Abstract
In temperate cereals, such as wheat (Triticum aestivum) and barley (Hordeum vulgare), the transition to reproductive development can be accelerated by prolonged exposure to cold (vernalization). We examined the role of the grass-specific MADS box gene ODDSOC2 (OS2) in the vernalization response in cereals. The barley OS2 gene (HvOS2) is expressed in leaves and shoot apices but is repressed by vernalization. Vernalization represses OS2 independently of VERNALIZATION1 (VRN1) in a VRN1 deletion mutant of einkorn wheat (Triticum monococcum), but VRN1 is required to maintain down-regulation of OS2 in vernalized plants. Furthermore, barleys that carry active alleles of the VRN1 gene (HvVRN1) have reduced expression of HvOS2, suggesting that HvVRN1 down-regulates HvOS2 during development. Overexpression of HvOS2 delayed flowering and reduced spike, stem, and leaf length in transgenic barley plants. Plants overexpressing HvOS2 showed reduced expression of barley homologs of the Arabidopsis (Arabidopsis thaliana) gene FLOWERING PROMOTING FACTOR1 (FPF1) and increased expression of RNase-S-like genes. FPF1 promotes floral development and enhances cell elongation, so down-regulation of FPF1-like genes might explain the phenotypes of HvOS2 overexpression lines. We present an extended model of the genetic pathways controlling vernalization-induced flowering in cereals, which describes the regulatory relationships between VRN1, OS2, and FPF1-like genes. Overall, these findings highlight differences and similarities between the vernalization responses of temperate cereals and the model plant Arabidopsis.
Many plants from temperate climates require prolonged exposure to low temperatures to become competent to flower; a phenomenon known as vernalization. The requirement for vernalization is often combined with daylength sensitivity. For example, many ecotypes of Arabidopsis (Arabidopsis thaliana) are vernalized during winter and then flower as daylength increases during spring (Imaizumi and Kay, 2006; Jaeger et al., 2006; Zeevaart, 2006; Turck et al., 2008). Similar seasonal flowering responses are found in economically important cereal crop species, including wheat (Triticum aestivum) and barley (Hordeum vulgare; Trevaskis et al., 2007a; Distelfeld et al., 2009; Greenup et al., 2009).
In Arabidopsis, the promotion of flowering by increasing daylength is mediated by FLOWERING LOCUS T (FT; Kardailsky et al., 1999; Kobayashi et al., 1999). FT encodes a mobile florigen that is produced in the leaves in long days and travels to the shoot apex, where it promotes floral development (Corbesier et al., 2007). Long-day induction of FT in the leaves is controlled by the CONSTANS (CO) protein (Onouchi et al., 2000; An et al., 2004). Expression of the CO transcript follows a diurnal rhythm, peaking in the late afternoon (Valverde et al., 2004; Jang et al., 2008). In long days, the peak in CO expression occurs in light, which stabilizes the CO protein, allowing activation of FT (Valverde et al., 2004; Jang et al., 2008).
Winter-annual ecotypes of Arabidopsis do not flower rapidly in long days unless plants have been vernalized. This requirement for vernalization is mediated by the MADS box floral repressor FLOWERING LOCUS C (FLC; Michaels and Amasino, 1999; Sheldon et al., 1999), which represses FT, and a second floral promoter, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1; Michaels and Amasino, 1999; Sheldon et al., 1999; Hepworth et al., 2002; Michaels et al., 2005). Vernalization down-regulates FLC, allowing long-day induction of FT and SOC1 to accelerate flowering. Vernalization-induced repression of FLC is mediated by protein complexes that chemically modify histones at the FLC locus to promote an inactive chromatin state (Schubert et al., 2006; Wood et al., 2006; De Lucia et al., 2008). The repressive histone modifications deposited at FLC chromatin during vernalization are stable, so repression of FLC is maintained postvernalization; this allows long-day induction of FT to occur in spring in vernalized plants (Sheldon et al., 2000).
The molecular mechanisms that promote flowering in response to long days in Arabidopsis are conserved in temperate cereals. For instance, CO and FT-like genes have been identified in barley and related grasses (Turner et al., 2005; King et al., 2006; Yan et al., 2006; Faure et al., 2007). The barley FT-like1 gene (FT1 or VRN3) is induced by long days and appears to be the functional equivalent of FT in cereals (Turner et al., 2005; King et al., 2006; Yan et al., 2006; Faure et al., 2007). As is the case for FT in vernalization-requiring Arabidopsis ecotypes, vernalization is a prerequisite for long-day induction of FT1 in vernalization-responsive barleys (Hemming et al., 2008). No homologs of FLC have been identified in cereals. Instead, VERNALIZATION2 (VRN2) is expressed in long days to suppress the induction of FT1 and delay flowering prior to vernalization (Takahashi and Yasuda, 1971; Yan et al., 2004; Trevaskis et al., 2006). Vernalization induces expression of VRN1 (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003; von Zitzewitz et al., 2005), which down-regulates VRN2 and promotes expression of FT1 in long days (Trevaskis et al., 2006; Yan et al., 2006; Hemming et al., 2008; Sasani et al., 2009). VRN1 also promotes inflorescence (spike) initiation at the shoot apex, irrespective of daylength (Trevaskis et al., 2006; Hemming et al., 2008; Sasani et al., 2009). Like FLC, changes in chromatin state at the VRN1 locus might provide a mechanism for a memory of vernalization in cereals by allowing stable activation of VRN1 by vernalization (Oliver et al., 2009).
The vernalization response has probably evolved independently in Arabidopsis and the temperate cereals (grasses). Here, we examine the function of grass-specific MADS box genes previously identified by gene expression analyses as potential components of the vernalization response in cereals: T. aestivum MADS box gene 23 (TaMx23; Trevaskis et al., 2003) and the closely related sequence T. aestivum AGAMOUS-like 33 (TaAGL33; Winfield et al., 2009). We show that these genes repress flowering and cell elongation by down-regulating a group of genes related to the FLOWERING PROMOTING FACTOR1 (FPF1) gene of Arabidopsis.
RESULTS
ODDSOC2 Is a Truncated MADS Box Gene Found in Cereals and Related Grasses
Two barley homologs of TaMx23 (Trevaskis et al., 2003) were identified among barley ESTs deposited in the GenBank database. These genes have no direct equivalent in Arabidopsis but show weak similarity to SOC1 (Supplemental Table S1; Supplemental Fig. S1). These genes were designated ODDSOC1 (HvOS1) and ODDSOC2 (HvOS2). ODDSOC-like genes also occur in a range of cereals other than barley, including wheat, rice (Oryza sativa), maize (Zea mays), and sorghum (Sorghum bicolor), and in the model grass Brachypodium distachyon (Table I; Supplemental Table S2). All genes share a high degree of sequence identity (Supplemental Fig. S2). A feature common to the predicted ODDSOC-like protein sequences is their short length compared with other plant MADS box proteins (152–167 versus 200 or more amino acids). No ODDSOC-like genes were identified outside the grasses.
