Abstract
Transposons are major components of all eukaryotic genomes. Although traditionally regarded as causes of detrimental mutations, recent evidence suggests that transposons may play a role in host gene diversification and evolution. For example, host gene transduction by retroelements has been suggested to be both common and to have the potential to create new chimeric genes by the shuffling of existing sequences. We have previously shown that the maize (Zea mays subsp. mays) retrotransposon Bs1 has transduced sequences from three different host genes. Here, we provide evidence that these transduction events led to the generation of a chimeric new gene that is both transcribed and translated. Expression of Bs1 is tightly controlled and occurs during a narrow developmental window in early ear development. Although all Bs1-associated transduction events took place before Zea speciation, a full uninterrupted open reading frame encoding the BS1 protein may have arisen in domesticated maize or in the diverse populations of its progenitor Z. mays subsp. parviglumis. We discuss potential functions based on domain conservation and evidence for functional constraints between the transduced sequences and their host gene counterparts.
Transposons are ubiquitous and fundamental components of prokaryotic and eukaryotic genomes. For example, they make up to 85%, 45%, 40%, and 12% of the maize (Zea mays), human, rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) genomes, respectively (Arabidopsis Genome Initiative, 2000; International Human Genome Sequencing Consortium, 2001; Goff et al., 2002; Yu et al., 2002; Schnable et al., 2009). Some recent studies suggest that over evolutionary time scales, transposons have contributed to the evolution of genes and genomes by providing means for gene and genome diversification (Kazazian, 2004). For example, an analysis of the human genome sequence reveals more than 1,000 predicted and known proteins to contain sequences derived from transposons, especially long interspersed nuclear elements of the L1 family and short interspersed nuclear elements such as Alu elements (Li et al., 2001). In addition, some mobility-related proteins may have evolved to contribute a cellular function. For example, Drosophila telomeres are composed of the telomere-specific retroposons HeT-A and TART (for review, see Pardue et al., 1996), the RAG1 and RAG2 proteins required for V(D)J recombination and the maturation of B- and T-cells are derived from an ancient DNA transposon (Hiom et al., 1998; for review, see Roth and Craig, 1998); and Syncytin, a protein involved in placental morphogenesis, is encoded by the envelope gene of the human endogenous retrovirus HERV-W (Mi et al., 2000). In plants, the transposase genes of Mutator-like elements (MULEs) may have provided ancestral sequences for two genes involved in red light signaling (FAR1 and FHY1; Hudson et al., 2003; Lin et al., 2007) as well as the MUG1 gene family (Cowan et al., 2005).
Other mechanisms, however, all based on transposon activity, have also been implicated. For example, “gene acquisition” by DNA transposons and “gene transduction” by retroelements have the potential to mediate gene diversification and the emergence of novel cellular functions (Goodier et al., 2000). Whereas gene acquisition by DNA transposons is likely to be mediated by recombination, gene transduction occurs by read-through transcription of retroelements (Goodier et al., 2000; Bennetzen 2005). Gene acquisition by MULEs in rice (Jiang et al., 2004; Juretic et al., 2005; Hanada et al., 2009) and Arabidopsis (Hoen et al., 2006) reveals that MULEs acquired gene fragments and duplicated or amplified them into gene families. A recent study suggests that 22% of MULE-generated duplications are transcribed in rice (Hanada et al., 2009), whereas the only case of an Arabidopsis duplicate that is transcribed (KI) behaves like the associated transposase gene, suggesting that the function of KI may be selfish rather than cellular (Hoen et al., 2006).
Like MULE-mediated gene acquisition in plants, cellular gene transduction by retroelements seems to be a common feature during L1 retrotransposition in human (Goodier et al., 2000). However, only transduction by infectious retroviruses has been shown to generate hybrid open reading frames (ORFs) that could modulate cell function, although in this case expression of the hybrid ORF leads to neoplastic transformation (Cooper, 1995). Thus, the significance of cellular gene transduction for gene diversification in the absence of a disease phenotype remains to be determined. A recent study suggests that the rate of chimeric gene formation by retroposition is 50-fold higher among grass genomes than in primates and that retroposition has kept grass genomes in constant flux of new chimeric retrogenes (Wang et al., 2006). In addition to transposon activity, several other mechanisms contribute to the origination of new genes, including exon shuffling caused by illegitimate recombination and retroposition, gene duplication, retroposition of a gene transcript, lateral gene transfer, gene fusion, and de novo formation of new genes from previously noncoding sequences (for review, see Long et al., 2003). In tomato (Solanum lycopersicum), a recent mutation fused exons from the gene encoding the β-subunit of inorganic pyrophosphate-dependent phosphofructokinase to those of the homeobox gene LeT6, leading to elevated levels and an altered pattern of expression of the latter (Chen et al., 1997). This mutation (called mouse ear) arose spontaneously in an isogenic tomato cultivar and leads to excessively proliferated leaves, consistent with the altered expression pattern of LeT6 (Rick and Harrison, 1959; Chen et al., 1997) and suggesting that the emergence of new gene variants may lead to phenotypic differences. Another tomato gene, SUN, was duplicated together with 24.7 kb of flanking sequences through gene transduction by the retrotransposon Rider (Xiao et al., 2008). This duplication event brought SUN into a new genomic context that increased its expression, leading to an elongated fruit shape. The identification of young genes such as these provides tools to study the origin and evolution of new genes, since many details on the origin of a gene are lost over long periods of time (Long et al., 2003).
We and others have previously reported the first case, outside of oncogenic retroviruses, of a retrotransposon that has transduced host cellular sequences (Bureau et al., 1994; Jin and Bennetzen, 1994; Palmgren, 1994; Elrouby and Bureau, 2001). The maize long terminal repeats (LTR) retrotransposon Bs1 has transduced sequences from three different maize cellular genes, namely, proton-dependent membrane ATPase (c-pma), xylan endohydrolase (c-xe), and β-1,3-glucanse (c-bg; where c corresponds to the cellular genes, whereas their retroelement-associated counterparts are designated r-pma, r-xe, and r-bg; Elrouby and Bureau, 2001). The transduction events generated a hybrid ORF (ORF1, 740 amino acids) that contains sequences corresponding to the Bs1 gag domain fused to the transduced sequences. Here, we report that the Bs1-associated transduction events and subsequent mutations led to the emergence of a novel gene by the shuffling of existing sequences. We show that Bs1 is both transcribed and translated in reproductive tissues, and specifically in ears. The BS1 protein is not detected in extracts obtained from sterile ears, suggesting that Bs1 expression may be associated with normal reproductive development in maize. Characterization of Bs1 from several maize landraces and inbred lines as well as from the wild relatives of maize, the teosintes, reveals that different large and small deletions/insertions mediated the formation of one uninterrupted ORF (ORF1) following the initial transduction events. A sequence highly related to maize ORF1 is first seen in Z. mays subsp. parviglumis, which has been shown by independent lines of evidence to be the progenitor of domesticated maize (Z. mays subsp. mays; Doebley, 2004). Collectively, the Bs1-associated transduction events generated a novel chimeric gene whose function, if any at this point of its evolution, may be involved in reproductive development.
