Summary
Viral genetic studies often focus on large open reading frames (ORFs) identified during genome annotation (ORF-based annotation). Here we provide a tool and software set for defining gene expression by murine gammaherpesvirus 68 (γHV68) nucleotide-by-nucleotide across the 119,450 basepair (bp) genome. These tools allowed us to determine that viral RNA expression was significantly more complex than predicted from ORF-based annotation, including over 73,000 nucleotides of unexpected transcription within 30 expressed genomic regions (EGRs). Approximately 90% of this RNA expression was antisense to genomic regions containing known large ORFs. We verified the existence of novel transcripts in three EGRs using standard methods to validate the approach and determined which parts of the transcriptome depend on protein or viral DNA synthesis. This redefines the genetic map of γHV68, indicates that herpesviruses contain significantly more genetic complexity than predicted from ORF-based genome annotations, and provides new tools and approaches for viral genetic studies.
Introduction
Murine gammaherpesvirus 68 is a useful model organism for studying the pathogenesis and immunology of oncogenic gammaherpesviruses, which include the human Kaposi’s sarcoma-associated virus (KSHV) and Epstein-Barr virus (EBV). A complete list of viral genes is essential for the application of genetic approaches to understanding viral pathogenesis and immunology. The 119,450 bp γHV68 genome has been fully sequenced and the initial annotation identified the presence of 80 genes (Virgin et al., 1997). This original annotation identified genes as regions of the genome encoding a methionine-initiated open reading frame containing more than 100 amino acids. In keeping with nomenclature conventions then in effect, conserved open reading frames were numbered based on prior sequenced gammaherpesvirus genomes while non-conserved open reading frames were numbered M1 to M14. The γHV68 homolog of the KSHV K3 gene was named K3. This ORF-based approach to genome annotation is limited in its ability to identify non-coding RNAs, spliced exons, and small protein-coding genes. In fact, small and large non-coding RNAs have been identified in EBV, KSHV, and γHV68. Such examples include small miRNAs and the large polyadenylated nuclear (PAN) RNA (Cai et al., 2006; Cai et al., 2005; Diebel et al., 2010; Pfeffer et al., 2005; Conrad and Steitz, 2005). Additionally, several spliced viral genes have been identified in γHV68 including versions of ORF73 with introns greater than 10kb (Allen et al., 2006). Therefore, the initial annotation of the γHV68 genome does not encompass all of the genetic complexity of this virus (Virgin et al., 1997; Nash et al., 2001; May et al., 2005; DeZalia and Speck, 2008; Gray et al., 2009).
A number of recent studies have demonstrated that the transcriptional complexity of a broad range of organisms is considerably higher than previously believed (Birney et al., 2007; He et al., 2008; Carninci et al., 2008; David et al., 2006). These studies have identified numerous novel antisense transcripts, large non-coding RNAs, and chimeric protein-coding transcripts derived from exons of different genes. These findings indicate that the transcription of eukaryotic genomes encompasses more forms and types of transcripts than previously assumed. Since herpesviruses have co-evolved with their hosts for millions of years, we hypothesized that the γHV68 genome would display this same level of transcriptional complexity. If so, the interpretation and future design of γHV68 genetic studies might be affected. Further, if the transcriptional complexity of the virus mimics the host, then the virus might provide a tractable model to understand aspects of the newly appreciated complexity of eukaryotic transcription. Current technologies, which require genome segment by genome segment analysis, are quite laborious for pan-genomic assessment of gene expression. Therefore, we designed a high-density oligonucleotide array with probes that tile across the γHV68 genome, and developed the computational methods for normalization and analysis of the tiled array data, to provide a tool that allows us to detect gene expression with high resolution across the entire genome in a manner not biased by prior ORF-based annotation. Additionally, the tiled array provides strand-specific information allowing the identification of regions of antisense transcription. Use of this newly developed tool allowed us to identify multiple unexpected regions of transcription across the viral genome.
Results
Design of a high-density tiled array to detect viral gene expression
We designed an oligonucleotide array with 42,545 overlapping 60mer probes complementary to each strand of the γHV68 genome, such that each nucleotide of the viral genome was covered by an average of 10 probes. These probes were designed using the eArray software package and arrays were produced by Agilent Technologies. In addition to the γHV68 probes, 10 probes were designed specific for Stratagene Spot Report Arabidopsis control RNAs. These probes were replicated 50 times across the array and were used in normalization. Total RNA was prepared from infected or mock-infected 3T12 fibroblast cells and analyzed for quality using Agilent Bioanalyzer DNA chips. Spike-in Arabidopsis RNA was added to these samples prior to reverse transcription, labeling with Cy5 dye, and array hybridization. Probe signal from mock-infected cells (Figure S1A), was normalized to and subtracted from that present in the infected cells using average spike-in RNA signal intensity. The region of highest mock signal was near the terminal repeat region, M12-14, and was most likely due to the high GC content of this region. However, the expression signal in infected cells was still 64 fold higher in this region. No significant signal was detected when using RNA prepared from infected cells that had been treated with RNAse (data not shown). To visualize the tiled array data, we wrote software that averaged the fluorescence signal from every probe overlapping a given nucleotide in the genome and graphed the data as mean fluorescence intensity on a log2 scale.
Detection of viral gene expression during lytic infection
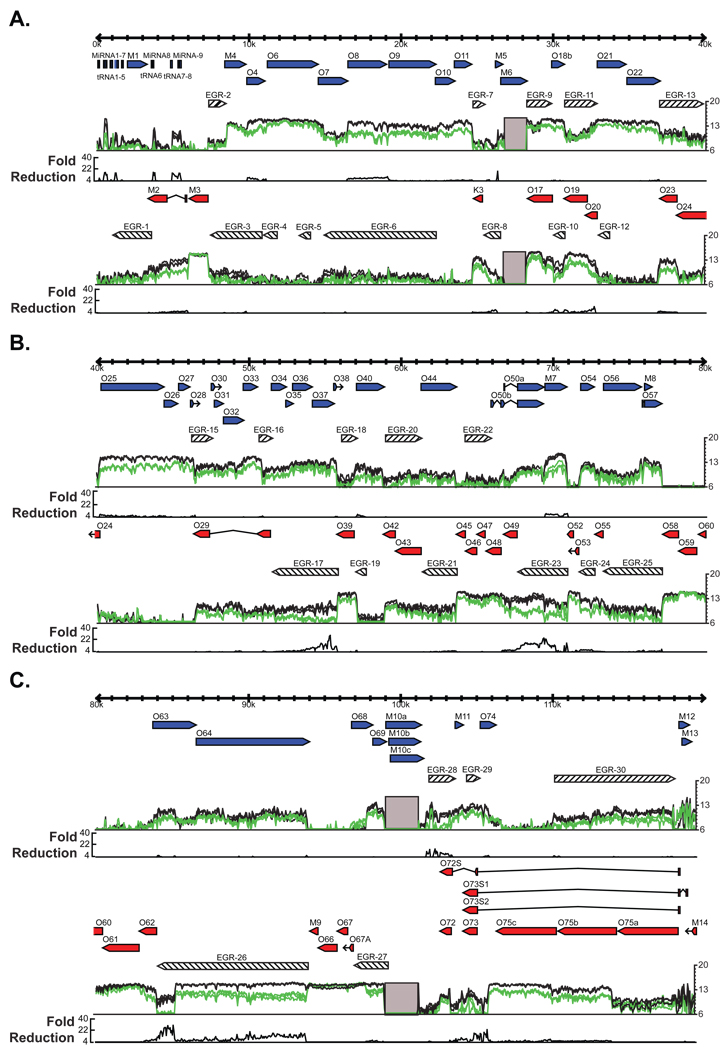
We first examined the expression of viral RNAs from infected (multiplicity of infection, MOI=10) 3T12 fibroblast cells 18 hours after infection. This is a late time point in the viral life cycle at which we expect the majority of viral genes to have been transcribed, including genes of the three standard classes of herpesvirus gene expression (immediate early, IE; early, E; and late, L). Figure 1 shows the expression of viral RNA at each nucleotide position from three independent experiments compared to the ORFs previously annotated in the genome (Virgin et al., 1997; Nash et al., 2001; May et al., 2005; Gray et al., 2009; Hwang et al., 2008). The data were highly reproducible. Approximately 87% of the nucleotide positions demonstrated less than 2 fold variation between the 3 biological replicates. We observed substantial RNA expression throughout the γHV68 genome, with many broad signal peaks in agreement with previously annotated ORF boundaries, such as ORF-23, ORF-40, and ORF-57 (Virgin et al., 1997; Mackett et al., 1997). This indicates that transcripts corresponding to known ORFs are detected by the tiled array. The known spliced intron of ORF-57 was clearly identified by the tiled array as a drop in signal corresponding to the expected splice site coordinates (Mackett et al., 1997) (Figure S1B). Interestingly, this gene expression signal does not disappear 5’ to the start of ORF-57, which parallels studies in KSHV identifying a second large transcript containing both ORF-56 and ORF-57 (Majerciak et al., 2006).