Table I. Nonredundant BLASTP results for HvOS2.
| Protein Description | Accession No. | Organism | Identity | BLAST Score | E Value |
| TaAGL33 | ABF57950 | T. aestivum | 93% (147/158) | 297 | 2e-79 |
| TaAGL41 | ABF57941 | T. aestivum | 84% (126/149) | 255 | 1e-66 |
| TaAGL42 | ABF57942 | T. aestivum | 73% (114/155) | 225 | 1e-57 |
| OsMADS51 (Os01g69850) | NP_001045235 | O. sativa | 74% (111/149) | 224 | 2e-57 |
| Hypothetical protein (Sb03g044170) | XP_002456860 | S. bicolor | 70% (110/157) | 219 | 6e-56 |
| Hypothetical protein (LOC100272251) | NP_001140218 | Z. mays | 69% (108/155) | 197 | 2e-49 |
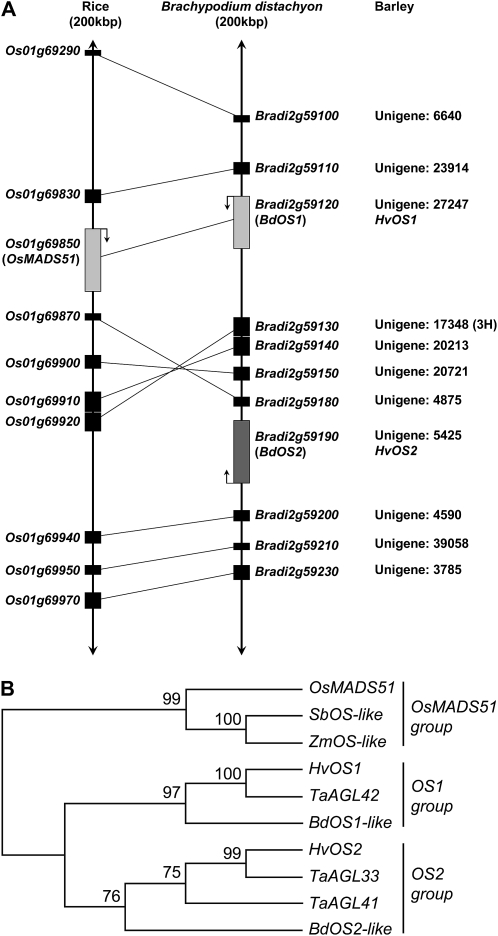
The two ODDSOC genes from Brachypodium are closely linked to one another in a region syntenous to barley chromosome 3H (long arm; 134 centimorgan; Fig. 1A). A single ODDSOC gene is found in the rice genome (OsMADS51; Kim et al., 2007), located in a syntenous region (Fig. 1A). Phylogenetic analysis showed that OS-like genes can be classified into three groups, OS1 and OS2 groups in the temperate cereals/grasses plus a third group corresponding to maize, sorghum, and rice (Fig. 1B), suggesting that these genes have undergone gene duplication during the evolution of the temperate grasses.
Figure 1.
HvOS1 and HvOS2 are members of a grass-specific class of MADS box genes. A, Diagrammatic representation of the syntentic region in rice and B. distachyon that contains the ODDSOC-like genes (OsMADS51, BdOS1, and BdOS2), and the corresponding barley Unigene numbers and map locations. Arrows indicate the direction of transcription. B, Phylogenetic relationships between the ODDSOC-like genes of rice (OsMADS51), maize (ZmOS-like), sorghum (SbOS-like), barley (HvOS1 and HvOS2), wheat (TaAGL33, TaAGL41, and TaAGL42), and B. distachyon (BdOS1-like and BdOS2-like) based on a sequence alignment of the coding sequence for each gene.
HvOS2 Is Repressed by Vernalization
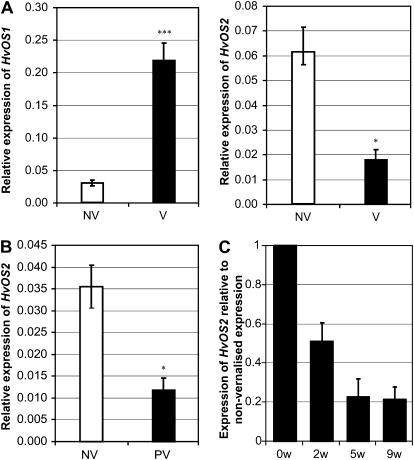
HvOS1 and HvOS2 transcript levels were monitored in seedlings during and after vernalization (Fig. 2A). HvOS1 transcript levels increased during vernalization, whereas HvOS2 transcript levels decreased (Fig. 2A). Since down-regulation of the MADS box floral repressor FLC plays a central role in the vernalization response of Arabidopsis, the role of HvOS2 in the vernalization response of barley was examined further. To determine if changes in HvOS2 expression were maintained after vernalization, transcript levels were assayed in leaves 2 weeks after plants were removed from the cold. Expression of HvOS2 remained low in plants that had been vernalized compared with nonvernalized controls (Fig. 2B). HvOS2 transcript levels were also assayed in the shoot apices, showing that expression of this gene decreases in the apices of vernalized plants (Fig. 2C).
Figure 2.
Vernalization-induced changes in HvOS1 and HvOS2 transcript levels. A, Expression of HvOS1 and HvOS2 in barley (cv Sonja) seedlings germinated in darkness at 20°C (NV, nonvernalized, white bars; n = 4) versus seedlings germinated in darkness at 4°C for 49 d (V, vernalized, black bars; n = 3) harvested at an equivalent stage of development. B, Expression levels of HvOS2 in fully expanded second leaves of nonvernalized plants (white bar; n = 3) versus vernalized plants (PV, postvernalization, black bar; n = 3) harvested in long days at the three-leaf stage, 10 d after the end of vernalization. C, Expression levels of HvOS2 in shoot apices (30–50 individual apices) from nonvernalized plants (0) or plants vernalized for 2, 5, or 9 weeks (w) and then grown in long days (n = 2). Plants were harvested at the three-leaf stage. All expression levels were assayed by qRT-PCR and are shown relative to ACTIN. Error bars show se (A and B) or range (C). Asterisks indicate P values of ANOVA: * P < 0.05, *** P < 0.001.
