Lignin is the generic term for a large group of aromatic polymers resulting from the oxidative combinatorial coupling of 4-hydroxyphenylpropanoids (Boerjan et al., 2003; Ralph et al., 2004). These polymers are deposited predominantly in the walls of secondarily thickened cells, making them rigid and impervious. In addition to developmentally programmed deposition of lignin, its biosynthesis can also be induced upon various biotic and abiotic stress conditions, such as wounding, pathogen infection, metabolic stress, and perturbations in cell wall structure (Caño-Delgado et al., 2003; Tronchet et al., 2010). Because lignin protects cell wall polysaccharides from microbial degradation, thus imparting decay resistance, it is also one of the most important limiting factors in the conversion of plant biomass to pulp or biofuels. The removal of lignin from plant biomass is a costly process; hence, research efforts are now aimed at designing plants that either deposit less lignin or produce lignins that are more amenable to chemical degradation (Sticklen, 2008; Weng et al., 2008a; Mansfield, 2009).
The main building blocks of lignin are the hydroxycinnamyl alcohols (or monolignols) coniferyl alcohol and sinapyl alcohol, with typically minor amounts of p-coumaryl alcohol (Fig. 1). The monolignols are synthesized from Phe through the general phenylpropanoid and monolignol-specific pathways. Phe is derived from the shikimate biosynthetic pathway in the plastid (Rippert et al., 2009). Certain enzymes of the lignin biosynthetic pathway, namely the cytochrome P450 enzymes CINNAMATE 4-HYDROXYLASE (C4H), p-COUMARATE 3-HYDROXYLASE (C3H), and FERULATE 5-HYDROXYLASE (F5H), are membrane proteins thought to be active at the cytosolic side of the endoplasmic reticulum (Chapple, 1998; Ro et al., 2001). Although metabolic channeling has been shown between PHENYLALANINE AMMONIA-LYASE (PAL) and C4H (Rasmussen and Dixon, 1999; Achnine et al., 2004), it remains unknown whether the other pathway enzymes are also part of metabolic complexes at the endoplasmic reticulum.
Figure 1.
The main biosynthetic route toward the monolignols p-coumaryl, coniferyl, and sinapyl alcohol (Boerjan et al., 2003). PAL, PHENYLALANINE AMMONIA-LYASE; C4H, CINNAMATE 4-HYDROXYLASE; 4CL, 4-COUMARATE:CoA LIGASE; C3H, p-COUMARATE 3-HYDROXYLASE; HCT, p-HYDROXYCINNAMOYL-CoA:QUINATE/SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE; CCoAOMT, CAFFEOYL-CoA O-METHYLTRANSFERASE; CCR, CINNAMOYL-CoA REDUCTASE; F5H, FERULATE 5-HYDROXYLASE; COMT, CAFFEIC ACID O-METHYLTRANSFERASE; CAD, CINNAMYL ALCOHOL DEHYDROGENASE.
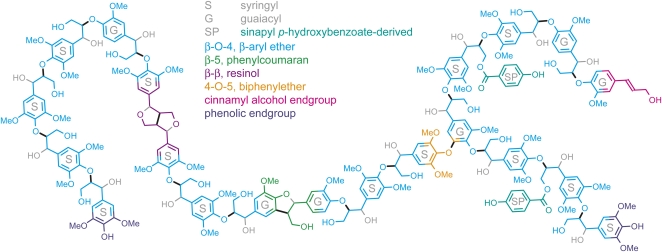
The units resulting from the monolignols, when incorporated into the lignin polymer, are called guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H) units (Figs. 1 and 2). With some notable exceptions (Novo Uzal et al., 2009), lignins from gymnosperms are composed of G-units only (with minor amounts of H-units), whereas angiosperm dicot lignins are composed of G- and S-units. H-units are elevated in softwood compression wood and may be slightly higher in grasses (Boerjan et al., 2003). A variety of less abundant units have been identified from diverse species, and these may be incorporated into the polymer at varying levels (Ralph et al., 2004). Some units, such as those derived from the monomer sinapyl acetate, can make up to 85% of all S-units in the polymer (Lu and Ralph, 2008; Martínez et al., 2008). In addition to differences in lignin composition among taxa and species, lignin composition can also differ among cell types, as readily visualized by the histochemical Mäule staining, which is indicative of S-units, or when chemically analyzed by laser capture microdissection followed by microanalysis of lignin (Nakashima et al., 2008; Ruel et al., 2009). Lignins can even be dissimilar at the level of individual cell wall layers, as revealed by Raman, IR, and UV microspectroscopy, or by immunolabeling of the secondary wall with antibodies cross-reacting with specific lignin substructures (Shi et al., 2006; Gierlinger and Schwanninger, 2007; Gou et al., 2008; Ruel et al., 2009).
Figure 2.
Representation of a lignin polymer from poplar, as predicted from NMR-based lignin analysis (adapted from Stewart et al., 2009).
EVOLUTIONARY ASPECTS
It is commonly accepted that lignin evolved together with the adaptation of plants to a terrestrial life to provide them with the structural support needed for an erect growth habit (Fig. 3). Comparative genomics studies based on available plant genomes indicate that the complete biosynthetic pathway, except for F5H, first appeared in moss (based on the Physcomitrella genome sequence; Xu et al., 2009) but was absent from green algae. However, recent studies have also detected secondary walls and apparent lignin in the marine red alga Calliarthron, which diverged from vascular plants more than 1 billion years ago (Martone et al., 2009), indicating either a very strongly conserved evolutionary history of the relevant pathways or convergent evolution. In support of the latter hypothesis, parallels between Calliarthron and angiosperms are obvious; lignin in the secondary cell walls of Calliarthron may have evolved to resist the bending stresses imposed by breaking waves, similar to lignin in the walls of vascular plants that provides biomechanical support (Martone et al., 2009). In the lycophyte species Selaginella, convergent evolution has been demonstrated with the identification of a new F5H that is capable of functioning in S-lignin biosynthesis but that is structurally unrelated to F5H characterized in angiosperms (Weng et al., 2008b, 2010). It is also noteworthy that cell cultures, but not woody cell walls of Ginkgo biloba, are able to synthesize S lignin; the S-lignin pathway in gymnosperms is generally thought to be absent (Novo Uzal et al., 2009). Clearly, the complex evolutionary puzzle regarding S lignin has yet to be resolved.
