Abstract
The arbuscular mycorrhiza (AM) brings together the roots of over 80% of land plant species and fungi of the phylum Glomeromycota and greatly benefits plants through improved uptake of mineral nutrients. AM fungi can take up both nitrate and ammonium from the soil and transfer nitrogen (N) to host roots in nutritionally substantial quantities. The current model of N handling in the AM symbiosis includes the synthesis of arginine in the extraradical mycelium and the transfer of arginine to the intraradical mycelium, where it is broken down to release N for transfer to the host plant. To understand the mechanisms and regulation of N transfer from the fungus to the plant, 11 fungal genes putatively involved in the pathway were identified from Glomus intraradices, and for six of them the full-length coding sequence was functionally characterized by yeast complementation. Two glutamine synthetase isoforms were found to have different substrate affinities and expression patterns, suggesting different roles in N assimilation. The spatial and temporal expression of plant and fungal N metabolism genes were followed after nitrate was added to the extraradical mycelium under N-limited growth conditions using hairy root cultures. In parallel experiments with 15N, the levels and labeling of free amino acids were measured to follow transport and metabolism. The gene expression pattern and profiling of metabolites involved in the N pathway support the idea that the rapid uptake, translocation, and transfer of N by the fungus successively trigger metabolic gene expression responses in the extraradical mycelium, intraradical mycelium, and host plant.
The arbuscular mycorrhizal (AM) symbiosis brings together the roots of the majority of land plant species and fungi of the phylum Glomeromycota to great mutual advantage (Smith and Read, 2008). AM fungi improve the acquisition of phosphate, nitrogen (N), sulfur, and trace elements such as copper and zinc (Clark and Zeto, 2000; Allen and Shachar-Hill, 2008) and increase the biotic and abiotic stress resistance of their host (Smith et al., 2010). In return, the host transfers up to 20% of its photosynthetically fixed carbon to the AM fungus (Jakobsen and Rosendahl, 1990), which depends on its host plant for its carbon supply (Bago et al., 2000).
N is the nutrient whose availability most commonly limits plant growth in natural ecosystems. AM fungi can take up NO3−NH4+ and can also increase access to organic N sources from the soil (Ames et al., 1983; Johansen et al., 1993; Bago et al., 1996; Hodge et al., 2001). The translocation by the fungus can represent a significant route for N uptake by the plant (Johansen and Jensen, 1996). For example, Toussaint et al. (2004) showed that in an in vitro mycorrhiza at least 21% of the total N uptake in the roots came from the fungal extraradical mycelium (ERM); for other mycorrhizal systems, even larger proportions have been described (more than 30% and 50%; Govindarajulu et al., 2005; Jin et al., 2005). Tanaka and Yano (2005) reported that 75% of the N in leaves of mycorrhizal maize (Zea mays) was taken up by the ERM of Glomus aggregatum.
A mechanism of N transfer from the fungus to the plant has been proposed (Bago et al., 2001) that involves the operation of a novel metabolic route in which N was translocated from the ERM to the intraradical mycelium (IRM) as Arg but transferred to the plant without carbon as inorganic N. This mechanism has been supported by labeling experiments (Johansen et al., 1996; Govindarajulu et al., 2005; Jin et al., 2005), enzyme activity analysis (Cruz et al., 2007), and limited gene expression data (Govindarajulu et al., 2005; Gomez et al., 2009; Guether et al., 2009). Nevertheless, our molecular knowledge of the metabolic and transport pathways involved and how they are regulated is still rudimentary. A better understanding of the mechanism and regulation of N uptake assimilation, translocation, and transfer to the host is important for potential applications of AM fungi as biofertilizers, bioprotectors, and bioregulators in sustainable agriculture and restoration as well as for understanding the role of AM fungi in natural ecosystems (Bruns et al., 2008).
In this study, we postulate that the uptake, translocation, and transfer of N by the fungus triggers the metabolic gene expression responses successively in the ERM, IRM, and host plant, which will result in the synthesis and accumulation of Arg in the ERM, the turnover of Arg to release ammonium in the IRM, and the assimilation of ammonium by the host plant via the glutamine synthetase (GS)/glutamate synthase (GOGAT) pathway inside the root (Fig. 1). To test these predictions, 11 genes involved in the N primary assimilation and metabolism were cloned and verified from Glomus intraradices; six enzymes with full-length coding sequences (CDSs) were functionally characterized by yeast knockout mutant complementation. Two GS proteins were found to have different substrate affinities and expression patterns, suggesting that they have different roles in N assimilation. The time courses of gene expression and N movement in fungal and host tissues were analyzed following nitrate supply to the ERM of a mycorrhiza grown under N-limited conditions. The results substantially increase our knowledge of the identity and regulation of most of the metabolic and transport genes involved in N movement through the AM symbiosis.
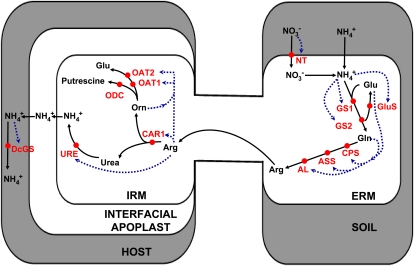
Figure 1.
Working model of N transport and metabolism in the symbiosis between plant roots and arbuscular mycorrhizal fungi. N moves (black arrows) from the soil into the fungal ERM, through a series of metabolic conversion reactions into Arg, which is transported into the IRM within the root (host) and there is broken down; N is transferred to and assimilated by the host as ammonia. Red circles refer to the sites of action of the genes identified and analyzed in this study. Blue arrows indicate mechanisms hypothesized to regulate gene expression by N metabolites involved in the pathway.
RESULTS
Gene Cloning and Functional Characterization
Based on data from high-throughput sequencing of cDNA from ERM (P.J. Lammers and Y. Shachar-Hill, unpublished data) and sequences that were previously deposited in public databases (GenBank), the putative sequences for 10 enzymes and one nitrate transporter of G. intraradices were identified. Seven full-length and four partial CDSs were obtained that show high sequence similarities to known genes involved in N uptake and metabolism in fungi (Table I; Supplemental Figs. S1–S10). The pathway for N metabolism in which these genes have been proposed to operate is shown in Figure 1, which includes the possible regulation of the transcriptional levels of genes by the N metabolites involved in the pathway. For the functional characterization, several full-length CDSs were expressed in yeast, and the antibody directed against the polyhistidine region fused within each Gi sequence was used to detect the expression of the proteins in yeast (Fig. 2A; Supplemental Fig. S11).
Table I. Gene identification from G. intraradices.
| Gene Name | CDS Length | Closest Homologa | Domains, Families, or Functional Sitesb |
| bp | |||
| Nitrate transporter (GiNT) | 990c | BAG70346 (Porphyra yezoensis), identity = 39% | Major facilitator superfamily |
| Glutamine synthetase 1 (GiGS1) | 1,065d | AAY62524 (Glomus intraradices), identity = 99% | GLNA_1, Gln synthetase signature 1; GLNA_ATP, Gln synthetase putative ATP-binding region signature |
| Glutamine synthetase 2 (GiGS2) | 1,065d | AAR11485 (Glomus mosseae), identity = 88% | GLNA_1, Gln synthetase signature 1; GLNA_ATP, Gln synthetase putative ATP-binding region signature |
| Glutamate synthase (GiGluS) | 759c | EED14920 (Talaromyces stipitatus), identity = 68% | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| Carbamoyl-phosphate synthase glutamine chain (GiCPS) | 990c | XP961982 (Neurospora crassa), identity = 55% | GATASE_TYPE_1, Gln amidotransferase type 1 domain profile |
| Argininosuccinate synthase (GiASS) | 1,239d | AAW43079 (Cryptococcus neoformans), identity = 71% | ARGININOSUCCIN_SYN_1, argininosuccinate synthase signature 1; ARGININOSUCCIN_SYN_2, argininosuccinate synthase signature 2. |
| Arginosuccinate lyase (GiAL) | 849c | XP002172712 (Schizosaccharomyces japonicus), identity = 66% | FUMARATE_LYASES, fumarate lyase signature |
| Arginase (GiCAR1) | 945d | XP001875445 (Laccaria bicolor), identity = 63% | ARGINASE_2, arginase family profile; ARGINASE_1, arginase family signature |
| Ornithine aminotransferase (GiOAT1) | 1,329d | XP002171771 (Schizosaccharomyces japonicus), identity = 69% | AA_TRANSFER_CLASS_3, aminotransferase class III pyridoxal-phosphate attachment site |
| Ornithine decarboxylase (GiODC) | 1,335d | CAB61758 (Mucor circinelloides), identity = 57% | ODR_DC_2_1, Orn/diaminopimelate/Arg decarboxylase family 2 pyridoxal-phosphate attachment site |
| Urease (GiURE) | 2,499d | XP964986 (Neurospora crassa), identity = 66% | UREASE_3, urease domain profile; UREASE_2, urease active site |
BLAST of the amino acid sequence against the National Center for Biotechnology Information protein database.