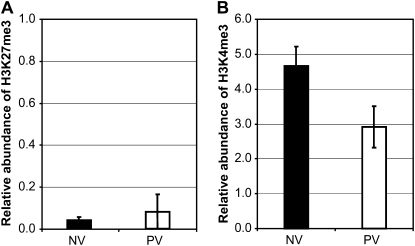
The Promoter of HvOS2 Is Not Enriched for H3K27 Trimethylation
In Arabidopsis, histone 3 Lys-27 trimethylation (H3K27me3), a histone modification associated with long-term gene repression, mediates vernalization-induced repression of FLC (Bastow et al., 2004; Sung and Amasino, 2004; Finnegan and Dennis, 2007; Schmitz et al., 2008). We examined whether a similar mechanism might mediate vernalization-induced repression of HvOS2. H3K27me3 levels were assayed at HvOS2 before and after vernalization. The level of H3K27me3 near the presumed transcriptional start site at HvOS2 chromatin was low irrespective of vernalization treatment (Fig. 3A), suggesting that this modification does not play a role in mediating the down-regulation of HvOS2 during vernalization. The level of H3K4me3, a modification associated with active gene transcription, was lower in vernalized leaves than nonvernalized leaves, consistent with the reduction in HvOS2 expression following vernalization (Fig. 3B).
Figure 3.
Analysis of histone modifications at HvOS2 during vernalization. A, Relative abundance of H3K27me3 at the start of transcription for HvOS2 in nonvernalized plants (NV, black bar) and vernalized plants (PV, postvernalization, white bar; cv Sonja). B, Relative abundance of H3K4me3 at the start of transcription for HvOS2 in nonvernalized plants (black bar) and vernalized plants (white bar; cv Sonja). Error bars show sd.
Expression of HvVRN1 Is Associated with Down-Regulation of HvOS2
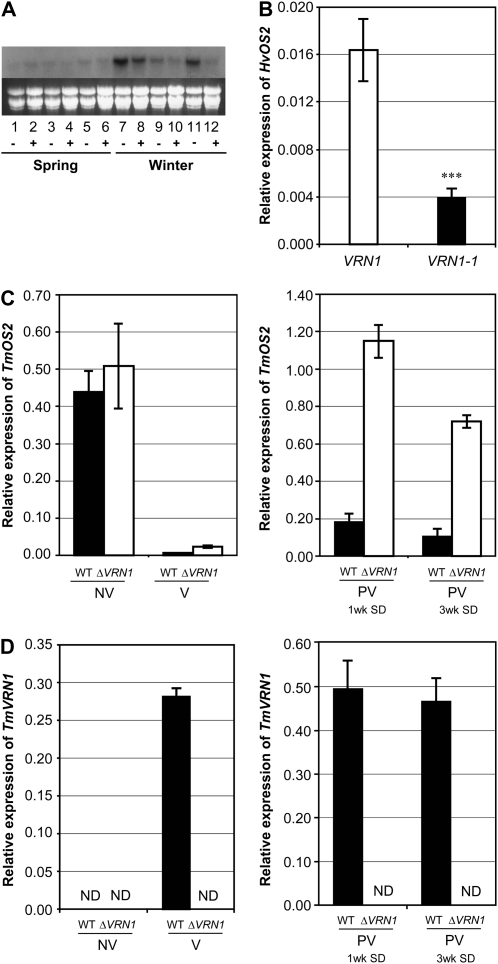
HvOS2 expression was compared between “winter” barleys that respond to vernalization and “spring” barleys that flower without vernalization. Expression of HvOS2 was strongest in winter barleys grown without vernalization treatment, and vernalization caused a decrease in HvOS2 expression in these barleys. Expression of HvOS2 was low in barleys that flower without vernalization, irrespective of vernalization treatment (Fig. 4A).
Figure 4.
Expression of OS2 in different genotypes of wheat and barley. A, HvOS2 expression levels in nonvernalized (−) versus vernalized (+) plants (2 weeks old, grown in long days) from different barley cultivars, including three spring barleys that flower without vernalization, Morgenrot (lanes 1 and 2), Randolph (lanes 3 and 4), and Malta (lanes 5 and 6), and three vernalization-responsive winter barleys, Sonja (lanes 7 and 8), Hudson (lanes 9 and 10), and Mirra (lanes 11 and 12). Expression was assayed by high-stringency hybridization of RNA gel blots with a HvOS2-specific riboprobe. Ethidium bromide staining of ribosomal RNA is shown as a loading comparison. B, HvOS2 expression levels assayed by qRT-PCR in RNA from barley seedlings of a doubled haploid barley population (Sloop × Halcyon). Expression of HvOS2 was assayed in individual lines, relative to ACTIN, and the average expression levels of the different HvVRN1 genotypic classes were compared (VRN1, n = 22; VRN1-1, n = 19). C, Relative expression levels of TmOS2-like (TmAGL33) in the TmVRN1 deletion mutant (ΔVRN1; white bars) versus the wild-type parent strain (black bars). Expression was assayed in vernalized (V; n = 4) and nonvernalized (NV; n = 4) seedlings and in the leaves of plants grown for 1 week or 3 weeks (wild type [WT], n = 3; ΔVRN1, n = 2) in short days (SD) postvernalization (PV). Expression is shown relative to ACTIN. D, Relative expression levels of TmVRN1 in the conditions described in C. Error bars show se. Asterisks indicate P values of ANOVA: *** P < 0.001. ND, Not detected.
To further examine the relationship between vernalization requirement and HvOS2 expression, HvOS2 transcript levels were assayed in lines from the Sloop × Halcyon doubled haploid barley population (Read et al., 2003) grown without vernalization. This population segregates for different alleles of HvVRN1: a wild-type allele that is activated by vernalization (VRN1) and an allele with a deletion in the first intron that is active without vernalization and reduces the vernalization requirement (VRN1-1; Trevaskis et al., 2006; Hemming et al., 2008). Expression of HvOS2 was lower in lines carrying VRN1-1 (Fig. 4B), suggesting that HvVRN1 down-regulates HvOS2.
OS2 expression was then examined in the maintained vegetative phase mutant of the diploid einkorn wheat (Triticum monococcum), which lacks the VRN1 gene (Shitsukawa et al., 2007; hereafter referred to as the ΔVRN1 mutant). When seedlings were germinated in darkness, without vernalization, expression of VRN1 was not detected in either the wild-type parent or the ΔVRN1 mutant and expression of the T. monococcum OS2 gene (TmOS2) did not differ (Fig. 4, C and D).
