 ± 0.0 | 55.4 ± 22.6 |
| 21 | Kil-0 | 46.9 ± 17.4 | 0.0 ± 0.0 | 1.1 ± 0.4 | n.d. | 0.3 ± 0.1 | 0.0 ± 0.0 | 1.2 ± 0.5 | n.d. | 49.5 ± 18.4 |
| 22 | JI-3 | 51.2 ± 20.1 | n.d. | 2.8 ± 1.2 | 0.1 ± 0.1 | 1.3 ± 0.3 | 0.0 ± 0.0 | 1.2 ± 0.4 | n.d. | 56.4 ± 22.1 |
| 23 | Ang-0 | 64.4 ± 21.9 | 0.8 ± 0.1 | 2.1 ± 0.8 | 0.1 ± 0.1 | 5.1 ± 1.4 | 0.2 ± 0.1 | 1.9 ± 0.9 | 0.0 ± 0.0 | 73.5 ± 25.0 |
| 24 | Ws | 64.6 ± 16.5 | 0.5 ± 0.3 | 5.4 ± 1.8 | 0.0 ± 0.0 | 7.2 ± 1.7 | 0.2 ± 0.1 | 2.2 ± 0.7 | 0.1 ± 0.1 | 79.3 ± 20.8 |
| 25 | Condara | 80.4 ± 19.4 | 0.3 ± 0.1 | 3.6 ± 0.8 | 0.3 ± 0.3 | 3.5 ± 0.8 | 0.1 ± 0.1 | 1.1 ± 0.3 | 0.0 ± 0.0 | 88.6 ± 21.4 |
| 26 | Ty-0 | 127.3 ± 19.4 | 0.1 ± 0.1 | 9.6 ± 1.8 | 0.1 ± 0.0 | 0.4 ± 0.2 | 0.1 ± 0.0 | 5.7 ± 3.3 | 0.0 ± 0.0 | 143.0 ± 24.6 |
| 27 | Kas-1 | 160.6 ± 12.2 | 0.2 ± 0.1 | 2.7 ± 0.2 | 0.0 ± 0.0 | 5.4 ± 0.9 | n.d. | 3.1 ± 0.4 | 0.1 ± 0.0 | 171.8 ± 13.6 |
Previous feeding experiments with the specialist P. xylostella on Col-0 had shown that insect feeding induces a volatile response similar to that observed with coronalon (Herde et al., 2008). In this study, when P. xylostella larvae were applied to rosette leaves of accession Ws, a high-(E)-β-ocimene emitter, emission of (E)-β-ocimene, TMTT, and MeSA was induced upon 21 to 30 h of feeding (Supplemental Fig. S1), with a compound ratio similar to that obtained upon coronalon treatment. Emission rates were substantially lower for all compounds [30-fold lower for (E)-β-ocimene] than after application of coronalon but in the range of those observed for Col-0 upon P. xylostella damage (Herde et al., 2008; Supplemental Fig. S1). Because of this overall lower response, no (E,E)-α-farnesene was detected in Ws in response to P. xylostella feeding.
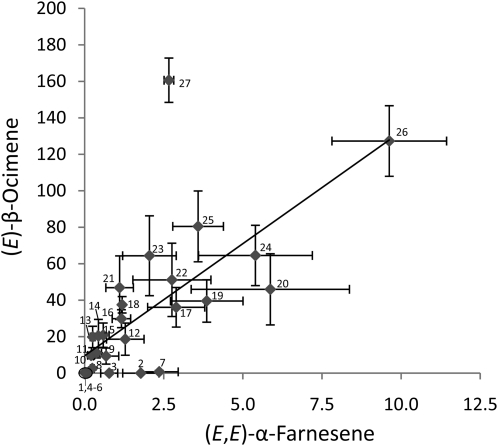
We investigated possible correlations among coronalon-induced emissions of the different volatile compounds in all accessions. A high correlation coefficient was found between emission of (E)-β-ocimene and (E,E)-α-farnesene (Fig. 1; Supplemental Table S1), indicating a common biosynthetic origin of both compounds. However, the formation of (E,E)-α-farnesene in Col-0 and other ecotypes that do not produce (E)-β-ocimene (Table I) suggested the existence of more than one biosynthetic route to (E,E)-α-farnesene in the Arabidopsis genome.
Figure 1.
Correlation of (E)-β-ocimene and (E,E)-α-farnesene emission from coronalon-treated leaves of 27 Arabidopsis accessions. Treatment with coronalon and volatile collection were conducted as described in “Materials and Methods.” Emissions are in ng g−1 fresh weight h−1. Numbers indicate individual accessions according to Table I. Each value represents the mean ± se of three replicates.
The Tandem TPS Genes TPS02 and TPS03 Differ in Expression between the Col-0 and Ws Accessions
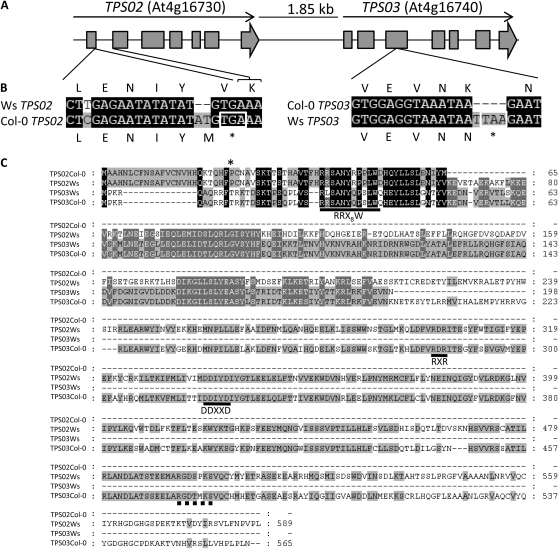
From the analyzed accessions, we selected Ws and Col-0 for further investigation since these two accessions showed clearly different elicitor-induced terpene volatile profiles and therefore seemed suitable for determining the molecular mechanisms underlying these differences. We reasoned that the variation of (E)-β-ocimene emission between the two accessions could be due to the differential expression or function of TPS03 (At4g16740) or a monoterpene synthase closely related to TPS03. A TPS03-encoded recombinant enzyme from Arabidopsis accession C24 was previously shown to catalyze the conversion of the ubiquitous precursor GPP to (E)-β-ocimene (Fäldt et al., 2003). Moreover, transcription of TPS03 was shown to be induced in leaves of the (E)-β-ocimene-emitting ecotype No-0 by treatment with jasmonic acid and wounding. Besides TPS03, only one other monoterpene synthase (TPS10) has been found to be induced in Arabidopsis leaves (Bohlmann et al., 2000). However, this enzyme produces myrcene as the primary product with minor amounts of (E)-β-ocimene in vitro and seems to have negligible activity in the investigated ecotypes, since no emission of myrcene was detected. In the Arabidopsis genome, TPS03 is positioned on chromosome 4 in close proximity to TPS02 (At4g16730; Fig. 2A). Both genes are 51.5% identical at the nucleotide sequence level and share similar structures, with seven exons and six introns, indicating that they likely emerged by gene duplication. Therefore, we analyzed both TPS02 and TPS03 alleles and their expression in the Col-0 and Ws accessions.
Figure 2.
Molecular nature of the TPS02 and TPS03 alleles in accessions Col-0 and Ws. A, Schematic representation of the structures of TPS02 and TPS03. Exons are represented by the gray boxes, and flanking regions and introns are represented by the lines between boxes. B, Alignment of nucleotide and amino acid sequence regions of the TPS02 (left) and TPS03 (right) alleles from accessions Col-0 and Ws indicating frame shift mutations caused by base pair insertions in the Col-0 TPS02 and Ws TPS03 genes. The white box marks premature stop codons. The gene-specific positions of the sequences are indicated. C, Amino acid sequence alignment of the full-length and truncated proteins of the Col-0 and Ws TPS02 and TPS03 alleles. Amino acids shaded in black are conserved in all sequences, and gray shading indicates amino acids conserved in two or three sequences. Dashes indicate gaps inserted for optimal alignment. Horizontal lines mark the highly conserved DDXXD, RXR, and RRX8W motifs. A motif similar to the H-α1 loop region of apple MdAFS1 is marked by a thick dashed line. The asterisk indicates the putative cleavage site for a 25-amino acid plastidial transit peptide of the TPS02 protein.
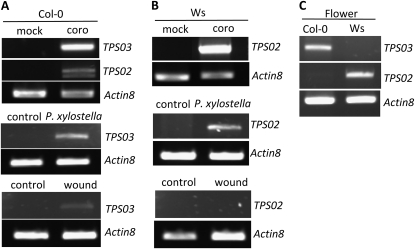
In the Col-0 ecotype, which does not emit (E)-β-ocimene, TPS02 does not encode a full-length TPS protein because of a two-base (AT) insertion 184 nucleotides downstream of the start codon leading to a frame shift and premature translational termination (Fig. 2, B and C). Despite the apparent loss of function of this allele, transcription of TPS02 was induced upon treatment with coronalon (Fig. 3A). A splice variant of the TPS02 transcript was found that lacks a part of the first exon and the entire second exon (Supplemental Fig. S2) and does not code for a functional protein. In contrast to TPS02, the Col-0 TPS03 gene encodes a full-length TPS protein of 565 amino acids (Fig. 2C). Transcription of TPS03 was induced by coronalon treatment as well as in response to 24 h of P. xylostella feeding, while only low levels of TPS03 transcript were observed upon mechanical wounding (Fig. 3A). However, the induced expression of TPS03 [reported as an (E)-β-ocimene synthase in ecotype C24 (Fäldt et al., 2003)] was hard to reconcile with the lack of (E)-β-ocimene emission in the Col-0 ecotype, indicating possible differences in the regulation or function of the Col-0 TPS03 protein compared with that in ecotype C24.
Figure 3.
Semiquantitative RT-PCR analysis of TPS02 and TPS03 transcript levels in Col-0 and Ws tissues. Actin8 transcripts were analyzed as a control. Results are representative for at least three independent experiments. A, TPS02 and TPS03 transcript analysis from rosette leaves of accession Col-0 treated with coronalon (coro; top panel), with P. xylostella larvae (middle panel), and after mechanical wounding (bottom panel). The two amplicons obtained for TPS02 represent splice variants (Supplemental Fig. S2). No TPS02 transcript was detected upon insect feeding and wounding. Treatments were conducted as described in “Materials and Methods.” B, Transcript levels of TPS02 in leaves of accession Ws in response to treatments as described for A. No mRNA of TPS03 was detected. C, Analysis of TPS02 and TPS03 transcripts in flowers of Col-0 and Ws.
