Abstract
Background
Physical maps employing libraries of bacterial artificial chromosome (BAC) clones are essential for comparative genomics and sequencing of large and repetitive genomes such as those of the hexaploid bread wheat. The diploid ancestor of the D-genome of hexaploid wheat (Triticum aestivum), Aegilops tauschii, is used as a resource for wheat genomics. The barley diploid genome also provides a good model for the Triticeae and T. aestivum since it is only slightly larger than the ancestor wheat D genome. Gene co-linearity between the grasses can be exploited by extrapolating from rice and Brachypodium distachyon to Ae. tauschii or barley, and then to wheat.
Results
We report the use of Ae. tauschii for the construction of the physical map of a large distal region of chromosome arm 3DS. A physical map of 25.4 Mb was constructed by anchoring BAC clones of Ae. tauschii with 85 EST on the Ae. tauschii and barley genetic maps. The 24 contigs were aligned to the rice and B. distachyon genomic sequences and a high density SNP genetic map of barley. As expected, the mapped region is highly collinear to the orthologous chromosome 1 in rice, chromosome 2 in B. distachyon and chromosome 3H in barley. However, the chromosome scale of the comparative maps presented provides new insights into grass genome organization. The disruptions of the Ae. tauschii-rice and Ae. tauschii-Brachypodium syntenies were identical. We observed chromosomal rearrangements between Ae. tauschii and barley. The comparison of Ae. tauschii physical and genetic maps showed that the recombination rate across the region dropped from 2.19 cM/Mb in the distal region to 0.09 cM/Mb in the proximal region. The size of the gaps between contigs was evaluated by comparing the recombination rate along the map with the local recombination rates calculated on single contigs.
Conclusions
The physical map reported here is the first physical map using fingerprinting of a complete Triticeae genome. This study demonstrates that global fingerprinting of the large plant genomes is a viable strategy for generating physical maps. Physical maps allow the description of the co-linearity between wheat and grass genomes and provide a powerful tool for positional cloning of new genes.
Background
Although wheat is a major food for the world population and the most extensively grown crop, progress in genomics had been slowed due to the size and the complexity of the genome. The hexaploid genome of common wheat (Triticum aestivum) contains 16,000 Mb of DNA organized into three genomes, A, B and D, with 7 chromosomes each. This makes the wheat genome far larger than the sequenced rice genome at 430 Mb [1] and Brachypodium genome at 271 Mb [2]. T. aestivum evolved via two hybridization events. The first event combined the A and B genomes into tetraploid wheat while the second hybridization event, which took place only about 8,500 years ago [3] and involved tetraploid wheat and diploid Ae. tauschii [4], contributed the D genome. Due to the recent origin of T. aestivum and limited gene-flow from Ae. tauschii [5], the D genome shows low polymorphism, and the genetic maps of the D genome chromosomes still tend to be poor.
Physical mapping strategies employing libraries of bacterial artificial chromosome (BAC) clones can potentially generate maps of a genome without extensive polymorphism and facilitate studying the structure of the genome, comparison with other genomes and map-based cloning of genes. Physical maps are also the prerequisite step for the sequencing and assembly of large and repetitive genomes such as those of wheat. Most physical mapping in wheat has generally targeted small regions covered by a few BAC clones. Two recent studies presenting physical map of the wheat chromosome 3B [6] and the short arm of chromosome 3AS and 3DS [7] exemplify a strategy adopted by the International Wheat Genome Sequencing Consortium http://www.wheatgenome.org/ for physical mapping of the 21 wheat chromosomes. The strategy makes use of BAC libraries constructed from DNA of single chromosomes or single chromosome arms isolated by chromosome or chromosome arm flow sorting [8]. A complementary approach is to construct physical maps of wheat diploid relatives and use the maps for the construction of the physical maps of the wheat chromosomes. Of the three diploid ancestors of T. aestivum, this strategy is the most applicable to Ae. tauschii because of high homology between its chromosomes and those of the wheat D genome. To further enhance the utility of Ae. tauschii physical map for using it as a resource for wheat D-genome genomics, an accession from the presumptive area of the origin of T. aestivum [5] showing a short genetic distance to the wheat D genome was used for the construction of Ae. tauschii BAC libraries [9] and BAC contigs [10]. This accession has also been used recently for the construction of extensive comparative map of Ae. tauschii based on ESTs (expressed-sequence-tags) with the rice and sorghum genome sequences [11].