Based on the databases http://www.expasy.ch/prosite and http://www.ebi.ac.
Partial CDS.
Full-length CDS.
Figure 2.
Heterologous expression, purification, and enzymatic kinetics of the two GS isoforms identified, GiGS1 and GiGS2. A, Western blot of the expressed GiGS1 and GiGS2 in yeast. Whole cell protein extracts from the yeast transformed with GiGS1 and GiGS2 were fractionated on a denaturing 12% polyacrylamide gel, transferred to polyvinylidene fluoride membranes, and detected with Anti-His(C-term)-HRP antibody. The yeast knockout mutants were independently transformed with the empty pYES2.1 vector or the vector fused with GiGS1 or GiGS2. Lanes a and c, The expression of GiGS on the medium without Gal; lanes b and d, the expression of GiGS on the medium with Gal; lane e, the expression of the empty vector on the medium with Gal (as a negative control). B, The purification stages of GiGS1 and GiGS2. Purified proteins were subjected to SDS-PAGE (12% NuPAGE Novex Bis-Tris Gel) and silver stained. Low-Mr markers are in lane f. Crude yeast extracts before purification were loaded in lane e, and successive elution fractions were loaded in lanes a to d. Each lane contained 1 μg of protein. C, Enzymatic kinetics for GiGS1 and GiGS2 detected by synthetase assay using the purified proteins. Biosynthetic GS activity assays were carried out by varying the concentration of Glu in the reaction mixture. Plots of 1/V versus [S] are shown. Velocity data, expressed as proportions of Vmax, were based on the three biological replicates; absolute Vmax values were 3.68 and 4.01 μm min−1 mg−1 for GiGS1 and GiGS2, respectively. Lineweaver-Burk plots of the data were obtained with GiGS1 (Km = 1.87 mm, r2 = 0.99) and GiGS2 (Km = 3.80 mm, r2 = 0.999).
Identification and Enzymatic Analysis for N Uptake and Assimilation Genes
GS (EC 6.3.1.2) belongs to a multigene family in most plants and some fungi (Stanford et al., 1993; Ishiyama et al., 2004). Of the two putative GiGS genes, each has a 1,062-bp full-length CDS corresponding to proteins with 354 amino acids with a predicted molecular mass of 39 kD. Analysis for the deduced proteins revealed two conserved domains for the GS family: GS signature 1 and a putative ATP-binding region signature; the corresponding amino acids were 5′-FDGSSAPGDDSDVLL-3′ and 5′-KPIKGDWNGAGCHTNYS-3′ for GiGS1 and 5′-FDGSSAPGHDSDILL-3′ and 5′-KPIKGDWNGAGCHTNYS-3′ for GiGS2, respectively (Supplemental Fig. S1A). GiGS1 is nearly identical to the GS sequence reported by Govindarajulu et al. (2005) except that the stop codon was not identified in the earlier study. Besides the striking similarity with GS from Glomus mosseae, GiGS1 and GiGS2 also possess a high similarity with GSs from Saccharomyces pombe and Filobasidiella neoformans, especially for the conserved region of GS family member signature 1 and the putative ATP-binding region (Supplemental Fig. S2A). The alignments of the deduced GiGS1 and GiGS2 sequences were used to calculate a phylogenetic tree (Supplemental Fig. S2B), which shows that GiGS1 and GiGS2 were clustered with the GSs from ectomycorrhizae into one large group, with the identities ranging from 70% to 90%.
The full-length CDSs of GiGS1 and GiGS2 fused with C-terminal polyhistidine tags were used for complementation of Saccharomyces cerevisiae strain ΔGLN1, which is auxotrophic for Gln (Bernard et al., 2008). The four cell types tested were (1) the corresponding wild-type strain BY4743, (2) the GLN1-deficient knockout mutant ΔGLN1 transformed with empty vector, and (3 and 4) ΔGLN1 transformed with GiGS1 or GiGS2. The haploid mutant ΔGLN1 could not grow on the medium without Gln, while it grew well on the plates with 2% Gln. The transformants expressing GiGS1 or GiGS2 grew well even on the plate without Gln added, which means that GiGS1 and GiGS2 could complement the auxotrophic mutant by the functional production of Gln (Fig. 3A).
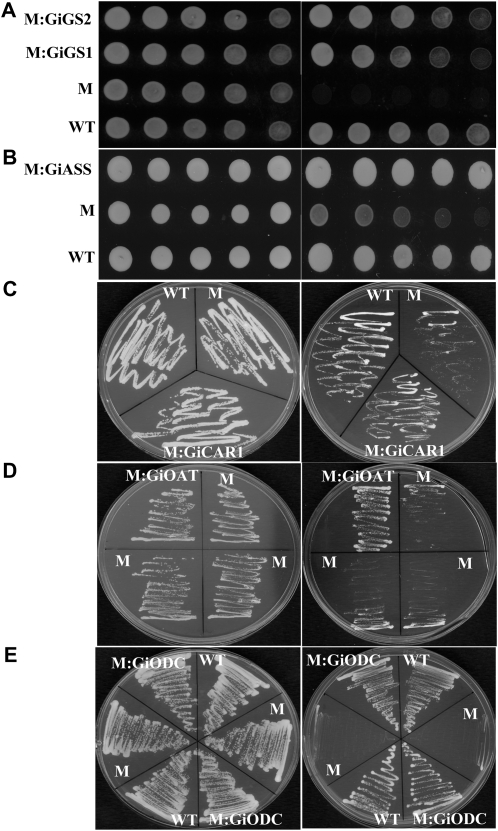
Figure 3.
Functional complementation of S. cerevisiae knockout mutants using the genes identified from G. intraradices. M, The knockout mutant expressing the empty vector; M:GiXXX, the knockout mutant expressing the GiXXX gene; WT, the wild type. A, GiGS1 and GiGS2. Left, Growth of colonies on rich medium plus 2% Gln; right, medium without Gln. B, GiASS. Left, Minimal medium plus all essential amino acids including 0.01% l-Arg; right, minimal medium with all essential amino acids excluding l-Arg. In A and B, 5 μL of cells was used for spreading on the plate. The optical density at 600 nm values for the cells are 1, 0.1, 0.01, 0.001, and 0.0001 from the left to the right columns. C, GiCAR1. Left, Minimal medium with all essential amino acids including 0.01% l-Arg; right, medium with 0.01% l-Arg as the sole N source. D, GiOAT1. Left, Medium with all essential amino acids; right, medium with only 0.01% Orn and Leu as N sources. E, GiODC. Left, Medium with 0.001% Leu, His, Lys, and putrescine as N sources; right, medium with 0.001% Leu, His, and Lys as N sources.
The catalytic reaction of GS is NH3 + Glu + ATP → Gln + ADP + inorganic phosphate. Figure 2 shows the expression of the two GiGS isozymes in yeast, their purification, and in vitro kinetics (Mitchell and Magasanik, 1983). GiGS1 and GiGS2 have a very similar Vmax of 3.7 and 4.0 μm min−1 mg−1, respectively, but differ in their Km. The Km value of GiGS2 was 3.8 mm, twice that of GiGS1 (1.9 mm).
In addition, two partial sequences (GiNT and GiGluS) involved in the primary N assimilation were identified here. Nitrate transporter plays an important role for transporting NO3− to the ERM. A 990-bp partial cDNA sequence for a putative high-affinity nitrate transporter was amplified from the total approximately 1,500-bp gene, sharing 39% identity with the nitrate transporter of Porphyra yezoensis (Table I; Supplemental Fig. S1). The next step of N assimilation via the GS/GOGAT pathway is catalyzed by glutamate synthase (GluS), which converts Gln and 2-oxoglutarate to two molecules of Glu. Approximately 800 bp of a putative GluS (EC 1.4.1.14) was identified with a 68% similarity to the Talaromyces stipitatus gene at the protein level (Table I; Supplemental Fig. S3). GiGluS includes a small subunit conserved for the GOGAT family, but the highly conserved NADPH-binding motif between residues in the N-terminal sequence could not be found in the partial C-terminal sequence (Pelanda et al., 1993).