We further analyzed the expression of the TPS02 and TPS03 alleles in the (E)-β-ocimene-emitting accession Ws. Semiquantitative reverse transcription (RT)-PCR analysis demonstrated that TPS02 was transcribed upon coronalon treatment (Fig. 3B). Transcription of Ws TPS02 was also induced by P. xylostella feeding, while no transcript was detected upon mechanical wounding (Fig. 3B). A full-length 1,770-bp TPS02 cDNA was amplified from coronalon-treated Ws leaves encoding a 589-amino acid protein (Fig. 2C). The TPS02 coding sequence differs from the Col-0 allele in the position of 18 nucleotides corresponding to 11 amino acid differences and does not show the frame shift mutation due to the AT insertion (Fig. 2B). The Ws TPS02 protein shares 62% amino acid sequence identity with the TPS03 (E)-β-ocimene synthase protein from C24. Thus, it seemed possible that a functional TPS02 enzyme could produce (E)-β-ocimene or very similar monoterpene compounds.
No transcript was found for TPS03 in response to any of the treatments of Ws leaves. To investigate the absence of the TPS03 transcript in Ws in more detail, we amplified the TPS03 gene including the 5′ and 3′ untranslated regions (UTRs) from genomic DNA of Ws. The nucleotide sequences of the Ws TPS03 gene and the Col-0 TPS03 gene were 99.5% identical. An insertion of four nucleotides (TTAA) was found in the third exon, causing a frame shift mutation and premature translational termination (Fig. 2B). We then performed RT-PCR with gene-specific primers designed to amplify small fragments of a putative TPS03 transcript. These experiments resulted in only two amplicons, 240 and 170 bp, situated consecutively at the 5′ end of the gene, indicating the instability and posttranscriptional degradation of the TPS03 mRNA (Supplemental Fig. S3).
We also analyzed the transcription of TPS02 and TPS03 in flowers of the Col-0 and Ws accessions to determine a possible correlation between the expression of one or the other gene and the floral emission of (E)-β-ocimene. Previous analysis of terpene volatiles from flowers of different Arabidopsis accessions demonstrated that inflorescences of Ws but not Col-0 emit (E)-β-ocimene, which is similar to the difference observed for elicitor-induced (E)-β-ocimene emission from both accessions. Inflorescences of both accessions also emit small amounts of (E,E)-α-farnesene. In Ws flowers, TPS02 but not TPS03 was found to be expressed (Fig. 3C). In flowers of the Col-0 ecotype, transcripts of TPS03 were detected, but, in contrast to elicitor-treated leaves, no or only traces of full-length TPS02 mRNA was amplified (Fig. 3C).
Based on the transcriptional differences of TPS02 and TPS03 between the two accessions, we hypothesized that TPS02 is responsible for the formation of (E)-β-ocimene in Ws, while the TPS02 allele is inactive in the Col-0 ecotype. We further presumed that the actively transcribed TPS03 gene in Col-0 might encode a protein that produced (E,E)-α-farnesene, the C15 analog of (E)-β-ocimene from FPP, instead of (E)-β-ocimene itself derived from GPP. Several enzymes with (E)-β-ocimene synthase activity from GPP have been shown to also catalyze the formation of (E,E)-α-farnesene in vitro when supplied with FPP (Pechous and Whitaker, 2004; Nieuwenhuizen et al., 2009; see below). As an intermediate of terpene biosynthesis, GPP is thought to be largely restricted to the plastids, while FPP is restricted mainly to the cytosol (Aharoni et al., 2005). Analysis of putative protein-targeting sequences via computer algorithms indicated that the Col-0 TPS03 protein does not carry a plastidial transit peptide while the TPS02 protein does.
TPS2 and TPS3 Loss-of-Function Plants Lack Induced Emission of (E)-β-Ocimene and/or (E,E)-α-Farnesene
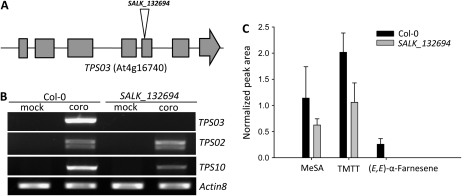
To investigate the in planta function of the Ws TPS02 and Col-0 TPS03 genes, we analyzed elicitor-induced volatile profiles of the respective gene knockout lines. In line Salk_132694, a T-DNA is located in the fifth exon of Col-0 TPS03 (Fig. 4A). In contrast to Col-0 wild-type plants, the mutant line did not accumulate any TPS03 mRNA after coronalon treatment as determined by semiquantitative RT-PCR (Fig. 4B). No (E,E)-α-farnesene was found to be emitted from Salk_ 132694 upon treatment with coronalon (Fig. 4C). Although MeSA and TMTT were released at rates somewhat lower than those of the Col-0 wild type, emission of these two compounds indicated that the elicitor had been successfully administered (Fig. 4C). Moreover, no changes were observed for the induced transcription of the TPS02 pseudogene and the myrcene/(E)-β-ocimene synthase TPS10 in comparison with wild-type plants. Since no other TPSs with putative (E,E)-α-farnesene or (E)-β-ocimene synthase activities are expressed in Col-0 leaves upon coronalon treatment (Herde et al., 2008), these results strongly suggested that the TPS03 gene is responsible for the induced formation of (E,E)-α-farnesene in the Col-0 accession.
Figure 4.
Coronalon-induced expression of TPS03 and volatile emission in detached leaves of Col-0 wild-type plants and the TPS03 T-DNA insertion line SALK_132694. A, Position of the T-DNA insertion in the TPS03 gene. Gray boxes represent exons, and flanking regions and introns are shown by the black line. B, Semiquantitative RT-PCR analysis of transcripts of TPS03 in comparison with genes TPS02 and TPS10. Transcript levels of Actin8 were analyzed as a control. coro, Coronalon. C, Emission of MeSA, TMTT, and (E,E)-α-farnesene as measured between 21 and 30 h of coronalon treatment of Col-0 wild-type plants and the T-DNA insertion line. Normalized peak areas are shown for each compound as analyzed by GC-MS (see “Materials and Methods”). No (E)-β-ocimene could be detected from wild-type or mutant plants under these conditions. Results are average values ± se (n = 3). None of the volatiles was detected in mock controls.
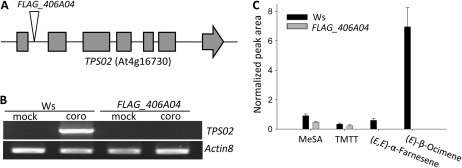
Next, we analyzed line FLAG_406A04, which carries a T-DNA in the first intron of the Ws TPS02 allele (Fig. 5A). When leaves of this mutant were treated with coronalon, no induced TPS02 transcript could be detected in comparison with wild-type Ws plants (Fig. 5B). The absence of the TPS02 transcript correlated with the loss of (E)-β-ocimene and (E,E)-α-farnesene emission in the FLAG_406A04 mutant (Fig. 5C). We also investigated two transgenic Ws lines, in which transcription of TPS02 was severely reduced or completely abolished by RNA interference (RNAi) in response to application of the fungal elicitor alamethicin (Supplemental Fig. S4). As expected, traces or no emission of (E)-β-ocimene and (E,E)-α-farnesene were found in the RNAi lines, confirming the result obtained for the FLAG_406A04 mutant (Supplemental Fig. S4, A and B). MeSA and TMTT were released from all lines at rates similar to those of wild-type Ws, showing that elicitor treatment was effective (Supplemental Fig. S4B). These results demonstrated that formation of (E)-β-ocimene and (E,E)-α-farnesene is dependent on the expression of TPS02 in the Ws ecotype.
Figure 5.
Coronalon-induced expression of TPS02 and volatile emission in detached leaves of Ws wild-type plants and the TPS02 T-DNA insertion line FLAG_406A04. A, Schematic presentation of the position of the T-DNA insertion in the TPS02 gene. Exons are represented by gray boxes, and flanking regions and introns are shown by the black line. B, Semiquantitative RT-PCR analysis of transcripts of TPS02. Transcript levels of Actin8 were analyzed as a control. No full-length transcripts were found for TPS03 and TPS10 in wild-type and mutant plants. coro, Coronalon. C, Emission of MeSA, TMTT, (E,E)-α-farnesene, and (E)-β-ocimene between 21 and 30 h of coronalon treatment of Ws wild-type plants and the T-DNA insertion line. Normalized peak areas are shown for each compound as analyzed by GC-MS (see “Materials and Methods”). Results represent mean values ± se (n = 3). None of the volatiles was detected from mock control leaves.
Recombinant TPS02 and TPS03 Proteins Both Produce (E)-β-Ocimene and (E,E)-α-Farnesene in Vitro
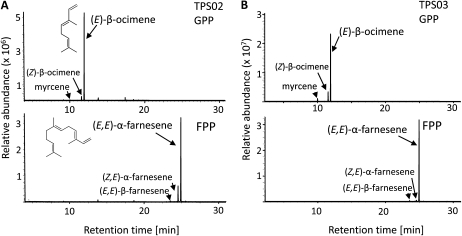
To further confirm the catalytic activities of the TPS02- and TPS03-encoded enzymes in Ws and Col-0, respectively, cDNAs of both genes were cloned into the Escherichia coli expression vector pET101 in fusion with a C-terminal His tag, and the resulting proteins were partially purified by affinity chromatography and assayed for TPS activity. For TPS02, a truncated 1,647-bp cDNA was used that was amplified from the isolated full-length Ws sequence. This truncation removed 41 amino acids containing a predicted plastidial transit peptide upstream of the conserved RR motif at the N terminus of the TPS02 protein (Fig. 2C) in an effort to enhance the activity of the recombinant enzyme. It has previously been shown that monoterpene synthases often have higher specific activity when expressed in E. coli as mature proteins rather than as full-length preproteins (Williams et al., 1998). The affinity-purified, recombinant Ws TPS02 protein converted the substrate GPP into (E)-β-ocimene as the major product, with (Z)-β-ocimene and myrcene as minor products (Fig. 6A). In assays with FPP as the substrate, the TPS02 enzyme produced primarily (E,E)-α-farnesene and small amounts (Z,E)-α-farnesene and (E,E)-β-farnesene (Fig. 6A). No activity was observed with GGPP as the substrate, and none of the terpene products was found in purified extracts of E. coli carrying the empty pET101 vector (data not shown).
Figure 6.
GC-MS analysis of monoterpene and sesquiterpene products of recombinant Ws TPS02 and Col-0 TPS03 enzymes. Recombinant proteins were expressed in E. coli, extracted, partially purified, and applied for TPS assays using the substrates GPP and FPP. A, Total ion GC-MS chromatograms of monoterpenes (top) and sesquiterpenes (bottom) produced by recombinant Ws TPS02 protein from GPP and FPP, respectively. The molecular structures of (E)-β-ocimene and (E,E)-α-farnesene are shown. B, Monoterpene products (top) and sesquiterpene products (bottom) of recombinant Col-0 TPS03 enzyme detected in assays with GPP and FPP, respectively. Terpene products were identified by comparison with authentic standards or by library suggestion [for (Z,E)-α-farnesene]. No products were found in purified extracts from E. coli carrying the empty expression vector.