Figure 3.
Phylogenetic tree showing the distribution of lignin monomer composition across major lineages. *, S-units are only found in cell cultures of Ginkgo, not in wood (Novo Uzal et al., 2009). **, Lignin-like structures are reported in some mosses and green algae, but the presence of real lignin in these nonvascular species remains questionable; red algae have been barely studied (Weng et al., 2008b; Martone et al., 2009).
PATHWAY PERTURBATIONS
Genes have been cloned for each of the steps of the lignin biosynthetic pathway and the impact on lignin amount and composition has been studied through mutants or reverse genetics in various species, mostly Arabidopsis (Arabidopsis thaliana), maize (Zea mays), alfalfa (Medicago sativa), poplar (Populus spp.), and tobacco (Nicotiana tabacum) but more recently also in flax (Linum usitatissimum; CCoAOMT; Day et al., 2009), the gymnosperms Pinus radiata (HCT and 4CL; Wagner et al., 2007, 2009) and Picea abies (CCR; Wadenbäck et al., 2008), and in rice (Oryza sativa; CAD; Zhang et al., 2006; Li et al., 2009). (For full gene/protein names not provided in the text, see legend of Fig. 1.) From these studies, it can be concluded that reducing the expression of each gene, except F5H, lowers lignin amount in the cell wall to varying extents, depending on the gene, the species, and the level of gene redundancy (Li et al., 2008; Nakashima et al., 2008; Vanholme et al., 2008). Secondary walls with decreased lignin amounts are often characterized by collapsed vessels. This “irregular xylem” phenotype, indicative of reduced mechanical cell wall strength, is not unique to reductions in lignin, as it is also noted in mutants with defects in the biosynthesis of other cell wall polymers (Brown et al., 2005). Strongly reduced lignin amounts result in altered plant development, but more importantly, modest reductions can lead to normal development, as nicely demonstrated by the allelic series of c4h (ref3) mutants in Arabidopsis; the strongest allele causes small and sterile plants, and the weaker alleles result in relatively normal plants with still less lignin (Ruegger and Chapple, 2001; Schilmiller et al., 2009). This implies that, for applications in crops, fine-tuning of the lignin level or targeting the transgene expression to only specific cell types will be an important endeavor.
In addition to lignin content, lignin H/G/S composition also appears to be rather flexible. Silencing of HCT or C3H leads to lignin with H-unit levels as high as 100% of the total thioacidolysis lignin monomers (Franke et al., 2002; Abdulrazzak et al., 2006; Ralph et al., 2006; Besseau et al., 2007; Coleman et al., 2008). Down-regulation of F5H or COMT strongly reduces S-unit content; in contrast, up-regulation of F5H increases the S-unit content (Franke et al., 2000; Stewart et al., 2009). Furthermore, down-regulation of CAD increases the incorporation of cinnamaldehydes into the polymer (Baucher et al., 1998; Lapierre et al., 1999, 2004; Kim et al., 2003). The most remarkable shifts in lignin composition are seen when COMT is down-regulated; in these lignins, 5-hydroxyconiferyl alcohol, derived from the COMT substrate, is incorporated, although it is below the detection limit in wild-type lignin (Van Doorsselaere et al., 1995; Lapierre et al., 1999; Fig. 5). It is interesting that shifts in G and S levels, as well as shifts toward more aldehydes, generally have only minor effects on plant development. Obviously, because the relative ratio of the different monomers determines the frequency of the different bonds in the polymer, all these compositional shifts have repercussions on the structure of the polymer and may thus alter the cell wall properties.
Figure 5.
2D 13C-1H correlation (HMQC, HSQC) NMR spectrum of an acetylated Arabidopsis cellulolytic enzyme lignin that contains the whole lignin fraction from the cell wall but is depleted in polysaccharides after their removal via treatment with crude cellulases. The spectrum provides a convenient profile of aspects of the lignin structure. A, The aromatic region allows the S/G ratio to be readily determined via integration of the well-separated S2/6 and G2 correlation peaks. B, The aliphatic region of the same spectrum allows the individual structures (at right, colored the same as their associated contours), and therefore the bonding patterns, to be visualized and (in principle) quantified. Dibenzodioxocin units (D) are only seen at lower contour levels (data not shown), like the other correlations from the spirodienones (S). Benzodioxane units (H) from the incorporation of 5-hydroxyconiferyl alcohol are below the detection limit in wild-type Arabidopsis lignin but are seen in lignins from COMT-down-regulated plants.
The effects of lignin pathway perturbations often go beyond alterations in lignin amount, composition, and cell wall structure. Indeed, several studies have now demonstrated that perturbing individual steps of the lignin biosynthetic pathway affects the expression level of other lignin pathway genes and also the expression of genes involved in a multitude of other, seemingly unrelated biological processes. These wider effects are also reflected at the metabolite level (Rohde et al., 2004; Sibout et al., 2005; Shi et al., 2006; Dauwe et al., 2007; Leplé et al., 2007). Uncovering their molecular basis might help mitigate the adverse effects on plant growth and development that often accompany lignin modifications.
THE REGULATORY CASCADE
In the past few years, significant progress has been made in understanding the regulation of lignification. New data indicate a regulatory cascade of upstream transcription factors that control the formation of secondary walls by activating a range of other transcription factors. Some of these downstream transcription factors are then able to induce the expression of genes of the lignin biosynthetic pathway (Zhong and Ye, 2007). The regulatory cascade explains why several of the currently described transcription factors lead to enhanced or reduced lignification when misexpressed in plants while they do not directly regulate the lignin biosynthetic genes by binding to their promoters (Zhong et al., 2006, 2008). Therefore, yeast one-hybrid assays, protoplast transient expression assays, and electrophoretic mobility shift assays have been essential to prove the direct binding of a given transcription factor to the promoters of lignin genes (Table I).