Enzymes Involved in Arg Synthesis
Sequences for argininosuccinate synthetase (GiASS; EC 6.3.4.5) and argininosuccinate lyase (GiAL; EC 4.3.2.1), which catalyze the last two steps in Arg biosynthesis, as well as a sequence for a putative carbamoyl-phosphate synthetase (GiCPS [Arg-specific small chain]; EC 6.3.5.5) were obtained.
GiCPS is involved in both Arg and pyrimidine biosynthesis and catalyzes the ATP-dependent formation of carbamoyl phosphate from Gln and carbon dioxide. This subunit promotes the hydrolysis of Gln to ammonia, which is subsequently used by the ammonia chain of CPS to synthesize carbamoyl phosphate. The amplified GiCPS shows high homology with the small subunit of CPS-A (an Arg biosynthetic enzyme), and a Gln amidotransferase type 1 domain was found (KIAVVDCGVK……….IDQIRSEKVK). A deduced 330-amino acid protein from an expected total approximately 1,350 bp has shown 55% identity to Neurospora crassa, which suggests that this enzyme should be located within the mitochondrion, since the N. crassa enzyme is known to be mitochondrially localized (Table I; Supplemental Fig. S4; Davis, 1986), even though the characteristic mitochondrial signal sequence of enriched Arg and Ser residues in the N terminus could not be found due to the lack of approximately 300 bp at the 5′ end.
GiASS catalyzes the ATP-dependent ligation of citrulline to Asp to form argininosuccinate, releasing AMP and pyrophosphate. ASS is distributed widely, from bacteria to vertebrates, and its primary structure is highly conserved. For ASS family members, some highly conserved regions have been identified, such as the presence of a Gly-rich motif and three other conserved regions involved in Asp and citrulline binding (Lemke and Howell, 2001). The amplified full-length CDS for a putative GiASS encodes a 413-amino acid protein with a predicted molecular mass of 46 kD and has two signature regions for the ASS family: the first is a highly conserved stretch of nine residues located in the N-terminal extremity of these enzymes (AYSGGLDTS), and the second is derived from a conserved region that contains one of the conserved Arg residues (GcTgKGNDqvRF; Table I; Supplemental Fig. S5). The deduced amino acid sequence for GiASS also shows some other highly conserved regions, including a Gly-rich motif A (AYSGGLDTS), common to a subset of N-type ATP pyrophosphatases (Tesmer et al., 1996); motif B (GCTGKGNDQVRF), the site involved in Asp binding (Lemke and Howell, 2001); and motifs C (STDENLFHISYE) and D (ENRFIGIKSRGCYE) that are responsible for citrulline binding by forming β-hairpins enabling the formation of a four-stranded β-sheet (Flint and Wilkening, 1986; Wagemaker et al., 2007). An Arg-auxotroph mutant, YOL058W, of S. cerevisiae deficient for the gene ARG1, which encodes an argininosuccinate synthetase, which grows poorly on medium without Arg, was used for the complementation analysis. As shown in Figure 3B, cells harboring GiASS survived well on medium without Arg, while the ARG1-deficient strain that was transformed with an empty vector was incapable of sustained growth, even though it grew well on Arg-containing medium. These results indicate that the GiASS-encoded protein can functionally substitute for S. cerevisiae ARG1.
AL catalyzes the formation of Arg and fumarate from argininosuccinate, the last step in the biosynthesis of Arg. A number of enzymes belonging to a lyase class that catalyze reactions that release fumarate, including AL, have been shown to share a short conserved sequence around a Met that is probably involved in the catalytic activity. An 849-bp-long partial sequence of a putative AL was also amplified that shared 66% identity with the respective enzyme from Schizosaccharomyces japonicus and also had a fumarate lyase signature domain, besides a highly conserved region denoted as C3 (Table I; Supplemental Fig. S6; Sampaleanu et al., 2001).
Enzymes Involved in Arg Breakdown
The proposed transport model for N in the AM symbiosis assumes the breakdown of Arg and the release of NH4 in the IRM in colonized roots (Fig. 1). A putative arginase (GiCAR1; EC 3.5.3.1) sequence that encodes a protein with 315 amino acids was identified. GiCAR1 was more than 60% identical at the amino acid level to previously described arginases of the ectomycorrhizal fungi Laccaria bicolor and Agaricus bisporus and the yeast Schizosaccharomyces pombe and also showed a region that is highly conserved in the arginase family (SFDVDaldPtvaPStgtpvrgG; Supplemental Fig. S7A). As shown in Supplemental Figure S7B, GiCAR1 was clustered with most of the yeast species into one group. The multiple regulation mechanisms for Schizosaccharomyces cerevisiae Arg metabolism has been studied in great detail, such as feedback inhibition, repression of the anabolic pathway, and induction of the catabolic pathway (Messenguy et al., 2000), which indicated the complicated regulation mechanism for Arg metabolism at the transcriptional, posttranscriptional, and translational levels (Werner et al., 1987; Crabeel et al., 1988; Boonchird et al., 1991).
In S. cerevisiae, the induction of arginase requires the presence of four proteins, ArgRIp (ArgSOp), ArgRIIp (Arg-8lp), ArgRIIIp (ArgRIIIp), and Mcmlp (Messenguy and Dubois, 1993), and a CAR1 mutation affecting the expression of the Arg catabolic gene encoding arginase results in the loss of the ability of the cells to use Arg as the N source. The mutant 12T7cI ura3, ΔCAR1, and the corresponding wild-type 2T7cI ura3 were used for functional complementation with GiCAR1. As shown in Figure 3C, GiCAR1 functionally complemented the knockout auxotrophic phenotype, allowing better growth than the mutant on minimal medium with 0.01% Arg as the sole N source (VanHuffel et al., 1994).
Ornithine aminotransferase (GiOAT; EC 2.6.1.13) and ornithine decarboxylase (GiODC; EC 4.1.1.17) are two enzymes that catalyze the breakdown of Orn (Borsuk et al., 1999; Dzikowska et al., 2003; Wagemaker et al., 2005). OAT catalyzes the transamination of α-ketoglutarate with Orn or N-acetylornithine and of Glu-5-semialdehyde with Glu and Ala. GiOAT1 has a deduced 442-amino acid sequence with a predicted molecular mass of 49 kD and an aminotransferase class III pyridoxal-phosphate attachment site (LIADEVLTGLARTGKLLCQEHDEVRADIVILGKALSGG) and differs from a partial sequence reported recently for a putative GiOAT2 (Gomez et al., 2009; Table I; Supplemental Fig. S8). The S. cerevisiae mutant strain O2463d Δarg81, which is unable to utilize Orn as the sole source of N, was used in a complementation study for GiOAT1 (Wagemaker et al., 2007). As shown in Figure 3D, the yeast strain O2463d Δarg81, which lacks OAT activity and is unable to utilize Orn as the sole N source, grew better on minimal medium with only 0.01% Orn and Leu as N sources than the vector control, which showed very limited growth in 2 d at 28°C. Thus, GiOAT1 could complement the O2468d ΔAg81 auxotrophic phenotype using Orn as the N source.
ODC is the key regulatory enzyme of the polyamine biosynthetic pathway and catalyzes the decarboxylation of Orn to 1,4-diaminobutane (putrescine). The predicted amino acid sequence of GiODC consists of 445 amino acids, and the highest degree of amino acid similarity was to the ODC sequence of Mucor circinelloides (57%). GiODC belongs to the Orn/Lys/Arg decarboxylase class II family, with a typical Orn/diaminopimelate/Arg decarboxylases family 2 pyridoxal-phosphate attachment site (YAVKCNGDPMLLRLLAALG; Table I; Supplemental Fig. S9). In general, active growth and cell division are associated with substantial polyamine biosynthesis in plants, animals, and microorganisms (Bagni et al., 1980; Palavan and Galston, 1982; Pegg and McCann, 1982). The yeast knockout mutant disrupted in Orn decarboxylase (SPE1) is auxotrophic for putrescine and biotin, shows putrescine-dependent growth, and could not grow on the medium without polyamines (Balasundaram et al., 1991; Mangold and Leberer, 2005). Since ODC catalyzes the decarboxylation of Orn to putrescine, the yeast SPE1 deletion strain YKL184W that lacks ODC is not able to grow without putrescine. Cells expressing GiODC could grow without exogenously supplied putrescine, while the vector control shows no growth, showing that GiODC functionally complements yeast putrescine auxotrophy resulting from the SPE1 mutation (Fig. 3E).