Expression of TmOS2 and VRN1 was then examined in seedlings at the end of a 7-week vernalization treatment. Expression of VRN1 was induced in vernalized seedlings of the wild-type parent but was not detected in the ΔVRN1 mutant (Fig. 4D). Compared with nonvernalized control seedlings, expression of TmOS2 was lower in vernalized seedlings, irrespective of VRN1 genotype (wild type, P < 0.001; ΔVRN1, P < 0.002; Fig. 4C). VRN1 and TmOS2 transcript levels were then examined in plants grown at normal temperatures for 1 or 3 weeks after vernalization treatment. VRN1 expression remained high in wild-type plants but was not detected in the ΔVRN1 mutant (Fig. 4D). In wild-type plants, expression of TmOS2 remained low but repression of TmOS2 was not maintained in the ΔVRN1 mutant (Fig. 4C).
Overexpression of HvOS2 Delays Flowering and Inhibits Leaf and Stem Elongation
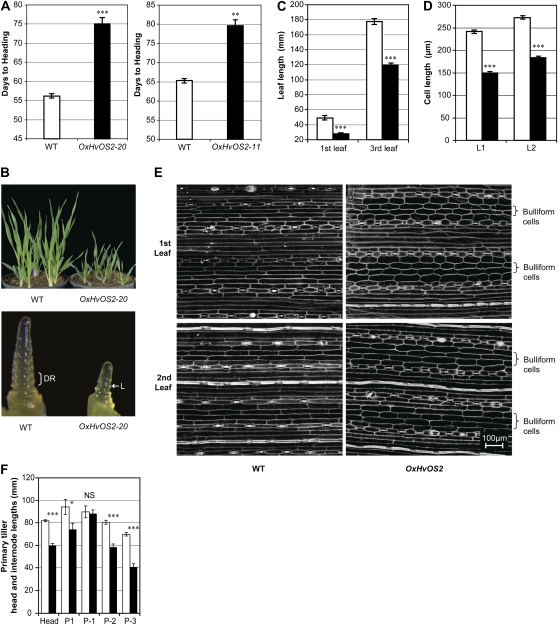
To further investigate the function of HvOS2, a spring barley that flowers without vernalization and has low levels of HvOS2 expression (cv Golden Promise; see “Materials and Methods”) was transformed with a transgene construct that placed HvOS2 under the control of the maize UBIQUITIN promoter. Approximately 50 independent transgenic lines were generated with this construct. The majority were late flowering compared with nontransformed plants, supporting the hypothesis that HvOS2 is a repressor of flowering. Two independent transgenic lines, the progeny of which showed segregation for the transgene construct, were characterized in detail: OxHvOS2-11 and OxHvOS2-20. In both these lines, expression of HvOS2 was higher in plants that inherited the transgene (Supplemental Fig. S3) and a late-flowering phenotype segregated with the transgene in both transgenic families (Fig. 5A). Plants from the OxHvOS2-11 line that inherited the transgene flowered on average 14 d later than siblings lacking the transgene (null siblings), which flowered at a similar time to wild-type Golden Promise plants. Similarly, OxHvOS2-20 transgenic plants flowered on average 18 d later than null siblings (Fig. 5A). Comparison of apex morphology at the third leaf stage, the developmental stage when inflorescence initiation typically occurs in Golden Promise plants under these growth conditions, showed that overexpression of HvOS2 delays the transition to reproductive development (Fig. 5B).
Figure 5.
Phenotypes of transgenic plants that overexpress HvOS2. A, Average days to head emergence (heading date) of transgenic barley lines overexpressing HvOS2 (black bars) versus sibling null segregant controls (plants from the same transgenic line that did not inherit the transgene; wild type [WT], white bars). B, Transgenic barley plants overexpressing HvOS2 versus null segregants at the fourth leaf stage (top), and apex images from the same stage (bottom). DR, Double ridges, the first sign of floral development; L, leaf primordia. C, Average length of the first and third leaves of plants overexpressing HvOxO2 (OxHvOS2-20; black bars) versus the null controls (white bars) at the sixth leaf stage (wild type, n = 5; OxHvOS2-20, n = 5). D, Average length of bulliform cells on the adaxial surface of mature leaves (first leaves [L1] and second leaves [L2]) from plants overexpressing HvOxO2 (OxHvOS2-20; black bars) versus wild-type siblings (white bars). L1, Wild type, n = 367 (cells) and OxHvOS2-20, n = 467; L2, wild type, n = 344 and OxHvOS2-20, n = 478. E, Scanning electron microscopy images of epidermal cells from the adaxial surface of mature leaves (first and second leaves). F, Average head and internode lengths of the primary tiller of plants overexpressing HvOxO2 (black bars) versus null segregants (white bars). P1, Peduncular internode; P-1, below P1; P-2, below P-1; P-3, below P-2. n = 15. Error bars show se. Asterisks indicate P values of ANOVA: * P < 0.05, ** P < 0.01, *** P < 0.001.
In addition to influencing flowering time, overexpression of HvOS2 inhibited leaf elongation. The length of the first and third leaves was reduced in plants overexpressing HvOS2 (Fig. 5C). This reduction in length was due to decreased cell length; the average length of bulliform cells was significantly reduced in the first and second leaves of the transgenic plants (Fig. 5, D and E). The final length of the primary spike was also reduced in transgenic plants overexpressing HvOS2, as were primary tiller (stem) internode lengths (Fig. 5F).
The effects of reducing HvOS2 expression levels was also investigated using a gene-specific RNA interference (RNAi) construct. One of the lines analyzed showed a reduction in expression levels for HvOS2 (Supplemental Fig. S4A). There were no observable phenotypic abnormalities or any change in heading date/final leaf number in any of the RNAi lines analyzed (Supplemental Fig. S4, B and C).
Overexpression of HvOS2 Down-Regulates Barley Homologs of FPF1
Microarray analysis of gene expression was used to investigate the molecular basis for the phenotype of plants overexpressing HvOS2. RNA from seedlings that overexpress HvOS2 was hybridized to the Affymetrix Barley1 chip (Close et al., 2004) and compared with RNA from null sibling control lines. A total of 90 genes were differentially expressed between plants that overexpress HvOS2 and sibling null controls (2-fold change in expression; P < 0.05; Supplemental Table S3). Of the 94 genes differentially expressed, 65 (69%) were up-regulated in plants that overexpress HvOS2 and 25 (27%) were down-regulated. Table II shows the top five up-regulated and down-regulated genes that were differently regulated between plants that overexpress HvOS2.
