Abstract
Grass carp reovirus (GCRV) is a member of the aquareovirus genus in the Reoviridae family and has a capsid with two shells—a transcription-competent core surrounded by a coat. We report a near-atomic-resolution reconstruction of the GCRV virion by cryo-electron microscopy and single-particle reconstruction. A backbone model of the GCRV virion, including seven conformers of the five capsid proteins making up the 1500 molecules in both the core and the coat, was derived using cryo-electron microscopy density-map-constrained homology modeling and refinement. Our structure clearly showed that the amino-terminal segment of core protein VP3B forms an ~120-Å-long α-helix-rich extension bridging across the icosahedral 2-fold-symmetry-related molecular interface. The presence of this unique structure across this interface and the lack of an external cementing molecule at this location in GCRV suggest a stabilizing role of this extended amino-terminal density. Moreover, part of this amino-terminal extension becomes invisible in the reconstruction of transcription-competent core particles, suggesting its involvement in endogenous viral RNA transcription. Our structure of the VP1 turret represents its open state, and comparison with its related structures at the closed state suggests hinge-like domain movements associated with the mRNA-capping machinery. Overall, this first backbone model of an aquareovirus virion provides a wealth of structural information for understanding the structural basis of GCRV assembly and transcription.
Keywords: aquareovirus, dsRNA virus, backbone model, Reoviridae, cryo-electron microscopy
Introduction
Viruses in the Reoviridae family represent one of the largest and most structurally divergent groups of viruses. These viruses have a segmented double-stranded RNA (dsRNA) genome, enclosed by single, double or triple proteinaceous capsid shells, and hosts ranging from vertebrates, invertebrates, plants, fungi and prokaryotes.1,2 In order to circumvent the antiviral defense mechanisms inside the cytoplasm of the host cell, these viruses conduct the entire viral RNA transcriptional process within the inner capsid in a characteristic process called endogenous RNA transcription.
The inner capsid (i.e., the core) of these viruses serves as a compartment for endogenous mRNA transcription and is a conserved architecture among all members of the Reoviridae,1 as well as other dsRNA viruses, such as totiviruses3,4 and some bacteriophages.5 Among viruses in different genera of the Reoviridae, the genera aquareovirus and orthoreovirus are considered to be the most similar; both are “turreted” reoviruses with double-layered capsids and have relatively high sequence identities between protein homologs (with sequence identities ranging from 15% to 31%).6,7 However, there are three significant differences between aquareoviruses and orthoreoviruses: First, the numbers of dsRNA segments in their viral genome are different (11 and 10 for aquareovirus and orthoreovirus, respectively).6 Second, aquareo-virus does not have a homolog of the orthoreovirus cell attachment protein σ1 (Ref. 7). Third, each aquareovirus core only contains 120 copies of its clamping protein VP6, in contrast to the 150 copies of clamping protein σ2 per orthoreovirus core, including 30 across its 2-fold axes.7 Orthoreoviruses have been subjected to extensive structural analyses by both X-ray crystallography8–10 and cryo-electron microscopy (cryo-EM).11–13 In contrast, there is no crystal structure of aquareovirus proteins and the highest-resolution cryo-EM structure reported to date is that of grass carp reovirus (GCRV) at ~9-Å resolution.7 At this resolution, it is not possible to build a backbone model from the cryo-EM density map, hindering our understanding of the molecular interactions critical to aquareovirus assembly and RNA transcription.
Recently, advancements in cryo-EM have pushed the resolution limit to near-atomic level,14–17 permitting separation of β-strands and identification of bulky side chains of amino acid residues in cryo-EM density maps. In this article, we report the first near-atomic-resolution structure of GCRV virion by cryo-EM and single-particle analysis and the derivation of a backbone model of the virion. We discovered an extended domain of the core protein VP3B in the virion that forms one bridge across near the 2-fold axis and interacts with genomic RNA. The bulk of this extended domain becomes disordered inside transcription-competent cores. We show the multiple enzymatic domains of VP1 and suggest plausible hinge-like domain movements between the open and closed states of the turret. Amino acids involved in inter-trimer interactions on the coat are also revealed.
Results and Discussion
Three-dimensional reconstruction and modeling of the GCRV virion
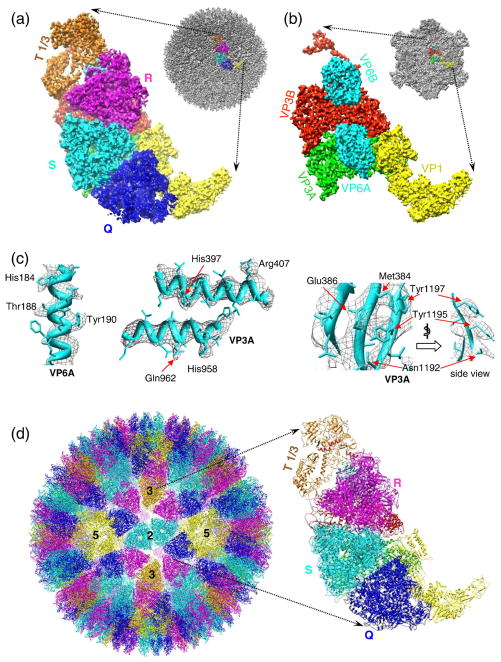
We reconstructed a near-atomic-resolution structure of the GCRV virion by merging 15,000 close-to-focus particle images recorded on a 16-megapixel CCD camera in a 300-kV cryo-electron microscope. The mass density maps for individual proteins in each asymmetric unit of the virion were segmented out and are shown in different colors in Fig. 1a and b. The GCRV virion structure is organized in two layers: a coat and a core. The coat is made of 200 triangular densities (“trimers”) arranged on an incomplete T=13 lattice. Each asymmetric unit of the coat layer contains one Q trimer, one S trimer, one R trimer and one-third of a T trimer (Fig. 1a), following the established nomenclature.12,18 Each asymmetric unit of the core consists of one turret protein (VP1), two conformers of the inner-capsid shell protein (VP3A and VP3B) and two conformers of the cementing protein (VP6A and VP6B) (Fig. 1b). Our map shows the pitches of α-helices, separation of β-strands in β-sheets and some densities for bulky side chains (Fig. 1c; Supplementary Fig. 1 and Movie 1c). The validity of our structure can also be judged by the fact that many side-chain densities were found in equivalent positions in the two conformers of VP3, which are not related by symmetry operation. The visibility of these features in core proteins (see Supplementary Movie 2) suggests that the resolution of the core proteins is about 4.5–5.0 Å (Refs. 14 and 19). The coat proteins have a slightly lower resolution, probably due to more flexibility of the coat protein and the resolution-limiting effect of the Ewald sphere curvature.20,21
Fig. 1.