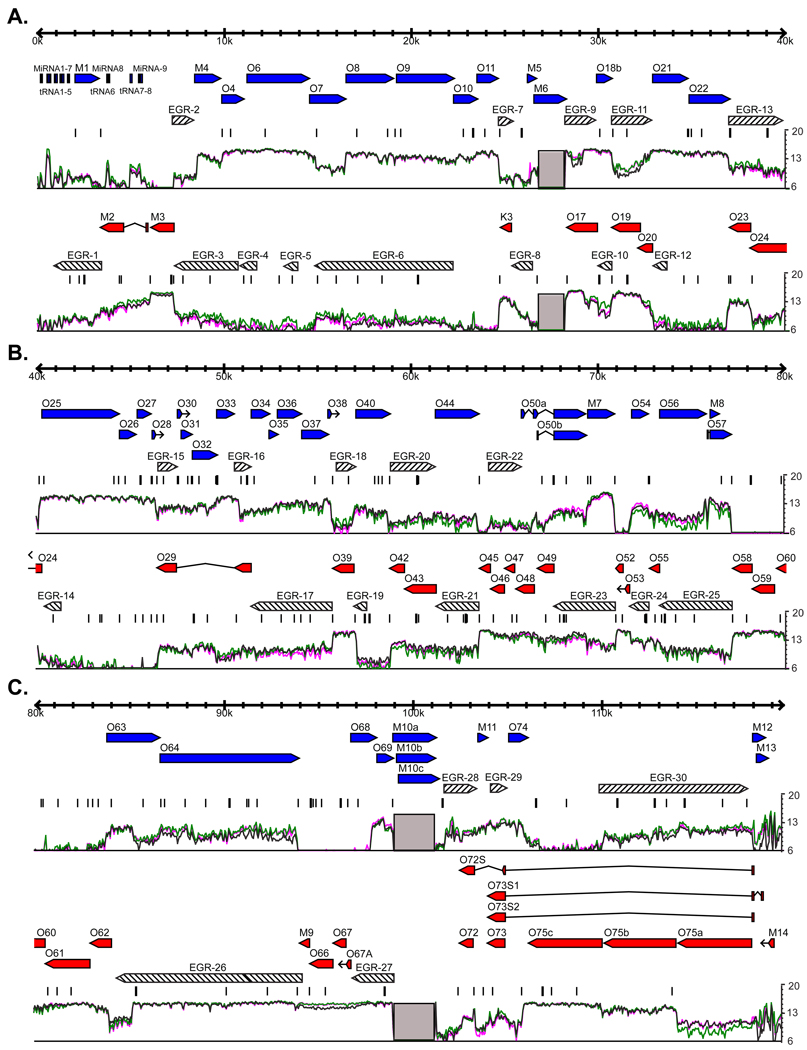
Figure 1. Genome-wide RNA expression in γHV68.
3T12 cells were infected with γHV68 at an MOI=10. Total RNA was harvested at 18 hpi, labeled, and hybridized to the γHV68 tiled array. The mean fluorescence of probes overlapping each nucleotide position is plotted on a log2 scale underneath the annotated gene positions. The three colored lines represent the data from three replicate infections. Blue or red arrows represent annotated genes on the positive or negative strand of the genome, respectively. Gray boxes represent the two internal repeat regions of the γHV68 genome that are excluded from our analysis. EGRs 1–30, which are greater than 700 nt and expressed one standard deviation above the mean mock-infected signal in at least two replicates, are shown as diagonal-filled arrows. Consensus polyadenylation signal sequences, AAUAAA and AUUAAA, are represented as black bars above the fluorescence signal. A) Genomic positions 1–40 kb. B) Genomic positions 40–80 kb. C) Genomic positions 80–119.45 kb. See also Figure S1.
RNA expression from unexpected γHV68 genome regions
Though many signal peaks detected by the tiled array agree with the boundaries of previously annotated genes or ORFs, a considerable amount of RNA expression was detected well outside of known genetic loci. To designate regions of RNA expression which are detected by the tiled array, but that were unexpected from ORF-based genomic annotation, we have elected the term ‘Expressed Genomic Region’ (EGR). Each of these regions may contain overlapping transcripts from multiple different loci and thus would not be accurately categorized as genes. We numbered the EGRs sequentially from the left end of the genome (Figure 1).
To identify EGRs, we established several criteria for annotating the genome based on RNA expression rather than the presence of ORFs. First, we focused on regions for which RNA expression was detected extending at least 700 nucleotides (nt) either 5' or 3' from an annotated ORF. Since there has been no systematic analysis of the average length of 5' and 3' non-coding regions of herpesvirus RNAs, we selected this arbitrary, but conservative, criterion to eliminate the designation of small untranslated regions of previously identified ORFs as 'unexpected'. For those regions that were not extensions of an annotated ORF, we required that the region of expression be at least 700 nt. Second, we considered insignificant any signal that was lower than one standard deviation above the mean signal obtained from mock-infected cells. We also analyzed the data with a baseline set at two standard deviations above the mean mock signal (Figure S1C). However, we found that using the one standard deviation criterion accurately predicted the structure of novel transcripts as defined by standard methods (see below), and therefore utilized this as our standard approach. Finally, we required that a significant signal be detected at a nucleotide in at least two of three independent experiments.
Using these criteria, we identified over 73 kilobases (kb) of unexpected RNA expression in 30 EGRs from the γHV68 genome (Table 1; Figure 1, diagonal-filled arrows). This represents about 31% of the 238,900 nucleotides that could theoretically be transcribed from the 119,450 bp γHV68 genome. EGRs range from a minimum of 700 nt to ~10,000 nt in length (Figure 1). It is notable that the entire genome is not transcribed, suggesting that EGRs represent significant features of the γHV68 genome rather than detection of unrestricted transcription across the genome.
Table 1.