RESULTS
Bs1 ORF1 Is Expressed in Reproductive Organs
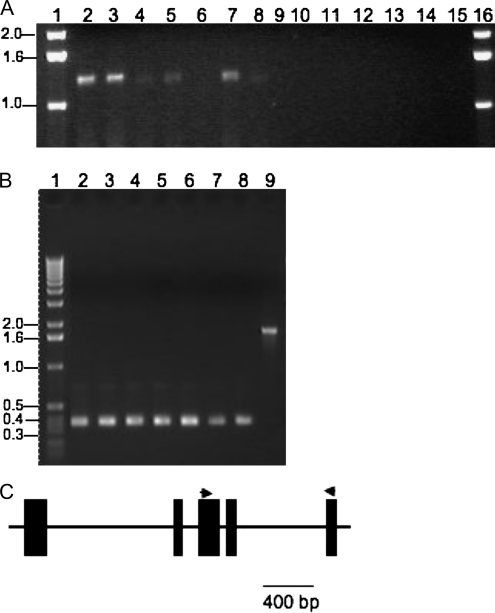
We have previously identified an EST that shares identity with the Bs1 3′ LTR and a part of the internal sequence (Elrouby and Bureau, 2001). This EST was isolated from mixed stages of anther and pollen, suggesting that Bs1 may be expressed in the germ line. To properly assess the Bs1 expression pattern, we performed reverse transcription (RT)-PCR with RNA extracted from different tissue types (Fig. 1) in two maize inbred lines, W22 and Oh43. As seen in Figure 1, Bs1 is specifically expressed in stage R1 or early stage R2 ears (when silk starts to be visible outside the husks; Hanway and Ritchie, 1984) and tassels of both inbred lines (Fig. 1A, lanes 2–5, 7, and 8) but not in any of the vegetative tissues tested (husk, root, 1-week-old seedling, 2-week-old seedling leaf, mature leaf; Fig. 1A, lanes 6 and 9–12).
Figure 1.
Bs1 is expressed in reproductive tissues. A, RT-PCR analysis of Bs1 mRNA extracted from Z. mays subsp. mays. Lanes 1 and 16 contain a Mr marker. Lanes 2 to 8 contain young ear (stage R1 or early stage R2, when silk starts to be visible outside the husks [Hanway and Ritchie 1984]; W22), young ear (Oh43), young tassel (W22), young tassel (Oh43), husk (Oh43), silk-free ear (Oh43), and silk (Oh43), respectively. Lanes 9 to 12 contain Oh43 root, 1-week-old seedling, 2-week-old seedling leaf, and mature leaf, respectively. Lanes 13 to 15 contain controls in which reverse transcriptase was omitted during first-strand cDNA synthesis; lane 13 contains young ear (Oh43), lane 14 contains young ear (W22), and lane 15 contains young tassel (Oh43). B, RT-PCR analysis of Abp1 mRNA extracted from Z. mays subsp. mays (Oh43) using the primer pair shown in the schematic in C (represented by arrowheads). Lane 1 contains a Mr marker and lanes 2 to 8 contain young ear, young tassel, root, seedling leaf, 8-week-old leaf, silk, and husk, respectively. Lane 9 contains a PCR amplification product using the same primer pair used for RT-PCR but with genomic DNA as a template. C, Schematic depiction of the Abp1 gene showing primers (arrowheads) used in B. The primers used to amplify the Bs1 transcript are indicated by arrows above the Bs1 structure depicted in Figure 4.
Since the Bs1 chimeric sequence was generated by retroelement-mediated gene transduction, it is devoid of any introns (Elrouby and Bureau, 2001). It is thus important to confirm that the RT-PCR products obtained were truly amplified from cDNA rather than contaminating genomic DNA. First, when reverse transcriptase was omitted from the reaction, no amplification products were obtained at all (Fig. 1A, lanes 13–15). Second, we used the same cDNA to amplify transcripts of an intron-containing gene. For this purpose, we used the gene coding for Abp1 (for auxin-binding protein 1; Elrouby and Bureau, 2000). Abp1 primers anchored in exons 3 and 5 yield an RT-PCR product consistent with amplification from cDNA only (Fig. 1B, lanes 2–8). The same primer pair amplified a genomic fragment containing the intervening introns when genomic DNA was used instead as a template (Fig. 1B, lane 9). Third, the fact that no Bs1 amplification products were obtained in vegetative tissues (Fig. 1A, lanes 6 and 9–12) whereas the same tissues supported amplification from Abp1 transcripts (Fig. 1B, lanes 4–8) suggests that the Bs1 amplification products obtained in reproductive tissues were derived from reverse-transcribed Bs1 mRNA expressed differentially in these tissues.
Bs1 ORF1 Is Translated
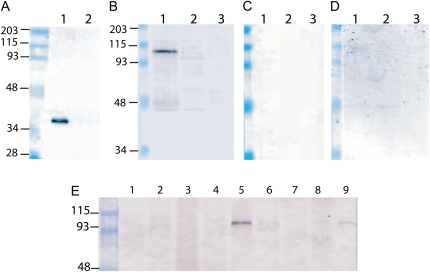
To determine whether the Bs1 transcript is translated, we raised polyclonal antibodies against ORF1. In ORF1, both r-xe and r-pma but not r-bg maintained the reading frame of their cellular gene counterparts (Elrouby and Bureau, 2001). To avoid cross-reactivity against cellular proteins encoded by c-xe and c-pma, only the sequence encoding the N-terminal 301 amino acids of ORF1 was used to raise the antibodies (anti-BS301). This sequence spans the Bs1 gag and r-bg domains (Elrouby and Bureau, 2001). Another antibody was raised against a synthetic peptide spanning residues 196 to 215 (anti-BS196). Both antisera recognize the BS1 protein expressed in Escherichia coli (Fig. 2A, shown for anti-BS196 antiserum), and a polypeptide of approximately 100 kD in extracts from maize young ears but not from sterile ears (see below) or leaves (Fig. 2B). The size of this polypeptide is consistent with a translational product encoded by Bs1 ORF1. Additionally, the approximately 100-kD protein is not recognized by preimmune sera (Fig. 2C) and is competed out when the antiserum is preincubated with the BS1 synthetic peptide (Fig. 2D). We conclude that the approximately 100-kD polypeptide we see in maize ear extracts is most likely encoded by Bs1 ORF1.
Figure 2.
Bs1 is translated. Immunoblot analysis using total bacterial (A) or plant (B–E) extracts. In A, extracts from a bacterial strain containing a plasmid expressing the N-terminal 301 amino acids (lane 1) or the empty plasmid (lane 2) were probed with anti-BS1 antiserum. In B to D, extracts from young ears, sterile ears, or leaves (lanes 1–3, respectively) were probed with the anti-BS1 antiserum (B), preimmune serum (C), or anti-BS1 antiserum that had been incubated with a BS1 synthetic peptide (D). In E, proteins extracted from mature leaf, 2-week-old seedling leaf, 1-week-old seedling, silk, young ear, young tassel, endosperm, pericarp, and embryo (lanes 1–9, respectively) were probed with anti-BS1 antibody. In all panels, the left-most lane contains a Mr marker. [See online article for color version of this figure.]
We performed immunoblot analyses using the anti-BS1 antibodies to test Bs1 expression in different maize tissues. Bs1 is translated primarily in young ears and to a much lesser extent in young tassels and mature embryos (Fig. 2E, lanes 5, 6, and 9). In tassels, two bands of approximately 97 and 100 kD are sometimes seen (Fig. 2E, lane 6). The nature of the smaller band is unclear. In vitro translation experiments have previously indicated that, in addition to ORF1, a longer polypeptide predicted for the frameshift fusion of ORF1 and ORF2 can be generated (Jin and Bennetzen, 1989). We do not see any evidence of such a fusion in plant extracts.