Cryo-EM density map and backbone models of GCRV. (a) Density map of an asymmetric unit (left panel) segmented out from the GCRV virion reconstruction (upper right). The trimers (Q, R, S, T) are labeled using the nomenclature of bluetongue virus and are colored differently.18 (b) Density map of an asymmetric unit of the core (left). The core map (upper right) was computationally extracted from the GCRV virion map shown in (a). (c) Cryo-EM densities (wire frames) and models (ribbons) of α-helices (left and middle) and a β-sheet (right) selected from VP6A and VP3A. Densities corresponding to some bulky side chains and partially resolved β-strands are resolved. (d) Backbone models of the virion (left) and an asymmetric unit (right). (e) Backbone models of the core (left) and an asymmetric unit (right). Different proteins (VP1, VP3A, VP3B, VP6A, VP6B and VP1) are shown in different colors.
The value of a three-dimensional structure can be substantially enhanced by deriving three-dimensional models with atomic coordinates. However, the limited resolution of the cryo-EM map does not permit atomic model building from the density map alone. For the GCRV virion, fortunately, model building through an integrative approach is possible due to the availability of both our near-atomic-resolution cryo-EM map and the experimentally determined atomic structures of the orthoreovirus core (λ1, λ2 and σ2) and coat (μ1 and σ3) proteins that are homologous to GCRV core and coat proteins.8,9 By means of a novel method of cryo-EM density-constrained homology modeling and refinement (Supplementary Fig. 2),22 we built backbone models for seven conformers (VP1, VP3A, VP3B, VP5, VP6A, VP6B and VP7) of the five GCRV virion structural proteins. The approach we used integrates a modeling-based protocol to build, modify and refine the local structures of initial homology models iteratively to match the cryo-EM maps, followed by a molecular-dynamics-based method to adjust the structural details to further improve the model.22 Because only some bulky side chains were resolved in our map, the side-chain densities were only used as additional constraints for determining backbone conformations during the refinement. For this reason, the geometries of the side chains have not been explicitly refined and our final models can only be considered backbone models. The models for the entire virion and the core were generated by imposing icosahedral symmetry of the asymmetric unit (Fig. 1d and e and Supplementary Movie 3). The complex patterns of molecular interactions on the core and coat and between the two layers are revealed by these models. We now focus on structural features that are of significant biological implications.
Two conformers of VP3 and inter-VP3 interactions involved in the core assembly
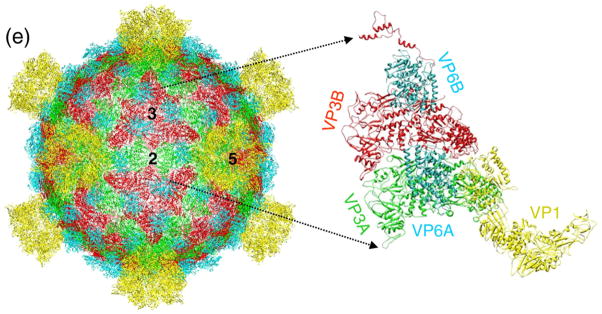
The core protein VP3 has two structural conformers: VP3A near the 5-fold axis (Fig. 2a) and VP3B near the 3-fold axis (Fig. 2b). These two conformers differ mainly in their amino-terminal domain and peripheral regions (cf. Fig. 2a and b). VP3A has a plate-like structure with two subdomains: subdomain I (amino acid residues 410–858) and subdomain II (amino acid residues 188–409 and 859–1214), which are similar to those of mammalian reovirus (MRV) λ1A (Ref. 8). VP3B has subdomain III (i.e., amino acid residues 19–187), in addition to subdomains I and II (Fig. 2b). Residues 1–18 of VP3B and residues 1–187 of VP3A (Fig. 2a) were not resolved, suggesting that these amino-terminal segments might be flexible and/or inserted into the non-icosahedrally organized RNA densities. In the crystal structure of MRV, fragments of the amino-terminal sequence of the core protein λ1B are modeled, but how the fragments are connected is not uniquely determined due to a missing loop (residues 168–180) (Ref. 8). In our cryo-EM density map of GCRV, the densities for the most part of the amino-terminal region of VP3B were clearly resolved, allowing us to directly trace the corresponding amino-terminal up to Thr19 (Fig. 2c).