Table II. Top five up-regulated and down-regulated genes in HvOS2 overexpression line.
| Probe Set | Best Match | Fold Change | P Value |
| Up-regulated | |||
| Contig 5058_x_at | RNase S-like (T. aestivum) | 37.6 | 5.3E-09 |
| Contig 5059_s_at | RNase S-like (T. aestivum) | 36.8 | 3.9E-09 |
| Contig 5185_at | rsh1, RNase S-like (H. vulgare) | 16.2 | 1.1E-07 |
| Contig 12031_at | HvODDSOC2 (H. vulgare) | 15.4 | 7.8E-10 |
| Contig 1568_x_at | THION9, plant thionin family protein (O. sativa) | 8.2 | 5.5E-04 |
| Down-regulated | |||
| Contig 3810_at | Galactinol synthase (T. aestivum) | −2.9 | 1.6E-05 |
| HVSMEm0003G16r2_at | Cytochrome P450 (O. sativa) | −3.4 | 5.7E-03 |
| HVSMEb0010F06r2_at | No description | −3.6 | 3.1E-06 |
| Contig 18182_at | FPF1-like (Arabidopsis) | −3.7 | 3.8E-06 |
| HU14G14r_s_at | FPF1-like (Arabidopsis) | −6.0 | 5.2E-05 |
To verify the results of microarray analysis, the expression levels of several differentially expressed genes were quantified by quantitative reverse transcription (qRT)-PCR (Supplemental Fig. S3). Consistent with the results of microarray analysis, barley orthologs of FPF1, designated HvFPF1-like1 (contig HU14G14r) and HvFPF1-like2 (contig 18182), were down-regulated in plants overexpressing HvOS2 (Supplemental Fig. S3, B and C). Conversely, expression levels of two RNase S-like genes, Hvrsh1 (Gausing, 2000; contig 5185) and Hvrsh2 (contig 5058/9), were elevated in plants overexpressing HvOS2 (Supplemental Fig. S3, D and E).
FPF1-Like Genes Are Regulated by Vernalization and Daylength in Barley
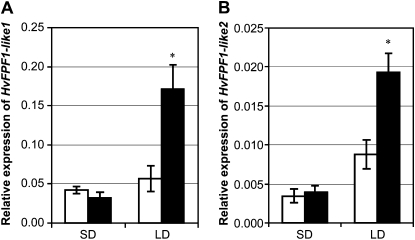
We examined whether expression of FPF1-like genes is influenced by vernalization in barley, a response predicted for genes regulated by HvOS2. Expression of HvFPF1-like1 and HvFPF1-like2 was higher in leaves of vernalized plants compared with nonvernalized plants, when plants were grown in long days postvernalization (Fig. 6). Vernalization did not influence the expression of FPF1-like genes when vernalized plants were grown in short days, where expression of FPF1-like genes was lower (Fig. 6). These data suggest that, in barley, down-regulation of HvOS2 by vernalization allows increased expression of FPF1-like genes when plants are exposed to long days.
Figure 6.
Influence of vernalization on the expression of FPF1-like genes in short or long days. Expression of HvFPF1-like1 (HU14G14r; A) and HvFPF1-like2 (contig 18182; B) in the fully expanded second leaf (harvested at the third leaf stage), nonvernalized (white bars) versus vernalized plants (black bars), grown in long days (LD) or short days (SD). Expression was assayed by qRT-PCR and is shown relative to ACTIN. Error bars show se. Asterisks indicate P values of ANOVA: * P < 0.05 (minimum of three biological repeats).
DISCUSSION
Reproductive development and stem elongation are closely coordinated in temperate cereals. In varieties that require vernalization, both processes are delayed until after winter and begin in spring, when temperature and daylength increase. We have identified a MADS box gene from barley, HvOS2, that potentially delays the transition to reproductive development and impedes cell elongation in stems and leaves but is down-regulated by vernalization (Fig. 2). We suggest that HvOS2 acts in a pathway that delays the transition to reproductive development and inhibits stem elongation prior to winter.
A single ODDSOC gene is found in rice, OsMADS51 (Kim et al., 2007). OsMADS51 promotes flowering and appears to be a component of the molecular pathway that promotes flowering in response to short days (Kim et al., 2007). It is possible that HvOS1 and the equivalent wheat gene (TaAGL42), which are induced by vernalization (Fig. 2A; Winfield et al., 2009), also act as floral promoters. Equally, it is possible that these genes acquired novel functions after the divergence of the ancestors of rice and the temperate cereals. HvOS2, which acts as a floral repressor, might have evolved from a duplication of OsMADS51/OS1 during the evolution of the vernalization response pathway in temperate cereals.
HvOS2 is quantitatively down-regulated by cold (longer cold treatments cause stronger down-regulation), and repression of HvOS2 is maintained in the shoot apex and leaves after vernalization (Fig. 2B; Supplemental Fig. S5). This pattern of gene expression is similar to FLC in Arabidopsis, but unlike FLC, repression of HvOS2 does not appear to involve the deposition of H3K27me3 (Fig. 3A). Vernalization-induced repression of TmOS2 does not require VRN1 (Fig. 4C), but VRN1 is required to maintain repression of TmOS2 after vernalization (Fig. 4C). HvVRN1 also down-regulates HvOS2 in barleys that flower without vernalization (Fig. 4B). Similarly, VRN1 down-regulates TmOS2 when T. monococcum plants (lacking VRN2) are grown in long days (Supplemental Fig. S6). Thus, we suggest that OS2 is down-regulated by cold independently of VRN1 but VRN1 represses OS2 during development at normal growth temperatures, both following vernalization and in barleys that flower without vernalization. Consistent with this hypothesis, microarray analysis shows that HvOS2 is repressed by cold treatments that are not long enough to induce HvVRN1, but repression is not maintained when plants are returned to normal growth temperatures (Supplemental Fig. S7; Plexdb accession no. BB81; Wise et al., 2008).