The barley diploid genome is only slightly larger than the wheat D genome and, like Ae. tauschii, genetic maps are well populated with molecular markers making it another good model for the Triticeae and T. aestivum. Barley and Ae. tauschii comparative mapping have shown that the grass genomes are highly collinear and gene order is conserved across chromosomes [11,12]. Gene co-linearity can be exploited by extrapolating the knowledge obtained on simpler model species to more complex species: from rice or sorghum to Ae. tauschii or barley, and then from them to wheat.
Here we report the construction of the physical map of a large portion of Ae. tauschii chromosome arm 3DS delineated by an X-ray induced deletion mutation called pairing homoeologous 2a (ph2a) and initially located on the distal 20 cM of the arm [13]. We describe here the assembly of Ae. tauschii BACs into contigs and their ordering based on the Ae. tauschii comparative genetic map [11]. By superimposing the physical map of the 3DS region on the Ae. tauschii recombination map, we estimate recombination rates along the chromosome arm and the gaps in the physical map. We also describe its synteny with Brachypodium and rice genomic sequences and the barley genetic map.
Results
Physical map of a distal region of chromosome 3DS in Ae. tauschii
Using the southern blot data from a previous study [13], we first identified the wheat deletion bins [14] overlapping the ph2a deletion. Among 53 ESTs mapped into the region deleted in ph2a, 8 EST mapped to 6 deletion bins of group 3 chromosomes: the telomeric bin 3AS4-0.45-1.00; the 3 bins 3BS8, 3BS9 and 3BS1 covering the chromosome arm from 0.33 to 1.00 (ratio of chromosome arm length); the bins 3DS6 and 3DS3 from position 0.24 to 1.00 [14]. The GrainGenes database http://wheat.pw.usda.gov contained a total of 591 ESTs that had been mapped to these 6 bins. These ESTs including 53 EST mapped within the ph2a deletion were assembled into 151 unigenes according to the DFCI wheat gene index database http://compbio.dfci.harvard.edu/tgi and used for BAC anchoring. To decrease the chance of hybridizing repeat elements, probes used for the BAC library screening contained only exon sequence. PCR products for 110 unigenes of wheat were verified by direct sequencing and used as markers. Eighty probes gave clean results by hybridization onto restricted DNA of nullisomic-tetrasomic wheat lines and were then used for hybridization to the Ae. tauschii BAC high density screening membranes.
Using 68 such probes identified 278 positive BAC clones. These clones had been previously assembled based on fingerprints into contigs containing 3,289 BACs http://wheatdb.ucdavis.edu/. We screened the 3,289 clones with 42 additional PCR markers which could not be used for hybridization because of the poor probe quality. After validation of marker presence on clones by PCR, 1,802 BAC clones were re-assembled into 24 contigs with a size that ranged from 188 to 3,935 kb (Table 1). The physical map covered 25.4 Mb which is about one third of the target region initially estimated to be about 80 Mb by comparison with rice genomic sequence [13]. The physical map included a total of 90 loci, composed of 85 unigenes and 5 genomic based markers (See Additional File 1). A contig contained an average of 3.7 markers (ranging from 1 to 17 markers). The genes were organized in 54 gene islands defined by the presence of at least 1 gene on one BAC. Among those, 19 different gene islands were identified with at least 2 genes and up to 5 genes on a single clone.
Table 1.