Genomic locations of novel Expressed Genomic Regions identified using a tiled array.
| Name | Start Coordinates | Stop Coordinates | Strand | Size |
|---|---|---|---|---|
| Expressed Genomic Region-1 | 847 | 3375 | − | 2528 |
| Expressed Genomic Region-2 | 7267 | 8409 | + | 1142 |
| Expressed Genomic Region-3 | 7277 | 10716 | − | 3439 |
| Expressed Genomic Region-4 | 10807 | 11718 | − | 911 |
| Expressed Genomic Region-5 | 13141 | 13916 | − | 775 |
| Expressed Genomic Region-6 | 14785 | 22194 | − | 7409 |
| Expressed Genomic Region-7 | 24654 | 25440 | + | 786 |
| Expressed Genomic Region-8 | 25335 | 26448 | − | 1113 |
| Expressed Genomic Region-9 | 28231 | 29917 | + | 1686 |
| Expressed Genomic Region-10 | 29957 | 30723 | − | 766 |
| Expressed Genomic Region-11 | 30771 | 32879 | + | 2108 |
| Expressed Genomic Region-12 | 32880 | 33672 | − | 792 |
| Expressed Genomic Region-13 | 37025 | 40100 | + | 3075 |
| Expressed Genomic Region-14 | 40253 | 41148 | − | 895 |
| Expressed Genomic Region-15 | 46360 | 47507 | + | 1147 |
| Expressed Genomic Region-16 | 50571 | 51465 | + | 894 |
| Expressed Genomic Region-17 | 51466 | 55799 | − | 4333 |
| Expressed Genomic Region-18 | 55999 | 57046 | + | 1047 |
| Expressed Genomic Region-19 | 56950 | 57664 | − | 714 |
| Expressed Genomic Region-20 | 58891 | 61303 | + | 2412 |
| Expressed Genomic Region-21 | 61334 | 63652 | − | 2318 |
| Expressed Genomic Region-22 | 64147 | 65909 | + | 1762 |
| Expressed Genomic Region-23 | 67643 | 70957 | − | 3314 |
| Expressed Genomic Region-24 | 71701 | 72741 | − | 1040 |
| Expressed Genomic Region-25 | 73313 | 77211 | − | 3898 |
| Expressed Genomic Region-26 | 84010 | 93959 | − | 9949 |
| Expressed Genomic Region-27 | 96675 | 98982 | − | 2307 |
| Expressed Genomic Region-28 | 101647 | 103418 | + | 1771 |
| Expressed Genomic Region-29 | 103933 | 105057 | + | 1124 |
| Expressed Genomic Region-30 | 109969 | 117992 | + | 8023 |
| 73478 |
Confirmation of Expressed Genomic Regions
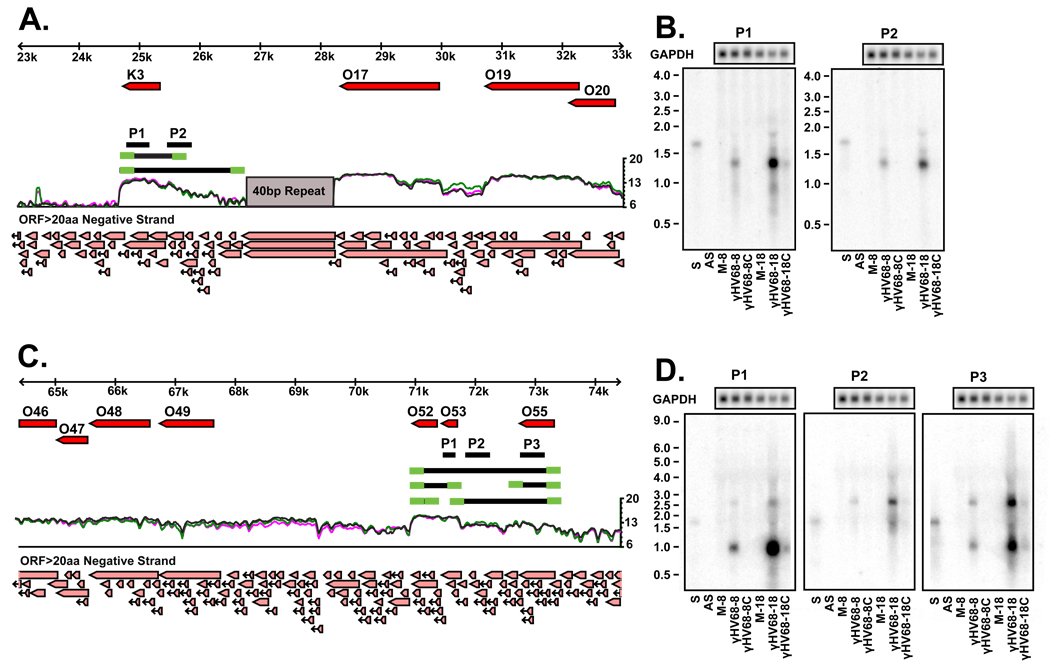
To understand how EGRs relate to viral transcripts detected by standard means, we elected to characterize regions that represent three different relationships between EGRs and already annotated ORFs in more detail. We selected (i) EGR 8 which appeared as a long 5' extension of the ORF K3, (ii) EGR 24 which was between ORF-53 and ORF-55, and (iii) EGR 26 which appeared as a very long 3' extension of the ORF M9 (Figure 2A and C; Figure 3A).
Figure 2. Expressed Genomic Regions 8 and 24.
A) Tiled array results for EGR 8 reveals a considerable 5’ extension to the K3 gene. Schematic representation of the transcripts revealed by Northern analysis and RACE are shown as black and green lines, respectively. Single stranded RNA probes used in the Northern analysis are shown as the black lines labeled P1 and P2. Light red arrows at the bottom of the figure represent predicted open reading frames of greater than 20 amino acids. B) Northern analysis of K3 and EGR 8 reveals the presence of 1.1 kb and 2 kb transcripts. C) Tiled array results for EGR 24 reveals transcriptional signal between ORF-53 and ORF-55. Schematic representation of the transcripts revealed by Northern analysis and RACE are shown as black and green lines, respectively. Single stranded RNA probes used in the Northern analysis are shown as the black lines labeled P1, P2 and P3. D) Northern analysis of ORFs 52, 53, 55 and EGR 25 reveals the presence of 800 nt, 860 nt, 1.8 kb, and 2.5 kb bands. S = sense control RNA; AS = antisense control RNA; M-8 = mock infected, collected at 8 hpi; γHV68-8 = infected at MOI=10, collected at 8 hpi; γHV68-8C = infected at MOI=10 and treated with 200ng/ml cycloheximide, collected at 8 hpi; M-18 = mock infected, collected at 18 hpi; γHV68-18 = infected at MOI=10, collected at 18 hpi; γHV68-18C = infected at MOI=10 and treated with 42 µg/ml cidofovir, collected at 18hpi.
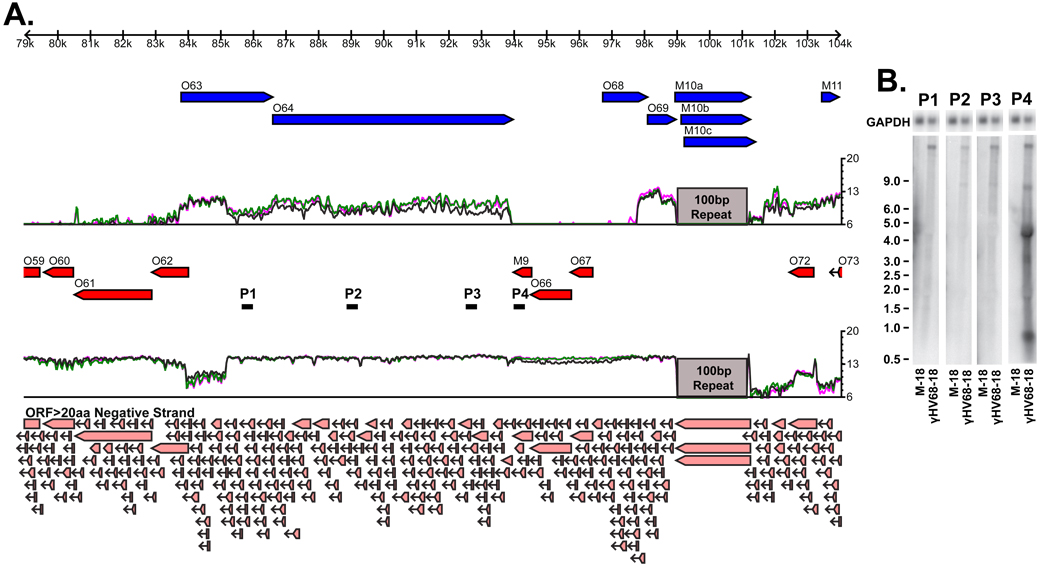
Figure 3. Expressed Genomic Region 26.