Table I. Overview of transcription factors with a role in the regulation of the phenylpropanoid pathway.
↑, ↓, or =, Target gene up-regulated, down-regulated, or not affected, respectively.
| Transcription Factor | PAL | C4H | 4CL | HCT | C3H | CCoAOMT | CCR | F5H | COMT | CAD | Methoda | Reference |
| Arabidopsis | ||||||||||||
| AtMYB4 | = | ↓b | ↓ | ↑ | = | = | qPCR, TEA | Jin et al.(2000) | ||||
| AtMYB46 | ↑b | ↑ | ↑ | ↑ | ↑b | ↑ | qPCR, TEA | Zhong et al. (2007); Ko et al. (2009) | ||||
| AtMYB58 | ↑ | ↑ | ↑b | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | qPCR, TEA, EMSA | Zhou et al. (2009) |
| AtMYB63 | ↑ | ↑ | ↑b | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | qPCR, TEA, EMSA | Zhou et al. (2009) |
| BREVIPEDICELLUS | ↓ | ↓ | ↓ | ↓b | ↓b | ↓ | qPCR, EMSA | Mele et al. (2003) | ||||
| Tobacco | ||||||||||||
| NtLIM1 | ↑ | ↑ | ↑ | qPCR, EMSAc | Kawaoka et al. (2000) | |||||||
| Maize | ||||||||||||
| ZmMYB31 | = | ↓ | = | ↓ | ↑ | qPCR | Fornalé et al. (2006) | |||||
| ZmMYB42 | ↓ | ↓ | = | ↓ | ↓ | qPCR | Fornalé et al. (2006) | |||||
| Poplar | ||||||||||||
| PttMYB21 | = | = | = | ↓ | = | = | qPCR | Karpinska et al. (2004) | ||||
| PtrMYB3 | ↑ | ↑ | qPCR, TEA | McCarthy et al. (2010) | ||||||||
| PtrMYB20 | ↑ | ↑ | qPCR, TEA | McCarthy et al. (2010) | ||||||||
| Eucalyptus | ||||||||||||
| EgMYB1 | ↓b | ↓b | TEA, EMSA | Legay et al. (2007) | ||||||||
| EgMYB2 | = | = | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑b | qPCR, EMSA | Goicoechea et al. (2005) |
| Antirrhinum majus | ||||||||||||
| AmMYB330 | ↓ | RNA blot | Tamagnone et al. (1998) | |||||||||
| AmMYB308 | = | ↓ | ↓b | ↓ | Y1H, RNA blot | Tamagnone et al. (1998) | ||||||
| Pinus taeda | ||||||||||||
| PtMYB4 | ↓ | = | = | ↑ | ↑ | ↑ | ↑ | ↑ | RNA blot, EMSAd | Patzlaff et al. (2003) | ||
| PtMYB1 | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑↓ | ↑ | qPCR | Bomal et al. (2008) | ||
| PtMYB8 | ↑ | ↑ | ↑ | ↑ | = | ↑ | ↓ | ↑ | qPCR | Bomal et al. (2008) | ||
| Vitis vinifera | ||||||||||||
| VvWRKY2 | ↑ | ↑b | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | qPCR, TEA | Guillaumie et al. (2010) | ||
qPCR refers to quantitative reverse transcription-PCR results from overexpressing lines, what does not necessarily imply direct binding on the respective promoters, except for Myb58, for which the qPCR was done in an estradiol-inducible system in the presence of the protein synthesis inhibitor cycloheximide (Zhou et al., 2009). EMSA, Electrophoretic mobility shift assay; TEA, protoplast transient expression assay; Y1H, yeast one-hybrid assay.
Direct promoter binding proven by TEA, EMSA, or Y1H.
EMSA shows binding to the Pal box.
EMSA shows binding to the AC-promoter element.
Microarray experiments on Arabidopsis cell suspension cultures that are induced to form tracheary elements have identified a set of transcription factors involved in this cell differentiation process (Kubo et al., 2005). Whereas VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 were shown to be key upstream regulators of the protoxylem and metaxylem formation, respectively (Kubo et al., 2005; Yamaguchi et al., 2008), SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1) was described as an upstream regulator of interfascicular fiber development, and overexpression of SND1 led to ectopic formation of secondary cell walls (Zhong et al., 2006). Moreover, SND1 was able to regulate at least 10 other transcription factors (Zhong et al., 2007, 2008; Zhou et al., 2009), some of which activated the phenylpropanoid pathway (e.g. Myb46, Myb63, and Myb58; Table I).
Engineering the expression of transcription factors has the potential to alter lignification with fewer adverse effects on plant development for two reasons. First, these transcription factors bind the promoters of multiple genes, thereby affecting the flux through the pathway in an orchestrated manner. Second, some might be specifically involved in the developmental lignification process and not in other processes, such as stress lignin formation, leaving the plant able to respond to environmental factors.
TRANSPORT AND POLYMERIZATION
After their biosynthesis, the monolignols are translocated to the cell wall by a mechanism that remains a mystery. In one model, the monolignols are translocated over the plasma membrane through their 4-O-glucosylated forms, coniferin and syringin, that are deglucosylated upon their arrival by glucosidases located in the cell wall. However, Arabidopsis mutants defective in the corresponding glucosyltransferases have normal lignin levels (Lanot et al., 2006; A. Chapelle, C. Lapierre, and L. Jouanin, unpublished data; A. Lanot, R. Dixon, and D. Bowles, personal communication). In another model, monolignols are transported to the plasma membrane by Golgi-derived vesicles. However, Kaneda et al. (2008), using [3H]Phe feeding experiments, did not find evidence for Golgi-derived vesicles in monolignol transport, nor did they find any significant label in the monolignol glucosides. Thus, at present, there is no convincing support for a role for monolignol glucosides or Golgi-derived vesicles in the transport of monolignols to the cell wall. Current working models assume that monolignols are translocated across the plasma membrane by transporters, but tests on several prime candidate transporters by reverse genetics have not yet revealed any effect on monolignol transport (Ehlting et al., 2005; L. Samuels, personal communication). Nevertheless, additional studies are needed. Alternatively, coniferyl and sinapyl alcohols might have the capability of unaided diffusion through the plasma membrane (Boija and Johansson, 2006).