Characteristics of the Ae. tauschii physical map of the distal region of chromosome 3DS
| Total | Average/contig | Minimum | Maximum | |
|---|---|---|---|---|
| Contigs | 24 | |||
| Clones | 1802 | 76 | 8 | 322 |
| Loci | 90 | 3.7 | 1 | 17 |
| CB* | 13,355 | 566.4 | 99 | 2,071 |
| kb | 25,374 | 1,057 | 188 | 3,935 |
* consensus band
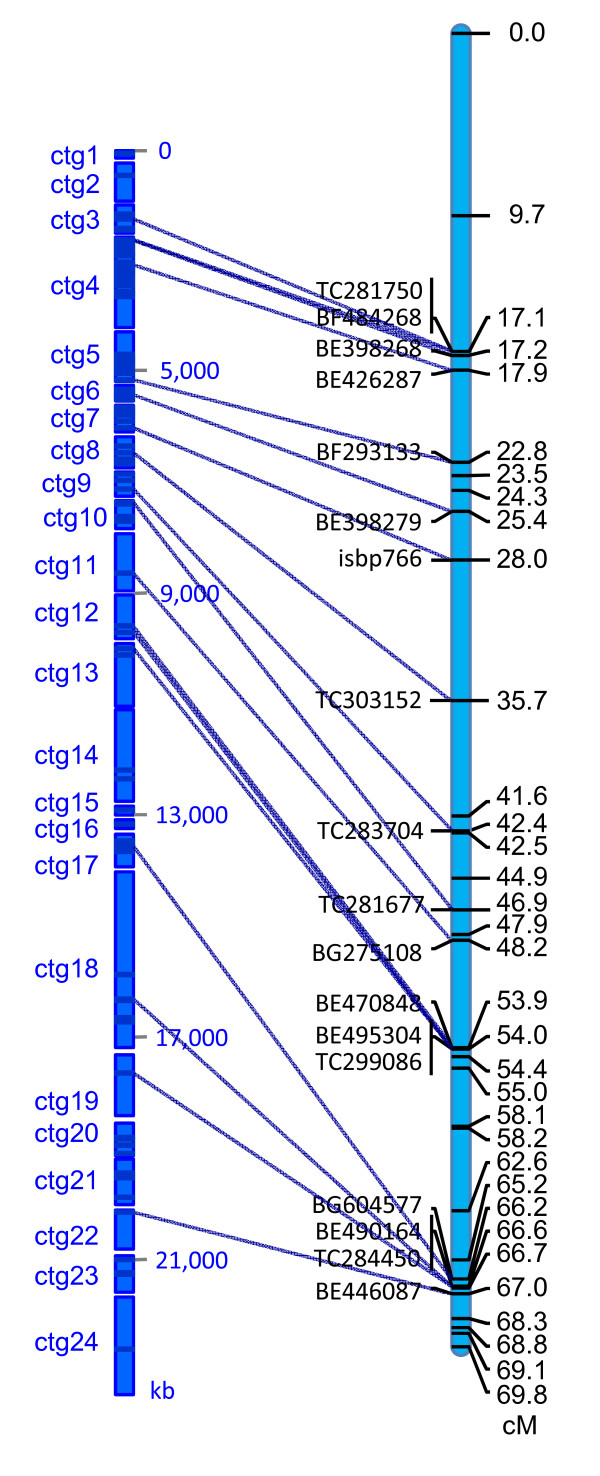
Due to the low polymorphism of the T. aestivum D genome, the wheat genetic maps are poorly populated with molecular markers and not informative enough for anchoring BAC contigs on a map. Therefore, whenever possible we have anchored the BAC contigs onto the Ae. tauschii genetic map built from AL8/78 × AS75 F2 mapping population [11]. The fact that one of the parents (AL8/78) was also used for the construction of the BAC libraries greatly assisted the anchoring of contigs on the genetic map. A contig was anchored only when a clone in the contig and a marker on the map contributed by AL8/78 had exactly the same phenotype. Eleven EST markers and one ISBP marker were used to align 14 contigs onto the Ae. tauschii genetic map (Figure 1 and Additional File 2). The remaining contigs were ordered as follows. Contigs (ctg) 1, 2, 21, 23 and 24 were ordered according to the 3B physical map [6]. Ctg 13, 14 and 20 were anchored and positioned in the order of the barley genetic map. Ctg 15 and 16 could not be anchored on a Triticeae map and were ordered according to the order of orthologous genes in the rice genome.