A) Tiled array results for EGR 26 reveals a 9.8 kb extension 3’ to the M9 gene. Single stranded RNA probes used in the Northern analysis are shown as the black lines labeled P1-4. Light red arrows at the bottom of the figure represent predicted open reading frames of greater than 20 amino acids. B) Northern analysis of EGR 26 reveals 0.7 kb, 3 kb, 4 kb, 8 kb, and 16 kb bands. M-18 = mock infected, collected at 18 hpi; γHV68-18 = infected at MOI=10, collected at 18hpi.
Transcripts within Expressed Genomic Region 8
EGR 8 appears as a possible 5’ extension to the start of the K3 ORF (Figure 2A). Northern blot analysis of this region using single-stranded RNA probes complementary to K3 (Figure 2B, probe P1) and 5’ to K3 (Figure 2B, probe P2) revealed the presence of a dominant ~1.1 kb and a fainter ~2.0 kb band (Figure 2B). Two transcriptional start sites were identified by 5’ RACE at genomic positions 25,735 and 26,749. A single 3’ transcript end was identified using 3’ RACE at position 24,698, approximately 23 nucleotides downstream of a consensus polyadenylation sequence (Figure 2A, green lines). A poly-A tail was detected at the expected site in the 3’ RACE product. These 5’ and 3’ ends define a 1,038 nt transcript at 25,735-24,698 and a 2,052 nt transcript at 26,749-24,698 (Figure 2A, black lines). These two transcripts are in good agreement with the two bands identified by Northern analysis and tiled array results that show expression from the edge of the 40 bp repeat region to the end of the annotated K3 ORF, thus confirming the presence of a previously unidentified γHV68 transcript that contains a 1448 nt extension 5' to the start of ORF K3. We further characterized the transcripts derived from this locus by examining their expression in the absence and presence of the protein synthesis inhibitor cycloheximide or the DNA synthesis inhibitor cidofivir (Figure 2B, see also Figures 4 and 5). Expression of both K3 transcripts were inhibited in the presence of either cycloheximide or cidofivir, indicating they are of the late class of viral genes. In KSHV, the K3 ORF is encoded in multiple transcripts, some of which are splice variants, which have been found to be expressed within different kinetic classes and during latency (Rimessi et al., 2001; Taylor et al., 2005). In γHV68, K3 has been described as an IE gene and is also expressed during latency (Ebrahimi et al., 2003; Rochford et al., 2001). In the conditions used in our study using a tiled array approach, we show that K3 transcripts can also follow a late gene class pattern of expression (see below).
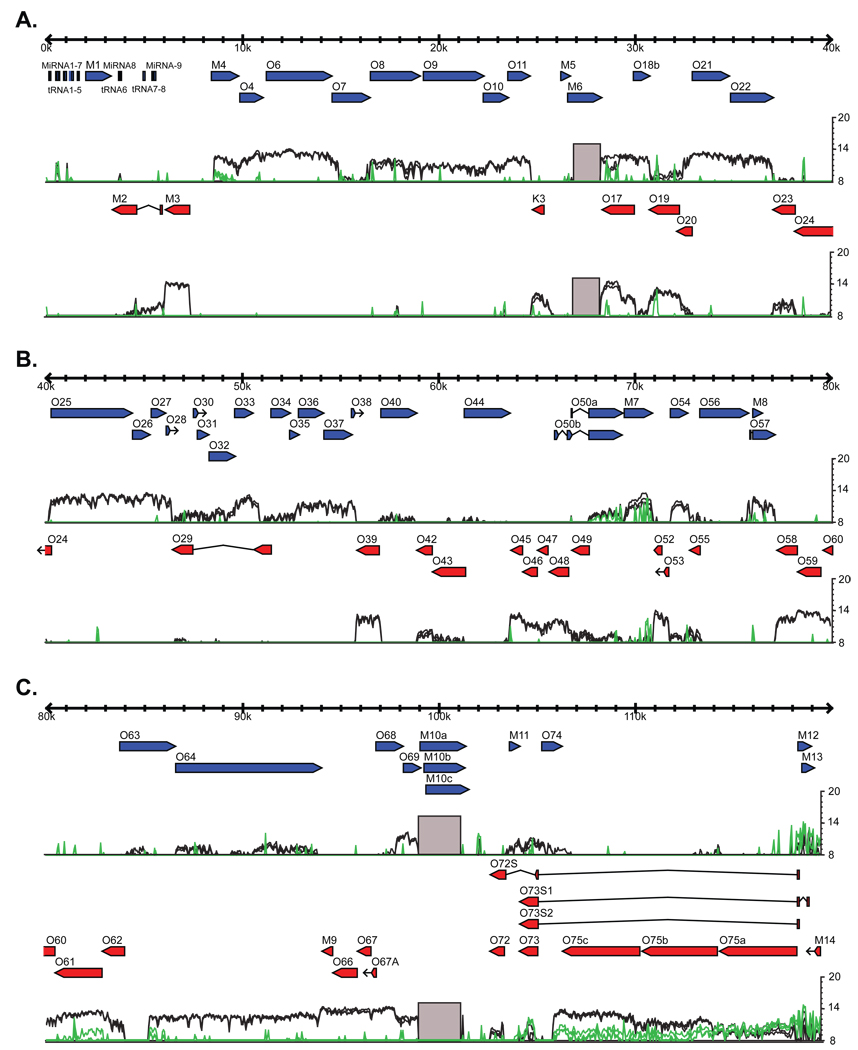
Figure 4. Tiled array identifies immediate early transcripts.
3T12 cells were infected in the presence, green lines, or absence, black lines, of 200 ng/ml cycloheximide and harvested 8 hpi. Three replicates of each condition were performed and are represented as individual lines. There is considerable disruption of transcription across the γHV68 genome with the exception of ORF-50, ORF-73, ORF-75a, M12, M13, and M14. Expression of ORF-61, M4, ORF-75b, and ORF-75c is decreased but still clearly detectable. A) Genomic positions 1–40 kb. B) Genomic positions 40–80 kb. C) Genomic positions 80–119.45 kb.
Figure 5. Genome-wide mapping of cidofovir sensitivity.
3T12 cells were infected in the presence, green lines, or absence, black lines, of 42 µg/ml cidofovir and harvested 18 hpi. Three replicates of each condition were performed and are represented as individual lines. Those regions that are greater than 4 fold down regulated in the presence of cidofovir are plotted on a linear scale on a nucleotide-by-nucleotide basis on the black line graph below the transcriptional signal. A) Genomic positions 1–40 kb. B) Genomic positions 40–80 kb. C) Genomic positions 80–119.45 kb. See also Figure S5.