Bs1 Expression Is Associated with Normal Reproductive Development
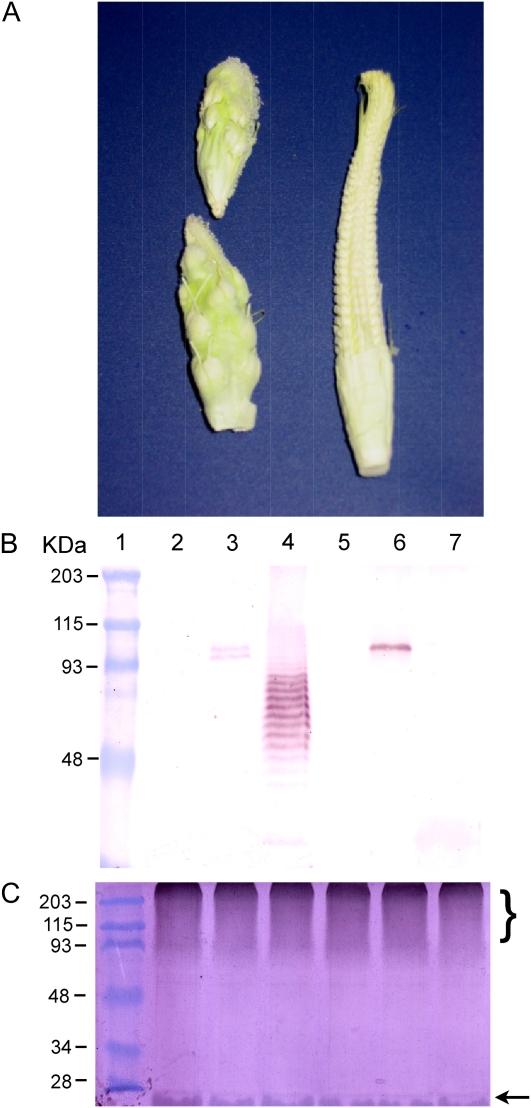
As a consequence of altered light and temperature conditions due to growth at elevated latitude, maize plants occasionally undergo normal vegetative development but produce sterile ears (Fig. 3A). These ears have a vegetative appearance, are stunted, light green in color, with aborted kernels that look like elevated swellings from the main axis of the cob, and arrested silk. When proteins prepared from these ears were tested by immunoblot analysis, we could not detect the Bs1 ORF1 polypeptide (Fig. 3B, lane 7). Furthermore, we could not detect the ORF1 polypeptide in proteins extracted from post-pollen tassel (Fig. 3B, lane 4). Instead, the anti-BS1 antibodies detected several bands, all of which are smaller than expected of the ORF1 polypeptide. Since the sum of the intensities of these bands is more than that seen in young tassel extracts (Fig. 3B, lane 3), they are unlikely to be degradation products. Thus, the nature of these bands remains to be determined. However, it is possible that they may correspond to shorter Bs1 translational products resulting from in-frame initiations at several internal ATG codons (ORF1 contains 20 internal Mets; data not shown). Immunoblots of protein extracts from both sterile ears and post-pollen tassels and probed with anti-ubiquitin antisera (Fig. 3C) suggest that both tissues contain a normal suite of proteins and that general protein degradation is not the case, as indicated by the presence of high molecular mass ubiquitinated proteins and free ubiquitin (Fig. 3C).
Figure 3.
Bs1 is involved during normal reproductive development. A, Photograph of normal (right) and sterile (left) maize ears. B, Immunoblot analysis using anti-BS1 antibody and proteins extracted from leaf, normal young tassel, post-pollen tassel, leaf, normal young ear, and sterile ear (lanes 2–7, respectively). C, Immunoblot analysis using anti-ubiquitin antibody and protein extracts as in B. The brace indicates high molecular mass ubiquitinated proteins, and the arrow indicates free ubiquitin. Lane 1 contains a molecular mass marker with sizes in kilodaltons. [See online article for color version of this figure.]
Structure and Evolution of Bs1 in Maize and the Teosintes
In order to study the structure and sequence evolution of Bs1 ORF1, we cloned and characterized Bs1 from domesticated maize and the teosintes (Zea luxurians, Zea diploperennis, Z. mays subsp. mexicana, Z. mays subsp. huehuetenangensis, Z. mays subsp. parviglumis). In these taxa, Bs1 copy number ranges from one to five (Johns et al., 1985; Elrouby and Bureau, 2001). To amplify all potential copies, the sense and antisense primers were anchored in the retroelement primer-binding site and polypurine tract, respectively, both of which are expected to be conserved. The number of copies isolated from the different taxa reflects expected copy numbers. We isolated five maize (inbred line W22) Bs1 copies (My1 to -5), two from Z. luxurians (L14, L15), three from Z. diploperennis (D12, D13, D18), four from Z. mays subsp. mexicana (Mx6, Mx16, Mx23, Mx24), three from Z. mays subsp. huehuetenangensis (H8, H10, H25), and one from Z. mays subsp. parviglumis (P22; Fig. 4; Table I). Although an intact ORF1 is only observed in two maize Bs1 copies (see below), all Bs1 copies contain all three transduced genes, suggesting that the transduction events took place before the speciation of the genus Zea.
Figure 4.
Structure and evolution of Bs1 in maize and the teosintes. Top, The Bs1 structure showing the three transduced genes (r-bg, r-xe, and r-pma), the gag and env domains, and the retroelement LTR. ORF1 and the hypothetical ORF2 are shown below. Primers used for RT-PCR are indicated by the arrows. Bottom, The different Bs1 copies in maize and the teosintes are represented by horizontal lines, deletions and insertions and their sizes in base pairs by triangles, and stop codons by asterisks.
Table I. Sequence analysis of Bs1 genes in maize and the teosintes.
| Codon Disrupteda | Nature of Disruption (Size in bp) | Bs1 Sequenceb |
| 6 | Stop | Mx16 |
| 12 | Stop | L14, L15, D13 |
| 16 | Indel (1) | H10 |
| 26 | Stop | D13, H8, H10 |
| 40 | Indel (2) | L14, L15 |
| 62, 63 | Indel (2) | L14, D13 |
| 79 | Indel (1) | L14, L15 |
| 88 | Indel (1) | L14, L15, D13, D18, H25, H10, Mx6, Mx16, Mx23, Mx24 |
| 138 | Indel (21) | D12, D18 |
| 171 | Indel (8) | D13 |
| 185 | Indel (1) | L14, L15, D12, D13, D18, H10, Mx16, P22, My3 |
| 194 | Indel (69) | L14, L15, D13 |
| 206 | Stop | L14, L15 |
| 210 | Stop | D13 |
| 234 | Indel (1) | Mx23 |
| 257 | Stop | Mx6, Mx23, Mx24 |
| 286 | Indel (1) | L14, L15 |
| 339 | Indel (1) | D13 |
| 344 | Indel (1) | D13 |
| 345 | Indel (1) | D18, H10, Mx23, My1, My3, My5 |
| 373 | Stop | Mx6, Mx23, Mx24 |
| 430 | Stop | D12, D18 |
| 435 | Indel (3) | Mx16 |
| 471 | Indel (1) | H25 |
| 473 | Indel (91) | Mx16 |
| 490–510 | Indel (60) | D12, D18 |
| 520 | Stop | D13 |
| 526 | Indel (1) | H8 |
| 548 | Indel (1) | H10 |
| 561 | Indel (187/183) | L14, L15, D13 |
| 619 | Indel (2) | D18 |
| 624 | Stop | My1 |
| 679–689 | Indel (30) | L14, L15 |
| 688 | Stop | D13 |
| 736 | Indel (2) | L14, L15, D12, D13, D18, H10, Mx6, Mx16, Mx23, Mx24, P22, My3 |
| 736, 737 | Indel (3) | H8 |
| 738 | Stop | L14, L15, D13 |
| Downstream of ORF1 | Indel (12) | L14, L15, D12, D13, D18 |
| Downstream of ORF1 | Indel (2/1) | L14, L15, D12, D18, P22 |
| Downstream of ORF1 | Indel (59) | My1, My3 |
Codon numbers are based on Bs1 ORF1 in Z. mays subsp. mays.