Figure 1.
Ae. tauschii physical and genetic maps of chromosome 3DS. Alignment of Ae. tauschii physical map onto Ae. tauschii genetic map of chromosome 3DS. The Ae. tauschii AL8/78 × AS75 F2 genetic map used EST based markers [11].
Recombination along the distal region of chromosome 3DS
Meiotic recombination activity was evaluated by calculating the recombination rate or coefficient of exchange (CE) in cM per Mb. Although the actual length of a contig can not be known without BAC sequencing, the physical size of a contig was evaluated based on the number of common bands (CB units) between BAC fingerprints. The average size of 1 CB was 1.9 kb for this Ae. tauschii library http://wheatdb.ucdavis.edu. The average CE along the map was 2.35 cM/Mb between the most distant markers TC281750 of ctg3 and BE446087 of ctg22 (49.9 cM and 11,187 CB) (Table 2). Local CE was also measured for two pairs of markers physically linked on the ctg4 and ctg12: respectively 2.19 and 1.42 cM/Mb. In the proximal region, we calculated the physical distance between the markers BG604577 and BE446087 by adding the size of ctg17, 18, 19, 20, 21 and 22 neglecting the gaps between them. With a genetic distance of 0.78 cM, the CE was 0.09 cM/Mb. The recombination rate decreased in Ae tauschii from the distal region of ctg4, through the middle region of ctg12 to the proximal ctg17-22.
Table 2.
Coefficient of exchange (CE) along the distal region of chromosome 3DS in Ae. tauschii
Synteny with B. distachyon, rice and barley
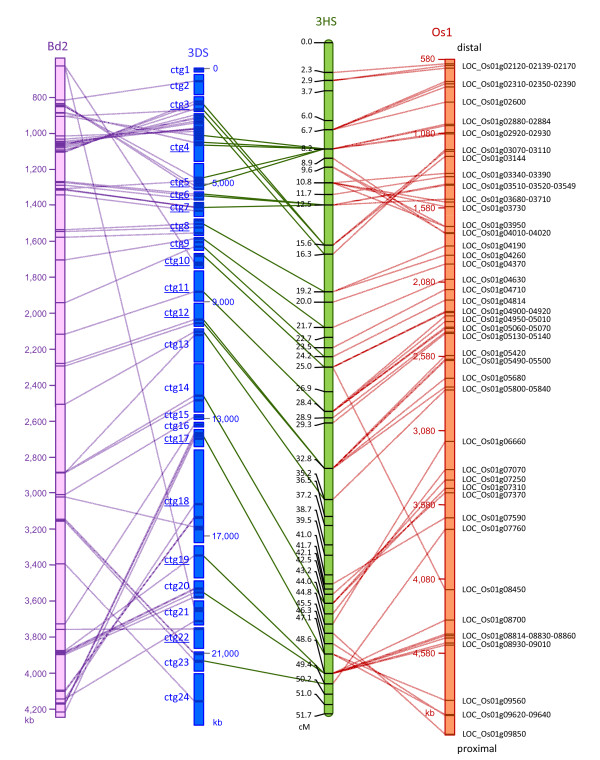
By comparing the Ae. tauschii physical map of 3DS to the B. distachyon 8× genome sequence, 53 wheat unigenes were orthologous to B. distachyon chromosome Bd2. A distal inversion occurred in the region between 834 and 1,105 kb of Bd2 matching the ctgs 3, 4 and 5 of Ae. tauschii (Figure 2). Separated by a large conserved block of about 2,035 kb in Brachypodium, a second large inversion has occurred in the proximal region between the ctgs 17 to 24 of Ae. tauschii and Bd2-3,140 and Bd2-4,211 kb. Compared to the rice genomic sequence, the Ae. tauschii map is highly collinear to chromosome 1 with 70 wheat unigenes of the Ph2 physical map orthologous to 59 rice genes of Os1 (See Additional File 1). They showed the same chromosomal rearrangements between 3DS and Os1 that have been previously described by using the same Ae. tauschii genetic map [11]. The synteny profile of Ae. tauschii-B. distachyon is almost identical to the one of Ae. tauschii-rice, showing the same inverted and conserved blocks (See Additional File 2).