Transcripts within Expressed Genomic Region 24
The tiled array revealed two peaks of expression that correspond to ORF52/53 and ORF55. Separating these peaks is a region of lower signal representing EGR 24 (Figure 2C). Transcripts emanating from this region were detected by Northern blot using single-stranded RNA probes complementary to ORF-53 and ORF-55 (Figure 4C, probes P1 and P3) as well as the intervening 1 kb unexpected region (Figure 4C, probe P2). Probes 1 and 3 detected bands of approximately 900 nucleotides, and all three probes detected bands of ~1.8 kb and ~2.7 kb, indicating that 4 discrete transcripts emanate from this region (Figure 2D).
We performed 5’ and 3’ RACE to further map transcripts from this region (Figure 2C, green lines). We identified four 5’ transcript ends slightly upstream of the start of ORF-52, ORF-53, and ORF-55 and within ORF-55, at genomic positions 71,399, 71,744, 73,420, and 72910. Three 3’ transcript ends were located downstream of ORF-52, downstream of ORF-55, and within ORF-53, at positions 70,937, 72,558, and 71,582. A poly-A tail was observed at the expected site in 3’ RACE products. There are canonical polyadenylation sequences directly at the ends of ORF-52 and ORF-55. While the canonical polyadenylation sequence of AAUAAA was not found upstream of the 3’ transcript end found within Orf-53, this region has several poly-A stretches possibly containing a non-canonical polyadenylation signal. These RACE products define four potential transcripts. The 808 nt (71,744-70,937) transcript contains ORF-52 and Orf-53 and is detected by probe 1. Transcripts of 2,484 nt (73,420-70,937) and 1,839 nt (73,420-71,582) are detected by probes 1, 2, and 3. The 863 nt (73,420-71,582) transcript is predicted to encode ORF-55 and is detected by probe 3. An additional transcript of 463 nt (71,399-70,937), which is predicted to encode ORF-52, lies downstream of the probes used in our Northern analysis and was not detected. Therefore, the Northern and RACE analysis confirm expression of several transcripts within EGR 24. Expression of all the transcripts from EGR 24 were disrupted by addition of either cycloheximide or cidofivir, and therefore these are late transcripts (Figure 2D).
Transcripts within Expressed Genomic Region 26
EGR 26 is a striking feature of the tiled array data in which RNA expression was detected extending 9,949 nt downstream of the M9 ORF. This transcript is antisense to the large capsid proteins ORF-63 and ORF-64 (Figure 3A). To define transcription in this region, Northern blot analysis was performed using four single-stranded RNA probes (Figure 3A, probes P1-4). An approximately 16 kb band was detected by all probes and an 8 kb band was detected by probes 2–4. Probe 4 detected several unique transcripts of approximately 0.7, 3, and 4 kb (Figure 3B). EGR 26 is an extremely large and complex region; however, these results confirm that our tiled array was truly detecting the expression of multiple unexpected transcripts arising from this genomic region. In addition to the EGR located in this region, it is notable that the region containing ORF-63 consistently demonstrates significantly higher expression signal for the N-terminal ca. 1.5 kb of the ORF. One possible explanation for this feature could be that ORF-63 gives rise to a truncated form in addition to the annotated gene.
Defining the gene class of Expressed Genomic Regions
Herpesvirus genes are classified as "immediate early" if their expression does not depend on new viral protein synthesis, as "late" if their expression is altered by inhibition of viral DNA synthesis, and as "early" if their expression depends both on new viral protein synthesis and viral DNA synthesis. These designations define class-specific mechanisms of transcriptional regulation and group genes with common functional characteristics. For example, many late genes encode structural proteins. We therefore expanded this classification approach to the γHV68 transcriptome. To identify immediate early RNA expression, we treated 3T12 cells during and after infection with cycloheximide. Cycloheximide significantly inhibited expression of most viral genes and EGRs (Figure 4). Two obvious exceptions were transcripts from the regions of the genome containing ORF-50 and ORF-73, both of which are known to be immediate early genes (Rochford et al., 2001). Additionally, ORF-75a, M12, M13, and M14 were expressed at comparable levels in the presence or absence of cycloheximide, indicating that these are immediate early genes. While expression of transcripts from most of the viral genome was reduced below detectable levels by treatment with cycloheximide, expression of transcripts from regions of ORF-61, M4, ORF-75b, and ORF-75c were decreased but still detectable. This could be due to multiple transcripts of differing kinetic classes emanating from the same region.
To identify regions of the genome of the late gene class, we measured viral RNA expression, on a nucleotide-by-nucleotide basis, in 3T12 fibroblast cells treated with cidofovir (Figure 5). It should be noted that the cidofovir concentration used in these experiments is over 500 fold higher than the reported IC50 value and effectively (>40-fold) inhibits viral DNA synthesis as determined by qPCR (Figure S5) (Neyts and De, 1998). The sensitivity of RNA expression to cidofovir treatment was highly variable across the genome, ranging from no change to regions with 32 fold decreases as measured on the tiled array. Regions for which cidofovir decreased RNA expression more than four-fold are plotted on the graph below the expression signal in Figure 5. Three of the most cidofovir-sensitive regions, with greater than 29 fold decreases, were located within EGRs 17, 23 and 26, indicating that these genome features are of the late gene class (Figure 5).
While expression from most regions of the γHV68 genome exhibited some degree of cidofovir sensitivity, some regions were notably unaffected by inhibition of viral DNA synthesis (Figure 5). These unaffected regions included the ORF for M3, which encodes a secreted viral chemokine binding protein (Parry et al., 2000), the region from 78,255 to 82,865 inclusive of ORFs 59, 60, and 61, and the region from 93,959 to 97,800 inclusive of ORFs M9, 66, and 67. These regions, whose expression is significantly inhibited by cycloheximide but not by cidofovir, are of the early class (Figures 4 and 5).
In addition, we compared RNA expression at eight hours after infection to expression 18 hours after infection (Figure S1D). The pattern of expression at the two time points is similar; however, the overall signal level is significantly higher at 18 hours after infection. One notable exception to this is Orf-75a which is expressed at the same level at both times indicating that, relative to other viral genes, it is expressed at a higher level at 8hpi than at 18hpi. Additionally, relative to neighboring regions, EGRs 26 and 27 demonstrate different expression patterns at eight and 18 hours after infection. In the case of EGR 27, the drop in signal in the middle of this EGR may indicate the boundary of a transcript in this region.
Computational analysis of Expressed Genomic Regions
To determine if the EGRs contain unannotated, but conserved, gammaherpesvirus protein-coding regions, we performed a comparative genomic analysis. We translated the γHV68, KSHV, EBV, and herpesvirus saimiri genomes in all possible reading frames and identified all open reading frames greater than 20 amino acids (Figure 2A,C; Figure 3A, light red arrows). Protein sequence conservation between the γHV68 open reading frames and those from the other gammaherpesvirus genomes was assessed by pair-wise sequence alignment (Wu-BLASTP) using a cutoff of P < 1×10−8. It should be noted that this analysis would miss conservation between proteins encoded by EGRs and proteins encoded by viruses not included in this analysis. Conservation of possible peptide sequences was detected in 8/30 EGRs (Table S1 and Figure S1F). However, these regions of conservation were quite small, averaging less than ~60 amino acids. Additionally, these regions were confined to portions of EGRs that had highly conserved protein-coding genes on the opposite strand. While it is possible these regions of conservation lie in novel protein-coding genes, it may also be that the conservation observed is due to sequence constraints due to these highly conserved protein genes. In addition to this comparative genome analysis, we analyzed the γHV68 genome using the eukaryotic virus gene prediction program GeneMarkS-EV (Besemer and Borodovsky, 2005). Using this approach, we were able to identify 69 of the 80 ORF annotations. Additionally, this analysis predicts the presence of 5 small novel protein-coding genes in the γHV68 genome. Three of these predictions were contained within EGRs 2, 7, and 28 (Figure 1SG).