Spring barleys that flower without vernalization express HvOS2 at low levels compared with vernalization-responsive winter barleys (Fig. 4, A and B). Increasing HvOS2 expression levels in transgenic spring barley (cv Golden Promise) delayed flowering, suggesting that HvOS2 functions as a repressor of flowering. The delay of flowering was caused by a delay of the transition to reproductive development, as evidenced by the impact of the HvOS2 overexpression on final leaf number (Supplemental Fig. S8). Increasing HvOS2 expression levels also influenced plant growth by inhibiting the elongation of cells in the leaves and stems but did not slow the rate of growth (Supplemental Fig. S8). The phenotypes observed in HvOS2 overexpression lines are different from those seen when other MADS box genes are overexpressed in barley (Trevaskis et al., 2007b), suggesting that these phenotypes are indicative of the actual function of HvOS2 and not simply an artifact of overexpressing a MADS box gene. Further reduction of the already low levels of HvOS2 expression in Golden Promise by RNAi did not influence flowering time, however. HvOS2 activity might be below a functional threshold in this spring barley, which flowers without vernalization. Equally, the level of reduction in HvOS2 expression by RNAi might not completely eliminate HvOS2 activity. Isolation of HvOS2 loss-of-function mutants could be used to further examine the role of this gene in future studies. Ideally, this will be done in a vernalization-responsive cultivar to allow the functional importance of HvOS2 to be assessed relative to other genes that delay flowering, such as HvVRN2.
The reduction of cell elongation and delay of flowering seen in HvOS2 overexpression lines was associated with reduced expression of FPF1-like genes (Table II; Supplemental Fig. S3, B and C). FPF1 promotes cell elongation and accelerates flowering when overexpressed in Arabidopsis (Kania et al., 1997; Melzer et al., 1999). These phenotypes mimic the effects of gibberellin (GA) application in Arabidopsis, and it has been suggested that FPF1 acts in a GA-dependent elongation pathway (Kania et al., 1997). Similar phenotypes have also been reported when FPF1-like genes were ectopically expressed in rice and tobacco (Nicotiana tabacum; Ge et al., 2004; Smykal et al., 2004), suggesting that the role of FPF1-like genes is conserved across divergent plant lineages. The reduction of cell elongation and delay of flowering seen in HvOS2 overexpression lines might be due primarily to reduced expression of FPF1-like genes. Many of the dwarfing phenotypes in transgenic plants overexpressing HvOS2 were abolished upon application of GA (Supplemental Fig. S9). This is consistent with the hypothesis that HvOS2 regulates HvFPF1-like genes, which may in turn alter GA responses.
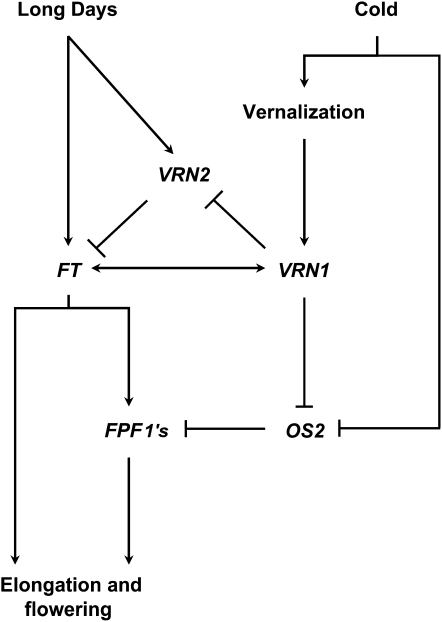
In Arabidopsis, expression of FPF1 increases rapidly at the shoot apex in response to long days (Kania et al., 1997). This long-day response is dependent on FT and CO (Schmid et al., 2003; Wise et al., 2008), suggesting that FPF1 acts downstream of FT in the long-day flowering response pathway. The FPF1-like genes of barley are also daylength responsive, with elevated expression in long days (Fig. 6), suggesting that this is a conserved feature of FPF1-like genes. Expression of FPF1-like genes is also determined by vernalization status in barley; the expression of these genes increases in vernalized plants where HvOS2 expression is reduced. The combined effects of vernalization and long daylength on FPF1 gene expression are consistent with a model where down-regulation of HvOS2 in vernalized plants derepresses FPF1-like genes, which are then further induced through a conserved CO-FT regulatory pathway as daylength increases during spring (Fig. 7).
Figure 7.
An extended model of the molecular genetic network that controls vernalization-induced flowering in temperate cereals. Low temperatures (cold) can transiently down-regulate OS2. Prolonged cold (vernalization) causes stable activation of VRN1. After vernalization, VRN1 down-regulates OS2, directly or indirectly. Consequently, FPF1s are derepressed. VRN1 also down-regulates VRN2 and allows activation of FT1 by long days (Trevaskis et al., 2007a; Distelfeld et al., 2009). Expression of FPF1s is induced as FT1 activity increases, and this promotes the transition to reproductive development at the shoot apex and cell elongation in the stem.
Microarray analysis identified other potential targets of HvOS2. For example, two RNase S-like genes were up-regulated in plants overexpressing HvOS2 (Table II). These RNases belong to a class that has only been identified in grasses and predicted to lack RNase activity due to amino acid substitutions at critical residues in the active site (Gausing, 2000). One of these RNases, rsh1, is expressed in leaves and is regulated by light and developmental cues (Gausing, 2000). Expression of both RNase genes decreases during cold treatment, when HvOS2 is down-regulated (Supplemental Fig. S7; Plexdb accession no. BB81; Wise et al., 2008). This supports the hypothesis that these genes are up-regulated by HvOS2. The function(s) of these RNases is not known (Gausing, 2000), so it is unclear whether these genes play a role in vernalization-induced flowering or other biological processes. Analysis of microarray comparisons of gene expression during barley development shows that HvOS2, the RNase S-like, and FPF1-like genes are all expressed during postvegetative development (Supplemental Fig. S10; Druka et al., 2006), suggesting broad roles for these genes during plant development in addition to potential roles in regulating flowering time.
In summary, we have identified a novel mechanism by which elongation and flowering are suppressed prior to vernalization in cereals. These findings further highlight the difference between the vernalization pathways of Arabidopsis and cereals, reinforcing the concept that the vernalization response has evolved independently in monocot and dicot plants.
MATERIALS AND METHODS
Plant Growth
Barley plants (Hordeum vulgare) were grown in glasshouses (18°C ± 2°C) in long days (16 h of light/8 h of dark) with supplementary light when natural levels dropped below 200 μE. For controlled growth conditions, plants were grown in growth chambers (20°C) with long days (16 h of light/8 h of dark) or short days (8 h of light/16 h of dark) under a mix of incandescent and fluorescent lighting. In instances where plants were vernalized, seeds were imbibed and germinated on moist filter paper for 4 to 7 weeks at 4°C in the dark.