Fig. 2.
Molecular interactions involved in the assembly of the core. (a and b) VP3A and VP3B models (ribbons) superimposed with their cryo-EM density maps (semitransparent gray). Their subdomains are shown in different colors. (c) Three copies of VP3B organized as a group of three around a 3-fold axis, as viewed from inside the virion. The icosahedral symmetry axes are designated by numbers (“2”, “3” and “5”, for 2-, 3- and 5-fold axes, respectively). The area enclosed in the red dashed box is enlarged and rotated 90° and shown in the box on the right. (d) The extended loops under the icosahedral 2-fold axis (indicated by “2”) and their zoomed-in views. In the zoomed-in view on the right, the atomic model is superimposed in the semitransparent density map and the amino acids contributing to the formation of the kinked bridge are indicated. (e) Organization of the core. Left: ribbon model of the core showing the organization of VP3A (green), VP3B (red) and VP6 (cyan); right: zoomed-in view of a region containing VP3A, VP3B, VP6A and VP6B. The backbone models are shown as ribbons and are superimposed with the cryo-EM density maps shown semitransparently.
The amino-terminal region (subdomain III) of VP3B begins with a segment of three three-turn α-helices connected by loops (Fig. 2b). This segment has the least structural similarities to its counterpart of λ1B in MRV and represents the most divergent sequence among VP3 homologs in other members of the Reoviridae family (Table 1). This segment is connected through a long loop with a kink (Pro103) to a highly conserved zinc finger motif (residues 119–140) having the characteristic structure of an α-helix and an anti-parallel β-sheet coordinated by a zinc ion (not seen due to limited resolution) (Fig. 2b). This zinc finger motif belongs to the “Cys–His–Cys–His” class, which is the best characterized class of zinc fingers and is known to bind RNA, as also suggested in MRV core.8
Table 1.
GCRV VP3 sequence identity with shell proteins of other reoviruses
| Full-length sequence identity/similarity (%) | Amino-terminal segment (up to the CCHH motif) sequence identity/similarity (%) | |
|---|---|---|
| American GCRV VP3 | 77/86 | 52/63 |
| MRV λ1 | 31/45 | 14/21 |
| Avian reovirus λA | 30/44 | 9/19 |
The National Center for Biotechnology Information accession numbers of GCRV VP3, American GCRV VP3, MRV λ1 and avian reovirus λA are AAG10437, YP_001837096, ABP48915 and AAT27445, respectively.
The extended subdomain III of VP3B may also play a structural role in core stability by strengthening the molecular contacts surrounding both 3-fold and 2-fold axes. When viewed from inside around the 3-fold axis, the three extended subdomain III's of the VP3B molecules form a triskelion joining three neighboring VP3B molecules together (Fig. 2c). When viewed from underneath the 2-fold axis (Fig. 2d), residues Pro103, Gln104 and Ser105 of this subdomain form a loop that attaches to the two adjacent VP3A molecules across the 2-fold axis. Therefore, this loop appears to serve as a rope tying two 2-fold-symmetry-related VP3A molecules to two neighboring 3-fold-symmetry-related VP3B molecules (Fig. 2c and d). Interestingly, GCRV does not have a cementing protein (VP6) situated above the connecting loop at the icosahedral 2-fold symmetry axis position (Fig. 2e), in contrast to that of MRV, which has a cementing protein σ2 at that position.8,23 Note that the corresponding λ1B amino terminal loop is not visible in MRV. Indeed, biochemical data have shown that the amino-terminal segment of λ1B is dispensable for MRV core assembly.24
Interactions between VP3 and RNA in the virion and transcription-competent core
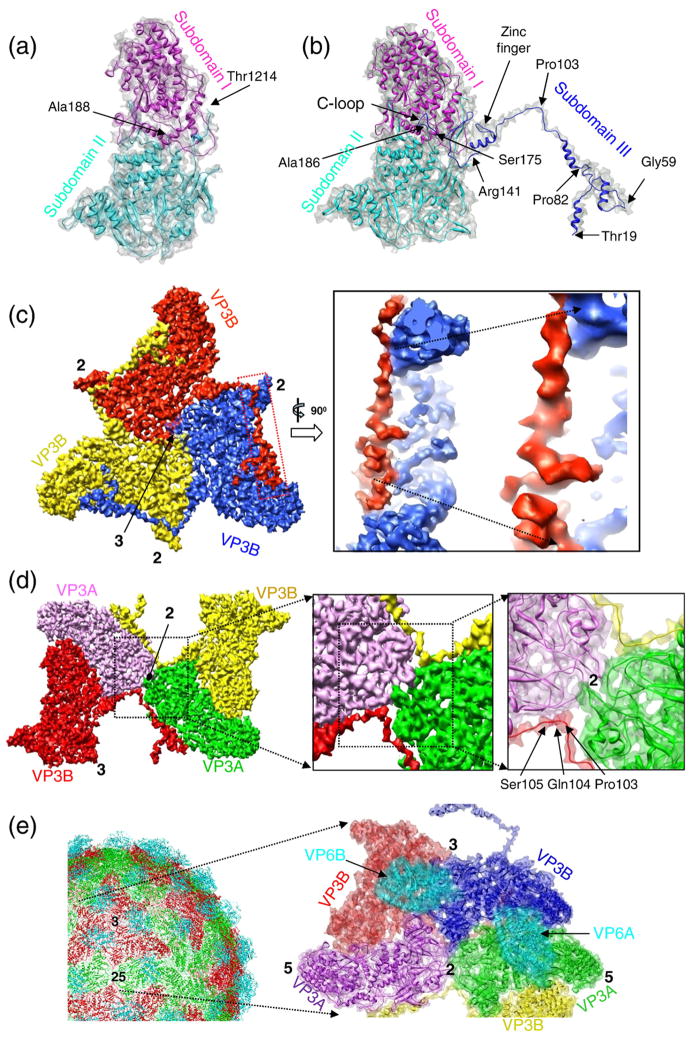
The ability to resolve not only protein densities but also RNA genome inside our cryo-EM maps allows us to examine the protein–RNA interactions and gain insight into the mechanism of endogenous RNA transcription. The amino-terminal segment (residues 19–29) of subdomain III of each VP3B interacts with the C-terminal loop (residues 175–186) of subdomain III of its neighboring VP3B (Fig. 3a and b). This part of the capsid is where the core protein interacts with the RNA inside (Fig. 3b). It is noteworthy that the loop in the C-terminal portion (residues 171–187; includes the C-terminal loop) of subdomain III does not share any sequence similarity with MRV λ1 (Table 1) and indeed exhibits a totally different structure.