Urease (EC 3.5.1.5) is a nickel-binding enzyme that catalyzes the hydrolysis of urea to carbon dioxide and ammonia. Two pathways are encountered for urea degradation: hydrolysis via urease or via an ATP-hydrolyzing urease (EC 3.5.1.45). In higher plants, some fungi, and many prokaryotes, urea is hydrolyzed by urease, which allows organisms to use externally and internally generated urea as a source of N (Mobley and Hausinger, 1989). In bacteria, three genes (ureA, ureB, and ureC) encode the structural subunits (α, β, and γ, respectively) of urease, which associate in an αβγ stoichiometry to form the urease apoenzyme (Jabri et al., 1995). In plants and fungi, the structural urease protein is encoded by one gene that comprises homologs of three bacterial genes that encode separate subunits of the functional enzyme. Here, the cloned urease from G. intraradices (GiURE) contains a CDS of 833 amino acids with a calculated molecular mass of 90 kD and a theoretical pI of 6.02. The encoded protein resembles α-, β-, and γ-subunits of the bacterial urease fused into one protein, as described before for other eukaryotic ureases (Jabri et al., 1995; Jabri and Karplus, 1996; Follmer, 2008). An alignment with fungal, plant, and bacterial ureases revealed a high similarity among all representative species included, with a high similarity to the eukaryotic ureases and to a lower extent to the bacterial enzymes. Several residues for the Klebsiella aerogenes urease were involved in substrate binding or catalysis (His-134, Lys-217, His-246, His-272, and Asp-360), and these residues were also found to be conserved in GiURE (Table I; Supplemental Fig. S10; Follmer, 2008). The yeast mutant lacking urease is also auxotrophic for several amino acids, which made it impossible to use urea as the sole N source for testing the function of GiURE by a complementation assay.
Gene Expression in Response to N Addition
Based on the current model, we hypothesized that the expression of genes required for N movement from soil through the fungus and into the host is temporally and spatially coordinated with the flux of N. The transcriptional levels for all the genes from G. intraradices identified here were measured in ERM and IRM tissues by quantitative real-time PCR. At the same time, plant GS is the initial enzyme for the assimilation of ammonium transferred by the fungus, so the carrot (Daucus carota) GS (DcGS) was also studied.
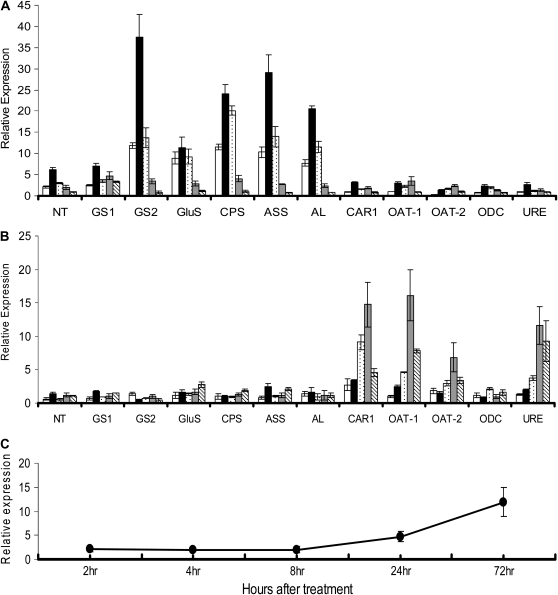
The transcript levels of several genes involved in N uptake and assimilation increased in the ERM within 2 h after 4 mm KNO3 was added to the fungal compartment (Fig. 4). The expression of a fungal GS, CPS (Arg-specific small chain), NADH-dependent GluS, ASS, AL, and a nitrate transporter (NT) increased up to 4 h after supply, increased to the highest levels after 8 h, and then returned to the initial values within 72 h. By contrast, the expression of CAR1, URE, OAT1 and OAT2, and ODC in the ERM was only slightly affected by the supply of nitrate. The expression of these genes (with the exception of ODC) was substantially up-regulated in the IRM within 24 h. The transcript levels of a plant GS (DcGS) began to increase 24 h after the supply of nitrate to the ERM and further increased up to the 72-h time point.
Figure 4.
The expression of primary N metabolic and transport genes in the arbuscular mycorrhizal symbiosis after the addition of 4 mm KNO3 to the fungal ERM. Bars are as follows: 2 h (white bars), 4 h (black bars), 8 h (dotted bars), 24 h (gray bars), and 72 h (hatched bars). Gene expression was measured by quantitative real-time PCR. A and B, ERM (A) and IRM (B) with fungal ribosomal protein S4 gene expression as the reference. C, Carrot GS expression in the colonized root tissue, with the expression of elongation factor 1-α from carrot used as the reference.
In the ERM, without the induction of N, the relative expression of GiGS1 (cycle threshold [CT] = 29.19 ± 1.00) is higher than GiGS2 (CT = 31.31 ± 1.07) with S4 ribosomal protein as reference gene (CT = 27.55 ± 0.60; P < 0.05). However, GiGS2 is much more highly induced than GiGS1 (Fig. 4) after the addition of N in the ERM. Thus, GiGS1 is likely the main functional enzyme for ammonium assimilation in the ERM at lower N levels, with GiGS2 being induced when N is more abundant (consistent with its higher Km). For OAT, GiOAT1 was found to be more up-regulated than GiOAT2 when N was added to the ERM, suggesting that this isoform is more important in catabolizing Orn released by arginase in the IRM. Compared with OAT, ODC expression is quite stable both in ERM and IRM, consistent with its main function being for polyamine synthesis rather than Orn breakdown.
N Movement through the AM Symbiosis
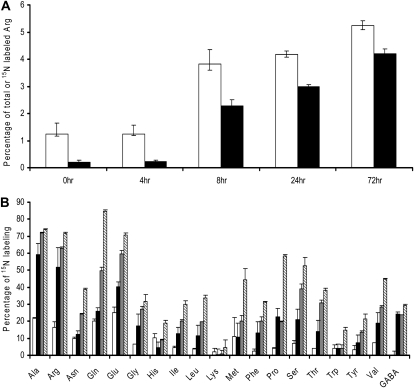
The levels and labeling of free amino acids in the IRM were measured after the ERM was exposed to 4 mm K15NO3 under the same conditions used for monitoring gene expression. The percentage of total free amino acids in the mycorrhizal roots represented by Arg and the percentage of labeled amino acid molecules represented by 15N-labeled Arg (see “Materials and Methods”) are shown as a function of time after 15N addition in Figure 5A, which indicated that Arg is the dominant amino acid in the pool. Figure 5B shows the time course of fractional enrichment in free amino acids of the mycorrhizal roots, which indicated the fast labeling ratio of Arg as the reaction of N addition.
Figure 5.
N movement from the ERM to the IRM in mycorrhizal carrot roots based on the timing of labeling free amino acids in the mycorrhizal roots after the addition of 4 mm K15NO3 to the fungal ERM. A, The percentage of total free amino acids represented by Arg (including 15N-labeled and unlabeled molecules; white bars) and the percentage of total free amino acids represented by only 15N-labeled Arg (black bars) in the colonized roots (plant plus IRM tissue) after 4 mm K15NO3 was applied to the ERM. B, The percentage labeling in the free amino acids in the mycorrhizal roots at 4 h (white bars), 8 h (black bars), 24 h (gray bars), and 72 h (hatched bars) after adding 4 mm K15NO3 to the ERM compartment. GABA, γ-Aminobutyrate.
Significant labeling in free amino acids was detectable in 4 h after 15N addition, and within 8 h, over half of the Arg molecules within the root were labeled, although Ala, Glu, and Gln also became rapidly labeled (Fig. 5B). The level of Arg in the colonized roots rose 3-fold and became the most abundant free amino acids within 8 h, whereas other amino acids remained at very similar relative levels throughout the time course. The percentage of the total amino acids represented by Arg increased rapidly in 8 h and more slowly from 8 to 72 h, consistent with the induction of arginase and the idea that Arg synthesis in the ERM is for N transport rather than longer term storage.