Seeds of the einkorn wheat (Triticum monococcum) maintained vegetative phase mutant, which lacks VRN1 (referred to here as ΔVRN1), and the wild-type parent strain were imbibed on moist filter paper. Vernalized samples were grown at 4°C for 7 weeks in the dark, and nonvernalized samples were grown in the dark at 22°C to a developmental stage equivalent to that of vernalized seedlings (4 cm coleoptile length, vegetative shoot apex). Individual seedlings were ground in liquid nitrogen, and a small sample of the ground material was used to extract DNA to determine the genotype of each individual seedling. The remaining material was used to extract RNA for gene expression studies. Genotyping of individual seedlings was carried out using two sets of primers that annealed to the TmVRN1 gene (Supplemental Table S4) in a PCR using Taq DNA polymerase (New England Biolabs). PCR products were run on a 1.2% agarose gel, and the absence of a visible band was considered an indication of a seedling that was homozygous for the ΔVRN1 mutation.
Apex Dissection and Flowering Time Measurements
Apices were isolated with a binocular dissecting microscope and then digitally photographed on a Leica M8 digital camera. Leaves were numbered sequentially, and plants were grown until the flag leaf emerged to determine final leaf number. Heading date was measured as the day when the head first emerged from the sheath on the main shoot (Z = 47; Zadoks et al., 1974).
Gene Expression Analysis
Total RNA was extracted using the method of Chang et al. (1993) or the Qiagen RNeasy Plant Miniprep kit. RNA gel blotting was performed as described previously (Trevaskis et al., 2003). cDNA was prepared for qRT-PCR using an oligo(T) primer (T18[G/C/A]) to prime first-strand cDNA synthesis from 1 to 5 μg of total RNA with SuperScript III reverse transcriptase enzyme (Invitrogen). qRT-PCR was performed on a Rotor-Gene 3000 real-time cycler (Corbett Research). The primers used for ACTIN have been described previously (Trevaskis et al., 2006), and additional primers are detailed in Supplemental Table S4. qRT-PCR was performed using Platinum Taq DNA polymerase (Invitrogen). Cycling conditions were 4 min at 94°C, 40 cycles of 10 s at 95°C, 15 s at 60°C, and 20 s at 72°C, followed by a melting-curve program (72°C–95°C with a 5-s hold at each temperature). Fluorescence data were acquired at the 72°C step and during the melting-curve program. Expression levels of genes of interest were calculated relative to ACTIN using the comparative quantification analysis method (Rotogene-5; Corbett Research), which takes into account the amplification efficiency of each primer set. Data presented are averages of a minimum of three biological replicates unless stated otherwise, and the error bars show se.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation of leaf tissue was performed as described by Oliver et al. (2009) using the third leaf from nonvernalized or postvernalized plants. Postvernalized plants were derived from seeds that had been germinated for 7 weeks under vernalizing conditions (4°C) and then transferred to normal glasshouse conditions and grown to the three-leaf stage. The results shown are means of two biological replicate experiments. Sequences for primers used in chromatin immunoprecipitation experiments are listed in Supplemental Table S4.
Plant Transformation
Overexpression constructs were made by introducing a full-length HvOS2 cDNA into a Gateway (Invitrogen)-adapted cloning vector described previously (Hemming et al., 2008), which uses the maize (Zea mays) UBIQUITIN promoter (Christensen et al., 1992) to drive transgene expression. RNAi constructs were made using the Gateway cloning system; the hairpin cassette from HELLSGATE12 (Wesley et al., 2001) was fused to the maize ubiquitin promoter and placed in the in the pWBVEC8 binary vector backbone (Wang et al., 1998). A map of the resulting vector (pSTARGATE) can be found at http://www.pi.csiro.au/RNAi/vectors.htm. Barley plants were transformed using Agrobacterium tumefaciens transformation of excised embryos of the variety Golden Promise (Tingay et al., 1997; Matthews et al., 2001). Golden Promise is a spring barley that flowers without vernalization (genotype HvVRN1-1, ΔHvVRN2) and is photoperiod insensitive. T1 and T2 plants were screened for segregation of the transgene using primers that amplify the hygromycin-selectable marker gene. Expression analysis was carried out on plants hemizygous or homozygous for the transgene and sibling null control lines that did not inherit the transgene.
Microarray Analysis
Plants were grown in glasshouse conditions and sampled at the two-leaf stage. RNA was extracted using the method of Chang et al. (1993) and then further purified using RNeasy columns (Qiagen). Probe synthesis, labeling, hybridization to the Barley1 gene chip (Close et al., 2004), and RNA quality were assessed at the Australian Genome Research Facilities, following the manufacturer's recommendations (Affymetrix). Microarray analyses were performed on three biological replicates of each sample. The resulting data set was analyzed in R version 2.7.0 and analyzed using packages from Bioconductor (Gentleman et al., 2004; http://www.bioconductor.org/) using default settings. Normalization was carried out by Robust Multichip Analysis, and differentially expressed genes were identified using the LIMMA package (Linear Models for Microarray Data; Smyth, 2005). Genes with P values higher than 0.01 or with a change in gene expression lower than 1.5-fold were excluded from further analysis.
Microscopy and Image Analysis
Leaf segments taken from positions at 33% and 66% of the total length of the leaf were fixed at room temperature in 70% ethanol for at least 2 h, then dehydrated to 100% ethanol in 10% steps (30 min each step). One hundred percent ethanol was replaced twice, and the tissue was critical point dried with CO2 and mounted on double-sided carbon tabs attached to scanning electron microscopy stubs, adaxial side up. Tissue was then viewed uncoated with a four-quadrant backscattered electron detector in a Zeiss EVO LS15 scanning electron microscope. Tissue was viewed using a 20-kV accelerating voltage under variable pressure mode, with 10-Pa chamber pressure. Images of the tissue were taken for analysis using the analySIS LS Professional (version 2.6). The length of bulliform cells (Wenzel et al., 1997) was measured manually with the line tool.
Sequence Database Searches
All sequence database searches (nucleotide and protein) were performed using BLAST at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic Analysis
Alignments of the full nucleotide coding sequences were performed using MUSCLE version 3.6 (Edgar, 2004) and were edited using the BioEdit interface (version 7.0.9.0; Hall, 1999; Supplemental Fig. S1, B and D). Phylogenetic analyses were conducted in MEGA4 using the neighbor-joining method (default settings; Saitou and Nei, 1987). Bootstrap values were calculated using 10,000 replicates.
Statistical Analysis
All statistical analysis was carried out using GenStat 11th edition (Payne et al., 2008) unless specified otherwise.
Sequence data from this article can be found in the GenBank data library under accession numbers HM130526 (HvOS1) and HM130525 (HvOS2). Microarray data have been deposited in the Plant Expression Database (www.plexdb.org) with accession number BB93.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic relationships between ODDSOC-like genes and other plant MADS box genes.
Supplemental Figure S2. Alignments of cDNA and predicted protein sequences.