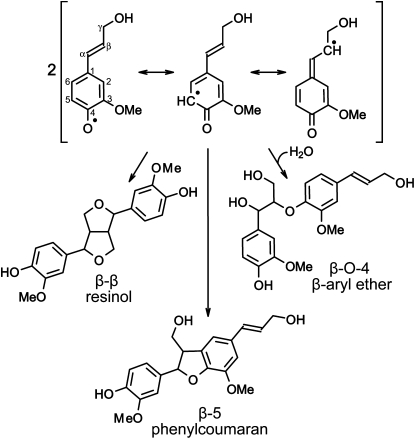
Lignin polymerization occurs via oxidative radicalization of phenols, followed by combinatorial radical coupling. In the first step, the monolignol phenol is oxidized (i.e. dehydrogenated). The resulting phenolic radical is relatively stable due to delocalization of the unpaired electron in the conjugated system (Fig. 4). Subsequently, two monomer radicals may couple to form a (dehydro)dimer, thereby establishing a covalent bond between both subunits. Monolignol radicals favor coupling at their β positions, resulting essentially in only the β-β, β-O-4, and β-5 dimers (Fig. 4). This radical-radical coupling occurs in a chemical-combinatorial fashion; thus, the ratio of each of the possible coupling products depends largely on the chemical nature of each of the monomers and the conditions in the cell wall (Ralph et al., 2004). Then, the dimer needs to be dehydrogenated, again to a phenolic radical, before it can couple with another monomer radical. This mode of action, in which a monomer (radical) adds to the growing polymer, is termed endwise coupling: the polymer grows one unit at a time. Coupling of two lignin oligomers is rare in S/G-lignins but relatively common in G-lignins, where 5-5 coupling accounts for approximately 4% of the linkages (Argyropoulos et al., 2002; Wagner et al., 2009). During each coupling reaction, two radicals are “consumed” (in a so-called “termination reaction”) as each single electron contributes to the newly formed bond, making this type of radical polymerization intrinsically different from the radical chain reactions that occur in the polymerization of several industrial polymers, such as polyethylene, polypropylene, and polystyrene. The average length of a linear lignin chain in poplar is estimated to be between 13 and 20 units (Reale et al., 2004; Stewart et al., 2009).
Figure 4.
Dimerization of two dehydrogenated coniferyl alcohol monomers. Resonance forms of dehydrogenated coniferyl alcohol with the unpaired electron “localized” at the C1 or C3 position are not shown because radical coupling (dimerization) reactions do not occur at these positions.
Monolignol dehydrogenation involves peroxidases and/or laccases. Whereas peroxidases use hydrogen peroxide as a substrate, laccases use oxygen to oxidize their metal centers to enable catalytical phenol oxidation. Both types of enzymes belong to large gene families of which the individual members have overlapping activities, making the process difficult to study in planta; a knockout may have little to no effect on lignification due to gene redundancy (McCaig et al., 2005; Sato and Whetten, 2006). Peroxidases may differ in their substrate specificities; whereas some almost exclusively accept coniferyl alcohol, others are highly specific toward sinapyl alcohol (Marjamaa et al., 2006; Gómez Ros et al., 2007). Because the structure of lignin depends on the availability of monolignol radicals, peroxidase specificity may determine in part the structure of the final lignin polymers, opening possibilities for altering lignin structure by modified expression of specific peroxidase isoforms. Although monolignols might be dehydrogenated via direct interaction with an electron-removing (oxidizing) enzyme, the radicals might alternatively be generated by radical transfer. Indeed, direct interaction of the growing lignin polymer with enzymes may be unlikely given the limited freedom of movement of both in the cell wall. The p-coumarate moiety in sinapyl p-coumarate has been proposed as a radical shuttle for lignification in grasses; p-coumarate, unlike sinapyl alcohol, is an excellent substrate for peroxidases found in grasses, but the p-coumarate radical efficiently transfers its unpaired electron to sinapyl alcohol and, presumably, also to lignin polymers (Hatfield et al., 2008).
In conclusion, the current model of lignification involves peroxidases and/or laccases to provide the oxidative capacity in the cell wall. All phenolic compounds that enter this region will eventually have the potential to radicalize and incorporate into the lignin polymer, subject to simple chemical oxidation and (cross-)coupling propensities. This lignification model also nicely explains why many other phenolic molecules can be integrated into the growing lignin polymer and opens up the possibility of tailoring lignins for industrial applications by regulating the influx and types of monolignols into the cell wall (Ralph, 2006; Grabber et al., 2008).
STRUCTURE
Various methods exist to reveal the molecular details of lignin structure. As lignin is a complex and heterogeneous mixture of polymers, the methods generally aim at estimating the average frequency of the main units and the main bond types in the polymer, from a conglomerate simply of the whole plant or in crudely fractionated major tissues. Nitrobenzene oxidation, pyrolysis-gas chromatography-mass spectrometry (GC-MS), and thioacidolysis and derivatization followed by reductive cleavage are degradative methods that reveal the H/G/S composition of the lignin polymer. All these methods liberate only a fraction of the polymer for analysis. Thioacidolysis and derivatization followed by reductive cleavage liberate monomers only from the so-called noncondensed fraction of the polymer (i.e. only those H-, G-, and S-units that are β-O-4-[ether] linked with other units are released from the polymer and quantified by GC-MS; Lapierre et al., 1985; Lu and Ralph, 1997). A streamlined thioacidolysis method now allows running some 50 samples per day (Robinson and Mansfield, 2009).