DISCUSSION
Gene Identification and Characterization Support the Proposed Pathway of N Transfer
The postulated pathway for N movement through the AM symbiosis (Fig. 1; Bago et al., 2001; Govindarajulu et al., 2005) has received experimental support from labeling and enzymatic activity measurements but so far has not been thoroughly tested at the molecular level. The operation of the proposed pathway implies the expression of fungal genes for N uptake, assimilation, and Arg synthesis in the ERM and the expression of genes for the breakdown of Arg and urea in the IRM. Here, we have identified candidate transcripts for all enzymes of the urea cycle pathway except those responsible for the synthesis of citrulline from Glu and its carbamylation. The presence of one or more high-affinity nitrate transporters in the ERM is implied by the ability of AM fungi to take up nitrate at low exogenous nitrate levels, and a candidate for this function was also identified. Functional characterization in a heterologous system and/or in vitro was employed because no system for knocking out or otherwise selectively suppressing gene expression has yet been developed in AM fungi. For those enzymes that we were unable to characterize by complementation or in vitro, sufficiently long sequences were obtained to test for the presence of conserved functional domains and for the expected phylogenetic relationships to known genes from other organisms. Those findings are consistent with the putative function of these genes.
GS is the first enzyme of the proposed N handling pathway, and its activity is highly regulated in other organisms. Therefore, GiGS1 and GiGS2 were studied in vitro and found to have lower Km values for Glu than those reported for GS from other organisms, such as 6.3 mm for S. cerevisiae, 5 mm for L. laccata, and 54 mm for Tuber borchii (Mitchell and Magasanik, 1983; Brun et al., 1992; Montanini et al., 2003). This may enable AM fungi to assimilate inorganic N and contribute to plant nutrition even under conditions of low N supply. The lower Km of GiGS1, together with its higher constitutive expression compared with GiGS2 under low-N conditions, suggest that GiGS1 serves at lower exogenous N and that GiGS2 is rapidly induced for assimilation when N is present at higher levels. The presence of two GS isoforms, with the more highly expressed gene being less transcriptionally regulated, has been observed in some but not all fungi (Benjamin et al., 1989; Margelis et al., 2001; Javelle et al., 2003). Previous measures of GS expression in G. intraradices by PCR probably detected both isoforms simultaneously due to the high level of similarity between the CDSs of the two transcripts. A previous report that GS in G. intraradices is not transcriptionally regulated in response to N addition was based on the levels of a GS transcript that has a closer similarity to GiGS1 (Breuninger et al., 2004; Supplemental Fig. S1). In addition, Gomez et al. (2009) reported the expression of a partial GS sequence that corresponds to the GiGS1 transcript characterized above in Medicago roots colonized by G. intraradices and suggested that this might be involved in N turnover within the fungal IRM.
Orn is released during Arg breakdown, and the proposed N handling scheme requires that it is stored, excreted, or broken down in the IRM or translocated to the ERM. Orn has not been found to be stored in substantial quantities in the IRM and is not transferred to the host (Govindarajulu et al., 2005). The lack of transfer to the plant of carbon from Arg or Orn is consistent with the dependence of the fungus on the plant for its carbon nutrition. A subsequent turnover of Orn and the release of inorganic N in the IRM would seem to be more efficient for N transfer to the host than its translocation to the ERM, which would return half of the N to its original location. The presence of two OAT and one ODC sequences raised the question of the fate of Orn: conversion to Glu or to putrescine. The increased expression of the two OAT sequences but not the putative ODC after N supply suggests that Orn is converted to Glu. We postulate that Glu is broken down to release more ammonium to the host, although a previously published putative Glu dehydrogenase (Govindarajulu et al., 2005) appears not to have this function, and we did not identify a better candidate.
Transcriptional Regulation
The regulation of N transport and metabolism in fungi includes a transcriptional regulatory network that responds to the levels and forms of available N (ter Schure et al., 2000). For the proposed mechanism of N transfer in the AM symbiosis to operate efficiently, the participating enzymes and transporters would be expected to be coordinately regulated in response to changes in the exogenous N supply. In earlier studies, the expression of several putative genes that are involved in the fungal N metabolism in ERM and IRM has been tested and interpreted as consistent with the proposed pathway (Govindarajulu et al., 2005; Gomez et al., 2009). However, the functional identity of several tested transcripts was still uncertain, their expression pattern in response to N treatment was difficult to interpret, and the sequences of several key enzymes of the proposed N pathway have not been identified. Enzymatic activity of ASS was shown to rise in the ERM but not in colonized roots, whereas arginase, urease, and GS activities rose in the roots, but not in the ERM, after ammonium supply to the ERM (Cruz et al., 2007). The evidence presented by Cruz et al. (2007) in support of the model of Bago et al. (2001) included measurements of total extractable enzyme activity from the colonized root and the ERM. To test the model thoroughly, however, it is necessary to distinguish between the fungus and host plant enzymes in mycorrhizal roots so as to distinguish host root from fungal activities. Since it is fungal arginase and urease genes that are induced, the model is supported, whereas if the enzyme activities had been of plant origin, then this would argue for a transfer of Arg and/or urea from fungus to host, which would have contradicted the original model. While consistent with the role for Arg in N movement in the AM symbiosis, the limited number of enzymes tested, the difficulty of distinguishing between host and fungal activities, and the lack of an activation of GS in the ERM in response to N supply make it important to investigate the dynamics of gene expression across the whole proposed pathway.
We predicted that activities for N uptake and assimilation and for Arg synthesis should be up-regulated in the ERM but not in the IRM after N addition and that the activities of genes involved in the breakdown of Arg and the release of ammonium should be increased in the IRM and not the ERM after N supply to the ERM. Furthermore, host GS levels should increase after ammonium is released in the IRM and transferred across the mycorrhizal interface. The timing and location of the observed transcriptional changes after nitrate was supplied to the ERM (Fig. 4) are completely consistent with these expectations. The timing of N transfer was followed with 15N labeling and measurements of amino acid levels, showing that N taken up in the ERM arrives in significant amounts at the colonized roots within hours (Fig. 5). 15N was detected in roots 4 h after N supply to the ERM, with high levels of labeling in amino acids and a 3-fold increase in Arg levels measured 4 h later. This coincides with the up-regulation of Arg breakdown genes, which occurs several hours after the expression changes in assimilatory genes in the ERM were observed. The level of plant GS transcript rose substantially after the response in fungal catabolic genes in the IRM was initiated. The pattern of gene expression in relation to N movement suggests that the arrival of N in significant quantities successively in the ERM, IRM, and host plant is the trigger for gene expression changes rather than signaling events that might precede significant N delivery.
A regulatory scheme is shown in Figure 1 that is based on our observations of the relative timing of N movement and gene expression and is consistent with findings in other fungi, particularly yeast. It is proposed that (1) nitrate availability stimulates the expression of the identified nitrate transporter (most likely together with other high- or low-affinity nitrate transporters), and presumably also of nitrate reductase, although we were unable to clone the enzyme sequence reported previously (Kaldorf et al., 1998; Montanini et al., 2006; Rekangalt et al., 2009); (2) ammonium stimulates the expression of the GS/GOGAT pathway, and increasing levels of Gln and Glu rather than ammonium could lead to an up-regulation of Arg synthesis genes (Hinnebusch, 1988; ter Schure et al., 2000); (3) Arg is then translocated from the ERM to the IRM (Cruz et al., 2007); (4) elevated Arg levels trigger an increase in arginase and urease activities in the IRM but not in the ERM (the difference between IRM and ERM may be due to N catabolite repression in the ERM [Hinnebusch, 1988; Wagemaker et al., 2007], as amino acid levels are substantial in the ERM in the presence of N [Jin et al., 2005]); (5) Arg or Orn that is produced in the IRM by the action of arginase stimulates the expression and activity of OAT (Messenguy et al., 2000; ter Schure et al., 2000); (6) the subsequent release of ammonium in the IRM could stimulate the passive efflux of ammonia into the mycorrhizal interface and the uptake of ammonium by the plant by specific transporters (Selle et al., 2005; Gomez et al., 2009; Guether et al., 2009); and (7) the subsequent increase in the availability of ammonium for the host causes an up-regulation of plant GS, especially near the sites of N transfer (Chalot et al., 2006).
The nature of signal(s) from the ERM to the IRM and host roots to regulate nutrient exchange when mineral nutrients become available to the ERM is unknown. The results above show that the rise in the expression of the fungal genes of Arg breakdown coincides with the arrival of new N (15N label) and a rise in Arg levels within the colonized root. Thus, it is likely that this is the signal for regulatory gene expression in the IRM, consistent with the observation that Arg induces arginase in S. pombe (Benitez and Farrar, 1980). However, Arg accumulation in the ERM does not repress Arg synthesis genes by a general amino acid control system seen in other fungi (Hinnebusch, 1988; ter Schure et al., 2000). This difference in transcriptional regulation is understandable if Arg in the ERM of AM fungi functions more as a transport molecule than a storage compound, as in saprophytes, and the transport into the fungal vacuole and its binding to polyphosphates keep the cytosolic levels low (Bücking et al., 1998). Although transcript levels for the Arg synthesis and catabolic genes fell after 24 h, these enzymatic activities must remain at substantial levels because high rates of N flow are sustained for extended periods following N addition (Govindarajulu et al., 2005; Jin et al., 2005; Cruz et al., 2007).