Discussion
In this paper we describe a new tool and software for the annotation of herpesvirus genomes using RNA expression, in addition to the presence of easily identified ORFs, as a basis for understanding genetic complexity. The transcriptome data provided by tiled array analysis reveals that we have underestimated the genetic potential of γHV68, and we speculate that the same may be true for other herpesviruses. Similar methodologies applied to other viruses may reveal a similar level of unexpected genetic complexity (Assarsson et al., 2008a; Satheshkumar and Moss, 2008; Assarsson et al., 2008b). Our tiled array approach is relatively fast, efficient, and is less expensive than the application of standard methods across the genome.
Implications of findings from tiled array analysis
The availability of transcriptome-based annotation may provide a basis for advances in five important areas. First, it will allow interpretation, and if necessary re-interpretation, of studies of gene knockouts and transposon mutant screens. Indeed, comparing the transcriptome from one time point in a single cell type to one transposon mutagenesis study, we found that many transposon insertions may alter more transcripts than previously appreciated (see below, Figure S1G). Second, these data serve as a starting point for assessing the function of RNAs emanating from across the viral genome. Third, comparison of tiled array data from cells infected with viral mutants will allow assessment of genome wide effects of viral regulatory genes. Fourth, comparison of tiled array data from different cell types will allow rapid identification of cell type-specific transcriptional patterns, a process that now must be accomplished on a transcript-by-transcript basis or by use of laborious, although sensitive, methods such as RNAse protection assays (Rochford et al., 2001). Lastly, tiled array data will serve as an essential component of the next step in herpesvirus genome annotation, which will undoubtedly come from deep sequencing of RNA from infected cells. In this latter application, tiled array data will provide important boundary conditions for transcription assemblies and a benchmark against which data on quantification of viral transcripts from deep sequencing data can be assessed.
The γHV68 transcriptome
In addition to detecting RNA expression from many previously annotated genes, we also detected over 73 kb of unexpected RNA expression from the γHV68 genome. We validated the accuracy and sensitivity of our tiled array for detecting previously unidentified transcripts by the application of standard RNA mapping methodologies to three selected EGRs. Approximately 120 kb of the potential 239 kb of the γHV68 genome was previously annotated as encoding ORFs of >100 amino acids. The newly identified 73 kb of transcription represents a 61% increase in the RNA coding potential of the γHV68 genome. This number assumes that the untranslated regions of previously annotated ORFs do not extend more than 700 bp in either direction. This assumption has not been rigorously tested. For this reason, our initial calculations of what is ‘unexpected’ may need to be reformed as further transcriptional mapping is done. Further, we do not know how the signal from EGRs relates to the processing of viral transcripts, alternative promoters, or 'read-through' transcription (i.e. polyadenylation signal suppression). These factors will likely impinge upon the interpretations of the source and importance of EGRs. However, it is important to note that the use of alternative promoters and polyadenylation signal suppression is well established in gammaherpesviruses. One such example is EBV's EBNA proteins which are vital for viral latency (Kieff and Rickinson, 2007).
It is not possible at this stage to determine what is encoded by the 30 EGRs detected here. However, computational analysis detected only minimal conservation of peptide fragments between small ORFs inside EGRs and other gammaherpesviruses. Further, the analysis of the γHV68 genome using the eukaryotic virus gene finder GeneMarkS-EV reveals three possible small protein-coding genes in EGRs 2, 7, and 28. These results possibly represent the identification of novel small γHV68-specific protein coding genes and indicate potential loci for future mutational studies. Thus, EGRs might encode small γHV68-specific or highly diverged protein-coding exons, non-coding RNAs, very long 5’ or 3’ untranslated regions of known ORFs, or unprocessed intronic sequences.
Interestingly, 90% of the unexpected transcription identified in our study is antisense to genes actively transcribed from the opposite strand. In particular, EGR 26 is complementary to over 9.8 kb of two large capsid proteins. Additionally, the three most sensitive regions of the γHV68 genome to a block in viral DNA synthesis, indicating that the transcription from these regions is expressed with 'late' gene properties, are within EGRs antisense to several viral protein coding genes including ORF-50, the main regulator of lytic replication. Such a large amount of overlapping transcription raises intriguing possibilities regarding widespread antisense regulation of viral messages in the γHV68 genome, or regulation of transcription by transcriptional interference as has been demonstrated in EBV (Puglielli et al., 1997).
The kinetic classes of Expressed Genomic Regions
Our study of γHV68 gene class revealed that none of newly detected EGRs fit within the classical definition of immediate-early genes. However, it should be noted that in cells treated with cycloheximide, we observe numerous small spikes of transcription spread throughout the genome. While many of these spikes do occur in regions that correspond to annotated genes, several are present only in cycloheximide-treated cells, for example, positions 42,590–42,670 on the negative strand. There are many possible explanations for these spikes. For example, they could be caused by cross-hybridization of host transcripts that are only expressed in infected cells. Alternatively, these could be small RNA transcripts that are de-repressed in the absence of host and/or viral de novo protein synthesis. This indicates the importance of deep sequencing of RNAs from infected cells. Additionally, these spikes could be explained in some cases if multiple transcripts of different viral gene classes arise from the same locus. For example, in the presence of cycloheximide, we detect a weak RNA expression signal that corresponds to the ORF-61 annotation, whereas in the absence of cycloheximide there is a large highly expressed block of signal that covers ORF-56 through ORF-62. This could be due to the presence of highly expressed immediate-early transcript(s) overlapping multiple viral genes and a smaller late transcript that primarily consists of coding sequences for ORF-61.
The impact of tiled array analysis on interpretation of genetic studies
A major advance in herpesvirus genetics has been the use of libraries of transposon mutants to identify and determine the function of herpesvirus genes. We therefore mapped the published transposon mutants of γHV68 from reference (Song et al., 2005) onto the γHV68 genome annotated with both ORFs (Virgin et al., 1997; Nash et al., 2001; May et al., 2005; Gray et al., 2009; Hwang et al., 2008) and EGRs (Figure S1G). Of 73 transposon mutants in the γHV68 genome, 40 (55%) fell within EGRs 1–30. Most EGRs (27/30, 90%) have a transposon mutant virus described. In particular, 22/40 transposon mutants contained within EGRs, with ORFs on the opposite strand, are characterized as essential for γHV68 lytic replication. This analysis indicates that interpretation of forward mutational screens in gammaherpesvirus genomes may be best interpreted in light of tiled array-based annotation of viral RNA expression.
γHV68 compared to eukaryotic cell gene expression
There has been growing number of studies examining the transcriptome of a wide variety of organisms (Birney et al., 2007; He et al., 2008; Carninci et al., 2008; David et al., 2006). One clear conclusion coming from this research is that these genomes are extensively transcribed well outside of the boundaries of previously annotated genes. A large number of antisense transcripts, transcripts with little protein coding potential, and unusual spliced RNA forms have been identified in these studies. Future studies that determine the transcriptional profile from nuclear and cytoplasmic fractions of the cell may give clues to the functions of these novel transcripts. For example, RNAs that are retained in the nucleus may have roles as non-coding RNAs or be leftovers from processing of transcripts destined for the cytoplasm. An important question arising from these findings is whether these transcripts possess biological function. Perhaps these transcripts reflect a previously unappreciated level of biological complexity. We believe that viruses, with their more tractable genomes and facile systems for analysis of biological function of transcripts provide a fundamental tool for future studies to address these central questions.