NMR, particularly two-dimensional (2D) 13C-1H-correlated (HSQC, HMQC) spectroscopy, which combines the sensitivity of 1H NMR with the higher resolution of 13C NMR, continues to be the best method to reveal the frequencies of the different lignin units and the interunit bonding patterns (Fig. 5; Ralph et al., 1999; Ralph and Landucci, 2010). Although the best quality spectra are obtained from purified isolated lignins, the entire lignin fraction can be analyzed when using cruder isolates, such as so-called “cellulolytic enzyme lignin,” in which most of the polysaccharides are removed by treatment with crude cellulases, or when using unfractionated whole cell wall material, either fully dissolved or simply swollen in the gel state (Yelle et al., 2008; Kim and Ralph, 2010). Analysis of the lignins in these preparations allows most of the details of the overall lignin structure to be determined. The 2D NMR “profile” of unfractionated walls now becomes a fingerprint of all of the components (the various polysaccharides and lignins) in the cell wall. A new development in 2D 13C-1H-correlated spectral data analysis is the use of multivariate analysis that provides even better peak assignment and, hence, a better characterization of the cell wall, as demonstrated by comparing the differences between tension wood and normal wood in poplar (Hedenström et al., 2009).
Although a fair number of the spectral data (contours) have been resolved (essentially those colored in Fig. 5), the molecular origins of many remain unknown. The comparative analysis of lignin from wild-type and transgenic or mutant plants by wet chemistry methods along with NMR analyses has helped significantly in revealing their origin. A striking example is that of COMT-down-regulated plants. Thioacidolysis and phenolic profiling of xylem extracts revealed the increased abundance of units derived from 5-hydroxyconiferyl alcohol; the resulting benzodioxane units, formed when this new monomer incorporates into the polymer, were revealed as new contours in the NMR spectra (Ralph et al., 2001; Marita et al., 2003; Morreel et al., 2004b). More recently, NMR of CCR-deficient plants (poplar, tobacco, and Arabidopsis) showed a low-level incorporation of ferulic acid into lignin. In addition to the previously anticipated β-O-4-ferulate structures, NMR and thioacidolysis evidence was presented for a novel bis-(β-O-4)-ether product arising from coupling of ferulic acid into lignin (Leplé et al., 2007; Ralph et al., 2008). These novel acetal units are now indicative of (free) hydroxycinnamic acid incorporation into lignins. These new units should not be confused with radical coupling products from lignification reactions of ferulate-polysaccharide esters, which are abundant in grasses.
NMR data also provide insight into other aspects of the lignin polymer, such as polymer length, as nicely demonstrated by the analysis of lignin from F5H-overexpressing poplar (Stewart et al., 2009). Lignins from these trees have the “world-record” S content, with an S/G ratio of greater than 35 versus approximately 2 for wild-type poplar lignin. As readily seen by NMR, the lignin is quite homogeneous, composed essentially only of syringaresinol and β-ether units (with small amounts of spirodienones; for bond types, see Fig. 5). Importantly, if the quantification is correct and the syringaresinol level is of the order of 10%, the average lignin chain can only be about 10 units, as it is a simple fact that a linear (S) lignin chain can only have a single resinol unit in that chain. Both factors (the high β-ether level and the short chain length) and the consequently high content of units with a free phenolic end probably contribute to this transgenic plant's remarkable pulping and biomass conversion performance (Huntley et al., 2003; Stewart et al., 2009).
TOWARD “LIGNIN OLIGOMER SEQUENCING” AND “LIGNOMICS”
As lignins have moderately high molecular weights and are highly heterogeneous polymers, deducing their primary structures (i.e. the sequence of H-, G-, and S-units in individual polymers and the bonds between them) is not possible using NMR methods that are typically based on lignin extracted from multiple cell types. In fact, because lignin is combinatorial, sequencing of lignin is meaningless, except for individual molecules. Nevertheless, understanding how monolignols couple (i.e. the propensity to couple a given way) provides insight into the factors that determine lignin structure. In an attempt to start determining the primary structures of low-Mr oligolignols, mixtures have been extracted from poplar xylem and analyzed by liquid chromatography-mass spectrometry (LC-MS; Morreel et al., 2004a). Forty of these structures were identified, and all were in agreement with combinatorial oxidative polymerization. When the relative abundances of these oligolignols were analyzed from transgenic plants with modified lignin, they nicely corresponded with the lignin structural data as obtained by NMR of the total lignin, advocating the value of oligolignol profiling as a high-throughput, easy-to-use, and more sensitive method than traditional NMR- or GC-based methods to reveal changes in incipient lignin structure. In addition, LC-MS-based oligolignol profiling has the potential to discover new lignin monomers. For example, sinapyl p-hydroxybenzoate was authenticated as a new monomer (or monomer conjugate) by identifying its cross-coupling product with sinapyl alcohol (Lu et al., 2004; Morreel et al., 2004a). Nonetheless, the identification of such oligolignols by LC-MS necessitates a profound knowledge of their fragmentation behavior in the gas phase. This laboriously gained knowledge has now allowed the development of an MS-based algorithm that allows sequencing of individual lignin oligomers (Morreel et al., 2010).
Finally, the MS analysis of xylem extracts reveals that many phenylpropanoid-derived molecules still have an unknown identity. Therefore, the structural elucidation and quantification of the full suite of these molecules by LC-MS, which we refer to in our laboratory as “lignomics,” may identify additional candidate lignin monomers and their possible biosynthetic routes.