MATERIALS AND METHODS
Spore Material and Mycorrhizal in Vitro Cultures for 15N Labeling and Gene Expression
The spore material of Glomus intraradices (DAOM 181602) was purchased from Premier Tech Biotechnologies in units of 106 and was stored at 4°C until further use. Ri T-DNA-transformed carrot (Daucus carota clone DCI) roots colonized by G. intraradices (DAOM 197198) were grown at 24°C in modified medium (Bécard and Fortin, 1988) gelled with 3.5 g L−1 Phytagel (Sigma) using the split-plate method of St-Arnaud et al. (1996). The medium of the root compartment was modified to limit the N concentration to 1 mm. The ERM was allowed to cross over the divider into the fungal compartment that contained modified medium with no N or Suc added and only 0.075 g L−1 Phytagel to form a semisolid medium. After 2 weeks, the ERM was supplied with 4 mm KNO3 for gene expression studies or K15NO3 to determine the labeling percentage of free amino acids. The colonized roots and ERM samples were collected after 0, 2, 4, 8, 24, and 72 h, rinsed with sterilized water, and immediately frozen in liquid N and stored at −80°C until RNA extraction.
RNA Extraction
The RNA used for gene sequencing was extracted from approximately 10,000 spores of G. intraradices germinated in M medium (Bécard and Fortin, 1988) without Suc for 4 to 5 d at 28°C using the TRIzol Plus RNA Purification Kit (Invitrogen), including the DNase treatment, according to the manufacturer's protocol. The RNA used for gene expression was extracted from approximately 100 mg fresh weight of mycorrhizal roots or 0.2 to 0.3 mg fresh weight of ERM tissue. The first strand of cDNA was synthesized using the SuperScript First-Strand Synthesis System for reverse transcription-PCR (Invitrogen) and random primers according to the manufacturer's instructions.
Gene Amplification and Sequence Analysis
The partial sequences used for cloning the full lengths are from 454 pyrosequencing of cDNA from the ERM of G. intraradices. The 5′ and 3′ untranslated regions, and when possible the entire coding region of the cDNA, were obtained by RACE using the cDNA from germinating spores as a template with the SMART RACE cDNA amplification kit (Invitrogen). To verify the start codon (ATG) of the amplified gene exactly, the promoter region of the gene was amplified using the GenomeWalker Universal Kit (Clontech). BLAST search and ClustalW 1.83 were used for the sequence comparisons and alignments (Thompson et al., 1994). The phylogenetic trees were constructed by the neighbor-joining method on the base of the alignments of our identified amino acid sequences with other available homologous sequences using MEGA 4.0.2. Secondary structure, protein family, domains, and functional sites were predicted using the ExPASy Proteomic tools (http://ca.expasy.org/tools/).
cDNA Cloning and Yeast Transformation
As the primers shown in Supplemental Table S1, seven full CDSs were cloned into the pYES2.1 TOPO TA (Invitrogen) vector was fused with a polyhistidine region for further analysis. The resulting plasmids were amplified in Escherichia coli and purified using the S.N.A.P Miniprep Kit (Invitrogen) and were used to transform Saccharomyces cerevisiae strains using Frozen-EZ Yeast Transformation II (Zymo Research) according to the manufacturer's protocol. The yeast strains used for the functional complementation are shown in Supplemental Table S2. Yeast colonies carrying recombinant plasmids (pYES2.1 gene) were screened using a selective Ura-drop medium (Clontech) and verified again by sequencing. As a negative control, the yeasts were also transformed with an empty vector (vector control).
The minimal medium (MM) composed of 0.175% yeast N base, 2.0% Gal (Q-BIOgene; Morgan) supplemented with Complete Supplement Mixture lacking uracil (Q-BIOgene), and 2.0% peptone agar was used for the yeast complementation for GS (GiGS1 and GiGS2). For the other enzymes, the MM for the complementation assays was modified as follows: GiASS, MM with all amino acids except Arg; GiOAT1, MM with 0.01% Orn and Leu as sole N sources; GiCAR1, MM with 0.01% Arg as sole N source; GiODC, the H medium (a polyamine-free defined medium) according to Balasundaram et al. (1991) and Mangold and Leberer (2005) was used. The H medium was sterilized by filtering the solutions through a 0.45-μm Millipore membrane or by boiling to prevent the growth of amine-deficient mutants resulting from impurities from the steam during the autoclaving. Glass vessels were washed with 6 m HCl, rinsed with 0.1 m HCl, and dried for 4 h at 170°C.
Enzyme Purification and Activity Detection
The total protein was extracted and the ProBond Purification System (Invitrogen) was used to purify the GS protein extended by six tandem His residues. The purified protein was desalted using Zeba Desalt Spin Columns (Thermo Scientific) and then kept at 4°C. The expression and size of the proteins in the transformed yeasts were confirmed using western blots based on the polyhistidine region using Novex chemiluminescence substrates (Invitrogen). Whole cell protein extracts from yeast transformants were fractionated on a denaturing 12% polyacrylamide gel, transferred to polyvinylidene fluoride membranes, and detected with Anti-His(C-term)-HRP antibody. Purified GiGS1 and GiGS2 were subjected to SDS-PAGE (12% NuPAGE Novex Bis-Tris Gel) and silver stained, and the cutting gels for two proteins were sequenced directly using gas chromatography-mass spectrometry to confirm. The protein level was measured using the BCA Protein Assay Kit-Reducing Agent Compatible (Pierce).
The GS activity was tested using the synthetase activity assay. The kinetic parameters for the substrate (Glu) were determined by varying the concentrations of individual reaction components in the presence of excess amounts of all the other components. Each enzyme activity assay was replicated independently at least three times (biological replicates); assays were always conducted in duplicate using parallel reaction mixtures lacking GiGS as blanks. The data were fitted to a Lineweaver-Burk plot ([S]/V versus [S]), and the regression analysis of the data was performed with Excel. A soybean (Glycine max) derivative of GS (Sigma) was used as the positive control. A total of 450 ng of the affinity-purified and desalted GiGS1 and GiGS2 was added in a 400-μL volume containing the following components at pH 7.0: 50 mm HEPES, 0.4 mm ethylene glycol bis(P-aminoethylether)-N,N,N′,N′-tetraacetic acid, 40 mm MgSO4, 0 to 32 mm potassium Glu, 10 mm hydroxylamine HC1, and 50 mm disodium potassium ATP. The reaction was incubated at 37°C for 10 to 30 min, then terminated by the addition of 1 mL of stop mix, consisting of 55 g of FeCl3·6H2O, 20 g of trichloroacetic acid, and 21 mL of concentrated HCl per liter. An A540 of 0.555 corresponds to 1 μm of γ-glutamylhydroxamate released into the 400-μL reaction mixture. One unit of synthetase activity catalyzes the formation of 1 μm of product per minute at 37°C.
Real-Time PCR
Gene expression was analyzed in DNA-free RNA samples by the SYBR Green fluorescence assay in an ABI Prism 7700 Sequence Detection System. Real-time PCR was performed on three independent biological samples and two to three technical replicates. The primers are shown in Supplemental Table S3. The comparative ΔΔCT method was used to measure changes in the expression of selected genes relative to untreated controls (Winer et al., 1999). For G. intraradices, the gene expression results were normalized using the S4 ribosomal protein as a reference gene due to its high expression stability (Govindarajulu et al., 2005). For host D. carota, the elongation factor 1-α gene was used as the reference (Clotault et al., 2008). Mean values were compared using Fisher's protected lsd test and were considered different at P < 0.01 using SPSS.