Bs1 copies were cloned from Z. luxurians (L), Z. diploperennis (D), Z. mays subsp. mexicana (Mx), Z. mays subsp. huehuetenangensis (H), Z. mays subsp. parviglumis (P), and Z. mays subsp. mays (My). A 696-bp deletion found in H25 is not represented here.
Except for the Z. mays subsp. parviglumis and four maize copies, all other Bs1 copies contain in-frame premature stop codons when compared with maize ORF1 (Jin and Bennetzen, 1989; copies My2, My4, Oh43 [this study]). The stop codons terminate the coding sequence at codons 6 (Mx16), 12 (L14, L15, D13), 26 (D13, H8, H10), 206 (L14, L15), 210 (D13), 257 and 373 (Mx6, Mx23, Mx24), 430 (D12, D18), 520 and 688 (D13), 624 (My1), and 738 (L14, L15, D13; Fig. 4; Table I). Likewise, all teosinte and three of the maize Bs1 copies sustain insertions/deletions (indels) that disrupt the Bs1 coding potential (Fig. 4; Table I). The teosinte Bs1 copies contain, in addition to 1- to 3-bp indels, larger indels such as those starting at codon 138 (21 bp, in D12, D18), codon 171 (8 bp, in D13), codon 194 (69 bp, in L14, L15, D13), codon 473 (91 bp, in Mx16), codon 490 to 510 (60 bp, in D12, D18), codon 561 (183/187 bp, in L14, L15, D13), and codon 679 to 689 (30 bp, in L14, L15). It is interesting that the 183/187-bp insertion seen in both Z. luxurians copies and in D13 corresponds to the 183 bp of c-pma that is later deleted in maize r-pma to form mature ORF1 (Bureau et al., 1994; Jin and Bennetzen, 1994; Palmgren, 1994; Elrouby and Bureau, 2001). Also, the 1-bp indel at codon 88 is conserved in all but three teosinte Bs1 copies, and the 2-bp indel at codon 736 is conserved in all teosinte copies as well as one copy from maize (My3). Another 1-bp indel that disrupts codon 185 is conserved in all perennial teosinte Bs1 copies, at least one copy from each annual teosinte (Mx16, H10, P22), and one copy from maize (My3). With the exception of one Z. mays subsp. huehuetenangensis copy (H25), Bs1 copies isolated from the annual teosintes contain only simple (1–3 bp) indel (Fig. 4; Table I). In H25, a large 696-bp deletion eliminates approximately two-thirds of the r-pma and all of the env regions and is likely to have occurred later after all transductions took place. In Z. mays subsp. parviglumis, P22 contains a 1-bp deletion at codon 185 as well as a 2-bp deletion at codon 736.
In maize (inbred line W22), five Bs1 copies were isolated (Fig. 4; Table I), but only two of them potentially encode an intact ORF1. The remaining three (My1, My3, My5) contain the same 1-bp deletion located at codon 345 (also present in D18, Mx23, H10). In addition, My3 contains the same 1-bp deletion present in all perennial Bs1 copies as well as in Mx16, H10, and P22 (disrupting codon 185) and the 2-bp deletion disrupting codon 736 in all perennial and annual teosinte copies. We also isolated Bs1 sequences from a number of maize exotic landraces and inbred lines and identified copies similar to maize (W22) and Z. mays subsp. parviglumis Bs1 (data not shown). The two maize Bs1 copies that potentially encode intact ORF1 (My2, My4) differ only by nucleotide substitutions (98% identical at the nucleotide level and 96% identical at the amino acid level). My4 is more similar to the published Bs1 sequence (isolated from maize inbred line 1s2p; Jin and Bennetzen, 1989). We also isolated a full-length Bs1 cDNA from another maize inbred line (Oh43). As in the case of My2 and My4, the Oh43 cDNA potentially encodes an intact ORF1 (Fig. 4).
Patterns of Nucleotide Sequence Evolution
The determination of the ratio of the nonsynonymous (leading to amino acid replacements) substitution rate per nonsynonymous sites and synonymous (leading to silent changes) substitution rate per synonymous sites (dN/dS) has been used extensively to infer the nature of selection operating on genes of interest (Yang, 2002). An excess of nonsynonymous over synonymous substitutions is an indication of positive selection, whereas a low dN/dS ratio is indicative of purifying selection (Yang, 2002). We calculated the dN/dS ratio for Bs1 ORF1 using the CODEML program of PAML (see “Materials and Methods”). Except for H25, all sequences described above were used in this analysis. H25 was eliminated because it contains a large (696 bp) deletion that is likely to skew the analysis. We obtained a dN/dS ratio not significantly different from 1 (0.89), possibly suggesting that, within the genus Zea, ORF1 is under neutral genetic drift. Since maize Bs1 sequences containing an intact ORF1 clustered in one clade (data not shown), we used CODEML to calculate a different dN/dS value for this cluster and assessed whether it is significantly different from that of the rest of the sequences. Likelihood ratio estimates suggest no significant difference (P = 0.59, 1 degree of freedom). Pairwise comparisons of the maize and Z. mays subsp. parviglumis Bs1 sequences, however, reveal dN/dS ratios of 0.60, 0.59, and 0.56 for Bs1(1s2p)/P22, My4/P22, and My2/P22, respectively (Table II). When the Bs1 copies that encode intact ORF1 were compared, we obtained ratios slightly higher than 1 when Bs1(1s2p) or My4 was compared with My2 (1.2 or 1.17, respectively) but lower than 1 (0.50) when Bs1(1s2p) and My4 were compared with each other (Table II). This confirms our earlier findings that My4 is more similar to the published Bs1 sequence (isolated from 1s2p) than it is to My2 and suggests that there may be two slightly different copies of Bs1 (1s2p/My4 type and My2 type) that are evolving differently.
Table II. dN/dS values of pairwise comparisons of potentially functional Bs1 copies and their parental teosinte copy.
| Bs1(1s2p) | My4 | My2 | P22 | |
| Bs1(1s2p) | – | |||
| My4 | 0.50 | – | ||
| My2 | 1.21 | 1.17 | – | |
| P22 | 0.60 | 0.59 | 0.56 | – |
We also used dN/dS ratio estimates to study the substitution patterns between the transduced genes and their parental host genes. We have previously reported that integration of both c-pma and c-xe but not c-bg occurred in a manner that preserved their open reading frames in Bs1 ORF1 (Elrouby and Bureau, 2001). We tested whether substitution patterns were also constrained to keep the cellular gene's amino acid composition (Table III). The dN/dS ratios for c-pma/r-pma, c-xe/r-xe, and c-bg/r-bg were 0.29, 0.18, and 0.91, respectively, suggesting that r-pma and r-xe are likely under pressure to maintain their cellular gene amino acid sequence. A high value for c-bg/r-bg is consistent with the random integration of r-bg and very similar to the value we determined for ORF1 comparisons (0.89), suggesting that r-bg and c-bg diverged at the same rate at which ORF1 diverged within the genus Zea.