Fig. 3.
VP3B–RNA interactions and conformational changes of VP3B between the virion and the transcription-competent core. (a) Stereo zoomed-in view of the loop region of VP3B facing the dsRNA genome inside the virion. The dashed region in the inset illustrates the region of the VP3B shown. The N-terminal extension is highlighted in blue, while the C-terminal loop (residues 175–186) of VP3B is highlighted in red. (b) Side view of a 12-Å-thick slab cut along the line drawn in the inset icon view of (a). (c) Negative-stain EM image of transcription-competent transcribing cores. The strands attached to the icosahedral vertices are nascent RNA molecules being released from the cores. (d) Negative-stain EM image of the intact non-transcribing virions. (e) A region of the core inside the virion, containing three copies of VP3B and a copy of VP3A, as viewed from outside of the virion. (f) Structure of the transcription-competent core (gray) superimposed with the virion molecules (color) shown in (e). (g) Zoomed-in view of the area marked by the red box in (f) after a rotation of 180°. (h) Top: Side view of the yellow boxed region in (g); bottom: backbone models of the same shell proteins (same view and color scheme). (i) Identical with (g) except that the blue virion VP3B was removed. The blue amino-terminal region is disordered and not visible in the transcription-competent core. (j) Top: Side view of the yellow boxed region in (i); bottom: backbone models of the same shell proteins (same view and color scheme).
Reovirus cores are transcriptional machines that produce viral mRNA. Biological data have indicated that MRV cores partially recoated with outer-capsid proteins can synthesize mRNA but that intact virions cannot.25 The levels of mRNA synthesized by partially recoated reovirus cores are inversely proportional to coat protein levels. The coat proteins are thus thought to prevent progeny virions to compete with transcribing particles for ribonucleo-side triphosphates.25 Our transcription experiments of GCRV also indicated that the cores can be activated for transcription (Fig. 3c) but that the intact GCRV virions (Fig. 3d) are incompetent to produce mRNA (Table 2). Upon GCRV entry into the cytoplasm, the coat proteins (VP5 and VP7) fall off during host cell membrane penetration,26,27 exposing the core proteins, which is considered to be necessary to activate the viral transcriptase inside the core. To further study the involvement of core proteins in the transcription, we compared the core proteins VP3A and VP3B computationally segmented from the virion with those in the transcription-competent core. By fitting the core protein structures from the virion into the 9-Å reconstruction of the transcription-competent core (Fig. 3e and f), we found that two α-helices in the amino-terminal region observed in VP3B of the virion became invisible in the transcription-competent core (Fig. 3g–j), while other helices of VP3A and VP3B in the virion fit well with those in the core, suggesting that the amino-terminal segment of VP3B is structurally flexible in the transcription-competent core. Superposition of the VP3B model into the core density map showed that this flexible segment roughly corresponds to the first 80 residues of VP3B (Figs. 2b and 3h).
Table 2.
Transcription activities of GCRV virions and cores
| Incorporated radioactivity CPMa (×103) | |
|---|---|
| Intact virions | 1.24±0.07 |
| Cores | 6.83±0.43 |
Incorporated radioactivity as measured in counts per minute (CPM). A low incorporated CPM value means a low rate of transcription.
It is likely that the amino-terminal segment of VP3B binds the genomic RNA and plays a regulatory role in RNA transcription. Using the sequence alignment program CLUSTALW,28 we found that the amino-terminal segments of the core proteins are the most divergent segments in the core proteins among aquareoviruses MRV and avian reovirus29,30 (Table 1). It was reported that the hydrophilic amino-terminal segment (sequence up to the zinc finger motif; see Fig. 2b) of MRV has dsRNA-binding property.31 In addition, residues 59–141 of the VP3B serve to block the clefts where VP3A and VP3B interact. Taken together, the observed conformational change of the VP3B amino-terminal segment from the GCRV virion to the transcription-competent core, the significant sequence divergence and the RNA-binding property of this segment all point to possible roles of subdomain III of VP3B in recognizing specific RNA, mediating RNA packing in the process of capsid assembly and regulating genomic RNA transcription.