Supplemental Figure S3. qRT-PCR analysis of gene expression in HvOS2 overexpression lines.
Supplemental Figure S4. Phenotypes and expression levels of HvOS2 in RNAi transgenic plants.
Supplemental Figure S5. qRT-PCR analysis of HvOS2 gene expression during development in leaf and crown tissue.
Supplemental Figure S6. qRT-PCR analysis of TmOS2 gene expression in the ΔVRN1 mutant grown in long days.
Supplemental Figure S7. Selected data from low-temperature stress microarray experiment (cv Dicktoo).
Supplemental Figure S8. Leaf appearance rate and final leaf number in HvOS2 overexpression lines versus null sibling control lines.
Supplemental Figure S9. Images of transgenic plants overexpressing HvOS2 and wild-type siblings with or without GA treatment.
Supplemental Figure S10. Selected data from microarray analysis of gene expression during barley development (cv Morex).
Supplemental Table S1. Nearest Arabidopsis homologs of HvOS1 and HvOS2, BLASTP.
Supplemental Table S2. Nearest Arabidopsis homologs of HvOS1 and HvOS2, BLASTN.
Supplemental Table S3. Microarray results for comparison of HvOS2 overexpression line versus a null sibling control.
Supplemental Table S4. Primers used in this study.
Note Added in Proof
The Brachypodium genome sequence, which was used in this study, has now been described by The International Brachypodium Initiative (The International Brachypodium Initiative [2010] Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768).
Supplementary Material
Acknowledgments
We thank our colleagues Steven Swain and Peter Chandler for helpful discussions and suggestions for improvement of the manuscript. We thank Sarah Fieg and Sandra Stops for technical support and K. Murai for the kind donation of the maintained vegetative phase seeds. We also thank Million Tadege for providing the cDNA clones used in this study.
References
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 [DOI] [PubMed] [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP. (2004) A new resource for cereal genomics: 22K Barley GeneChip comes of age. Plant Physiol 134: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signalling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AME, Greb T, Dean C. (2008) A PHD-Polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Druka A, Muehlbauer G, Druka I, Caldo R, Baumann U, Rostoks N, Schreiber A, Wise R, Close T, Kleinhofs A, et al. (2006) An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics 6: 202–211 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. (2007) The FLOWERING LOCUS T-Like gene family in barley (Hordeum vulgare). Genetics 176: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. (2007) Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17: 1978–1983 [DOI] [PubMed] [Google Scholar]
- Gausing K. (2000) A barley gene (rsh1) encoding a ribonuclease S-like homologue specifically expressed in young light-grown leaves. Planta 210: 574–579 [DOI] [PubMed] [Google Scholar]
- Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan K, Xu ZH, Chong K. (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135: 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A, Peacock WJ, Dennis ES, Trevaskis B. (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann Bot (Lond) 103: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. (2008) Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol 147: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Graf A, Wigge PA. (2006) The control of flowering in time and space. J Exp Bot 57: 3415–3418 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S. (1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9: 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G. (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Moritz T, Evans LT, Martin J, Andersen CH, Blundell C, Kardailsky I, Chandler PM. (2006) Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol 141: 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Matthews PR, Wang M-B, Waterhouse PM, Thornton S, Fieg SJ, Gubler F, Jacobsen JV. (2001) Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNAs’ on a standard Agrobacterium transformation vector. Mol Breed 7: 195–202 [Google Scholar]
- Melzer S, Kampmann G, Chandler J, Apel K. (1999) FPF1 modulates the competence to flowering in Arabidopsis. Plant J 18: 395–405 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. (2009) Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc Natl Acad Sci USA 106: 8386–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G. (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RW, Harding SA, Murray DA, Soutar DM, Baird DB, Glaser AI, Channing IC, Welham SJ, Gilmour AR, Thompson R, et al. (2008) GenStat Release 11 Reference Manual. Part 2. Directives. VSN International, Hemel Hempstead, England [Google Scholar]
- Read BJ, Raman H, McMichael G, Chalmers KJ, Ablett GA, Platz GJ, Raman R, Genger RK, Boyd WJR, Li CD, et al. (2003) Mapping and QTL analysis of the barley population Sloop × Halcyon. Aust J Agric Res 54: 1145–1153 [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sasani S, Hemming MN, Oliver SN, Greenup A, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi H-R, Dennis ES, Peacock WJ, et al. (2009) The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J Exp Bot 60: 2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Sung S, Amasino RM. (2008) Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N, Ikari C, Shimada S, Kitagawa S, Sakamoto K, Saito H, Ryuto H, Fukunishi N, Abe T, Takumi S, et al. (2007) The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet Syst 82: 167–170 [DOI] [PubMed] [Google Scholar]
- Smykal P, Gleissner R, Corbesier L, Apel K, Melzer S. (2004) Modulation of flowering responses in different Nicotiana varieties. Plant Mol Biol 55: 253–262 [DOI] [PubMed] [Google Scholar]
- Smyth G. (2005) LIMMA: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Sung S, Amasino RM. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Yasuda S. (1971) Genetics of earliness and growth habit in barley. Nilan RA, , Barley Genetics II: Proceedings of the Second International Barley Genetics Symposium. Washington State University Press, Pullman, WA, pp 388–408 [Google Scholar]
- Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thornton S, Brettell R. (1997) Agrobacterium tumefaciens-mediated barley transformation. Plant J 11: 1369–1376 [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. (2007a) The molecular basis of vernalisation-induced flowering in cereals. Trends Plant Sci 12: 352–357 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140: 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C. (2007b) Short Vegetative Phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol 143: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szucs P, Dubcovsky J, Yan LL, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS. (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59: 449–467 [DOI] [PubMed] [Google Scholar]
- Wang M, Li Z, Matthews PR, Upadhyaya NM, Waterhouse PM. (1998) Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic 461: 401–408 [Google Scholar]
- Wenzel CL, Chandler PM, Cunningham RB, Passioura JB. (1997) Characterization of the leaf epidermis of barley (Hordeum vulgare L. ‘Himalaya’). Ann Bot (Lond) 79: 41–46 [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Winfield M, Lu C, Wilson I, Coghill J, Edwards K. (2009) Cold and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biol 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Caldo RA, Hong L, Shen L, Cannon E, Dickerson JA. (2008) BarleyBase/PLEXdb. Edwards D, , Plant Bioinformatics. Humana Press, Totowa, NJ, pp 347–363 [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. (2006) The Arabidopsis thaliana vernalization response requires a Polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]
- Zeevaart JAD. (2006) Florigen coming of age after 70 years. Plant Cell 18: 1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.