We also tested whether a truncated 539-amino acid protein resulting from possible alternative translation at nucleotide 153 of the TPS02 gene from Col-0 had enzyme activity. However, this protein was shown to be inactive when expressed in E. coli. In addition, we investigated if removal of the frame shift mutation in the Col-0 TPS02 gene restored a functional TPS02 protein. A 1,772-bp cDNA of Col-0 TPS02 was amplified from RNA isolated from coronalon-treated Col-0 leaves, and site-directed mutagenesis was applied to remove the AT two-base nucleotide insertion (Fig. 2B). When the resulting cDNA clone was expressed in E. coli as described above, no TPS activity was detected, indicating that additional mutations in the Col-0 TPS02 gene contribute to the loss of enzyme activity.
To analyze the activity of the Col-0 TPS03-encoded protein, a 1,695-bp TPS03 cDNA corresponding to the full-length TPS03 protein was isolated from RNA of Col-0 leaves treated with coronalon. When enzyme assays were performed with GPP and FPP as substrates, the recombinant, partially purified TPS03 enzyme produced the same monoterpenes and sesquiterpenes as those synthesized by the TPS02 protein from Ws (Fig. 6B).
To analyze possible differences of the recombinant TPS proteins in conversion rates of GPP and FPP, we determined the catalytic properties of both enzymes for these substrates. Both enzymes had similar Vmax and kcat values for GPP and FPP, with a 7- to 8-fold higher catalytic activity for GPP than for FPP (Table II). The apparent Km values for GPP and FPP (1.7–6.7 μm) were low for both enzymes (Table II) and in the range of Km values reported previously for other plant monoterpene and sesquiterpene synthases (Cane, 1999), specifically other (E,E)-α-farnesene/(E)-β-ocimene synthases (Nieuwenhuizen et al., 2009). Despite these similarities, the Ws TPS02 protein had an approximately 4-fold lower Km for GPP and an approximately 1.5-fold higher Km for FPP in comparison with the Col-0 TPS03 enzyme, resulting in 4-fold higher and 1.5-fold lower corresponding kcat/Km values for GPP and FPP, respectively (Table II). Catalysis of both enzymes was dependent on Mg2+ for maximum activity. We also tested the dependency of both enzymes on K+ ions. Neither Ws TPS02 nor Col-0 TPS03 showed any change in activity in the presence of 40 mm K+ (data not shown).
Table II. Kinetic parameters of Ws TPS02 and Col-0 TPS03 recombinant enzymes.
Each value represents the average ± se of three replicates. Substrates were at 20 mm Mg2+.
| Enzyme | Substrate | Km | Vmax | kcat | kcat/Km |
| μm | pkat mg−1 | s−1 | s−1 mm−1 | ||
| Ws TPS02 | GPP | 1.7 ± 0.3 | 539.0 ± 40.0 | 3.5 × 10−2 ± 2.6 × 10−3 | 21.4 ± 2.35 |
| FPP | 4.0 ± 0.4 | 69.0 ± 3.1 | 4.5 × 10−3 ± 2 × 10−4 | 1.12 ± 0.05 | |
| Col-0 TPS03 | GPP | 6.7 ± 0.9 | 514.8 ± 49.8 | 3.4 × 10−2 ± 3.3 × 10−3 | 5.12 ± 0.18 |
| FPP | 2.6 ± 0.2 | 72.4 ± 1.2 | 4.8 × 10−3 ± 8.0 × 10−5 | 1.82 ± 0.09 |
Taken together, our analysis of the recombinant TPS02 and TPS03 proteins from Ws and Col-0, respectively, showed that both enzymes can produce (E)-β-ocimene and (E,E)-α-farnesene in vitro without major kinetic differences. However, analyses of the loss-of-function plants clearly indicated different product profiles of the encoded enzymes in vivo, with Ws TPS02 making both compounds (Fig. 5C) and Col-0 TPS03 producing predominantly (E,E)-α-farnesene (Fig. 4C). Thus, these findings supported the notion that TPS02 and TPS03 might be present in separate subcellular compartments with differential access to the substrates GPP and FPP.
Subcellular Localization of TPS02 in Plastids and TPS03 in the Cytosol
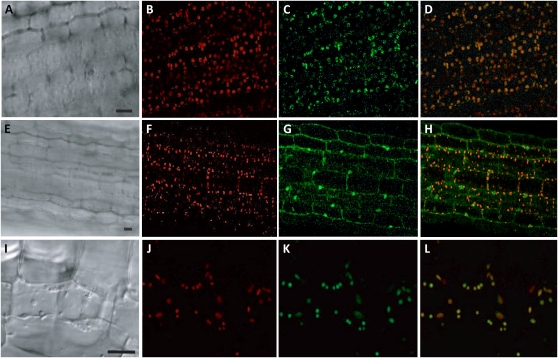
Analysis of putative targeting sequences using different algorithms (CHLOROP [http://www.cbs.dtu.dk/services/ChloroP], TARGETP [http://www.cbs.dtu.dk/services/TargetP], PWOLF PSORT [http://wolfpsort.seq.cbrc.jp], and Predator [http://urgi.versailles.inra.fr/predotar/predotar.html]) suggested that the TPS02 protein from Ws carries a plastidial transit peptide of approximately 25 amino acids and therefore is targeted to chloroplasts. To experimentally determine the subcellular localization of the Ws TPS02 protein, a 105-bp cDNA fragment beginning with the start codon of the TPS02 gene was inserted into the vector pCAMBIA 1302 under the control of the cauliflower mosaic virus 35S promoter, generating a 35-amino acid TPS02 peptide with a C-terminal fusion to GFP. GFP analysis of hypocotyl cells of several independent plant lines transformed with the TPS02-GFP construct showed green fluorescence located in plastids, which clearly indicated that the TPS02 protein is targeted to chloroplasts (Fig. 7, C and D).
Figure 7.
Confocal laser scanning microscopy of stably expressed Ws TPS02 and Col-0 TPS03 peptide-GFP fusion proteins. Microscopic images were taken from the hypocotyls of 2-week old seedlings. The first column (A, E, and I) shows light microscopic images of hypocotyl cells. Chlorophyll autofluorescence, detected in the red channel, is shown in the second column (B, F, and J). The third column (C, G, and K) shows GFP fluorescence, detected in the green channel. The fourth column (D, H, and L) shows merged green and red channel images. A 35-amino acid N-terminal TPS02 peptide (containing a putative 25-amino acid plastidial transit peptide) fused to GFP localizes to chloroplasts (A–D). No plastidial localization was detected for a fusion protein containing a 50-amino acid N-terminal peptide of Col-0 TPS03 (E–H). Ferredoxin N-reductase-eGFP carrying a plastidial target peptide was used as a chloroplast marker (I–L). Bars = 20 μm.
In contrast to TPS02, no consistent prediction for the subcellular localization of TPS03 was obtained with different algorithms, with suggestions of either a cytosolic/nuclear localization or the presence of a nine- to 28-amino acid mitochondrial targeting sequence. Longer transit peptides beginning at earlier start codons were not in frame with the TPS03 protein. We generated a Col-0 TPS03-GFP fusion construct as described for TPS02 by cloning of a 150-bp TPS03 cDNA fragment encoding a 50-amino acid N-terminal TPS03 peptide. In plants transformed with the TPS03-GFP construct, GFP fluorescence was not observed in plastids but appeared to reside in the cytosol with diffusion into nuclei (Fig. 7, G and H).
The GFP constructs clearly support the localization of TPS02 in plastids and TPS03 in the cytosol. Given the preponderance of GPP in the plastids, TPS02 thus seems responsible for synthesizing (E)-β-ocimene from GPP in the Ws accession. On the other hand, the cytosolic TPS03 is in a compartment thought to be supplied with FPP rather than GPP and thus is likely to form (E,E)-α-farnesene from FPP in Col-0. The formation of small amounts of (E,E)-α-farnesene in Ws is probably also attributable to TPS02 and demonstrates the existence of low levels of FPP in the chloroplast or low levels of TPS02 in a subcellular compartment provided with FPP.
Inhibitor Studies of the Methylerythritol Phosphate and Mevalonate Pathways Support the Subcellular Localization of TPS02 and TPS03
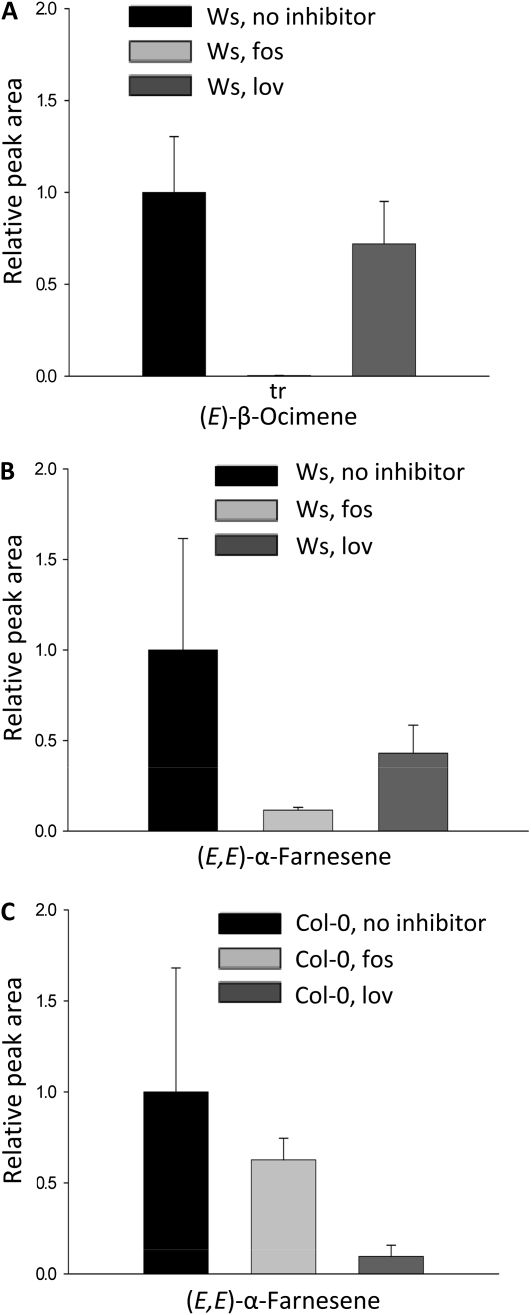
To further investigate the subcellular compartmentation of the Ws TPS02 and Col-0 TPS03 enzymes, inhibitors were employed that are specific for one of the two pathways of isopentenyl diphosphate/dimethylallyl diphosphate formation in plants, either the plastid-localized methylerythritol phosphate (MEP) pathway (Lichtenthaler, 1999) or the cytosol-localized mevalonate pathway. Inhibitors were applied together with coronalon to leaves of both ecotypes. Administration of fosmidomycin, which inhibits 1-deoxyxylulose-5-phosphate reductoisomerase in the MEP pathway, to Ws leaves caused an almost complete loss of coronalon-induced (E)-β-ocimene emission (Fig. 8A). By contrast, inhibition of the mevalonate pathway enzyme 3-hydroxy-3-methylglutaryl-CoA reductase by lovastatin led to only a 28% reduction in (E)-β-ocimene formation (Fig. 8A). The emission of (E,E)-α-farnesene was reduced by both inhibitors in a similar way as (E)-β-ocimene, indicating that both products are likely produced in the same compartment (Fig. 8B). Treatment of Col-0 leaves with one or the other inhibitor caused effects opposite to those observed for the Ws ecotype. While application of lovastatin severely reduced the release of (E,E)-α-farnesene in response to coronalon treatment, emission of (E,E)-α-farnesene was inhibited by only 37% upon administration of fosmidomycin (Fig. 8C). The fact that some reduction was observed in the emission of (E,E)-α-farnesene by treatment of Col-0 with fosmidomycin and in the emission of (E)-β-ocimene upon treatment of Ws with lovastatin can be attributed to an exchange of terpenoid precursors between the cytosol and plastid, as described previously (Hemmerlin et al., 2003; Laule et al., 2003; Schuhr et al., 2003; Dudareva et al., 2005). Overall, our results are in agreement with the biosynthesis of (E)-β-ocimene and (E,E)-α-farnesene by Ws TPS02 in plastids and the formation of (E,E)-α-farnesene by the Col-0 TPS03 enzyme in the cytosol.