Table III. dN/dS values of pairwise comparisons of transduced sequences and their parental maize cellular gene counterparts.
| r-pma | r-xe | r-bg | |
| c-pma | 0.29 | ||
| c-xe | 0.18 | ||
| c-bg | 0.91 |
DISCUSSION
Gene transduction by retroelements occurs frequently during the retrotransposition of L1 elements in human (Moran et al., 1999; Goodier et al., 2000). Approximately 23% of human L1 elements seem to have transduced host sequences. With approximately 400,000 L1 elements, transduction events like these have enlarged the diploid human genome by as much as 19 Mb, or 0.6% (Goodier et al., 2000). L1-mediated gene transduction has also been shown to occur in an experimental system involving a human cell culture and proposed to have the potential as a mechanism for the evolution of new genes by shuffling of already existing sequences (Moran et al., 1999; Goodier et al., 2000). In addition, the human PMCHL1 gene was created by retrotransposition of an antisense MCH mRNA coupled with the de novo creation of splice sites (Courseaux and Nahon, 2001). A computational survey of the rice genome identified 1,235 retrogenes, and 27 of these are located within LTR retrotransposons (Wang et al., 2006). Additionally, 380 of these retrogenes contain chimeric protein-coding sequences. Combined with an exceptionally high rate of chimeric gene formation by retroposition in grass genomes, gene transduction by retroelements is likely to contribute to the phenotypic diversity of grasses. In maize, Bs1 remains the best studied LTR retrotransposon with clear evidence for multiple gene transduction events (Elrouby and Bureau, 2001). In this study, we provide evidence that these transduction events may have resulted in the formation of a chimeric new gene. Whereas all transduction events took place before the speciation of the genus Zea, the formation of ORF1 may have happened in domesticated maize or in the diverse populations of its progenitor Z. mays subsp. parviglumis, suggesting a possible recent emergence of this new gene.
The Birth of a New Gene
Several findings suggest that Bs1 has evolved as a new gene. First, it is both transcribed and translated. This is remarkable, since gene transduction and integration of the transduced sequences into the retroelement genome is theoretically a random process. In Bs1, none of the transduced sequences were full-length ORFs (i.e. only fragments of the three genes were incorporated into the Bs1 genome; Elrouby and Bureau, 2001). Moreover, in the case of r-xe, a part of the transduced sequence is noncoding in c-xe (5′ untranslated region), and r-bg was integrated into Bs1 without maintaining the c-bg translational frame. Bs1 transductions were also associated with numerous mutations, including two major deletions. The first deletion removed 385 bp in r-xe compared with its cellular gene counterpart (c-xe). This deletion eliminated 44 bp from the first exon, all of the intervening intron, and 82 bp of the second exon (Elrouby and Bureau, 2001). The second deletion removed 183 bp in r-pma when compared with c-pma and hence eliminated most of exon 6 (Bureau et al., 1994; Jin and Bennetzen, 1994). Additionally, single point mutations account for a large number of amino acid changes, and the transduced sequences considerably diverged from their cellular gene counterparts (sequence identities are 81%, 86%, and 88% for r-bg/c-bg, r-xe/c-xe, and r-pma/c-pma, respectively; Elrouby and Bureau, 2001). Despite all these mutations, one long uninterrupted ORF (ORF1) formed in maize and is both transcribed and translated. The size of the protein product observed in our immunoblot analyses is consistent with a translational product of ORF1.
Second, analysis of the Bs1 sequence in maize and its wild relatives, the teosintes, suggests a process that guided the emergence of this new gene. Bs1 is likely to be a Zea-specific element. DNA hybridization experiments using DNA from a variety of monocotyledonous and dicotyledonous plants reveal its presence only in Zea species (Johns et al., 1985). Similarly, no Bs1 sequences were found in closely related grasses (rice, sorghum [Sorghum bicolor], barley [Hordeum vulgare]), since BLAST searches only identified short regions of similarity with cellular gene orthologs of the transduced genes (data not shown). In Bs1 copies from the different Zea species, several deletions/insertions were identified, mainly in the teosintes, and obviously led to the formation of one long uninterrupted ORF (ORF1) in maize. For example, we have previously noticed that, compared with c-pma, r-pma in maize Bs1 contained a deletion of 183 bp (Bureau et al., 1994; Elrouby and Bureau, 2001). In this study, we identified teosinte Bs1 copies (in Z. luxurians and Z. diploperennis) that still contain this 183-bp sequence. This further supports our hypothesis that c-pma was the last of the three genes to be transduced (Elrouby and Bureau, 2001). More importantly, it indicates that all three Bs1 transductions took place before the speciation of the genus Zea and that the formation of mature ORF1 most probably took place in the diverse populations of Z. mays subsp. parviglumis (the progenitor of maize) or during the domestication of maize. In addition, several deletions/insertions are conserved (identified in the same position) among the different taxa studied. In particular, the 1-bp deletion at codon 88 is seen in almost all teosinte copies but not in maize inbred lines or Z. mays subsp. parviglumis. Likewise, the 2-bp deletion at codon 736 is found in all teosinte sequences as well as in one maize sequence, and the 1-bp deletion at codon 185 is identified in one maize and eight teosinte copies (Fig. 4; Table I). It is likely that simple insertions at these positions were instrumental to the formation of ORF1.
Third, when the dN/dS ratio, a strict measure of selection, was calculated for teosinte Bs1 copies, a value not significantly different from 1 was obtained throughout the length of a reconstructed ORF1. This rules out purifying selection and suggests that in the genus Zea, ORF1 is under neutral drift, a result to be expected for pseudogenes (all teosinte Bs1 copies). However, when potentially “functional” copies of Bs1 obtained from maize inbred lines were compared with their presumed parental copy (from Z. mays subsp. parviglumis), we obtained dN/dS ratios significantly lower than 1, suggesting potential functional constraints on Bs1 ORF1 in domesticated maize. This is also confirmed by the finding that the dN/dS ratio of Bs1(1s2p) and My4, two copies isolated from two different inbred lines, is also lower than 1. Interestingly, the two potentially functional copies identified in maize seem to diverge slightly in sequence, and this is reflected in a slightly increased number of nonsynonymous over synonymous changes. This was evident from the fact that the two copies diverge more at the amino acid level than at the nucleotide level and also from a dN/dS ratio that is higher than 1.
Fourth, Bs1 is expressed only during a specific developmental window. This is evident again both at the transcript and the protein levels and both spatially and temporally. The Bs1 transcript is detected primarily in young ears and to a lesser extent in young tassels but not in any of the vegetative tissues tested. The BS1 protein mirrors its transcript localization; however, whereas it was detected in abundant levels in young ears, it was barely detectable in young tassels. Bs1 expression is also temporally regulated. Immunoblot analysis with protein extracts from young ears (stage R1 or early stage R2, when silk starts to be visible outside the husks; Hanway and Ritchie, 1984), whole mature kernels, mature embryos, endosperm, and pericarp reveals that the BS1 protein is detected only in young ears (Fig. 2). Embryos show very low expression levels, suggesting that, in young ears, Bs1 is probably expressed in ovules and young developing embryos and that expression is either down-regulated or shut down during later stages. This tight expression pattern suggests that Bs1 may have evolved as a new gene and may be involved in early aspects of maize reproduction and/or kernel development.
Fifth, Bs1 expression seems to correlate with normal reproductive development. The BS1 protein was undetectable in sterile ears collected from plants that developed abnormally, probably due to unfavorable growth conditions. These plants grew normal vegetative structures but produced very few pollen and sterile ears. Specifically, the ears were vegetative in appearance (i.e. light green in color with kernels replaced with structures that look like elevated swellings from the main axis of the cob). The cob axis itself was enlarged (in diameter) and constituted most of the mass of the ear. The average length of the cob was 30% to 50% of the length of an R1/R2-stage ear. Silk was arrested early and appeared only when husks were manually removed. These ears arrested at this stage and did not develop further. Whereas the BS1 protein was not detected in protein extracts prepared from these ears, high molecular mass ubiquitinated proteins as well as free ubiquitin (Fig. 3C) were unaffected, suggesting that general protein degradation is not the case and that Bs1 is expressed only during normal ear development. It remains to be determined whether normal reproductive development requires or is necessary for Bs1 expression.