Structural transition between open and closed turret states
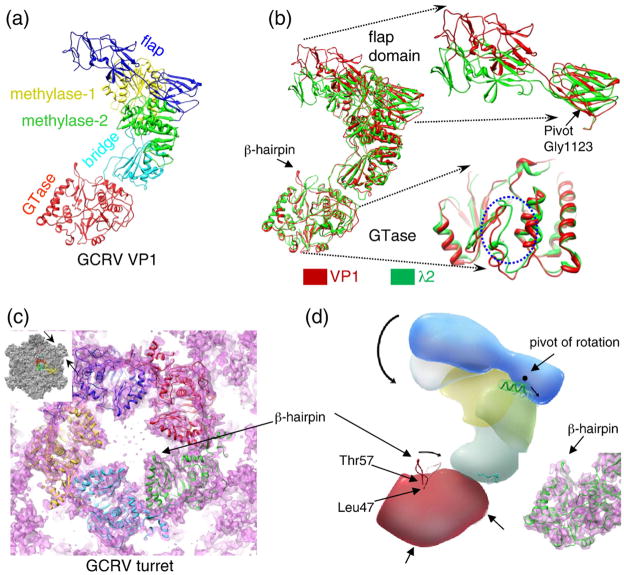
The turret protein VP1 and MRV λ2 share a sequence identity of 26% (Refs. 32 and 33) and have highly conserved enzymatic activities, including guanylyltransferase (GTase) and methyltransferase activities in mRNA capping.10,34 Consistently, our cryo-EM structure of the GCRV virion shows that VP1 has highly conserved domain folds (Fig. 4a and b) as those in MRV λ2. Simply superimposing VP1 onto MRV λ2 reveals a similar overall topological structure and conserved domains involved in RNA capping and release through the turret. Similar to MRV λ2 (Ref. 8), GCRV VP1 can be divided into seven domains (Fig. 4a): the GTase domain (residues 1–389), a bridge domain (residues 390–437 and 695–805), the first methyltransferase domain (methylase-1, residues 438–694), the second methyltransferase domain (methylase-2, residues 806–1029) and three immunoglobulin domains forming a flap (residues 1030–1299).
Fig. 4.
Comparison between GCRV VP1 and MRV λ2. (a) Ribbon diagram of VP1 with functional domains shown in different colors. (b) Superposition of VP1 (red) and MRV λ2 (green), with enlarged views of the flap and the GTase domains. The channel for nascent mRNA release is marked by a dotted blue circle. (c) A slab of the cryo-EM map of pentameric turret (transparent) superimposed with five copies of VP1 models shown as ribbons. The arrow points to a protruding β-hairpin (Leu47–Thr57). The inset shows the core and the position of the slab (pointed by the two arrows). (d) Schematic illustration of VP1 depicting putative conformational changes (arrows) associated with the transition from the open state to the closed state of the turret protein. The right inset shows the VP1 GTase domain density (pink) superimposed with its backbone model (green ribbon).
The pentameric turrets resolved in our GCRV virion and core reconstructions are in the open conformation. However, in both GCRV and MRV, turrets can adopt two conformational states: an open state and a closed state,13,24,35,36 suggesting the existence of a transition between the open state and the closed state in both viruses. A comparison of high-resolution structures representative of the open (VP1) and closed (λ2) states of the turrets thus reveals clues about this transition (Supplementary Movie 4). The structural changes appear to be localized in four regions—the immunoglobulin domains (i.e., the flap), the GTase domain and the methylase-2 and bridge domains (Fig. 4b). The tilt of the flap can be considered as a pivot type of movement characterized by a rotation of the flap around a pivot at residue Gly1123 of VP1 (Fig. 4b and d). In the GTase domain, the β-hairpin (Leu47–Thr57) making up the constriction inside the channel is tilted farther away from the 5-fold axis (Fig. 4b). Two major conformational changes occur at regions where VP1 and VP5 interact, suggesting a possible association of the coat protein trimers with control of the open/closed states. However, such a causal relationship is not firmly established yet. For example, in MRV, the removal of μ1 trimers triggered the conformational change of the flap in λ2, resulting in the open state of the turret.37 Such a relationship is supported by a number of cryo-EM studies of MRV cores13,24,35–39 but not by the crystal structure of the MRV core, in which the turret was closed.8
Coat proteins and molecular interactions contributing to virion assembly
The coat consists of 200 triangular densities (trimers of VP5/VP7 dimers) arranged on an incomplete T=13 icosahedral lattice, exposing the VP1 turrets on the icosahedral vertices (Fig. 1a and d). Each coat trimer is formed by three VP7 molecules sitting on top of three VP5 molecules (Fig. 5c) (Ref. 7). VP7 has the least structural similarity to its orthoreovirus homolog protein σ3 among all five capsid proteins. This structural divergence of VP7 is consistent with specific cell tropisms observed across different reoviruses.40
Fig. 5.
Molecular interactions involved in the assembly of the outer shell. (a) A copy of VP1 and adjacent trimer Q and a zoomed-in view of their contact sites. Key residues at the contact sites are indicated. (b) Top view of the trimers R and S. (c) Side view of trimers R and S. (d) Two slabs as indicated in (c). (e) Zoomed-in view of the contact sites between the VP5 molecules in two neighboring trimers.