TRENDS AND RESEARCH AVENUES
One new trend in lignin research is to steer biosynthetic pathways toward the biosynthesis of molecules that, upon incorporation into the lignin polymer, will improve lignin degradation. This idea stems from the growing list of newly discovered lignin monomers, from the observation that lignin is able to readily copolymerize alternative units that derive from incomplete monolignol biosynthesis in plants with pathway perturbations, and from examples in which plants with altered lignin structures are shown to be viable and more easily processed (Pilate et al., 2002; Huntley et al., 2003; Ralph et al., 2006; Leplé et al., 2007). This concept of copolymerizing alternative lignin monomers into the polymer has already been demonstrated in a biomimetic system by polymerizing coniferyl ferulate together with normal monolignols into maize primary cell walls (Grabber et al., 2008). The resulting lignin, now strongly enriched in easily breakable ester bonds in the lignin backbone, degrades at lower temperature and alkaline charges. If such dramatic improvement can also be achieved by genetic engineering, it may result in less energy-demanding biomass pretreatments to liberate the cellulose.
Additional insight into the factors that determine lignin structure and degradability can be obtained by mathematical modeling. According to Freudenberg's concept, coupling of monolignols is dependent on their speed of delivery to the cell wall, their chemical characteristics, and the oxidative environment in the wall, among other factors (Vanholme et al., 2008). A mathematical model has now been developed that predicts the frequency of the different bond types, the number of different polymers, their lengths, and the relative abundance of the different length polymers under varying parameters. This in silico tool can be used to define the parameters that influence lignin degradation in processes such as alkaline pulping or biomass pretreatment methods used in biofuel production (van Parijs et al., 2010). The model is such that it can accommodate the incorporation of alternative lignin monomers to evaluate the effect on lignin degradability.
Regarding more fundamental research, systems biology has revealed that lignin perturbations have wide-ranging consequences on the regulation of the pathway itself and on other metabolic pathways and biological processes (Vanholme et al., 2008). Understanding how this network functions, how it responds to lignin perturbations, and how it can be mathematically modeled to predict the effects of lignin perturbations while taking into account feedback loops, enzyme kinetics, and fluxes still has a long way to go.
Finally, although many studies have demonstrated the beneficial effects of lignin engineering on end-use applications, such as pulping and saccharification in the laboratory, only a few studies have evaluated modified plants in the field (Boerjan, 2005; Kawaoka et al., 2006; Hopkins et al., 2007; Leplé et al., 2007; Wadenbäck et al., 2008; Wei et al., 2008; Walter et al., 2010). However, in the field, plants are exposed to various conditions and stresses caused by weather and pests that are difficult or even impossible to mimic in a greenhouse. Field trials are thus an essential translation toward end-use application. Field trials to examine the effects of reduced lignin on the conversion of biomass to bioethanol have recently been initiated (Custers, 2009; Strauss et al., 2009). In addition to biotech plants, clear benefits have been shown years ago for a number of natural mutants, both in grasses (Vermerris et al., 2007) and pine (Pinus taeda; Dimmel et al., 2001; Yu et al., 2006). With the advent of the biobased economy, the easy release of sugars from the plant cell wall and their use as a carbon source for bacteria that have been engineered to biosynthesize a wide range of biobased products is becoming ever more important.
Acknowledgments
We thank Martine De Cock for help in preparing the manuscript.
References
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, et al. (2006) A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140: 30–48; erratum Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, et al (2006) Plant Physiol 141: 1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA. (2004) Colocalization of l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16: 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyropoulos DS, Jurasek L, Krištofová L, Xia Z, Sun Y, Paluš E. (2002) Abundance and reactivity of dibenzodioxocins in softwood lignin. J Agric Food Chem 50: 658–666 [DOI] [PubMed] [Google Scholar]
- Baucher M, Monties B, Van Montagu M, Boerjan W. (1998) Biosynthesis and genetic engineering of lignin. Crit Rev Plant Sci 17: 125–197 [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W. (2005) Biotechnology and the domestication of forest trees. Curr Opin Biotechnol 16: 159–166 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Boija E, Johansson G. (2006) Interactions between model membranes and lignin-related compounds studied by immobilized liposome chromatography. Biochim Biophys Acta 1758: 620–626 [DOI] [PubMed] [Google Scholar]
- Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JEK, Blais S, Tremblay L, Morency MJ, Pavy N, et al. (2008) Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot 59: 3925–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M. (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Chapple C. (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49: 311–343 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD. (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci USA 105: 4501–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custers R. (2009) First GM trial in Belgium since 2002. Nat Biotechnol 27: 506. [DOI] [PubMed] [Google Scholar]
- Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, Van Beeumen J, Ralph J, Boudet AM, Kopka J, Rochange SF, et al. (2007) Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Day A, Neutelings G, Nolin F, Grec S, Habrant A, Crônier D, Maher B, Rolando C, David H, Chabbert B, et al. (2009) Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem 47: 9–19 [DOI] [PubMed] [Google Scholar]
- Dimmel DR, MacKay JJ, Althen EM, Parks C, Sederoff RR. (2001) Pulping and bleaching of CAD-deficient wood. J Wood Chem Technol 21: 1–17 [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Sonbol FM, Maes T, Capellades M, Puigdomènech P, Rigau J, Caparrós-Ruiz D. (2006) Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol Biol 62: 809–823 [DOI] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C. (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Franke R, McMichael CM, Meyer K, Shirley AM, Cusumano JC, Chapple C. (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22: 223–234 [DOI] [PubMed] [Google Scholar]
- Gierlinger N, Schwanninger M. (2007) The potential of Raman microscopy and Raman imaging in plant research. Spectroscopy 21: 69–89 [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, et al. (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J 43: 553–567 [DOI] [PubMed] [Google Scholar]