Recent Birth
Our results suggest that the Bs1 transduction events occurred before the speciation of the genus Zea. We identified the transduced sequences in all Bs1 copies isolated from all five teosinte species tested (Fig. 4). However, all the teosinte copies of Bs1 contain deletions, insertions, or premature stop codons that disrupt ORF1. Although this is very clear for perennial (Z. luxurians and Z. diploperennis) and two of the annual (Z. mays subsp. mexicana and Z. mays subsp. huehuetenangensis) teosintes, Bs1 in Z. mays subsp. parviglumis has a structure more similar to that of domesticated maize (Z. mays subsp. mays). The Bs1 sequence in Z. mays subsp. parviglumis differs from maize Bs1 by only several nucleotide substitutions and three simple deletions. The first deletion is located at codon 185 (relative to ORF1) and is 1 bp long. The second deletion is located at position 736 and is 2 bp in length. The third deletion is also 2 bp in length but is located downstream of ORF1 (in the hypothetical ORF2). A single nucleotide insertion at codon 185 would restore ORF1 and produce an ORF (94% identical to maize ORF1, 904 amino acids in length) that terminates 7 bp upstream of the hypothetical ORF2 stop codon (Fig. 4). Although we cannot rule out the presence of such an ORF in Z. mays subsp. parviglumis, we sequenced two more independent PCR products and confirmed the presence of the deletion at position 185. Additionally, this 1-bp deletion was also identified in eight other Bs1 sequences isolated from perennial and annual teosintes as well as domesticated maize (see “Results”), ruling out an amplification error and suggesting that it descends from an ancestral Bs1 copy. Although it is possible that one or more Bs1 copies may have escaped PCR amplification in the taxa used in this study, it is unlikely that this copy encodes an intact ORF1. Given the high degree of sequence similarity between Bs1 copies that contain an intact ORF1 or an ORF1 that contains only simple (1- or 2-bp) indels, amplification would probably miss copies of considerable structural differences rather than copies that are more similar to maize Bs1. This is confirmed by the facts that all five maize Bs1 copies were cloned, that the number of copies cloned from the teosintes corresponds to copy number estimates reported previously (Johns et al., 1985; Elrouby and Bureau, 2001), and that some internal primers (anchored in maize ORF1) failed to amplify any Bs1 sequences from the teosintes.
The high degree of sequence and structural similarity between the Z. mays subsp. parviglumis and maize Bs1 copies makes it difficult to infer the time of emergence of ORF1. It is tempting to suggest that ORF1 formed at or immediately after the domestication of maize, since copies of Bs1 with intact ORF1 were only identified in domesticated maize. However, populations of Z. mays subsp. parviglumis exhibit a very high degree of diversity, and maize has maintained a substantial proportion (60%–70%) of this diversity (Tenaillon et al., 2001; for review, see Tian et al., 2009). Given our small sample size, we are not able to discern whether ORF1 formed in maize or in its progenitor populations. This may require a more detailed examination of Bs1 sequence in a large number of Z. mays subsp. parviglumis and maize populations.
The notion that ORF1 may have acquired, or is in the process of acquiring, a function is extraordinary, given how recently this must have taken place. Based on recent archaeological and molecular data, maize (Z. mays subsp. mays) was domesticated from its progenitor teosinte (Z. mays subsp. parviglumis) approximately 6,250 years ago (Riperno and Flannery, 2001; Matsuoka et al., 2002), although evidence derived from microsatellite analyses estimates the upper limit of the time of divergence of the two subspecies at 9,188 years ago (Matsuoka et al., 2002). Although it has been suggested that most domesticated crops are the products of multiple independent domestications, analysis of 99 microsatellite loci in a large maize and teosinte population suggests a single domestication for maize that is likely to have occurred in the central Balsas River Valley in Mexico (Matsuoka et al., 2002). In archaeological maize samples, analysis of three genes involved in the control of plant architecture, storage protein synthesis, and starch production (and hence that were major players during domestication) revealed that alleles of the three genes typical of contemporary maize were present in Mexican maize by 4,400 years ago, yet allelic selection at one of these genes may have not been completed by as recently as 2,000 years ago (Jaenicke-Despres et al., 2003). As mentioned earlier, maize Bs1 is most similar (based on structure and the degree of sequence identity) to Z. mays subsp. parviglumis Bs1. It is conceivable that the Z. mays subsp. parviglumis allele passed on to maize during domestication approximately 9,000 years ago and that this was followed by two simple insertions that created ORF1. The finding of intermediates containing intact codon 185 or intact codon 736 among maize exotic landraces supports this idea. Alternatively, ORF1 or an ORF1-related fusion of ORF1 and the hypothetical ORF2 may have arisen in Z. mays subsp. parviglumis, followed by selection during domestication to maintain ORF1 in postdomestication maize. The domestication of maize is thought to have involved strong selective sweeps that are likely to reduce genetic diversity in genes and genomic regions important during domestication (Vigouroux et al., 2002). The absence of teosinte-type Bs1 copies (with large indels and premature stop codons) among maize inbred lines is consistent with this idea.
Strong selection for traits that improved agronomic performance, palatability, or nutritional value was instrumental for the domestication of crop plants (Vigouroux et al., 2002). In maize, the ear received much of the attention. The morphological differences between the maize and teosinte ears suggest that traits unique to maize confer a selective disadvantage for surviving in the wild and more suitability as a cultivated crop. For example, maize has a rigid enlarged polystichous (multi-ranked) rachis with tenaciously attached grains that require human intervention for dispersal and propagation, whereas the teosinte ear is distichous (two-ranked) with a thin rachis that naturally disarticulates, aiding in seed dispersal. Teosinte grains are also protected inside fruit cases formed by an invaginated rachis and lower glume. As discussed before, we suggest that Bs1 ORF1 is expressed in the ear (especially young developing ears) and that its expression correlates with normal reproductive development. It is possible that the Bs1 ORF1 contributes some of the traits that farmers favored during domestication (for a discussion of potential functions, see below). Alternatively, the Bs1 ORF1 may have hitchhiked with some of the genes contributing these traits.
Potential Function
It is likely that both r-xe and r-pma contribute properties to the BS1 protein similar to those of their parental cellular proteins. This is based on several observations. First, both r-xe and r-pma integrated in ORF1 in a nonrandom manner that maintained the same reading frame of their cellular gene counterparts (Jin and Bennetzen, 1994; Elrouby and Bureau, 2001). Second, dN/dS ratio estimates reveal potential functional constraints between r-xe and r-pma and their cellular gene counterparts (see “Results”). Third, conserved domain analysis revealed that r-xe corresponds to the sequence encoding the xylan endohydrolase signal peptide (Banik et al., 1996; Elrouby and Bureau, 2001; data not shown) that potentially targets the enzyme to cell wall xylans, and r-pma encodes a slightly truncated ATP-binding and hydrolysis domain characteristic of membrane and vacuolar proton-dependent ATPases (Jin and Bennetzen, 1994; Michelet and Boutry, 1995; Elrouby and Bureau, 2001; data not shown).