Figure 8.
Effects of the MEP pathway inhibitor fosmidomycin (fos) and the mevalonate pathway inhibitor lovastatin (lov) on emission of (E)-β-ocimene and (E,E)-α-farnesene from leaves of accessions Ws and Col-0. Volatiles were collected for 8 h from detached rosette leaves treated with coronalon in the presence of a single inhibitor. Relative peak areas of compounds are shown. Peak areas from controls without the addition of inhibitors were arbitrarily set to 1.0. Results represent mean values ± se (n = 3).
Expression Profile of TPS02 and TPS03 Promoters
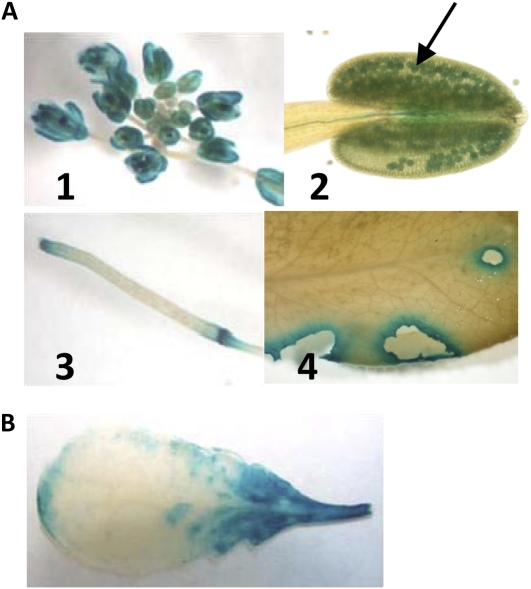
To gain a better understanding of the organ- and tissue-specific formation of (E)-β-ocimene and (E,E)-α-farnesene in leaves and flowers of the Col-0 and Ws ecotypes, a 2.1- and 1.8-kb intergenic fragment upstream of the start codon of the functionally active Ws TPS02 and Col-0 TPS03 gene, respectively, was cloned 5′ to the GUS reporter gene of the pDW137 vector, and the TPS promoter-GUS constructs were stably transformed into the respective Ws and Col-0 backgrounds. GUS activity driven by the Col-0 TPS03 promoter was detected in sepal, anther, and stigma of immature and mature flowers (Fig. 9A, 1). In anthers, GUS activity was found particularly in pollen (Fig. 9A, 2). Moreover, GUS staining was observed in the abscission zone of developing siliques (Fig. 9A, 3). In leaves, TPS03-GUS activity was induced locally around sites of feeding damage by P. xylostella (Fig. 9A, 4). No activity was found in undamaged leaves. GUS staining was also detected at mechanical wound sites (data not shown), in agreement with the transcription of TPS03 observed upon mechanical wounding (Fig. 3A). Analysis of transgenic Ws plants expressing GUS under the control of a TPS02 promoter fragment showed GUS staining only in response to treatment of leaves with the strong elicitor coronalon (Fig. 9B). GUS activity was strongest around the petiole after submersion in coronalon solution. No GUS activity was observed in Ws flowers or leaves upon P. xylostella feeding, in contrast to the detection of TPS02 transcripts in these organs, which suggested that the cloned promoter fragment lacked regulatory elements responsible for full activity in the intact plant.
Figure 9.
GUS activity in Col-0 ProTPS03:GUS and Ws ProTPS02:GUS plants. The results are representative for at least three independent lines. A, Histochemical GUS staining of an inflorescence (1), pollen grains (2, arrow), a silique (3), and a P. xylostella-damaged mature leaf from a Col-0 ProTPS03:GUS plant (4). In panel 4, GUS activity is induced locally around the sites of feeding damage. B, Induced GUS activity in a rosette leaf of a Ws ProTPS02:GUS plant treated for 24 h with coronalon through the petiole. The results are representative for at least three independent lines.
DISCUSSION
Elicitor- and Insect-Induced Volatile Emissions Vary among Arabidopsis Ecotypes
Intraspecific variation of herbivore-induced volatile emissions has been reported primarily from varieties of cultivated plants (Loughrin et al., 1995; Geervliet et al., 1997; Gouinguene et al., 2001; Degen et al., 2004; Lou et al., 2006) but has been the subject of only a few investigations of wild species such as solanaceous plants (Halitschke et al., 2000; Glawe et al., 2003; Hare, 2007; Delphia et al., 2009) and teosinte (Gouinguene et al., 2001). In this study, we conducted a survey of 27 Arabidopsis accessions for volatiles released upon treatment with coronalon, a synthetic mimic of octadecanoid plant hormones that induces volatile blends similar to those emitted upon insect feeding (Schuler et al., 2001; Herde et al., 2008). Most Arabidopsis accessions emitted blends of the same composition of volatiles, all of which are common constituents of herbivore-induced volatile mixtures (Turlings et al., 1990; McCall et al., 1994; Takabayashi and Dicke, 1996; Pichersky and Gershenzon, 2002; Ament et al., 2004).
Large ecotype-specific quantitative differences were observed for the emission of (E)-β-ocimene, with approximately one-third of the accessions, including Col-0, producing none or only traces of this monoterpene. Quantitative variation was also apparent in the emission of the other volatile compounds (MeSA, TMTT), resulting in ecotype-specific differences of compound ratio. Our results correspond to differences in herbivore-induced volatile blends observed among genotypes of other wild species (Halitschke et al., 2000; Gouinguene et al., 2001; Hare, 2007; Delphia et al., 2009). The ecological significance of intraspecific variation of insect-induced volatile production is still not well understood because the roles of these compounds in plants are not completely elucidated. Among the investigated Arabidopsis accessions, no definite correlation could be found between the geographical distribution and the total amount of volatiles emitted or their profiles. However, variability of herbivore-induced volatile emissions may reflect habitat-dependent, selective adaptations of ecotypes to specific populations of herbivores and their natural enemies. Natural selection is also assumed to be responsible for significant variation of other specialized metabolites among Arabidopsis accessions, such as glucosinolates (Kliebenstein et al., 2001). The insect-induced volatile blend emitted from Arabidopsis Col-0 has been shown to attract parasitic wasps such as Cotesia rubecula (Van Poecke et al., 2001), and preferences of parasitoid wasps for particular volatile mixtures have been demonstrated (Hoballah et al., 2002). Moreover, associative learning of parasitoids (De Boer and Dicke, 2006; Smid and Vet, 2006) may facilitate optimal use of the volatile cues emitted by a specific plant population. Since volatile terpenes exhibit antimicrobial activities, ecotype-specific variation of induced volatiles might also emerge under selective pressure by different microbial pathogens. Bacterial and fungal pathogens can elicit emission of volatile mixtures in Arabidopsis, which are very similar to those induced by insects, as shown for infections by Pseudomonas syringae (Attaran et al., 2008) and treatment with the fungal elicitor alamethicin (Herde et al., 2008). Interestingly, two accessions, Bla-1 and Bla-10, originating from the same region (Blanes/Genora, Spain), differed more than 100-fold in the emission of (E)-β-ocimene, indicating intraspecific variability even among populations occurring close together.
A significant correlation was apparent between emissions of (E)-β-ocimene and (E,E)-α-farnesene (Fig. 1; Supplemental Table S1), which suggested that in most accessions both terpene volatiles are produced either simultaneously by a single bifunctional (E)-β-ocimene/(E,E)-α-farnesene synthase or by coexpressed enzyme activities. Significant correlations were also found between emissions of MeSA and TMTT, on the one hand, and MeSA and (E,E)-α-farnesene, on the other (Supplemental Table S1), which reflect overlapping, coronalon-induced responses in the expression of biosynthetic enzymes in the formation of MeSA (benzoic acid/salicylic acid carboxyl methyltransferase 1; Chen et al., 2003a), TMTT, and (E,E)-α-farnesene.
Col-0 and Ws Accessions Show Allelic Differences for the Duplicated (E)-β-Ocimene/(E,E)-α-Farnesene Synthase Genes TPS02 and TPS03 That Lead to Variability in Terpene Biosynthesis
Within the Arabidopsis TPS family, the proteins encoded by genes TPS02 and TPS03 cluster together with five other monoterpene synthases (TPS10, TPS14, TPS23, TPS27, and TPS24), all of which belong to the plant TPS-b subfamily (Aubourg et al., 2002; Chen et al., 2003b; Supplemental Fig. S5). Based on their close proximity and similarity in gene structure and sequence, TPS02 and TPS03 are most likely the result of tandem gene duplication. Several other TPS genes are arranged as tandem pairs in the Arabidopsis genome, such as identical gene copies of the root-expressed 1,8-cineole synthase AtTPS-Cin (TPS23 and TPS27; Chen et al., 2004) and the (Z)-γ-bisabolene synthases TPS12 and TPS13 (Ro et al., 2006). Rapid radiation of genes by duplication and sequence divergence is common within the plant TPS superfamily and other gene families of plant specialized metabolism (Pichersky and Gang, 2000; Kliebenstein et al., 2001; Köllner et al., 2004) and is thought to contribute to generating the diversity of metabolites that function in plant-organism interactions (van der Hoeven et al., 2000; Aubourg et al., 2002).
The Ws and Col-0 accessions each maintain only one functional allele at the TPS02 and TPS03 locus, whose recombinant proteins exhibit both (E)-β-ocimene and (E,E)-α-farnesene synthase activities. The TPS03 allele in Ws and the TPS02 allele in Col-0 are inactive because of frame shift mutations, which cause the introduction of premature stop codons (Fig. 2B). Both alleles are still transcribed under induced conditions; however, in the case of the Ws TPS03 gene, most of the mRNA is degraded except for a short fragment upstream of the nonsense codon (Supplemental Fig. S3). Premature nonsense codons, particularly those occurring in early exons, can decrease mRNA stability by activating nonsense-mediated decay pathways (vanHoof and Green, 1996; Gutierrez et al., 1999; Hori and Watanabe, 2007). For the TPS02 transcript in Col-0, where the nonsense codon is positioned at the junction between the first and second exons, full-length mRNA transcripts were observed in elicitor-treated leaves. By contrast, no or only traces of full-length mRNA of TPS02 was found in Col-0 flowers, and only partial transcripts of this gene were previously amplified successfully from floral tissue (Chen et al., 2003b), which indicates a possible higher mRNA instability of the TPS02 pseudogene in Col-0 flowers than under induced conditions in leaves.