It is reported that the MRV outer capsid cannot assemble without the participation of turret protein λ2 (Ref. 24). Consistent with this observation, our structure shows that VP1 and VP5 have two areas of interaction (Fig. 5a)—one involving VP1 residues Val379–Asn380 in contact with VP5 residues Asp59–Leu61 and another involving VP1 residues Ala723–Val724 in contact with VP5 residues Lys100–Gly102.
In addition to their interactions with the turret protein VP1 and the cementing protein VP6 of the core,7 we identified two kinds of trimer–trimer interactions on the coat (Fig. 5b–e). One is between residue 379 and residue 381 of VP5 projecting toward their counterpart in a neighboring trimer (Fig. 5d), and the other is between the Cys103 residues of two neighboring VP5 molecules (Fig. 5e). The presence of connecting density between the two nearby Cys residues suggests a possible disulfide bond between the two Cys residues in the neighboring trimers. In addition to serving as a stabilizing factor, these VP5–VP5 interactions, particularly the putative disulfide bonds, lead to the formation of a VP5/VP7 trimer network. Such a network of trimers can collectively promote membrane perforation, thus facilitating viral entry and uncoating. Indeed, thiol/disulfide rearrangement is often necessary during membrane penetration and viral uncoating processes, as shown in a number of enveloped viruses.41
Location of VP2 and relationship to VP3A
The density underneath VP3A near each 5-fold axis is thought to be that of the RNA-dependent RNA polymerase VP2 (Ref. 7). Due to lack of icosahedral symmetry, the bulk of VP2 was unresolved except for an α-helix attached to VP3A at a location adjacent to a 5-fold hole (Supplementary Fig. 3b). The 5-fold hole is formed by five copies of four consecutive residues 502–505 (Glu, Ser, Thr and Thr) of VP3A, which are fully conserved among GCRV, MRV and avian reovirus. In addition, these conserved residues in VP3B are flexible and are located next to the putative channel for the pathway of nascent mRNA (the situation is the same for MRV)8 and point toward the RNA molecules inside. Conceivably, these four residues of VP3A might be involved in RNA transcription.
Implication to reovirus transcription
The structural differences revealed in VP1 and VP3 proteins of the GCRV virion and core particles and between GCRV and MRV may shed light onto reovirus transcription. The coat protein VP5/μ1, in complex with its “protector” protein VP7/σ3, plays a key role in cell membrane penetration by the non-enveloped reovirus.9,27 The interactions found between VP1 and VP5, and the turret comparisons between the virions and cores suggest that, under certain in vivo conditions, uncoating of coat protein trimers during membrane penetration can trigger the conformational change of the turret protein VP1/λ2. Given that VP3B/λ1B contacts the GTase domain of the turret protein VP1/λ2, it is possible that the conformational change of the turret protein might not only prepare VP1/λ2 for transcription but also serve as a signal to trigger the conformational change of VP3B subdomain III.
The genomic RNA inside GCRV capsid, similar to that of MRV, exhibits a statistically significant icosahedral order,8 indicating that there are obvious interactions between the RNA and icosahedral shell proteins. In GCRV, these interactions were observed and pinpointed to specific locations between the RNA and the amino-terminal subdomain III of the shell protein VP3B. The amino-terminal segment of VP3B probably plays a regulating role by switching the RNA from a static state to a dynamic state, as in the actively transcribing cores.
Methods
Preparation of GCRV virions and cores, cryo-EM imaging and reconstruction
GCRV virions and cores were purified using an established method as described previously.7,42 Focal pair images of GCRV particles were recorded on a TVIPS CCD 16-megapixel CCD camera in an FEI 300-kV Polara cryo-electron microscope at underfocus values of about 0.8 and 2.3 μm, respectively. More than 4000 focal pairs were recorded, from which about 30,000 particles were selected for in-depth single-particle reconstruction using the IMIRS package.43 The CTF (contrast transfer function) was determined by the ctfit program of the EMAN package44 based on incoherently averaged Fourier transforms of particle images.45 The final reconstruction was obtained by combining 15,000 particle images by including image data up to 1/4.3-Å spatial frequency. All data processing steps were carried out using two Dell personal computers running Windows XP. The resolution of the map was estimated to be around 4.5–5.0 Å based on the strand separation in β-sheets and the visibility of some bulky side chains in the density map. Protein subunit densities were segmented from the maps and visualized using UCSF Chimera.46
Atomic modeling and model refinement
The homology models for seven conformers of five GCRV proteins were refined using two protocols consecutively. Starting with the homology model and segmented cryo-EM map, modeling-based refinement was first carried out to obtain a new model in which the backbone conformation overall is consistent with the density. In this protocol, after fitting the atomic model into the cryo-EM map, regions that are inconsistent with the map were identified with visual inspection and refined using the EM-IMO method.22 The refined regions were then merged into a single model, which was refitted into the cryo-EM map and used as the starting model in the next iteration of refinement. EM-IMO, as the core component of this protocol, incorporates the cryo-EM map as a constraint in structure refinement. The structures obtained from the modeling-based protocol were further subjected to molecular-dynamics-based refinement, in which the structures were adjusted in greater detail. During molecular dynamics simulation, the pseudo-energies and forces derived from the grid representation of the cryo-EM map were used in conjunction with the standard force field energy function and implicit solvation model to guide the sampling in conformational space. After refinement, the conformation with the best density fitting score was selected as the final model from the ensemble of candidates.