Experimental Procedures
Virus and viral stocks
γHV68 clone WUMS (ATCC VR1465) was passaged, and titers were determined by plaque assay of NIH 3T12 cells (Weck et al., 1996).
Cell culture and viral infections
Wild-type NIH 3T12 cells (3T12) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, 10 mM HEPES, and 2 mM L-glutamine (complete DMEM). Cells were maintained in a 5% CO2 tissue culture incubator at 37°C. 3T12 cells were either mock-infected or infected with γHV68 WUMS at a multiplicity of infection (MOI) of 10. Virus was allowed to adsorb onto the cells at 37°C for 1 hour with occasional rocking. The cells were rinsed with DMEM and allowed to grow in standard conditions. RNA was harvested at 8 and 18 hours post infection (hpi). Mock-infected or infected 3T12 cells were also treated during infection and incubation with 200ng/ml cycloheximide for 8 hours or 42 µg/ml cidofovir for 18 hours. Cidofovir inhibition of viral DNA synthesis was assessed by treating 3T12 cells with 0, 262 pg/ml, 5 ng/ml, 105 ng/ml, 2.1 µg/ml, and 42 µg/ml of cidofovir at the viral adsorption stage. Viral and host DNA was harvested at 0 or 18 hours post infection and measured with quantitative PCR using primers for ORF-50 and GAPDH, respectively. Viral DNA amount of untreated infected 3T12 cells (No CID) was used to set the 0-fold baseline level. Fold inhibition was calculated as No CID ΔCt/Experimental ΔCt, where ΔCt was calculated as GAPDH Ct – ORF50 Ct.
RNA isolation and cDNA labeling
Total RNA was harvested from NIH 3T12 cells using Trizol (Invitrogen) according to manufacturer's instructions. This RNA was treated with Ambion Turbo DNAfree Dnase and subjected to phenol/chloroform extraction. Labeled cDNA was generated using Invitrogen’s SuperScript Plus Indirect cDNA Labeling Module using random hexamer priming. Stratagene's Spot Report Arabidopsis RNAs were added to cDNA labeling reactions as spike-in RNA controls.
Tiled array design and hybridization
Probes of 60 nucleotide length complementary to each strand of the γHV68 genome and advancing 6–10 nucleotides were designed using Agilent's eArray package. This package was also used to design probes complementary to our spike-in RNAs and several mouse genes. The paired and balanced cDNAs were suspended in Agilent 2× Gene Expression buffer, Agilent 10× Blocking agent, and water and then applied to custom Agilent 4×44K microarrays. Hybridization was carried out at 65C for 20 hours. Washing procedures were carried out according to Agilent gene expression protocols.
Tiled array data analysis
Local background signal was subtracted from all probe fluorescence signals. Probe signals from mock and γHV68-infected samples were normalized to each other by spike-in RNA ratios. The normalized mock-infected probe signals were subtracted from the respective γHV68 infected samples; this corrected signal is plotted in figures. Quantile normalization was performed on these subtracted γHV68-infected replicates. Average spike-in RNA fluorescence signal was used to normalize between cycloheximide and cidofovir-treated infected cells with untreated infected cells. Probe intensities were mapped back to the γHV68 genome and mean fluorescence of each nucleotide was calculated from all overlapping probes. The baseline used in graphs is set at one standard deviation above the mean fluorescence signal detected in mock-infected cells at the same time point post infection. This data was converted to gff3 file format and visualized using gBrowse (Stein et al., 2002). Raw and normalized microarray data have been deposited in the Gene Expression Omnibus (GEO) database under Series entry GSE21500. A tar archive of the perl and R scripts used in the computational analysis is available upon request.
Computational analysis of Expressed Genomic Regions
The γHV68, KSHV, EBV, and herpesvirus saimiri genomes were translated in all six reading frames using "translate" from Dr. Sean Eddy's SQUID library version 1.51 (ftp://selab.janelia.org/pub/software/squid/). Open reading frames were identified with a minimum length of 20 amino acids and no initiator methionine was required. Protein sequence conservation between the γHV68 open reading frames and those from the other gammaherpesvirus genomes was determined by Wu-BLASTP 2.0MP using default settings. Gene predictions were done by GeneMarkS-EV and coordinates were extracted from the VIOLIN database of phage and virus genomes (http://opal.biology.gatech.edu/GeneMark/VIOLIN/violin_display.cgi?accession=U97553) (Mills et al., 2003).
Cloning of Northern probes and positive control regions
Northern probe regions were amplified by PCR from viral genomic DNA. These PCR products were cloned into Invitrogen's Topo TA dual promoter vector and sequenced. The primers used in cloning the Northern probe regions were as follows: EGR 8: 5’-gatctgcatttgccacgtctgaag-3’ and 5’-tgtgtgggactccatattccatgaa-3’ for Probe 1, 5’-ttgtttaggttgggatcctgtggc-3’ and 5’-tgcaagattcaccgaaacccttc-3’ for Probe2; EGR 24: 5’-ggtagtaagaaattttgaaaagcatgcg-3’ and 5’- cgtggttctttttttgatgtgtgcc-3’ for Probe 1, 5’-tactcctttgtgcccaagcactttt-3’ and 5’-agaggctctgaaatcaaatcctgca-3’ for Probe 2, 5’-ggcttttttcagtgctatttggtcg-3’ and 5’-cgacctgattaggagcaccaaggat-3’ for Probe 3; EGR 26: 5’-cagaccgctgggcatgttctact-3’ and 5’-tgagggtctccaagtgaacttccag-3’ for Probe 1, 5’-agtaatcccccagctgcaagctg-3’ and 5’-cggacctcggatgtgataatcaagt-3’ for Probe 2, 5’-aggtcttgtcggtccacaccttca-3’ and 5’-gtggcgtcccctctgtaatgga-3’ for Probe 3, and 5’-tttccatttccactccttgttttgg-3’ and 5’-ccaaaatgatggaccctgtcttgg-3’ for Probe 4.
Sense and antisense control regions were amplified by PCR utilizing primers with T7 and Sp6 promoter sequences on their 5' ends. Control region PCR products were purified using spin columns. Sense and antisense RNAs were transcribed utilizing Ambion's Maxiscript T7/Sp6 kit. The PCR primers used in cloning the sense and antisense control regions are as follows (T7 and Sp6 promoter sequences are underlined): EGR 8: 5’-taatacgactcactatagggcaatgagcaggctagaattttggca-3’ and 5’-tcacacaccccagtctacaacagga-3' (sense) and 5’-caatgagcaggctagaattttggca-3’ and 5'-atttaggtgacactatagggtcacacaccccagtctacaacagga-3’ (antisense); EGR 24:, 5’-taatacgactcactatagggcatttgggaaacccggtgtacctta-3’ and 5’-tgccaacacactagagacactcccc-3’ (sense) and 5’-catttgggaaacccggtgtacctta-3’ and 5’-atttaggtgacactatagggtgccaacacactagagacactcccc-3’ (antisense); EGR 26: 5’-taatacgactcactatagggtgcgaggtttgggtctgatatgaga-3’ and 5’-atttaggtgacactatagggtacaacacactgatgctcgtcagcc-3’ for positive control 1, 5’-taatacgactcactatagggggtggaatgtgcgcaggtagtattt-3’ and 5’-atttaggtgacactatagggtctcatatcagacccaaacctcgca-3’ for positive control 2, 5’-taatacgactcactatagggtacgtcgctcccaacagtga-3’ and 5’-atttaggtgacactatagggggaggctttgcctgcttgttatca-3’ for positive control 3, 5’-taatacgactcactatagggcccttcctcaaggccctagatctgt-3’ and 5’-atttaggtgacactatagggctccattacagaggggacgcca-3’ for positive control 4.