Figure 2.
Comparative maps of Ae. tauschii 3DS with rice, Brachypodium and barley genome. Physical map of the distal region of Ae. tauschii 3DS compared to the orthologous genomic sequences of B. distachyon Bd2 (8× release) and rice Os2, and to the genetic map of barley 3HS. The underlined ctg names indicate the contigs which were aligned onto the Ae. tauschii AL8/78 × AS75 F2 genetic map [11]. The names to the right are rice genes as listed in MSU Rice Genome Annotation (Osa1) Release 6.0. The barley Steptoe x Morex genetic map used gene-based SNP [15,16].
A comparison was made with the genetic map of barley 3HS derived from the Steptoe x Morex doubled haploid population populated with gene-based SNP [15] and Transcript Derived Markers [16] loci. A total of 24 unigenes homologous between wheat and barley were used to integrate the Ae. tauschii physical map and the barley genetic map. Sixteen contigs were therefore anchored onto 15 barley genetic map positions (Figure 2). Synteny was conserved on a large block between the two species, from ctg 8 to 24. A region of 8.1 cM in length in barley has been rearranged in Ae. tauschii between ctgs 3, 4 and 5. This observation is supported by the anchoring of 3 markers of ctg4, which was oriented and positioned onto the Ae. tauschii genetic map. This rearrangement overlapped the distal rice-Ae. tauschii inversion previously described. It could either be due to an inversion between Ae. tauschii and barley leading to the translocation of ctg6 and 7 in a more distal region in barley, or to the proximal translocation of ctg 3-distal part of ctg4 in barley.
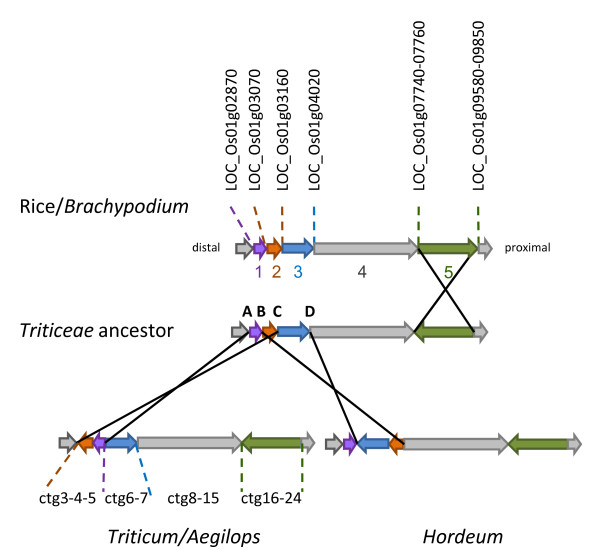
The disruption of co-linearity between genomes is due to chromosomal rearrangements which occurred after the divergence of the two species from their common ancestor [17]. With the hypothesis of minimum chromosomal rearrangements occurring before speciation, we propose a model explaining the discontinuities in rice, B. distachyon, barley and Ae. tauschii synteny along the distal region of chromosome 3DS (Figure 3). The segment 5 (corresponding to ctg 17-24) was conserved between Ae. tauschii and barley while it appeared inverted between Ae. tauschii and rice/B. distachyon, and between barley and rice/B. distachyon (Figure 2). This proximal inversion probably took place in the Triticeae ancestor after its divergence from a common ancestor of Oryzae and Brachypodium lineages.