Transcriptase reactions and in vitro RNA transcription assay
Purified virions of GCRV at a concentration of about 2 mg/ml in 10 mM phosphate-buffered saline were digested with 100 mg/ml of chymotrypsin at 37 °C to generate transcription-competent cores for transcriptase reaction. Chymotrypsin digestion was stopped by placing the reaction mixtures on wet ice, and cores were then recovered by CsCl gradient centrifugation.27 The final pellet was resolved in 10 mM phosphate-buffered saline and stored at 4 °C. We followed previous publications47–49 for our GCRV transcriptase reactions. Briefly, the reactions were performed in 250-μl volumes containing 100 mM Tris–HCl, pH 8.0; 10 mM MgCl2; 3.3 mM phosphoenolpyruvate; 100 pg/ml of pyruvate kinase; 0.4 μmol each of ATP, GTP, CTP and UTP; 2.0 μCi of [α-32P]ATP; and 50 μl of digestion mixture or purified virus sample. The reactions were incubated at 28 °C for 90 min, after which 25-μl portions were removed and precipitated by adding 500 μl of 5% trichloroacetic acid. Precipitates were collected by filtration through Whatman GF/C filters, and radioactivity was measured by counting with a Beckman liquid scintillation counter.
The complete RNA transcriptase reaction mixture contained the following components in a volume of 1.5 ml: 30 μmol Tris–HCl buffer, pH 8.5; 5.0 μmol MgCl2; 3 pmol phosphoenolpyruvate; 1 pmol pyruvate kinase; 3 μmol each of ATP, GTP, CTP and UTP; and approximately 0.5 mg each of purified GCRV virion and core. After incubation at 28 °C for 30 min, samples were removed from the reaction mixture with a chilled pipette and synthesis was terminated by placing the sample on wet ice. Reaction cores were further purified by CsCl gradients as described elsewhere27 and further diluted with components of a cytochrome c spreading mixture to give a final concentration of 100 μg of cytochrome c per milliliter and 1% formaldehyde. Following surface spreading was performed as described previously.50,51 The samples were negatively stained and imaged in a transmission electron microscope.
Data deposition and accession numbers
The electron density maps and associated backbone models reported here have been deposited with the Electron Microscopy Data Bank at the European Bioinformatics Institute under accession code EMD-1653 and with the Protein Data Bank under accession code 3K1Q.
Supplementary Material
Acknowledgments
This research was supported in part by grants from the National Institutes of Health (GM071940 and AI069015 to Z.H.Z. and GM30518 to B.H.), the National Science Foundation (NSF HRD-0420407 to Z.H.Z. and MCB-0416708 to B.H.), the National Basic Research Program of China (973 Program, 2009CB118701 to Q.F.), the National Natural Scientific Foundation of China (30671615 and 30871940 to Q.F.) and the Chinese Academy of Sciences (KSCX2-YW-N-021 to Q.F.). We thank Xing Zhang and Lei Jin for discussions, Ivo Atanasov for assistance in cryo-EM and Xiaorui Zhang for assistance in editing and graphics illustration.
Abbreviations used
- GCRV
grass carp reovirus
- dsRNA
double-stranded RNA
- EM
electron microscopy
- MRV
mammalian reovirus
- GTase
guanylyltransferase
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.12.027
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit:
References
- 1.Reinisch KM. The dsRNA viridae and their catalytic capsids. Nat Struct Biol. 2002;9:714–716. doi: 10.1038/nsb1002-714. [DOI] [PubMed] [Google Scholar]
- 2.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Caston JR, Luque D, Trus BL, Rivas G, Alfonso C, Gonzalez JM, et al. Three-dimensional structure and stoichiometry of Helmintosporium victoriae 190S totivirus. Virology. 2006;347:323–332. doi: 10.1016/j.virol.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 4.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 5.Huiskonen JT, De Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Attoui H, Fang Q, Mohd Jaafar F, Cantaloube JF, Biagini P, De Micco P, De Lamballerie X. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of golden shiner reovirus, grass carp reovirus, striped bass reovirus and golden ide reovirus (genus Aqua-reovirus, family Reoviridae) J Gen Virol. 2002;83:1941–1951. doi: 10.1099/0022-1317-83-8-1941. [DOI] [PubMed] [Google Scholar]
- 7.Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH. Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol. 2008;382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 9.Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage–structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Ji Y, Zhang L, Harrison SC, Marinescu DC, Nibert ML, Baker TS. Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 angstrom resolution. Structure. 2005;13:1545–1557. doi: 10.1016/j.str.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dryden KA, Wang GJ, Yeager M, Nibert ML, Coombs KM, Furlong DB, et al. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformations. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 18.Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 19.Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derosier DJ. Correction of high-resolution data for curvature of the Ewald sphere. Ultramicroscopyc. 2000;81:83–98. doi: 10.1016/s0304-3991(99)00120-5. [DOI] [PubMed] [Google Scholar]
- 21.Wan Y, Chiu W, Zhou ZH. Full contrast transfer function correction in 3D cryo-EM reconstruction. 2004. pp. 960–964. [Google Scholar]