In some cases, transcripts of pseudogenes have been shown to regulate the stability of their homologous coding gene transcripts (Hirotsune et al., 2003). By analyzing the Ws FLAG_406A04 mutant and TPS02 RNAi lines, we did not find evidence for this regulatory mechanism, since the absence of the TPS02 transcript did not positively affect the mRNA stability of the TPS03 allele. In addition, no microRNAs have been identified for both TPS02 and TPS03 genes.
Examples of pseudogene transcripts have been described in other defense metabolic pathways in Arabidopsis (Kliebenstein et al., 2001). In particular, stress-responsive gene families such as the TPS family have elevated levels of pseudogenization, which can be interpreted as a rapid turnover of genes under varying selection pressures (Thibaud-Nissen et al., 2009; Zou et al., 2009). Analysis of the TPS02-TPS03 gene pair in Col-0 and Ws demonstrated that either one of the duplicated genes can lose function. However, since most of the investigated accessions emit (E)-β-ocimene, selection pressure seems to support the expression of an active (E)-β-ocimene synthase. Moreover, the presence of a full-length although inactive TPS02 transcript in elicitor-treated leaves of the Col-0 ecotype indicates a more recent loss of function of the TPS02 allele than the TPS03 allele.
Allelic variation of TPS genes contributing to terpene diversity has also been described in varieties of different crop plants such as basil, tomato (Solanum lycopersicum), and maize (van der Hoeven et al., 2000; Iijima et al., 2004; Köllner et al., 2004). For example, in maize, inactivation of alleles of the insect-induced, duplicated sesquiterpene synthase genes tps4 and tps5, caused by frame shift mutation, was observed in a comparison of two different cultivars (Köllner et al., 2004). While extrapolating from these results to wild species is possible, the findings presented here provide a direct demonstration that allelic diversification of TPS genes in wild gene pools contributes to the natural variation in terpene formation.
Subcellular Compartmentalization of the TPS02 and TPS03 Proteins Contributes to Ecotype Variation of Induced (E)-β-Ocimene and (E,E)-α-Farnesene Emission
Subcellular localization experiments demonstrated that the Ws TPS02 protein is targeted to plastids (Fig. 7, C and D), where it converts GPP to (E)-β-ocimene. Inactivation of TPS02 in Ws caused the loss of emission of both (E)-β-ocimene and (E,E)-α-farnesene (Fig. 5; Supplemental Fig. S4), which clearly showed that the TPS02 enzyme not only converts GPP to (E)-β-ocimene in plastids but also catalyzes FPP to (E,E)-α-farnesene conversion in the same organelle. The presence of plastidial FPP was also inferred when a strawberry (Fragaria species) linalool/nerolidol synthase carrying a plastid-targeting peptide was expressed in Arabidopsis, causing the emission of a small amount of the sesquiterpene nerolidol together with linalool (Aharoni et al., 2003). A plastidial FPP pool might be the result of an import of FPP from the cytosol as a result of metabolic cross talk between both compartments (Schuhr et al., 2003) and/or the biosynthesis of FPP inside the organelle. The latter explanation is supported by the lower emission of (E,E)-α-farnesene in the presence of fosmidomycin, an inhibitor of the plastid-localized MEP pathway, than after treatment with lovastatin, an inhibitor of the cytosol-localized mevalonate pathway (Fig. 8, A and B). Although an isoform of the Arabidopsis FPP synthase 1 has been shown to be targeted to mitochondria, a specific FPP synthase activity in plastids has not been demonstrated. Instead, FPP might be produced as a side product of plastidial GPP synthase activity.
In comparison with TPS02, the TPS03 protein lacks 14 amino acids at its N terminus (Fig. 2C) and is not targeted to plastids but resides instead in the cytosol (Fig. 7, G and H). The TPS03 enzyme produces (E,E)-α-farnesene mostly from cytosolic FPP, which is evident from a severe reduction of (E,E)-α-farnesene emission by treatment with lovastatin in comparison with the application of fosmidomycin (Fig. 8C). Formation of only trace amounts of (E)-β-ocimene in the Col-0 ecotype indicates the absence of a high GPP level in the cytosol and rules out an efficient export of GPP from plastids to the cytosol. Experimental evidence for small cytosolic levels of GPP has recently been provided from tobacco (Nicotiana tabacum) and kiwifruit (Actinidia deliciosa; Wu et al., 2006; Nieuwenhuizen et al., 2009).
A previous analysis of TPS03 from accession C24 assumed that the TPS03 enzyme is responsible for the in planta formation of (E)-β-ocimene based on the characterization of the overexpressed protein in vitro (Fäldt et al., 2003). However, since the N-terminal region of the C24 TPS03 protein is identical to that of the Col-0 enzyme and because of negligible GPP pools in the cytosol of Arabidopsis leaf cells, we can now hypothesize that (E)-β-ocimene, if emitted from this ecotype, is probably produced by a functional TPS02 protein rather than a TPS03 enzyme activity. No (E,E)-α-farnesene synthase activity was found for the TPS03 recombinant enzyme from C24 (Fäldt et al., 2003), despite only a single amino acid difference at position 267 (Ser in Col, Phe in C24). It will be interesting to determine to what extent this amino acid change is indeed responsible for the loss of (E,E)-α-farnesene synthase activity of the TPS03 protein.
Subcellular segregation of homologous bifunctional TPSs in plastids and the cytosol has also been documented in cultivated plants, such as for two linalool/nerolidol synthases in snapdragon (Nagegowda et al., 2008) and in cultivated strawberry (Aharoni et al., 2004). Our results on enzymes from a noncultivated species expand these findings by showing that differential subcellular targeting of dual-function TPSs is a molecular mechanism of general importance in the natural evolution of intraspecific volatile terpene diversity.
TPS02 and TPS03 Expression Is under Constitutive Control in Arabidopsis Flowers and Stress Induced in Leaves
Transcript analysis of the Ws TPS02 and Col-0 TPS03 genes showed that expression of both genes is induced in leaves upon treatment with coronalon and in response to feeding by P. xylostella (Fig. 3, A and B). Moreover, histochemical assays of Col-0 TPS03 promoter activity demonstrated local expression of this gene at the site of P. xylostella feeding damage (Fig. 9A, 4). The fact that no expression of Col-0 TPS03 was found in undamaged leaves of P. xylostella-treated plants supports the notion of the lack of a systemic response in herbivore-induced terpene volatile emission in Arabidopsis. Induction of the TPS03 transcript was also demonstrated in leaves of Arabidopsis ecotype No-0 in response to jasmonate treatment and mechanical wounding but not to feeding by the specialist herbivore P. rapae (Fäldt et al., 2003), suggesting ecotype- and/or insect-specific differences in herbivore-induced responses of the TPS03 gene.
Both Ws TPS02 and Col-0 TPS03 are also expressed constitutively in flowers (Fig. 3C), which is consistent with the formation of (E)-β-ocimene and (E,E)-α-farnesene as common constituents of floral volatile blends in plants (Dudareva et al., 2003; Nieuwenhuizen et al., 2009). However, for the TPS03 gene, the strong promoter-GUS activity and transcript abundance in Col-0 flowers (Fig. 9A, 1 and 2) do not correlate with the low emissions of its enzymatic product (E,E)-α-farnesene (approximately 0.3 ng h−1 per 70 inflorescences) reported from Col-0 floral tissues (Tholl et al., 2005). This discrepancy might be caused by differences in TPS03 enzyme activity and/or substrate availability in floral tissue in comparison with elicitor-treated leaves, where (E,E)-α-farnesene is readily detected. The specific expression profile of TPS03 in the stigma and sepal of Col-0 flowers may reflect the functions of (E,E)-α-farnesene in pollinator attraction or florivore/antimicrobial defense, as discussed for the flower-specific genes TPS21 [encoding a (E)-β-caryophyllene synthase; Tholl et al., 2005] and TPS24 (a multiproduct monoterpene synthase), which exhibit similar expression patterns in Arabidopsis flowers (Chen et al., 2003b). Interestingly, TPS03 promoter activity was observed in pollen grains and resembles the recently reported expression of a valencene sesquiterpene synthase in the pollen of grape (Vitis vinifera) flowers (Martin et al., 2009). A pattern of constitutive expression in reproductive organs and induced expression in vegetative tissues has also been observed for Arabidopsis geranyllinalool synthase (TPS04/GES; Herde et al., 2008) in the formation of the volatile homoterpene TMTT and has been demonstrated for other genes involved in plant defense (Pollak et al., 1993; Hoegen et al., 2002; Stotz et al., 2009).
In contrast to (E,E)-α-farnesene, (E)-β-ocimene is the predominant volatile in Ws flowers (Tholl et al., 2005), consistent with the expression of an active TPS02 gene (Fig. 3C). All of the accessions that emit (E)-β-ocimene from their foliage under induced conditions also release (E)-β-ocimene from floral tissue (Tholl et al., 2005), while nonemitters or trace emitters of (E)-β-ocimene from leaves also lack emissions from flowers. Therefore, we assume that (E)-β-ocimene is produced primarily by the TPS02 enzyme in both leaves and flowers of most of the investigated ecotypes. Despite amplification of a 2.1-kb intergenic promoter region, we were unable to detect TPS02 promoter activity in flowers and in response to insect feeding. An expanded analysis of the TPS02 promoter region may reveal regulatory elements necessary for full promoter activity and allow a more detailed comparison of organ-specific expression and function for the TPS02 and TPS03 genes.
Formation of (E)-β-Ocimene and (E,E)-α-Farnesene by Bifunctional TPSs Evolved Several Times in Flower, Fruit, and Herbivore-Induced Volatile Biosynthesis
A phylogenetic comparison of the Arabidopsis Ws TPS02 and Col-0 TPS03 proteins with other plant TPSs shows that both enzymes cluster in the TPS-b subfamily together with the bifunctional (E,E)-α-farnesene/(E)-β-ocimene synthases from apple (Malus domestica; MdAFS1) and pear (Pyrus communis; Pechous and Whitaker 2004; Supplemental Fig. S5). Several other TPSs producing (E)-β-ocimene or (E,E)-α-farnesene or both of these terpenes have been identified previously from gymnosperms (TPS-d; Martin et al., 2004) and in the TPS-a, -b, -f, and -g subfamilies of angiosperms (Dudareva et al., 2003; Arimura et al., 2004; Mercke et al., 2004; Nieuwenhuizen et al., 2009; Supplemental Fig. S5). The sequence variation among these enzymes demonstrates that the formation of (E,E)-α-farnesene and (E)-β-ocimene has arisen independently several times in the evolution of higher plants, suggesting repeated selection for their roles in pollinator attraction, fruit dispersal, and plant defense.