Liquid Chromatography-MS
Liquid chromatography (LC)-MS analyses were carried out using a Quattro micro (Waters) mass spectrometer system bundled with an electrospray ionization source and a Shimadzu HPLC system. Underivatized amino acids were separated using a Waters Symmetry C18 (100 × 2.1 mm, 3 μm) column with 1 mm perflurohetanoic acid in a water/acetonitrile gradient at ambient temperature and a flow rate at 0.3 mL min−1. The 15N content of Arg was determined using a full scan of the protonated amino acid [i.e. mass-to-charge ratio (m/z) 175 (15N0), 176 (15N1), 177 (15N2), 178 (15N3), and 179 (15N4)]. Arg content was determined by stable isotope analysis based on 15N2-Arg. Arg isotopomers (i.e. m/z 175, 176, 177, 178, and 179) were investigated by LC-MS. Mass spectrometric peak intensities were integrated for labeled and unlabeled molecules of each free amino acid and used to obtain total levels (the sum of all the labeled and unlabeled molecules of an amino acid) as well as fractional labeling (the sum of labeled molecules as a proportion of the total number of amino acids). Thus, for example, the amount of Arg is the sum of peak intensities for Arg 15N-labeled molecules (m/z of 176, 177, 178, and 179 representing molecules with one, two, three, and four 15N atoms, respectively) and unlabeled molecules (m/z 175). Samples from unlabeled tissues were used to account for heavy isotopes at natural abundance (mainly 13C). Values are means of three biological replicates. The 4-h Arg spectra contained greater background signals for the most labeled molecules, which increased the uncertainty of label quantification. The error bars in Figure 5 for labeling of Arg at this time point represent mean maximal and mean minimal integrated values, which are larger than the sd of the biological replicates.
The gene sequences are deposited in GenBank as follows: GiGS1 (GU111909), GiGS2 (GU111910), GiASS (GU111911), GiOAT1 (GU111912), GiCAR1 (GU111913), GiODC (GU111914), GiURE (GU111915), GiGluS (GU111916), GiCPS (GU111917), GiAL (GU111918), and GiNT (GU111919).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment for GiNT.
Supplemental Figure S2. Amino acid alignment and phylogenetic analysis for GiGS1 and GiGS2.
Supplemental Figure S3. Amino acid alignment for GiGluS.
Supplemental Figure S4. Amino acid alignment for GiCPS.
Supplemental Figure S5. Amino acid alignment for GiASS.
Supplemental Figure S6. Amino acid alignment for GiAL.
Supplemental Figure S7. Amino acid alignment and phylogenetic analysis for GiCAR1.
Supplemental Figure S8. Amino acid alignment for GiOAT1.
Supplemental Figure S9. Amino acid alignment for GiODC.
Supplemental Figure S10. Amino acid alignment for GiURE.
Supplemental Figure S11. Western blot of the expressed proteins in yeast.
Supplemental Table S1. Primers for gene cloning.
Supplemental Table S2. Yeast strains for functional complementation.
Supplemental Table S3. Primers for real-time PCR.
Supplementary Material
Acknowledgments
We thank Kyaw Aung and Yani Chen for assistance with the gene amplification and functional complementation assays; Mintu Kumar Desai for help with the enzyme activity tests; Sara J. Glimour for advice on real-time PCR; Lijun Chen and Dan Jones for help with LC-MS; James Allen and Yugandhar Beesetty for advice on mycorrhizal culture and inoculation; Taghleb Al-Deeb and Cheng Zou for help on the figures; and Doug Allen, Ana Paula Alonso, Inga Krassovskaya, and Weiqing Zeng for the suggestion of this project.
References
- Allen JW, Shachar-Hill Y. (2008) Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol 149: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames RN, Reid CP, Porter LK, Cambardella C. (1983) Hyphal uptake and transport of nitrogen from two 15N-labelled sources by Glomus mosseae, a vesicular-arbuscular mycorrhizal fungus. New Phytol 95: 381–396 [Google Scholar]
- Bagni N, Malucelli B, Torrigiani P. (1980) Polyamines, storage substances and abscisic acid-like inhibitors during dormancy and very early activation of Helianthus tuberosus tuber tissues. Physiol Plant 49: 341–345 [Google Scholar]
- Bago B, Pfeffer P, Shachar-Hill Y. (2000) Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol 124: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Pfeffer P, Shachar-Hill Y. (2001) Could the urea cycle be translocating nitrogen in the arbuscular mycorrhizal symbiosis? New Phytol 149: 4–8 [DOI] [PubMed] [Google Scholar]
- Bago B, Vierheilig H, Piché Y, Ázcon-Aguilar C. (1996) Nitrate depletion and pH changes induced by the extraradical mycelium of the arbuscular mycorrhizal fungus Glomus intraradices grown in monoxenic culture. New Phytol 133: 273–280 [DOI] [PubMed] [Google Scholar]
- Balasundaram D, Tabor CW, Tabor H. (1991) Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 88: 5872–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G, Fortin JA. (1988) Early events of vesicular arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108: 211–218 [DOI] [PubMed] [Google Scholar]
- Benitez T, Farrar L. (1980) Induction of arginase and ornithine transaminase in the fission yeast Saccharomyces pombe. J Bacteriol 144: 836–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin PM, Wu JL, Mitchell AP, Magasanik B. (1989) Regulatory systems control expression of glutamine synthetase in Saccharomyces cerevisiae at the level of transcription. Mol Gen Genet 217: 370–377 [DOI] [PubMed] [Google Scholar]
- Bernard SM, Moller ALB, Dionisio G, Kichey T, Jahn TP, Dubois F, Baudo M, Lopes MS, Terce-Laforgue T, Foyer CH, et al. (2008) Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol Biol 67: 89–105 [DOI] [PubMed] [Google Scholar]
- Boonchird C, Messenguy F, Dubois E. (1991) Determination of amino-acid-sequences involved in the processing of the Arg 5 Arg6 precursor in Saccharomyces cerevisiae. Eur J Biochem 199: 325–335 [DOI] [PubMed] [Google Scholar]
- Borsuk P, Dzikowska A, Empel J, Grzelak A, Grzeskowiak R, Weglenski P. (1999) Structure of the arginase coding gene and its transcript in Aspergillus nidulans. Acta Biochim Pol 46: 391–403 [PubMed] [Google Scholar]
- Breuninger M, Trujillo CG, Serrano E, Fischer R, Requena N. (2004) Different nitrogen sources modulate activity but not expression of glutamine synthetase in arbuscular mycorrhizal fungi. Fungal Genet Biol 41: 542–552 [DOI] [PubMed] [Google Scholar]
- Brun A, Chalot M, Botton B, Martin F. (1992) Purification and characterization of glutamine-synthetase and NADP-glutamate dehydrogenase from the ectomycorrhizal fungus Laccaria laccata. Plant Physiol 99: 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns TD, Arnold AE, Hughes KW. (2008) Fungal networks made of humans: UNITE, FESIN, and frontiers in fungal ecology. New Phytol 177: 586–588 [DOI] [PubMed] [Google Scholar]
- Bücking H, Beckmann S, Heyser W, Kottke I. (1998) Elemental contents in vacuolar granules of ectomycorrhizal fungi measured by EELS and EDXS: a comparison of different methods and preparation techniques. Micron 29: 53–61 [Google Scholar]
- Chalot M, Blaudez D, Brun A. (2006) Ammonia: a candidate for nitrogen transfer at the mycorrhizal interface. Trends Plant Sci 11: 263–266 [DOI] [PubMed] [Google Scholar]
- Clark RB, Zeto SK. (2000) Mineral acquisition by arbuscular mycorrhizal plants. J Plant Nutr 23: 867–902 [Google Scholar]
- Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E. (2008) Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot 59: 3563–3573 [DOI] [PubMed] [Google Scholar]
- Crabeel M, Seneca S, Devos K, Glansdorff N. (1988) Arginine repression of the Saccharomyces cerevisiae arg1 gene: comparison of the arg1 and arg3 control regions. Curr Genet 13: 113–124 [DOI] [PubMed] [Google Scholar]
- Cruz C, Egsgaard H, Trujillo C, Ambus P, Requena N, Martins-Loucao MA, Jakobsen I. (2007) Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiol 144: 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH. (1986) Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev 50: 280–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowska A, Kacprzak M, Tomecki R, Koper M, Scazzocchio C, Weglenski P. (2003) Specific induction and carbon/nitrogen repression of arginine catabolism gene of Aspergillus nidulans-functional in vivo analysis of the otaA promoter. Fungal Genet Biol 38: 175–186 [DOI] [PubMed] [Google Scholar]
- Flint HJ, Wilkening J. (1986) Cloning of the arg-12 gene of Neurospora crassa and regulation of its transcript via cross-pathway amino-acid control. Mol Gen Genet 203: 110–116 [DOI] [PubMed] [Google Scholar]