Figure 3.
Model of chromosomal rearrangements of the Ph2 locus in rice, Brachypodium, Ae. tauschii and barley. 1-5 indicate chromosomal segments, A-D, the putative breakpoints. The breakpoint B would be located within the ctg5 of Ae. tauschii physical map. The names on top are rice genes as listed in MSU Rice Genome Annotation (Osa1) Release 6.0.
Additional rearrangements seemed to have occurred in the distal region after the divergence of the Triticum/Aegilops lineage from the Hordeum lineage. The ctg3-4-5 of the map showed inversions between Ae. tauschii and rice (LOC_Os01g02870-LOC_Os01g03160) (See Additional File 2). It partially overlapped the inversion observed between the barley region 8.9 cM-16.3 cM and the rice sequence LOC_Os01g03070-LOC_Os01g04020 (Figure 2). Thus, the segment 3 (ctg6-7) was collinear between rice/Brachypodium and Ae. tauschii while it was inside the inverted block between rice and barley. A translocation of the ctg3-distal ctg4 segment in a more proximal region in barley wouldn't explain the difference of synteny of the segment 3 between Ae. tauschii, barley and rice. A possible explanation would be some overlapping inversions with different chromosomal breakpoints in the Triticum/Aegilops and the Hordeum radiations (Figure 3). The block formed by segments 1 and 2 (corresponding to ctg3-4-5) would be inverted in the Triticum/Aegilops radiation, with the breakpoints A and C, while in Hordeum, the breakpoints may have occurred in B (within ctg4) and D leading to the inversion of a block formed by segments 2 and 3 (corresponding to a part of ctg4 and ctg5-6-7-8).
Discussion
Beside the Ph2 locus, which is a suppressor of homoeologous chromosome pairing [13], other genes of interest have been previously mapped on chromosome 3DS including a resistance to tan spot [18], the brittle rachis gene [19], the Gigantea gene [20] and the developmental gene Sphaerococcoid S1 [21]. Some QTLs associated to yield components [22-24], spike length [25], black point resistance [26], resistance to Fusarium head blight Fhb [27], and grain protein content [28] have also been identified on 3DS. In addition, genes coding for several metabolic enzymes are located on 3DS: esterase 1, hexokinase-1,2, malic enzyme, peroxydase and triosphosphate isomerase [29]. Although a total of 57 molecular markers were found on chromosome 3D of the consensus genetic map of hexaploid wheat [30], the linkage group 3D usually shows only 10 to 20 markers for a specific segregating population [19,22,23,25]. This low density of markers makes the isolation of specific genes on the 770 Mb of chromosome 3D almost impossible. The difficulty could be overcome by using physical and genetic maps of Ae. tauschii [31].
We have used the Ae. tauschii genomic resources and a specific deletion bin (ph2a) to make the first physical map of part of a large plant genome based on global fingerprinting of the whole genome. The gaps in the map were evaluated by comparing the average CE along the region with the CE previously reported in wheat. If the average recombination is similar between wheat chromosomes, the difference observed would be due to a difference in physical distance. The average CE of the map was 2.35 cM/Mb, which is much higher than the maximum CE observed for all wheat chromosomes (0.87 cM/Mb) and for wheat chromosome 3BS (0.85 cM/Mb) [32,33]. The rate is about triple of previous reports, meaning that the actual physical distance would be 73 Mb, which is close to the estimated size of the ph2a deletion (80 Mb).