Several scenarios are possible for a potential function for Bs1 ORF1. For example, ORF1 may contribute a novel function that may or may not require the r-xe and/or r-pma domains, although amino acid and domain conservation would argue for a function that utilizes one or both domains. Alternatively, ORF1 may alter already existing functions (e.g. those encoded by c-xe and/or c-pma). In this case, Bs1 may down-regulate c-xe and/or c-pma functions in the ear. This is conceivable both at the transcript and the protein levels. Bs1 transcripts share a high degree of sequence identity with those of c-xe and c-pma and may thus mediate their down-regulation by posttranscriptional gene silencing. The ORF1 protein may also compete with proteins encoded by c-xe and/or c-pma for xylan binding (for c-xe) and/or localization to the membrane or the cell wall (for c-pma and c-xe, respectively). One of the main differences between the maize and the teosinte ear is cob size (both in length and diameter). The maize cob is much larger in size and is multi-ranked (polystichous) with tenaciously attached grains that require human intervention for dispersal and propagation (or harvest), traits that are suitable for a field crop. Xylans constitute more than 60% of cell wall polysaccharides in the maize cob (Ebringerova et al., 1997). In the teosinte ear, on the other hand, grains attach to a thin rachis that is mostly cellulose in nature. Reduction of xylan endohydrolase activity in the maize ear could result in a larger xylan content and cob size, traits that might have appealed to farmers during domestication. Elucidating the exact function of Bs1 will require the generation of Bs1 null alleles or knockdown lines as well as detailed molecular and biochemical characterization.
Retroelement-mediated gene transduction has been previously proposed as a general mechanism to generate new genes with the potential to modulate host cellular functions. In this report, we show that gene transduction events mediated by the maize LTR retrotransposon Bs1 led to the formation of a chimeric new gene whose function may be implicated during reproductive development. Given the high frequency with which gene transduction takes place (Moran et al., 1999; Wang et al., 2006) and the frequent occurrence in genomes (Goodier et al., 2000; Wang et al., 2006), other cases similar to Bs1 are likely to be identified, and the full extent of how chimeric genes produced by retroposition may contribute to genetic and phenotypic diversity may be elucidated.
MATERIALS AND METHODS
Plant Material
The teosinte germplasm was a gift from Dr. John Doebley (University of Wisconsin, Madison). Zea luxurians (accession G-38), Zea diploperennis (accession 2549), Zea mays subsp. huehuetenangensis (accession G-120), Z. mays subsp. mexicana (accession 178), and Z. mays subsp. parviglumis (Chilpancingo, Guerrero, Mexico) were previously described (Wang et al., 1999; White and Doebley, 1999). Z. mays subsp. mays landraces Nal-Tel (accession YUC 7), Zapalote Chico (accession OAX 70), Conico (accession PUE 32), Gordo (CHH 160), Assiniboine (accession P1213793), and Serrano (accession GUA 14) and inbred lines (W22, W23, A188, Oh43, K55) were obtained from Dr. Brandon Gaut (University of California, Irvine) and the North Central Regional Plant Introduction Station (Iowa State University, Ames) and were described previously (Tenaillon et al., 2001).
PCR Amplification, RT-PCR, Plasmid Cloning, and Sequencing
The Bs1 sequence in maize and the teosintes was amplified using primer Bs-296S (5′-GCTAACAATTGGTATCAAAGG-3′) and primer Bs-3202A (5′-GTTAGCAACCCAATACCAGTG-3′). The thermocycle was as follows: 95°C for 1 min, 55°C for 2 min, 72°C for 2 min (40 cycles). To achieve fidelity, we used a mixture of a high-fidelity DNA polymerase (Pwo; Roche Diagnostics) and AmpliGold Taq DNA polymerase (Perkin-Elmer) in a 1:10 ratio as recommended by the supplier.
For RT-PCR, total RNA was isolated as before (Elrouby and Bureau, 2000) and treated with RQ1 RNase-free DNase (Promega) to eliminate any contaminating genomic DNA. mRNA was then purified using the Qiagen Oligotex mRNA mini kit (Qiagen). For first-strand cDNA synthesis, approximately 50 ng of mRNA was incubated with an oligo(dT) primer and 200 units of SuperScript II reverse transcriptase (Invitrogen) as recommended by the supplier, or with 1 μL of water (as a negative control for cDNA synthesis). To amplify the Bs1 cDNA shown in Figure 1, the following primers were used: Bs-1881S (5′-GCCAGTGGGCTGAGGAGG-3′) and Bs-3202A. Full-length Bs1 cDNA was amplified from Z. mays subsp. mays (Oh43) using primers Bs-296S and Bs-3202A. To amplify the Abp1 cDNA and genomic controls, we used primer Abp-3442S (5′-CAATAAGTCCAGGTCAAAGGACGCCAATC-3′) and Abp-5490A (5′-GGAAACACTTGTGACCTAGAG-3′; Elrouby and Bureau, 2000). All PCR and RT-PCR products were cloned into pCR2.1 using the TA cloning kit (Invitrogen) and sequenced as before (Elrouby and Bureau, 2001).
Antibody Production, Protein Extraction, and Immunoblot Analyses
The sequence coding for the N-terminal 301 amino acids of Bs1 ORF1 was amplified (as described above) using primer Bs-338S (5′-GGGGGATCCGAGCCCACCCTGCAGTCC-3′) and primer Bs-1237A (5′-GGGAAGCTTTCATGCGCGGCTAAGCCAGC-3′). Primer Bs-338S starts at the second codon of ORF1 and contains an engineered BamHI site at its 5′ end, whereas primer Bs-1237A contains an engineered termination codon (TGA) followed by a HindIII site at its 5′ end. The PCR product was cloned into pCR2.1 and sequenced (as described above). The insert in pCR2.1 was then subcloned into the BamHI-HindIII sites of the bacterial expression vector pQE30 (Qiagen) to generate a translational fusion with six N-terminal His (6×His) residues. The expression of recombinant pQE30, in Escherichia coli (XL1-Blue) cells, was then induced using 200 μm isopropyl β-d-1-thiogalactopyranoside for 5 h at 37°C. Purification of the 6×His-tagged BS1 protein on nickel-nitrilotriacetic acid agarose was performed according to the manufacturer's (Qiagen) instructions. This BS1 antigen was called BS301 and used to raise anti-BS301 antibodies as described below. We also raised a second antibody against a synthetic peptide. The peptide CAETQRQGPRQARRQCRLRV spanning residues 196 to 215 of ORF1 was synthesized and coupled to a carrier protein (keyhole limpet hemocyanin) at the Sheldon Biotechnology Centre of McGill University. This antigen was called BS196. Anti-BS1 antibodies were raised in rabbits at Pocono Rabbit Farm and Laboratory (Canadensis, PA) by injecting 100 μg of purified protein or coupled peptide mixed with Complete Freund's Adjuvant intradermally, followed by three booster injections of 50 μg of purified protein mixed with Incomplete Freund's Adjuvant.
For immunoblot analyses, total protein extracts were prepared from the different tissue types as follows. Plant material was frozen in liquid nitrogen, ground to a fine powder, and then extracted in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, and 0.1% Triton X-100. The suspension was centrifuged at 3,000g for 10 min to pellet tissue debris, the supernatant was recovered and concentrated using CentriPreps YM-10 (Millipore), and a protease inhibitor cocktail (Complete, Mini; Roche Diagnostics) was added as recommended by the supplier. Protein concentration was estimated using the Bio-Rad Laboratories protein assay reagent. Fifty micrograms of total proteins was separated on 10% SDS-polyacrylamide gels and blotted onto nitrocellulose membranes (Bio-Rad Laboratories) using standard methods. Blots were rinsed briefly in 1× phosphate-buffered saline (PBS) and then incubated for 2 h at room temperature in 1× PBS and 5% (w/v) low-fat dry milk with agitation. Anti-BS1 (1:1,000 dilution) or anti-ubiquitin (1:100 dilution; Upstate/Millipore) antibody was then added, and the blots were allowed to agitate for 2 h. The blots were then washed twice (10 min each) in 1× PBS/5% milk, and a secondary antibody (goat anti-rabbit IgG-alkaline phosphatase conjugate; Sigma) was added according to the supplier's instructions. After incubation for 2 h, the blots were washed as before and the signal was visualized by the addition of a solution of 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Sigma) as recommended by the supplier. For peptide competition, the anti-BS1 antibody was incubated with the BS1 peptide (10 μg mL−1 in PBS) shaking for 2 h at 4°C and then used for immunoblot analysis.