- Follmer C. (2008) Insights into the role and structure of plant ureases. Phytochemistry 69: 18–28 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang YH, Blancaflor EB, Udvardi MK, Harrison MJ. (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin HR, Abubaker J, Douds DD, Allen JW, Bucking H, Lammers PJ, Shachar-Hill Y. (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435: 819–823 [DOI] [PubMed] [Google Scholar]
- Guether M, Neuhauser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P. (2009) A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol 150: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. (1988) Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev 52: 248–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge A, Campbell CD, Fitter AH. (2001) An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413: 297–299 [DOI] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Tabuchi M, Yamaya T, Takahashi H. (2004) Biochemical background and compartmentalized functions of cytosolic glutamine synthetase for active ammonium assimilation in rice roots. J Biol Chem 279: 16598–16605 [DOI] [PubMed] [Google Scholar]
- Jabri E, Carr MB, Hausinger RP, Karplus PA. (1995) The crystal structure of urease from Klebsiella aerogenes. Science 268: 998–1004 [PubMed] [Google Scholar]
- Jabri E, Karplus PA. (1996) Structures of the Klebsiella aerogenes urease apoenzyme and two active-site mutants. Biochemistry 35: 10616–10626 [DOI] [PubMed] [Google Scholar]
- Jakobsen I, Rosendahl L. (1990) Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plants. New Phytol 115: 77–83 [Google Scholar]
- Javelle A, Andre B, Marini AM, Chalot M. (2003) High-affinity ammonium transporters and nitrogen sensing in mycorrhizas. Mol Microbiol 47: 411–430 [DOI] [PubMed] [Google Scholar]
- Jin H, Pfeffer PE, Douds DD, Piotrowski E, Lammers PJ, Shachar-Hill Y. (2005) The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol 168: 687–696 [DOI] [PubMed] [Google Scholar]
- Johansen A, Finlay RD, Olsson PA. (1996) Nitrogen metabolism of external hyphae of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 133: 705–712 [Google Scholar]
- Johansen A, Jakobsen I, Jensen ES. (1993) Hyphal transport by a vesicular-arbuscular mycorrhizal fungus of N applied to the soil as ammonium or nitrate. Biol Fertil Soils 16: 66–70 [Google Scholar]
- Johansen A, Jensen ES. (1996) Transfer of N and P from intact or decomposing roots of pea to barley interconnected by an arbuscular mycorrhizal fungus. Soil Biol Biochem 28: 73–81 [Google Scholar]
- Kaldorf M, Schmelzer E, Bothe H. (1998) Expression of maize and fungal nitrate reductase genes in the arbuscular mycorrhiza. Mol Plant Microbe Interact 11: 439–448 [DOI] [PubMed] [Google Scholar]
- Lemke CT, Howell PL. (2001) The 1.6 angstrom crystal structure of E. coli argininosuccinate synthetase suggests a conformational change during catalysis. Structure 9: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Mangold U, Leberer E. (2005) Regulation of all members of the antizyme family by antizyme inhibitor. Biochem J 385: 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margelis S, D'Souza C, Small AJ, Hynes MJ, Adams TH, Davis MA. (2001) Role of glutamine synthetase in nitrogen metabolite repression in Aspergillus nidulans. J Bacteriol 183: 5826–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E. (1993) Genetic evidence for a role for mcm1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol 13: 2586–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Vierendeels F, Scherens B, Dubois E. (2000) In Saccharomyces cerevisiae, expression of arginine catabolic genes CAR1 and CAR2 in response to exogenous nitrogen availability is mediated by the Ume6 (CargRI)-Sin3 (CargRII)-Rpd3 (CargRIII) complex. J Bacteriol 182: 3158–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP, Magasanik B. (1983) Purification and properties of glutamine-synthetase from Saccharomyces cerevisiae. J Biol Chem 258: 119–124 [PubMed] [Google Scholar]
- Mobley HLT, Hausinger RP. (1989) Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev 53: 85–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanini B, Betti M, Marquez AJ, Balestrini R, Bonfante P, Ottonello S. (2003) Distinctive properties and expression profiles of glutamine synthetase from a plant symbiotic fungus. Biochem J 373: 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanini B, Gabella S, Abba S, Peter M, Kohler A, Bonfante P, Chalot M, Martin F, Ottonello S. (2006) Gene expression profiling of the nitrogen starvation stress response in the mycorrhizal ascomycete Tuber borchii. Fungal Genet Biol 43: 630–641 [DOI] [PubMed] [Google Scholar]
- Palavan N, Galston AW. (1982) Polyamine biosynthesis and titer during various developmental stages of Phaseolus vulgaris. Physiol Plant 55: 438–444 [Google Scholar]
- Pegg AE, McCann PP. (1982) Polyamine metabolism and function. Am J Physiol 243: C212–C221 [DOI] [PubMed] [Google Scholar]
- Pelanda R, Vanoni MA, Perego M, Piubelli L, Galizzi A, Curti B, Zanetti G. (1993) Glutamate synthase genes of the diazotroph Azospirillum brasilense: cloning, sequencing, and analysis of functional domains. J Biol Chem 268: 3099–3106 [PubMed] [Google Scholar]
- Rekangalt D, Pepin R, Verner MC, Debaud JC, Marmeisse R, Fraissinet-Tachet L. (2009) Expression of the nitrate transporter nrt2 gene from the symbiotic basidiomycete Hebeloma cylindrosporum is affected by host plant and carbon sources. Mycorrhiza 19: 143–148 [DOI] [PubMed] [Google Scholar]
- Sampaleanu LM, Vallee F, Thompson GD, Howell PL. (2001) Three-dimensional structure of the argininosuccinate lyase frequently complementing allele Q286R. Biochemistry 40: 15570–15580 [DOI] [PubMed] [Google Scholar]
- Selle A, Willmann M, Grunze N, Geßler A, Weiß M, Nehls U. (2005) The high-affinity poplar ammonium importer PttAMT1.2 and its role in ectomycorrhizal symbiosis. New Phytol 168: 697–706 [DOI] [PubMed] [Google Scholar]
- Smith SE, Facelli E, Pope S, Smith FA. (2010) Plant performance in stressful environments: interpreting new and established knowledge of the roles of arbuscular mycorrhizas. Plant Soil 326: 3–20 [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis, Ed 3. Elsevier and Academic, New York [Google Scholar]
- Stanford AC, Larsen K, Barker DG, Cullimore JV. (1993) Differential expression within the glutamine synthetase gene family of the model legume Medicago truncatula. Plant Physiol 103: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Fortin JA. (1996) Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res 100: 328–332 [Google Scholar]
- Tanaka Y, Yano K. (2005) Nitrogen delivery to maize via mycorrhizal hyphae depends on the form of N supplied. Plant Cell Environ 28: 1247–1254 [Google Scholar]
- ter Schure EG, van Riel NAW, Verrips CT. (2000) The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev 24: 67–83 [DOI] [PubMed] [Google Scholar]
- Tesmer JG, Klem TJ, Deras ML, Davisson VJ, Smith JL. (1996) The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat Struct Biol 3: 74–86 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint JP, St-Arnaud M, Charest C. (2004) Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T-DNA roots of Daucus carota L., in an in vitro compartmented system. Can J Microbiol 50: 251–260 [DOI] [PubMed] [Google Scholar]
- VanHuffel C, Dubois E, Messenguy F. (1994) Cloning and sequencing of Saccharomyces pombe car1 gene encoding arginase: expression of the arginine anabolic and catabolic genes in response to arginine and related metabolites. Yeast 10: 923–933 [DOI] [PubMed] [Google Scholar]
- Wagemaker MJM, Eastwood DC, Van Der Drift C, Jetten MSM, Burton K, Van Griensven LJLD, Den Camp HJMO. (2007) Argininosuccinate synthetase and argininosuccinate lyase: two ornithine cycle enzymes from Agaricus bisporus. Mycol Res 111: 493–502 [DOI] [PubMed] [Google Scholar]
- Wagemaker MJM, Welboren W, van der Drift C, Jetten MSM, Van Griensven LJLD, den Camp HJMO. (2005) The ornithine cycle enzyme arginase from Agaricus bisporus and its role in urea accumulation in fruit bodies. Biochim Biophys Acta 1681: 107–115 [DOI] [PubMed] [Google Scholar]
- Werner M, Feller A, Messenguy F, Pierard A. (1987) The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 49: 805–813 [DOI] [PubMed] [Google Scholar]
- Winer J, Jung CKS, Shackel I, Williams PM. (1999) Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270: 41–49 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.