The activity of the MdAFS1 enzyme from apple was shown to be dependent on K+, and the protein contains an H-α1 loop motif for optimal binding of K+ ions (Green et al., 2009). The TPS02 and TPS03 proteins were not K+ dependent, and in these as well as other TPS-b monoterpene synthases, a Ser residue of the H-α1 loop motif is replaced by a Lys residue at the corresponding position (Fig. 2C).
In contrast to the bifunctional (E,E)-α-farnesene/(E)-β-ocimene synthases from apple and kiwifruit (AdAFS1, TPS-f subfamily), which preferably produce (E,E)-α-farnesene from FPP (Green et al., 2009; Nieuwenhuizen et al., 2009), the recombinant TPS02 and TPS03 enzymes exhibit 19- and 3-fold higher catalytic efficiency for GPP than FPP, respectively (Table II), suggesting their original function as monoterpene synthases. The cytosolic Col-0 TPS03 enzyme is slightly more efficient in converting FPP to (E,E)-α-farnesene than the plastidic Ws TPS02 protein and less efficient in producing (E)-β-ocimene from GPP (Table II). These differences may reflect substrate preferences arising from the predominant substrate present in the subcellular environment of the enzyme.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds of all accessions with the exception of Col-0 and Ws were obtained from Tom Mitchell-Olds. Seeds of Arabidopsis T-DNA insertion mutant lines (TPS3KO [SALK_132694] and TPS2KO [FLAG_406A04]) were from the Arabidopsis Biological Resource Center stock center and INRA, respectively. Wild-type and transgenic/mutant plants were cultivated on soil (Sunshine Growing Mix No.1:sand, 8:1) for 5 to 6 weeks under controlled growth conditions (10-h-light/14-h-dark photoperiod with 150 μmol m−2 s−1 photosynthetically active radiation, 23°C, 55% relative humidity). Kanamycin- or hygromycin-resistant transgenic plants were preselected on 1× Murashige and Skoog (Duchefa) plates with 1% Suc and 100 μg mL−1 kanamycin or 30 μg mL−1 hygromycin prior to being transferred to soil. Hydroponic plants of various accessions were grown from seeds on rock wool support (Gibeaut et al., 1997) under the same light and temperature conditions as described above. All plants were used in the prebolting rosette stage.
Reagents and Radiochemicals
Unlabeled GPP and FPP were purchased from Echelon Biosciences. Tritium-labeled GPP and FPP ([1-3H]GPP and [1-3H]FPP, both approximately 0.74 TBq mmol−1) were purchased from American Radiolabeled Chemicals. All other reagents or solvents were obtained from Fisher Scientific, Sigma-Aldrich, Invitrogen, and Fluka, unless otherwise stated.
Plant Treatments
For treatment of various accessions with coronalon, hydroponically grown plants were placed with their roots in glass beakers containing 30 mL of hydroponic medium with 33 μg mL−1 (100 μm) coronalon, 0.1% ethanol (Schuler et al., 2004). Treatment was performed for 30 h. For treatments of detached Arabidopsis leaves with coronalon or alamethicin, 16 leaves were cut off from soil-grown plants and transferred with their petioles into glass beakers filled with 10-mL aqueous solutions of 16.5 μg mL−1 (50 μm) coronalon, 0.1% ethanol or 10 μg mL−1 alamethicin, 0.1% ethanol. Mock solutions for all treatments contained 0.1% ethanol. Treatments were performed for 24 h (coronalon) and 30 h (alamethicin). For inhibitor treatments of detached leaves, leaves were placed in aqueous solutions of fosmidomycin (50 μm) and lovastatin (50 μm) and incubated for 24 h prior to the addition of coronalon (50 μm) and an additional 24-h treatment. Mock treatments were done with 0.2% ethanol. For wounding experiments, fully expanded rosette leaves of intact soil-grown plants were evenly penetrated 20 times by needles and harvested after 30 h. Insect feeding experiments were performed by placing an average of six third- to fourth-instar Plutella xylostella larvae on each fully expanded rosette leaf of soil-grown plants and allowing them to feed for 24 h (for RNA extraction and GUS staining) or 30 h (for volatile collection). P. xylostella larvae were from a G88 colony and reared on artificial diet with a wheat germ base at 27°C with an 18-h-light/6-h-dark cycle.
Volatile Collection and Analysis
Volatile collection from detached leaves or intact hydroponic plants was performed in 1-L bell jars using the closed-loop stripping method (Donath and Boland, 1995) under controlled growth conditions as described previously (Chen et al., 2003b). If not indicated otherwise in the figure legends, volatiles were collected in the light for 8 h during 22 to 30 h from the beginning of the treatment. Collections were performed during this time period because of higher volatile emissions in comparison with earlier time points of treatment. Volatiles emitted from inhibitor-treated rosette leaves were collected between 40 and 48 h of incubation with the inhibitor (i.e. between 16 and 24 h of coronalon treatment). For the P. xylostella treatment, plants were placed with their root balls wrapped in aluminum foil in 3-L bell jars and volatiles were collected during 21 to 30 h from the start of larval feeding. Volatiles were trapped on 25 mg of Super-Q (hydroponic plants, insect feeding; Tholl et al., 2006) or 5 mg of activated charcoal (detached leaves) and eluted with 100 μL (Super-Q) or 40 μL (charcoal) of CH2Cl2 containing 120 ng of nonyl acetate or 80 ng of 1-bromodecane, respectively, as an internal standard. No major differences in the volatile profiles were observed between the different trapping materials.
The eluted samples (1 μL) were injected in splitless mode into a GC-2010 gas chromatograph (Shimadzu) coupled with a QP2010S mass spectrometer (Shimadzu). Separation was performed on an Rxi-XLB column (Restek) of 30 m × 0.25 mm i.d. × 0.25 μm film thickness. Helium was used as the carrier gas (1.4 mL min−1 flow rate), and a temperature gradient of 5°C min−1 from 40°C (hold for 2 min) to 220°C was applied. Samples collected from hydroponically grown accessions were analyzed in 2-μL volumes on a Hewlett-Packard 6890 gas chromatograph coupled to a Hewlett-Packard 5973 quadrupole mass detector. Compounds were separated at a flow rate of 2 mL min−1 on a 5% phenyl-methyl-polysiloxane (DB5) column (J&W Scientific) of the same dimensions as described above. Qualitative analysis of volatile products of the TPS02 and TPS03 recombinant enzymes was performed using an AOC-5000 Shimadzu autosampler with automated solid-phase microextraction. Compounds were thermally desorbed in a 2:1 split for 5 min at 240°C in the gas chromatograph injector.
The identities of volatile compounds were confirmed by comparison of their retention times and mass spectra with those of authentic standards (individual compounds or components of essential oils) and with mass spectra in the National Institute of Standards and Technology and Wiley libraries. For absolute quantification of MeSA and TMTT, the primary ion peaks of each compound were integrated (single ion method) and the amounts were calculated in relation to the response of nonyl acetate at mass-to-charge ratio (m/z) 69. Response curves for the quantified compounds relative to the internal standard were generated by injecting a mixture of equal amounts of authentic standards and internal standard. Absolute quantification of (E)-β-ocimene and (E,E)-α-farnesene was performed using a flame ionization detector. For relative quantification of volatiles, primary ion peaks [m/z 93 for MeSA, (E)-β-ocimene, and (E,E)-α-farnesene; m/z 69 for TMTT] were integrated and normalized against 1-bromodecane (at m/z 135).
Genotyping of Plant Material
Homozygous mutants in the Col-0 and Ws backgrounds carrying a T-DNA insertion in the TPS03 and TPS02 genes, respectively, were identified in the insertional mutant population (Sessions et al., 2002). Genomic DNA was isolated using the method of Edwards et al. (1991). The T-DNA insertion in Ws TPS02 and Col-0 TPS03 was confirmed by PCR, using primers P1 to P3 and P4 to P6, respectively (Supplemental Table S2).
Generation of Ws TPS02 RNAi Lines
To prepare an RNAi construct for targeting Ws TPS02, first, a DNA fragment spanning a portion of the last exon of TPS02, the downstream intergenic region, and a portion of the first exon of TPS03 was amplified by PCR using primers P7 and P8 and Ws genomic DNA. The PCR product was cloned into the pCR2.1-TOPO vector (Invitrogen) and used as a template for a second PCR to amplify a 106-bp fragment spanning 29 bp of the last exon and 77 bp of the 3′ UTR of Ws TPS02 with primers P9 and P10. The amplicon was cloned into pENTR-TOPO-D (Invitrogen) and transferred to pHELLSGATE8 (Wesley et al., 2001) using the LR recombination reaction (Invitrogen). The resulting construct was transformed into Agrobacterium tumefaciens (strain GV3101) using chemical transformation (An, 1987), and Ws plants were transformed with the vacuum infiltration method (Bechtold et al., 1993). Transgenic plants were selected on kanamycin resistance as described above. Experiments were performed with plants in the T3 generation.
Determination of TPS Gene Expression by Semiquantitative RT-PCR
Total RNA was isolated from treated and untreated leaves of Col-0 and Ws wild-type and mutant lines as well as from roots and flowers of both accessions using the Trizol reagent (Invitrogen). One microgram of total RNA was treated with RQ1 RNase-free DNase (Promega). The DNA-free total RNA was reverse transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen) in a total volume of 20 μL following the manufacturer's instructions. PCRs were performed with primers P11 and P12 for TPS02 (At4g16730), P13 and P14 for TPS03 (At4g16740), P15 and P16 for TPS10 (At2g24210), and P17 and P18 for Actin8 (At1g49240), as listed in Supplemental Table S2. PCRs were conducted with 0.5 μm of each primer, 0.2 mm of each deoxynucleotide triphosphate, 0.5 units of Taq DNA polymerase (New England Biolabs), and 30 cycles for each reaction. Reactions for Actin8 were applied to judge the equality of cDNA template concentrations. RT-PCRs were performed in three replicates with similar results using RNA extracted from three individual plants. Expression of the TPS03 gene in Ws was probed with seven pairs of gene-specific primers (P19–P32; Supplemental Table S2) designed to amplify small fragments of a putative TPS03 transcript.
Genomic and cDNA Cloning of TPS02 and TPS03 by RT-PCR
To obtain genomic sequence information of the TPS03 gene from accession Ws, PCR primers P19 and P33 were used to amplify a TPS03 genomic fragment, and 5′ and 3′ UTRs were amplified using primer pairs P34/P35 and P36/P37, respectively. The TPS03 amplicon was cloned into the pGEM-T Easy vector (Promega) and sequenced.