Nucleotide and Amino Acid Sequence Analysis
To infer Bs1 structure in maize and the teosintes, multiple sequence alignments were generated using ClustalW (Higgins et al., 1994) with minor manual editing. To determine dN/dS, we used the CODEML program of the PAML (for Phylogenetic Analysis by Maximum Likelihood) suite of programs (Goldman and Yang, 1994; version 4.2a, January 2009). To obtain intact ORFs for all sequences, stop codons and codons with simple deletions (1–3 bp) were replaced by question marks, insertions were manually removed, and larger deletions (30 bp in one case and 60 bp in another) were replaced by a reconstructed consensus sequence (based on the remaining sequences). Maximum parsimony trees were generated using the PHYLIP suite of programs. CODEML default parameters (codon Freq = 2, clock = 0) were used to estimate omega for all branches (model = 0, NSsites = 0) using branch lengths of the tree generated by PHYLIP or independent of branch length. We also allowed CODEML to estimate different values for omega for the clade that contains intact ORF1 sequences and the rest of the Bs1 copies (model = 2, NSsites = 0) and used likelihood ratio test statistics (χ2 with 1 degree of freedom) to assess whether the difference was significant. For pairwise comparisons, we used runmode = −2.
Acknowledgments
We thank Ilkka Kronholm and Asis Hallab for assistance with PAML and Dr. Daniel Schoen for critical reading of the manuscript.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Banik M, Garrett TPJ, Fincher GB. (1996) Molecular cloning of cDNAs encoding 1,4-β-xylan endohydrolases from the aleurone layer of germinated barley (Hordeum vulgare). Plant Mol Biol 31: 1163–1172 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15: 621–627 [DOI] [PubMed] [Google Scholar]
- Bureau TE, White SE, Wessler SR. (1994) Transduction of a cellular gene by a plant retroelement. Cell 77: 479–480 [DOI] [PubMed] [Google Scholar]
- Chen JJ, Janssen BJ, Williams A, Sinha N. (1997) A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell 9: 1289–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM. (1995) Oncogenes, Ed 2. Jones and Bartlett, Sudbury, MA, pp 21–65 [Google Scholar]
- Courseaux A, Nahon JL. (2001) Birth of two chimeric genes in the Hominidae lineage. Science 291: 1293–1297 [DOI] [PubMed] [Google Scholar]
- Cowan RK, Hoen DR, Schoen DJ, Bureau TE. (2005) Mustang is a novel family of domesticated transposase genes found in diverse angiosperms. Mol Biol Evol 22: 2084–2089 [DOI] [PubMed] [Google Scholar]
- Doebley J. (2004) The genetics of maize evolution. Annu Rev Genet 38: 37–59 [DOI] [PubMed] [Google Scholar]
- Ebringerova A, Hromadkova Z, Hribalova V, Mason TJ. (1997) Effect of ultrasound on immunogenic corn cob xylan. Ultrason Sonochem 4: 311–315 [DOI] [PubMed] [Google Scholar]
- Elrouby N, Bureau TE. (2000) Molecular characterization of the Abp1 5′ flanking region in maize and the teosintes. Plant Physiol 124: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Bureau TE. (2001) A novel hybrid ORF formed by multiple cellular gene transductions by a plant LTR-retroelement. J Biol Chem 276: 41963–41968 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11: 725–736 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr. (2000) Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9: 653–657 [DOI] [PubMed] [Google Scholar]
- Hanada K, Vallejo V, Nobuta K, Slotkin RK, Lisch D, Meyers BC, Shiu SH, Jiang N. (2009) The functional role of Pack-MULEs in rice inferred from purifying selection and expression profile. Plant Cell 21: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanway JJ, Ritchie SW. (1984) How a Corn Plant Develops: Special Report No. 48. Iowa State University, Ames, IA [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94: 463–470 [DOI] [PubMed] [Google Scholar]
- Hoen DR, Park KC, Elrouby N, Yu Z, Mohabir N, Cowan RK, Bureau TE. (2006) Transposon-mediated gene expansion and diversification of a family of ULP-like genes. Mol Biol Evol 23: 1254–1268 [DOI] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH. (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by phytochrome A-signaling pathway. Plant J 34: 453–471 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Jaenicke-Despres V, Buckler E, Smith BD, Gilbert MTP, Cooper A, Doebley J, Paabo S. (2003) Early allelic selection in maize as revealed by ancient DNA. Science 302: 1206–1208 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jin YK, Bennetzen JL. (1989) Structure and coding properties of Bs1, a maize retrovirus-like retrotransposon. Proc Natl Acad Sci USA 86: 6235–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YK, Bennetzen JL. (1994) Integration and nonrandom mutation of plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns MA, Mottinger J, Freeling M. (1985) A low copy number copia-like transposon in maize. EMBO J 4: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res 15: 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr (2004) Mobile elements: drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Li WH, Gu Z, Wang H, Nekrutenko A. (2001) Evolutionary analysis of the human genome. Nature 409: 847–849 [DOI] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. (2003) The origin of new genes: glimpses from the young and old. Nat Rev Genet 4: 865–875 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez J, Buckler E, Doebley J. (2002) A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99: 6080–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li XP, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403: 785–789 [DOI] [PubMed] [Google Scholar]
- Michelet B, Boutry M. (1995) The plasma membrane H+-ATPase: a highly regulated enzyme with multiple physiological functions. Plant Physiol 108: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr (1999) Exon shuffling by L1 retrotransposition. Science 283: 1530–1534 [DOI] [PubMed] [Google Scholar]
- Palmgren MG. (1994) Capturing of host DNA by a plant retroelement: Bs1 encodes plasma membrane H(+)-ATPase domains. Plant Mol Biol 25: 137–140 [DOI] [PubMed] [Google Scholar]
- Pardue ML, Danilevskaya ON, Lowenhaupt K, Slot F, Traverse KL. (1996) Drosophila telomeres: new views on chromosome evolution. Trends Genet 12: 48–52 [DOI] [PubMed] [Google Scholar]
- Rick CM, Harrison AL. (1959) Inheritance of five new tomato seedling characters. J Hered 50: 91–98 [Google Scholar]
- Riperno DR, Flannery KV. (2001) The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications. Proc Natl Acad Sci USA 98: 2101–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Craig NL. (1998) V(D)J recombination: a transposase goes to work. Cell 94: 411–414 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98: 9161–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Stevens NM, Buckler ES., IV (2009) Tracking footprints of maize domestication and evidence for a massive selective sweep on chromosome 10. Proc Natl Acad Sci USA 106: 9979–9986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux Y, McMullen M, Hittinger CT, Houchins K, Schulz L, Kresovich S, Matsuoka Y, Doebley J. (2002) Identifying genes of agronomic importance in maize by screening microsaellites for evidence of selection during domestication. Proc Natl Acad Sci USA 99: 9650–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. (1999) The limits of selection during maize domestication. Nature 398: 236–239 [DOI] [PubMed] [Google Scholar]
- Wang W, Zheng H, Fan C, Li J, Shi J, Cai Z, Zhang G, Liu D, Zhang J, Vang S, et al. (2006) High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SE, Doebley JF. (1999) The molecular evolution of terminal ear1, a regulatory gene in the genus Zea. Genetics 153: 1455–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2002) Inference of selection from multiple species alignments. Curr Opin Genet Dev 12: 688–694 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]