- Gómez Ros LV, Gabaldón C, Pomar F, Merino F, Pedreño MA, Ros Barceló A. (2007) Structural motifs of syringyl peroxidases predate not only the gymnosperm-angiosperm divergence but also the radiation of tracheophytes. New Phytol 173: 63–78 [DOI] [PubMed] [Google Scholar]
- Gou JY, Park S, Yu XH, Miller LM, Liu CJ. (2008) Compositional characterization and imaging of “wall-bound” acylesters of Populus trichocarpa reveal differential accumulation of acyl molecules in normal and reactive woods. Planta 229: 15–24 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Hatfield RD, Lu F, Ralph J. (2008) Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9: 2510–2516 [DOI] [PubMed] [Google Scholar]
- Guillaumie S, Mzid R, Méchin V, Léon C, Hichri I, Destrac-Irvine A, Trossat-Magnin C, Delrot S, Lauvergeat V. (2010) The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol 72: 215–234 [DOI] [PubMed] [Google Scholar]
- Hatfield R, Ralph J, Grabber JH. (2008) A potential role for sinapyl p-coumarate as a radical transfer mechanism in grass lignin formation. Planta 228: 919–928 [DOI] [PubMed] [Google Scholar]
- Hedenström M, Wiklund-Lindström S, Öman T, Lu F, Gerber L, Schatz P, Sundberg B, Ralph J. (2009) Identification of lignin and polysaccharide modifications in Populus wood by chemometric analysis of 2D NMR spectra from dissolved cell walls. Mol Plant 2: 933–942 [DOI] [PubMed] [Google Scholar]
- Hopkins DW, Webster EA, Boerjan W, Pilate G, Halpin C. (2007) Genetically modified lignin below ground. Nat Biotechnol 25: 168–169 [DOI] [PubMed] [Google Scholar]
- Huntley SK, Ellis D, Gilbert M, Chapple C, Mansfield SD. (2003) Significant increases in pulping efficiency in C4H-F5H-transformed poplars: improved chemical savings and reduced environmental toxins. J Agric Food Chem 51: 6178–6183 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Rensing KH, Wong JCT, Banno B, Mansfield SD, Samuels AL. (2008) Tracking monolignols during wood development in lodgepole pine. Plant Physiol 147: 1750–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, Bhalerao R, Wingsle G. (2004) MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Biol 56: 255–270 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H. (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J 22: 289–301 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Nanto K, Ishii K, Ebinuma H. (2006) Reduction of lignin content by suppression of expression of the LIM domain transcription factor in Eucalyptus camaldulensis. Silvae Genet 55: 269–277 [Google Scholar]
- Kim H, Ralph J. (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Lu F, Ralph SA, Boudet AM, MacKay JJ, Sederoff RR, Ito T, Kawai S, Ohashi H, et al. (2003) NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org Biomol Chem 1: 268–281 [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim WC, Han KH. (2009) Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 60: 649–665 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanot A, Hodge D, Jackson RG, George GL, Elias L, Lim EK, Vaistij FE, Bowles DJ. (2006) The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J 48: 286–295 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C. (1985) Thioacidolysis of lignin: comparison with acidolysis. J Wood Chem Technol 5: 277–292 [Google Scholar]
- Lapierre C, Pilate G, Pollet B, Mila I, Leplé JC, Jouanin L, Kim H, Ralph J. (2004) Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65: 313–321 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leplé JC, Boerjan W, Ferret V, De Nadai V, et al. (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have opposite impact on the efficiency of industrial Kraft pulping. Plant Physiol 119: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay S, Lacombe E, Goicoechea M, Brière C, Séguin A, Mackay J, Grima-Pettenati J. (2007) Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci 173: 542–549 [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Weng JK, Chapple C. (2008) Improvement of biomass through lignin modification. Plant J 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu X. (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69: 685–697 [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. (1997) Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J Agric Food Chem 45: 2590–2592 [Google Scholar]
- Lu F, Ralph J. (2008) Novel tetrahydrofuran structures derived from β-β-coupling reactions involving sinapyl acetate in Kenaf lignins. Org Biomol Chem 6: 3681–3694 [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J, Morreel K, Messens E, Boerjan W. (2004) Preparation and relevance of a cross-coupling product between sinapyl alcohol and sinapyl p-hydroxybenzoate. Org Biomol Chem 2: 2888–2890 [DOI] [PubMed] [Google Scholar]
- Mansfield SD. (2009) Solutions for dissolution—engineering cell walls for deconstruction. Curr Opin Biotechnol 20: 286–294 [DOI] [PubMed] [Google Scholar]
- Marita JM, Ralph J, Hatfield RD, Guo D, Chen F, Dixon RA. (2003) Structural and compositional modifications in lignin of transgenic alfalfa down-regulated in caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase. Phytochemistry 62: 53–65 [DOI] [PubMed] [Google Scholar]
- Marjamaa K, Kukkola E, Lundell T, Karhunen P, Saranpää P, Farstedt KV. (2006) Monolignol oxidation by xylem peroxidase isoforms of Norway spruce (Picea abies) and silver birch (Betula pendula). Tree Physiol 26: 605–611 [DOI] [PubMed] [Google Scholar]
- Martínez AT, Rencoret J, Marques G, Gutiérrez A, Ibarra D, Jiménez-Barbero J, del Río JC. (2008) Monolignol acylation and lignin structure in some nonwoody plants: a 2D NMR study. Phytochemistry 69: 2831–2843 [DOI] [PubMed] [Google Scholar]
- Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J. (2009) Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol 19: 169–175 [DOI] [PubMed] [Google Scholar]
- McCaig BC, Meagher RB, Dean JFD. (2005) Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221: 619–636 [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Fowler S, Lyskowski D, Piyasena H, Carleton K, Spicer C, Ye ZH. (2010) The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol 51: 1084–1090 [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S. (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inzé D, Messens E, et al. (2010) Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W. (2004a) Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Lu F, Goeminne G, Busson R, Herdewijn P, Goeman JL, Van der Eycken J, Boerjan W, Messens E. (2004b) Phenolic profiling of caffeic acid O-methyltransferase-deficient poplar reveals novel benzodioxane oligolignols. Plant Physiol 136: 4023–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA. (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol 179: 738–750 [DOI] [PubMed] [Google Scholar]
- Novo Uzal E, Gómez Ros LV, Pomar F, Bernal MA, Paradela A, Albar JP, Ros Barceló A. (2009) The presence of sinapyl lignin in Ginkgo biloba cell cultures changes our views of the evolution of lignin biosynthesis. Physiol Plant 135: 196–213 [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, et al. (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36: 743–754 [DOI] [PubMed] [Google Scholar]
- Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leplé JC, Pollet B, Mila I, Webster EA, Marstorp HG, et al. (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20: 607–612 [DOI] [PubMed] [Google Scholar]
- Ralph J. (2006) What makes a good monolignol substitute? Hayashi T, , The Science and Lore of the Plant Cell Wall: Biosynthesis, Structure and Function. BrownWalker Press, Boca Raton, FL, pp 285–293 [Google Scholar]
- Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA. (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281: 8843–8853 [DOI] [PubMed] [Google Scholar]
- Ralph J, Kim H, Lu F, Grabber JH, Leplé JC, Berrio-Sierra J, Derikvand MM, Jouanin L, Boerjan W, Lapierre C. (2008) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ralph J, Landucci LL. (2010) NMR of lignins. Heitner C, Dimmel DR, , Lignins. Marcel Dekker, New York, pp 137–234 [Google Scholar]
- Ralph J, Lapierre C, Marita JM, Kim H, Lu F, Hatfield RD, Ralph S, Chapple C, Franke R, Hemm MR, et al. (2001) Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Ralph J, Marita JM, Ralph SA, Hatfield RD, Lu F, Ede RM, Peng J, Quideau S, Helm RF, Grabber JH, et al. (1999) Solution-state NMR of lignins. Argyropoulos DS, , Advances in Lignocellulosic Characterization. TAPPI Press, Atlanta, pp 55–108 [Google Scholar]
- Rasmussen S, Dixon RA. (1999) Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11: 1537–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale S, Di Tullio A, Spreti N, De Angelis F. (2004) Mass spectrometry in the biosynthetic and structural investigation of lignins. Mass Spectrom Rev 23: 87–126 [DOI] [PubMed] [Google Scholar]
- Rippert P, Puyaubert J, Grisollet D, Derrier L, Matringe M. (2009) Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiol 149: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Mah N, Ellis BE, Douglas CJ. (2001) Functional characterization and subcellular localization of poplar (Populus trichocarpa × Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol 126: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AR, Mansfield SD. (2009) Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J 58: 706–714 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, De Rycke R, Kushnir S, Van Doorsselaere J, Goeminne G, Joseleau JP, Vuylsteke M, Van Driessche G, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Chapple C. (2001) Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159: 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K, Berrio-Sierra J, Derikvand MM, Pollet B, Thévenin J, Lapierre C, Jouanin L, Joseleau JP. (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Sato Y, Whetten RW. (2006) Characterization of two laccases of loblolly pine (Pinus taeda) expressed in tobacco BY-2 cells. J Plant Res 119: 581–588 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C. (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Shi C, Koch G, Ouzunova M, Wenzel G, Zein I, Lübberstedt T. (2006) Comparison of maize brown-midrib isogenic lines by cellular UV-microspectrophotometry and comparative transcript profiling. Plant Mol Biol 62: 697–714 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A. (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JJ, Akiyama T, Chapple C, Ralph J, Mansfield SD. (2009) The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol 150: 621–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticklen MB. (2008) Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Genet 9: 433–443 [DOI] [PubMed] [Google Scholar]
- Strauss SH, Tan H, Boerjan W, Sedjo R. (2009) Strangled at birth? Forest biotech and the Convention on Biological Diversity. Nat Biotechnol 27: 519–527 [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Stacey N, Plaskitt K, Parr A, Chang CF, Lynn D, Dow JM, Roberts K, Martin C. (1998) Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell 10: 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchet M, Balagué C, Kroj T, Jouanin L, Roby D. (2010) Cinnamyl alcohol dehydrogenases C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol Plant Pathol 11: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier MT, Petit-Conil M, Leplé JC, Pilate G, Cornu D, Monties B, et al. (1995) A novel lignin in poplar trees with a reduced caffeic acid/5-hydroxyferulic acid O-methyltransferase activity. Plant J 8: 855–864 [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W. (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- van Parijs FRD, Morreel K, Ralph J, Boerjan W, Merks RMH. (2010) Modeling lignin polymerization. I. Simulation model of dehydrogenation polymers. Plant Physiol 153: 1332–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, Saballos A, Ejeta G, Mosier NS, Ladisch MR, Carpita NC. (2007) Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci 47: S142–S153 [Google Scholar]
- Wadenbäck J, von Arnold S, Egertsdotter U, Walter MH, Grima-Pettenati J, Goffner D, Gellerstedt G, Gullion T, Clapham D. (2008) Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR). Transgenic Res 17: 379–392 [DOI] [PubMed] [Google Scholar]
- Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J. (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L. (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter C, Fladung M, Boerjan W. (2010) Twenty years of GM tree field testing implies high environmental safety. Nat Biotechnol (in press) [DOI] [PubMed] [Google Scholar]
- Wei J, Wang Y, Wang H, Li R, Lin N, Ma R, Qu L, Song Y. (2008) Pulping performance of transgenic poplar with depressed caffeoyl-CoA O-methyltransferase. Chin Sci Bull 53: 3553–3558 [Google Scholar]
- Weng JK, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C. (2010) Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Li X, Bonawitz ND, Chapple C. (2008a) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol 19: 166–172 [DOI] [PubMed] [Google Scholar]
- Weng JK, Li X, Stout J, Chapple C. (2008b) Independent origins of syringyl lignin in vascular plants. Proc Natl Acad Sci USA 105: 7887–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P, et al. (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics (Suppl 11) 10: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T. (2008) VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yelle DJ, Ralph J, Frihart CR. (2008) Characterization of nonderivatized plant cell walls using high-resolution solution-state NMR spectroscopy. Magn Reson Chem 46: 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Li B, Nelson CD, McKeand SE, Batista VB, Mullin TJ. (2006) Association of the cad-n1 allele with increased stem growth and wood density in full-sib families of loblolly pine. Tree Genet Genomes 2: 98–108 [Google Scholar]
- Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z. (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2007) Regulation of cell wall biosynthesis. Curr Opin Plant Biol 10: 564–572 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]