For cloning of TPS02 and TPS03 cDNAs from accessions Ws and Col-0, respectively, total RNA was extracted from alamethicin-treated leaves and reverse transcribed as described above. Primer P38, which binds 123 nucleotides downstream of the start codon of TPS02, and P39, which corresponds to the end of the TPS02 coding region, were used for RT-PCR amplification of the TPS02 cDNA from Ws. For amplification of a truncated version of TPS02 from Col-0, primer P40, binding 153 bp downstream of the start codon, and P39 were employed. To remove the AT insertion of the Col-0 TPS02 pseudogene, site-directed mutagenesis was performed with primer P41 and the Col-0 TPS02 full-length cDNA cloned in the pCR-T7/CT vector (Invitrogen) using the Stratagene QuikChange Site-Directed Mutagenesis Kit (Stratagene).
The entire open reading frame of TPS03 from Col-0 was amplified using primers P42 and P43. Both reverse primers (P39 and P43) allowed a translational fusion to a C-terminal His6 tag. The amplified TPS02 and TPS03 cDNA products were inserted by directional cloning into the protein expression vector pET101-TOPO (Invitrogen). Correctness of cDNA clones was confirmed by sequencing.
Heterologous Protein Expression and Purification of Ws TPS02 and Col-0 TPS03 in Escherichia coli
The pET101-TOPO plasmids containing Ws TPS02 or Col-0 TPS03 cDNA were transformed into E. coli BL21 DE3 competent cells (Invitrogen) for recombinant protein expression. Transformed cells were incubated in 500-mL cultures, started with an overnight culture, at 18°C until an optical density at 600 nm of 0.6 was reached. Protein expression was induced by adding 1 mm isopropyl-β-d-thiogalactopyranoside followed by additional incubation for 16 h at 18°C. Cells were extracted and the TPS proteins were partially purified on 0.5-mL nickel-nitrilotriacetic acid agarose columns (Qiagen) as described by Tholl et al. (2005) and according to the manufacturer's protocol. Following desalting into assay buffer (10 mm 3-N-morpholino-2-hydroxypropanesulfonic acid, pH 7.0, 1 mm dithiothreitol, and 10% [v/v] glycerol), fractions with the highest enzyme activity were used for determination of enzyme kinetic parameters. Protein concentrations were determined by the Bradford method (Bradford, 1976) using reagents obtained from Bio-Rad and bovine serum albumin as the calibration standard. The specific amount of TPS protein in the active fraction was determined by SDS-PAGE separation of the partially purified protein and quantitative comparison of the intensity of the Coomassie Brilliant Blue-stained TPS protein band with those of bovine serum albumin standards.
TPS Enzyme Assay and Kinetic Characterization
For qualitative analysis of TPS enzyme products via solid-phase microextraction, enzyme assays were performed in screw-capped 10-mL glass vials in a total volume of 250 μL containing 10 μL (TPS02) or 150 μL (TPS03) of partially purified enzyme, 20 mm MgCl2, 0.2 mm NaWO4, 0.1 mm NaF, 1 mm dithiothreitol, and 60 μm GPP or FPP. The assay was incubated in the presence of a polydimethylsiloxane fiber at 30°C for 10 min prior to volatile analysis by gas chromatography-mass spectrometry (GC-MS) as described above. For enzyme characterization, assays were carried out for 5 min (TPS03 with FPP) or 20 min in 50-μL reaction volumes with 0.35 to 4.5 μg of partially purified Ws TPS02 or Col-0 TPS03 enzyme under the same buffer conditions described above. Reaction products were extracted with 300 μL of hexane, and total radioactivity was determined by scintillation counting. For evaluation of the Km values for GPP and FPP, five different concentrations of [1-3H]GPP (35.2 MBq μmol−1) and [1-3H]FPP (35.2 MBq μmol−1) were applied. Assays were conducted in three replicates, and apparent Km values were determined by Hanes plot analysis using the Hyper 1.01 program (J.S. Easterby, University of Liverpool). To determine a possible activation of the TPS enzymes by K+ ions, assays were performed with 5 μm [1-3H]GPP or [1-3H]FPP in the presence or absence of 50 mm KCl.
Construction and Analysis of TPS02 and TPS03 Promoter-GUS Reporter Gene Fusions
A 2.1-kb promoter fragment of Ws TPS02 was isolated from genomic DNA of Ws via PCR using the forward primer P44, containing a HindIII site, and the reverse primer P45, containing the TPS02 start codon and a BamHI site. The PCR product was inserted into the HindIII and BamHI cloning sites of the uidA (GUS) gene-containing binary vector pDW137 (Blazquez et al., 1997) according to Chen et al. (2004). A 1.8-kb Col-0 TPS03 promoter-GUS fusion construct was obtained accordingly using primers P46 and P47. Transformation of the Ws TPS02 and Col-0 TPS03 promoter-fusion constructs into Ws and Col-0 plants, respectively, selection of transformed lines, and GUS enzyme assays were conducted as described previously (Chen et al., 2003b, 2004). At least four independent transformed lines were analyzed. For histochemical analysis of insect-induced GUS activity, P. xylostella larvae were allowed to feed for 24 h. Coronalon-induced GUS activity was determined after 24 h of treatment.
Construction of Ws TPS02- and Col-0 TPS03-GFP Reporter Fusions and Subcellular Localization
A 105-bp fragment encoding a 35-amino acid N-terminal peptide of Ws TPS02 was amplified by PCR with primers P48, carrying a NcoI site, and P49, which contains a SpeI site, and cloned into pCR-TOPO4 (Invitrogen). The fragment was cut out of the vector with NcoI and SpeI and subsequently cloned downstream of a cauliflower mosaic virus 35S promoter in the vector pCAMBIA 1302 (Hajdukiewicz et al., 1994) to generate a C-terminal fusion to mGFP5 (Siemering et al., 1996). The same procedure was applied to clone a 150-bp fragment encoding a 50-amino acid N-terminal peptide of the Col-0 TPS03 protein using primers P50 and P51 for initial PCR amplification. Following confirmation of error-free constructs by sequencing, constructs were transformed in A. tumefaciens, and Ws and Col-0 plants were transformed as described above. Transgenic plants were screened on agar plates for hygromycin resistance, and genomic insertion of the corresponding fusion constructs was confirmed by genomic PCR. Two-week-old plantlets of at least four independent lines were used for observation by confocal laser scanning microscopy using a LSM 510 microscope (Carl Zeiss) equipped with a HeNe laser. Tissue autofluorescence was excited at 458 nm and GFP fluorescence at 488 nm. Band pass was set to 500 to 550 nm, and long pass was set to 560 nm. Bright-field images were acquired with the differential interference contrast channel. Images were processed from optical sections taken along the optical axis and projected into one image using the Zeiss LSM Image Browser 3.2.0.
Phylogenetic Analysis
Amino acid sequence alignment of plant TPS proteins was produced with ClustalW (Lasergene 8) and exported as a Nexus file. Phylogenetic analysis of the data set was conducted using maximum parsimony in PAUP* (Swofford, 2002). Maximum parsimony analyses were conducted using heuristic tree searches with tree bisection-reconnection branch swapping and 1,000 random addition sequence replicates. Support for the clades was obtained by performing bootstrap (Felsenstein, 1985) searches with 1,000 replicates and 10 random sequence replicates. Trees were compiled using TreeGraph2 (Stover and Muller, 2010).
Statistical Analysis
Correlations of induced volatile emissions were determined with a Pearson correlation hypothesis test using the SAS program.
The deduced amino acid sequences of cited TPS genes from gymnosperms and angiosperms can be found in the GenBank database with the following accession numbers: Arabidopsis multiproduct monoterpene synthase (AtTPS24, At3g25810), NP_001031651; Arabidopsis 1,8-cineole synthase (AtTPS23 and AtTPS24, At3g25820 and At3g25830), NP_189212; Arabidopsis myrcene/(E)-β-ocimene synthase (AtTPS10, At2g24210), NP_179998; Malus domestica α-farnesene synthase, AAS01424; Pyrus communis α-farnesene synthase, ABC25002; Lotus japonicus (E)-β-ocimene synthase, AAT86042; Cucumis sativus (E,E)-α-farnesene synthase, AAU05951; Antirrhinum majus (E)-β-ocimene synthase, AAO42614; Pinus taeda α-farnesene synthase, AAO61226; Picea abies (E,E)-α-farnesene synthase, AAS47697; Actinidia deliciosa (E,E)-α-farnesene/(E)-β-ocimene synthase, ACO40485.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Induced emission of (E)-β-ocimene, MeSA, and TMTT from leaves of accession Ws upon insect feeding.
Supplemental Figure S2. Nucleotide sequence alignment of RT-PCR products of Col-0 TPS02 amplified from RNA isolated from coronalon-treated Col-0 leaves.
Supplemental Figure S3. RT-PCR analysis of partial transcripts of the TPS03 gene in coronalon-treated leaves of accession Ws.
Supplemental Figure S4. Expression of TPS02 and volatile emission in detached leaves of Ws wild-type plants and two TPS02 RNAi lines treated with the fungal elicitor alamethicin.
Supplemental Figure S5. Phylogenetic tree illustrating the relationship of Arabidopsis Ws TPS02 and Col-0 TPS03 to other selected Arabidopsis monoterpene synthases and to other plant α-farnesene and (E)-β-ocimene synthases of different TPS subfamilies.
Supplemental Table S1. Pearson correlation coefficients determined for emission of the four main volatile compounds induced in leaves of 27 Arabidopsis accessions.
Supplemental Table S2. Primer sequences.
Supplementary Material
Acknowledgments
We are thankful to Bettina Raguschke (Max Planck Institute for Chemical Ecology) for excellent technical assistance. We are grateful to Wilhelm Boland (Max Planck Institute for Chemical Ecology) for providing coronalon and Tom Mitchell-Olds (Duke University) for the seeds of all accessions except Col-0 and Ws. We thank Tobias Köllner (University of Halle-Wittenberg) for providing an (E,E)-α-farnesene-containing essential oil from ginger. We are thankful to Anthony Shelton and Hilda Collins (Cornell University) for providing P. xylostella larvae. We also thank Sheena Friend (Department of Biological Sciences, Virginia Tech) and Lucas Roberts (Statistics Department, Virginia Tech) for support with the phylogenetic and statistical analyses.