Northern blotting and probe labeling
Five micrograms of total RNA or 0.01 ng control RNA was loaded per lane on a 1% agarose-formaldehyde gel. Ambion’s RNA Millenium size markers were loaded to estimate RNA size. After electrophoresis, RNA was transferred to a positively-charged nylon membrane through downward capillary transfer and cross-linked to the membrane using an ultraviolet source. Pre-hybridization and hybridization were performed at 68°C in Ambion’s ULTRAhyb hybridization buffer. Single-stranded RNA P32-labeled probes were generated from linearized plasmid using Ambion’s Maxiscript Sp6/T7 kit. Unincorporated labeled nucleotides were removed using Ambion’s NucAway spin columns and specific activity of the probes was estimated by scintillation counting. Membranes were exposed to a phosphor storage plate and scanned using a Storm 840 Phosphor Imager.
RACE
RACE determination of 5’ and 3’ transcript ends was performed using Invitrogen's 5' and 3' RACE systems according to manufacturer's instructions. For 3’ RACE, oligo-dT primers were used to generate cDNA, while for 5’ RACE, cDNA was generated using a gene-specific reverse transcriptase primer. The PCR amplification step used Invitrogen’s adaptor primers and gene-specific primers. The following reverse transcriptase primers were used in 5’ RACE: EGR 8: RT1 5’-catgtccactcacgcaaag-3’, RT2 5’- cggcgctctagacacacttt -3’; EGR 24: RT1 5’- atgatactttctctcttggc -3’, RT2 5’- cagcgaaggaacaaaagtg -3’, RT3 5’-tgctgaagaaaagaataggc -3’, and RT4 5’- tttgactgctgtggctat -3’. The PCR primers used in 5’ RACE reactions were as follows: EGR 8: P1 5’-ttcatggaatatggagtcccacaca -3’, P2 5’ -tgacagcatacagcgtgggaatg -3’, N1 5’-tctccagccaaccacgtagacagt -3’, N2 5’- gcggagatacagctgtgctcacat -3’; EGR 24: P1 5’- catctgatgagacagcgcctgaa-3’, P2 5’- ttatccagaaaacaagcggatttcc-3’, P3 5’-agcaaacagaggccaggaaagc-3’, P4 5’- tcgagaagagccaccgtttcct-3’, N1 5’-gctttttggacgccatcgct-3’, N2 5’- cctaaatgcgatttgtgccaa-3’, N3 5’-gggtttgggtcaagtggggtgaat-3’, N4 5’- gctggaagtgtggcctcatctaca-3’. The PCR primers used in 3’ RACE reactions were as follows: EGR 8: 5’-ctgtgtgggactccatattccatga-3’; EGR 24: 1 5’- cgtggttctttttttgatgtgtgcc -3’, 2 5’-aattggcccggttgaatctc -3’, 3 5’- gctttcctggcctctgtttgct -3’, and 4 5’-ttaggagcaccaaggataagcag -3’. RT = reverse transcriptase primer, P= PCR primer, N= nested PCR primer.
Supplementary Material
Acknowledgements
We would like to thank the Washington University Microarray Core facility for running the tiled arrays, Seth Crosby's assistance on the original tiled array design, and members of the Virgin lab for reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Allen RD, Dickerson S, Speck SH. Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection. J. Virol. 2006;80:2055–2062. doi: 10.1128/JVI.80.4.2055-2062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. U. S. A. 2008a;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Reply to Satheshkumar and Moss: Poxvirus transcriptom analysis. Proc. Natl. Acad. Sci. U. S. A. 2008b;105:E63–E64. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic. Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 2008;20:274–280. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Steitz JA. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO. J. 2005;24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZalia M, Speck SH. Identification of closely spaced but distinct transcription initiation sites for the murine gammaherpesvirus 68 latencyassociated M2 gene. J Virol. 2008;82:7411–7421. doi: 10.1128/JVI.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebel KW, Smith AL, Van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA. 2010;16:170–185. doi: 10.1261/rna.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, Nash AA. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 2003;84:99–109. doi: 10.1099/vir.0.18639-0. [DOI] [PubMed] [Google Scholar]
- Gray KS, Allen RD, III, Farrell ML, Forrest JC, Speck SH. Alternatively initiated gene 50/RTA transcripts expressed during murine and human gammaherpesvirus reactivation from latency. J. Virol. 2009;83:314–328. doi: 10.1128/JVI.01444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Wu TT, Tong LM, Kim KS, Martinez-Guzman D, Colantonio AD, Uittenbogaart CH, Sun R. Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J Virol. 2008;82:12498–12509. doi: 10.1128/JVI.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E, Rickinson A. Epstein-Barr Virus and Its Replication. In: Knipe D, Howley P, editors. In Fields Virology. Philadelphia: Lippincott, Williams & Wilkins; 2007. pp. 2605–2654. [Google Scholar]
- Mackett M, Stewart JP, de V Pepper S, Chee M, Efstathiou S, Nash A, Arrand JR. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J. Gen. Virol. 1997;78:1425–1433. doi: 10.1099/0022-1317-78-6-1425. [DOI] [PubMed] [Google Scholar]
- Majerciak V, Yamanegi K, Zheng ZM. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J Virol. 2006;80:11968–11981. doi: 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JS, Coleman HM, Boname JM, Stevenson PG. Murine gammaherpesvirus-68 ORF28 encodes a non-essential virion glycoprotein. J. Gen. Virol. 2005;86:919–928. doi: 10.1099/vir.0.80661-0. [DOI] [PubMed] [Google Scholar]
- Mills R, Rozanov M, Lomsadze A, Tatusova T, Borodovsky M. Improving gene annotation of complete viral genomes. Nucleic. Acids Res. 2003;31:7041–7055. doi: 10.1093/nar/gkg878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash AA, Dutia BM, Stewart JP, Davison AJ. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond B Biol. Sci. 2001;356:569–579. doi: 10.1098/rstb.2000.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyts J, De CE. In vitro and in vivo inhibition of murine gamma herpesvirus 68 replication by selected antiviral agents. Antimicrob. Agents Chemother. 1998;42:170–172. doi: 10.1128/aac.42.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry CM, Simas JP, Smith VP, Stewart CA, Minson AC, Efstathiou S, Alcami A. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J Exp. Med. 2000;191:573–578. doi: 10.1084/jem.191.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, Van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Puglielli MT, Desai N, Speck SH. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp) J Virol. 1997;71:120–128. doi: 10.1128/jvi.71.1.120-128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimessi P, Bonaccorsi A, Sturzl M, Fabris M, Brocca-Cofano E, Caputo A, Melucci-Vigo G, Falchi M, Cafaro A, Cassai E, Ensoli B, Monini P. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J Virol. 2001;75:7161–7174. doi: 10.1128/JVI.75.15.7161-7174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R, Lutzke ML, Alfinito RS, Clavo A, Cardin RD. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 2001;75:4955–4963. doi: 10.1128/JVI.75.11.4955-4963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satheshkumar PS, Moss B. Poxvirus transcriptome analysis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:E62. doi: 10.1073/pnas.0804594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Hwang SM, Wong WH, Wu TT, Lee SM, Liao HI, Sun R. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3805–3810. doi: 10.1073/pnas.0404521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: a building block for a model organism system database. Genome. Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JL, Bennett HN, Snyder BA, Moore PS, Chang Y. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J Virol. 2005;79:15099–15106. doi: 10.1128/JVI.79.24.15099-15106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck KE, Barkon ML, Yoo LI, Speck SH, Virgin HW. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.