- 22.Zhu J, Cheng L, Fang Q, Zhou ZH, Honig B. Building and refining protein models within cryo-electron microscopy density maps based on homology modeling and multi-scale structure refinement. J Mol Biol. 2010;397:835–861. doi: 10.1016/j.jmb.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Tang J, Walker SB, O'Hara D, Nibert ML, Duncan R, Baker TS. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology. 2005;343:25–35. doi: 10.1016/j.virol.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Zhang X, Centonze VE, Bowman VD, Noble S, Baker TS, Nibert ML. The hydrophilic amino-terminal arm of reovirus core shell protein lambda1 is dispensable for particle assembly. J Virol. 2002;76:12211–12222. doi: 10.1128/JVI.76.23.12211-12222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farsetta DL, Chandran K, Nibert ML. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J Biol Chem. 2000;275:39693–39701. doi: 10.1074/jbc.M004562200. [DOI] [PubMed] [Google Scholar]
- 26.Rivas C, Bandin I, Cepeda C, Dopazo CP. Efficacy of chemical disinfectants against turbot aquareovirus. Appl Environ Microbiol. 1994;60:2168–2169. doi: 10.1128/aem.60.6.2168-2169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Q, Seng EK, Ding QQ, Zhang LL. Characterization of infectious particles of grass carp reovirus by treatment with proteases. Arch Virol. 2008;153:675–682. doi: 10.1007/s00705-008-0048-3. [DOI] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen PC, Chiou YF, Liu HJ, Song CH, Su YP, Lee LH. Genetic variation of the lambdaA and lambdaC protein encoding genes of avian reoviruses. Res Vet Sci. 2007;83:394–402. doi: 10.1016/j.rvsc.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Harrison SJ, Farsetta DL, Kim J, Noble S, Broering TJ, Nibert ML. Mammalian reovirus L3 gene sequences and evidence for a distinct amino-terminal region of the lambda1 protein. Virology. 1999;258:54–64. doi: 10.1006/viro.1999.9707. [DOI] [PubMed] [Google Scholar]
- 31.Bisaillon M, Lemay G. Molecular dissection of the reovirus lambda1 protein nucleic acids binding site. Virus Res. 1997;51:231–237. doi: 10.1016/s0168-1702(97)00092-0. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Tao Y, Reinisch KM, Harrison SC, Nibert ML. Orthoreovirus and aquareovirus core proteins: conserved enzymatic surfaces, but not protein–protein interfaces. Virus Res. 2004;101:15–28. doi: 10.1016/j.virusres.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Qiu T, Luongo CL. Identification of two histidines necessary for reovirus mRNA guanylyl-transferase activity. Virology. 2003;316:313–324. doi: 10.1016/j.virol.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 34.Fang Q, Attoui H, Cantaloube JF, Biagini P, Zhu Z, De Micco P, De Lamballerie X. Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (genus Aquareovirus, family Reoviridae) Biochem Biophys Res Commun. 2000;274:762–766. doi: 10.1006/bbrc.2000.3215. [DOI] [PubMed] [Google Scholar]
- 35.Nason EL, Samal SK, Venkataram Prasad BV. Trypsin-induced structural transformation in aquareovirus. J Virol. 2000;74:6546–6555. doi: 10.1128/jvi.74.14.6546-6555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandran K, Zhang X, Olson NH, Walker SB, Chappell JD, Dermody TS, et al. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J Virol. 2001;75:5335–5342. doi: 10.1128/JVI.75.11.5335-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandran K, Walker SB, Chen Y, Contreras CM, Schiff LA, Baker TS, Nibert ML. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins mu1 and sigma3. J Virol. 1999;73:3941–3950. doi: 10.1128/jvi.73.5.3941-3950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dryden KA, Farsetta DL, Wang G, Keegan JM, Fields BN, Baker TS, Nibert ML. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology. 1998;245:33–46. doi: 10.1006/viro.1998.9146. [DOI] [PubMed] [Google Scholar]
- 39.Mendez II, Weiner SG, She YM, Yeager M, Coombs KM. Conformational changes accompany activation of reovirus RNA-dependent RNA transcription. J Struct Biol. 2008;162:277–289. doi: 10.1016/j.jsb.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Chandran K, Nibert ML, Harrison SC. Reovirus mu1 structural rearrangements that mediate membrane penetration. J Virol. 2006;80:12367–12376. doi: 10.1128/JVI.01343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fenouillet E, Barbouche R, Jones IM. Cell entry by enveloped viruses: redox considerations for HIV and SARS-coronavirus. Antioxid Redox Signal. 2007;9:1009–1034. doi: 10.1089/ars.2007.1639. [DOI] [PubMed] [Google Scholar]
- 42.Fang Q, Shah S, Liang Y, Zhou ZH. 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China, Ser C: Life Sci. 2005;48:593–600. doi: 10.1360/062004-105. [DOI] [PubMed] [Google Scholar]
- 43.Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 44.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semi-automated software for high resolution single particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 45.Zhou ZH, Hardt S, Wang B, Sherman MB, Jakana J, Chiu W. CTF determination of images of ice-embedded single particles using a graphics interface. J Struct Biol. 1996;116:216–222. doi: 10.1006/jsbi.1996.0033. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Skehel JJ, Joklik WK. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology. 1969;39:822–831. doi: 10.1016/0042-6822(69)90019-1. [DOI] [PubMed] [Google Scholar]
- 48.Drayna D, Fields BN. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin P, Cheang M, Coombs KM. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J Virol. 1996;70:1223–1227. doi: 10.1128/jvi.70.2.1223-1227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gillies S, Bullivant S, Bellamy AR. Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science. 1971;174:694–696. doi: 10.1126/science.174.4010.694. [DOI] [PubMed] [Google Scholar]
- 51.Bartlett N, Gillies S, Bullivant S, Bellamy A. Electron microscopy study of reovirus reaction cores. J Virol. 1974;14:315–326. doi: 10.1128/jvi.14.2.315-326.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.