References
- Abel C, Clauss M, Schaub A, Gershenzon J, Tholl D. (2009) Floral and insect-induced volatile formation in Arabidopsis lyrata ssp petraea, a perennial, outcrossing relative of A. thaliana. Planta 230: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Giri AP, Deuerlein S, Griepink F, de Kogel WJ, Verstappen FWA, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ. (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Giri AP, Verstappen FWA, Bertea CM, Sevenier R, Sun ZK, Jongsma MA, Schwab W, Bouwmeester HJ. (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16: 3110–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Jongsma MA, Bouwmeester HJ. (2005) Volatile science? Metabolic engineering of terpenoids in plants. Trends Plant Sci 10: 594–602 [DOI] [PubMed] [Google Scholar]
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC. (2004) Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol 135: 2025–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. (1987) Binary Ti-vectors for plant transformation and promoter analysis. Methods Enzymol 153: 292–305 [Google Scholar]
- Arimura G, Ozawa R, Kugimiya S, Takabayashi J, Bohlmann J. (2004) Herbivore-induced defense response in a model legume. Two-spotted spider mites induce emission of (E)-β-ocimene and transcript accumulation of (E)-β-ocimene synthase in Lotus japonicus. Plant Physiol 135: 1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran E, Rostas M, Zeier J. (2008) Pseudomonas syringae elicits emission of the terpenoid (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene in Arabidopsis leaves via jasmonate signaling and expression of the terpene synthase TPS4. Mol Plant Microbe Interact 21: 1482–1497 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J. (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In-planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199 [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D. (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Martin D, Oldham NJ, Gershenzon J. (2000) Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-beta-ocimene synthase. Arch Biochem Biophys 375: 261–269 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cane DE. (1999) Sesquiterpene biosynthesis: cyclization mechanisms. Barton SB, Nakanishi K, Meth-Cohn O, , Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Steroids, Vol 2 Pergamon, Oxford, pp 155–200 [Google Scholar]
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E. (2003a) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36: 577–588 [DOI] [PubMed] [Google Scholar]
- Chen F, Ro DK, Petri J, Gershenzon J, Bohlmann J, Pichersky E, Tholl D. (2004) Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol 135: 1956–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, D'Auria JC, Farooq A, Pichersky E, Gershenzon J. (2003b) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer JG, Dicke M. (2006) Olfactory learning by predatory arthropods. Anim Biol 56: 143–155 [Google Scholar]
- Degen T, Dillmann C, Marion-Poll F, Turlings TCJ. (2004) High genetic variability of herbivore-induced volatile emission within a broad range of maize inbred lines. Plant Physiol 135: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Köllner TG, Gershenzon J. (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Delphia CM, Rohr JR, Stephenson AG, De Moraes CM, Mescher MC. (2009) Effects of genetic variation and inbreeding on volatile production in a field population of horsenettle. Int J Plant Sci 170: 12–20 [Google Scholar]
- Dicke M, van Poecke RMP, de Boer JG. (2005) Inducible indirect defence of plants: from mechanisms to ecological functions. Basic Appl Ecol 4: 27–42 [Google Scholar]
- Donath J, Boland W. (1995) Biosynthesis of acyclic homoterpenes: enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39: 785–790 [Google Scholar]
- Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J. (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci USA 102: 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Fäldt J, Miller B, Bohlmann J. (2003) (E)-β-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15: 1227–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Negre F, Nagegowda DA, Orlova I. (2006) Plant volatiles: recent advances and future perspectives. Crit Rev Plant Sci 25: 417–440 [Google Scholar]
- Edwards K, Johnstone C, Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäldt J, Arimura G, Gershenzon J, Takabayashi J, Bohlmann J. (2003) Functional identification of AtTPS03 as (E)-beta-ocimene synthase: a monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta 216: 745–751 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Geervliet JBF, Posthumus MA, Vet LEM, Dicke M. (1997) Comparative analysis of headspace volatiles from different caterpillar-infested or uninfested food plants of Pieris species. J Chem Ecol 23: 2935–2954 [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawe GA, Zavala JA, Kessler A, Van Dam NM, Baldwin IT. (2003) Ecological costs and benefits correlated with trypsin protease inhibitor production in Nicotiana attenuata. Ecology 84: 79–90 [Google Scholar]
- Gouinguene S, Degen T, Turlings TCJ. (2001) Variability in herbivore-induced odour emissions among maize cultivars and their wild ancestors (teosinte). Chemoecology 11: 9–16 [Google Scholar]
- Green S, Squire CJ, Nieuwenhuizen NJ, Baker EN, Laing W. (2009) Defining the potassium binding region in an apple terpene synthase. J Biol Chem 284: 8652–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, MacIntosh GC, Green PJ. (1999) Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci 4: 429–438 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile Ppzp family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Kessler A, Kahl J, Lorenz A, Baldwin IT. (2000) Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia 124: 408–417 [DOI] [PubMed] [Google Scholar]
- Hare JD. (2007) Variation in herbivore and methyl jasmonate-induced volatiles among genetic lines of Datura wrightii. J Chem Ecol 33: 2028–2043 [DOI] [PubMed] [Google Scholar]
- Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ. (2003) Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco Bright Yellow-2 cells. J Biol Chem 278: 26666–26676 [DOI] [PubMed] [Google Scholar]
- Herde M, Gärtner K, Köllner TG, Fode B, Boland W, Gershenzon J, Gatz C, Tholl D. (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C-16-homoterpene TMTT. Plant Cell 20: 1152–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A. (2003) An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91–96 [DOI] [PubMed] [Google Scholar]
- Hoballah MEF, Tamo C, Turlings TCJ. (2002) Differential attractiveness of induced odors emitted by eight maize varieties for the parasitoid Cotesia marginiventris: is quality or quantity important? J Chem Ecol 28: 951–968 [DOI] [PubMed] [Google Scholar]
- Hoegen E, Stromberg A, Pihlgren U, Kombrink E. (2002) Primary structure and tissue-specific expression of the pathogenesis-related protein PR-1b in potato. Mol Plant Pathol 3: 329–345 [DOI] [PubMed] [Google Scholar]
- Hori K, Watanabe Y. (2007) Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol 48: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Iijima Y, Davidovich-Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E. (2004) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol 136: 3724–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. (2001) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J. (2004) The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 16: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange BM. (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 50: 47–65 [DOI] [PubMed] [Google Scholar]
- Lou YG, Hua XY, Turlings TCJ, Cheng JA, Chen XX, Ye GY. (2006) Differences in induced volatile emissions among rice varieties result in differential attraction and parasitism of Nilaparvata lugens eggs by the parasitoid Anagrus nilaparvatae in the field. J Chem Ecol 32: 2375–2387 [DOI] [PubMed] [Google Scholar]
- Loughrin JH, Manukian A, Heath RR, Tumlinson JH. (1995) Volatiles emitted by different cotton varieties damaged by feeding beet armyworm larvae. J Chem Ecol 21: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Martin DM, Fäldt J, Bohlmann J. (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Toub O, Chiang A, Lo BC, Ohse S, Lund ST, Bohlmann J. (2009) The bouquet of grapevine (Vitis vinifera L. cv. Cabernet Sauvignon) flowers arises from the biosynthesis of sesquiterpene volatiles in pollen grains. Proc Natl Acad Sci USA 106: 7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall PJ, Turlings TCJ, Loughrin J, Proveaux AT, Tumlinson JH. (1994) Herbivore-induced volatile emissions from cotton (Gossypium hirsutum L) seedlings. J Chem Ecol 20: 3039–3050 [DOI] [PubMed] [Google Scholar]
- Mercke P, Kappers IF, Verstappen FWA, Vorst O, Dicke M, Bouwmeester HJ. (2004) Combined transcript and metabolite analysis reveals genes involved in spider mite induced volatile formation in cucumber plants. Plant Physiol 135: 2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N. (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J 55: 224–239 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Wang MY, Matich AJ, Green SA, Chen XY, Yauk YK, Beuning LL, Nagegowda DA, Dudareva N, Atkinson RG. (2009) Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). J Exp Bot 60: 3203–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous SW, Whitaker BD. (2004) Cloning and functional expression of an (E,E)-alpha-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219: 84–94 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR. (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gershenzon J. (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Pollak PE, Vogt T, Mo YY, Taylor LP. (1993) Chalcone synthase and flavonol accumulation in stigmas and anthers of Petunia hybrida. Plant Physiol 102: 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Ehlting J, Keeling CI, Lin R, Mattheus N, Bohlmann J. (2006) Microarray expression profiling and functional characterization of AtTPS genes: duplicated Arabidopsis thaliana sesquiterpene synthase genes At4g13280 and At4g13300 encode root-specific and wound-inducible (Z)-γ-bisabolene synthases. Arch Biochem Biophys 448: 104–116 [DOI] [PubMed] [Google Scholar]
- Schuhr CA, Radykewicz T, Sagner S, Latzel C, Zenk MH, Arigoni D, Bacher A, Rohdich F, Eisenreich W. (2003) Quantitative assessment of crosstalk between the two isoprenoid biosynthesis pathways in plants by NMR spectroscopy. Phytochem Rev 2: 3–16 [Google Scholar]
- Schuler G, Gorls H, Boland W. (2001) 6-Substituted indanoyl isoleucine conjugates mimic the biological activity of coronatine. Eur J Org Chem 1663–1668 [Google Scholar]
- Schuler G, Mithofer A, Baldwin IT, Berger S, Ebel J, Santos JG, Herrmann G, Holscher D, Kramell R, Kutchan TM, et al. (2004) Coronalon: a powerful tool in plant stress physiology. FEBS Lett 563: 17–22 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J. (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol 6: 1653–1663 [DOI] [PubMed] [Google Scholar]
- Smid HM, Vet LEM. (2006) Learning in insects: from behaviour to brain. Anim Biol 56: 121–124 [Google Scholar]
- Stotz HU, Spence B, Wang YJ. (2009) A defensin from tomato with dual function in defense and development. Plant Mol Biol 71: 131–143 [DOI] [PubMed] [Google Scholar]
- Stover BC, Muller KF. (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP* Beta Version. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- Takabayashi J, Dicke M. (1996) Plant-carnivore mutualism through herbivore-induced carnivore attractants. Trends Plant Sci 1: 109–113 [Google Scholar]
- Thibaud-Nissen F, Shu OY, Buell R. (2009) Identification and characterization of pseudogenes in the rice gene complement. BMC Genomics 10: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. (2006) Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol 9: 297–304 [DOI] [PubMed] [Google Scholar]
- Tholl D, Boland W, Hansel A, Loreto F, Rose USR, Schnitzler JP. (2006) Practical approaches to plant volatile analysis. Plant J 45: 540–560 [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. (2005) Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J 42: 757–771 [DOI] [PubMed] [Google Scholar]
- Turlings TCJ, Tumlinson JH, Lewis WJ. (1990) Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 250: 1251–1253 [DOI] [PubMed] [Google Scholar]
- Unsicker SB, Kunert G, Gershenzon J. (2009) Protective perfumes: the role of vegetative volatiles in defense against herbivores. Curr Opin Plant Biol 12: 479–485 [DOI] [PubMed] [Google Scholar]
- van der Hoeven RS, Monforte AJ, Breeden D, Tanksley SD, Steffens JC. (2000) Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12: 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanHoof A, Green PJ. (1996) Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J 10: 415–424 [DOI] [PubMed] [Google Scholar]
- Van Poecke RMP, Posthumus MA, Dicke M. (2001) Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: chemical, behavioral, and gene-expression analysis. J Chem Ecol 27: 1911–1928 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Williams DC, McGarvey DJ, Katahira EJ, Croteau R. (1998) Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37: 12213–12220 [DOI] [PubMed] [Google Scholar]
- Wu SQ, Schalk M, Clark A, Miles RB, Coates R, Chappell J. (2006) Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat Biotechnol 24: 1441–1447 [DOI] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thibaud-Nissen F, Prakash T, Buell CR, Shiu SH. (2009) Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol 151: 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.