Comparison of the size of rice and Ae. tauschii short arm chromosomes reveals a genomic inference between the two species. The Ph2 map spanned 4.3 Mb of the distal region of rice short arm chromosome 1, from LOC_Os01g02090 to LOC_Os01g09580 [13]. The rice Os1S and wheat 3DS arms are respectively 17.1 Mb and 321 Mb long [6,34]. The increase of genome size between rice and Ae. tauschii would be 17× for the distal half (4.3 Mb in rice, 73 Mb in Ae. tauschii) and 19× for the proximal half of chromosome (12.8 Mb in rice, 248 Mb in Ae. tauschii). When rice genome was compared to sorghum genome, the genome size increase was entirely accounted for by the increase in the proximal heterochromatic regions [35]; the distal euchromatic regions were of identical lengths in the two species. The increase in genome size in the Triticeae is nearly equivalent in the distal and proximal regions, suggesting that the growth of Triticeae genomes took place along the entire chromosomes, in contrast to small sorghum genome where the growth took place only in the proximal region.
In plants, the recombination rate is higher in gene-dense regions and towards the telomeres and lower in centromeric regions [36], although reversals of this pattern exist. In wheat and barley, meiotic recombination rate follows a gradient along the chromosome arms decreasing from the telomere to the centromere [32,37]. The recombination rates along the 3DS region in Ae. tauschii dropped from 2.19 cM/Mb in the distal region to 0.09 cM/Mb in the proximal region. The CE in the distal and middle regions (2.19 and 1.42 cM/Mb) were calculated to be much higher for single contigs than the rate in the distal 3BS8 and 3BS7 bins (respectively 0.85 and 0.37 cM/Mb) [33]. In the proximal region, although the CE is overestimated due to the gaps between the contigs, the value is lower (0.09 cM/Mb) than the one of the orthologous bin 3BS9 (0.17 cM/Mb) [33]. Considering that the ctgs 17-22 were anchored on 3DS6-0.55-1.00 deletion bin (See Additional File 1), it means that recombination is almost null in half of the chromosome arm. While the recombination followed a gradient from telomere to centromere in barley, the Ae. tauschii genetic map showed alternated low and high- recombination regions along the short arm chromosome 3S, showing that the pattern of recombination might be chromosome specific.
Comparative studies between wheat, barley and model genomes such as rice and Brachypodium are helpful in understanding the structure of grass genomes and developing strategies to overcome linkage drag, which limits the introgression of wild genes in elite varieties [17]. As shown in previous studies, wheat chromosome 3 is largely collinear to rice Os1 [29,38] and to B. distachyon Bd2 [39]. The mid-scale comparative maps presented here revealed chromosomal rearrangements between Brachypodium and wheat that haven't been revealed in whole genome study [39]. Although the rice and Brachypodium genomes diverged 40-53 million years ago (Mya) and were largely rearranged from their common ancestor [39], the rice- and Brachypodium-Ae. tauschii co-linearity were similar with disruption in the same distal and proximal regions of Ph2 and 2 major inversions (Figure 3). Both inversions differentiating the 3DS physical map and the Os1 sequence have been detected in the comparison of the Ae. tauschii genetic map and rice and sorghum genome sequences and their origins were assigned to the Triticeae lineage [11], in agreement with inferences reported here. Additional Ae. tauschii-rice/Brachypodium rearrangements probably occurred but could not be detected in our map probably because of the low density markers on some contigs. For example, two inversions between rice chromosome 1 and 3BS reported in [6] were not observed in the Ae. tauschii 3DS map due to the lack of markers on ctgs1-2-3.
The diploid genome of barley is usually considered as a good model for Triticeae genomics. Comparative studies using deletion bin, cytomolecular and low-resolution linkage maps showed a high level of synteny between wheat and barley chromosomes and revealed very few genomic rearrangements at the genome scale [40-43]. For example, the comparison of barley and T. monococcum revealed inversions and translocation between chromosomes 1H and 1A, and between chromosomes 5A and 4H [40]. As expected, the Ae. tauschii chromosome 3DS is largely collinear to the barley chromosome 3HS. However we reported here a disruption of the synteny for a distal region of 8.1 cM in barley, which has not been observed in previous studies.
The phylogeny of grasses suggests that the divergence of rice and Brachypodium from Triticeae occurred about 41-47 Mya and 32-39 Mya, respectively [39,44-46]. The structure of the orthologous Ph2 locus was almost identical in Brachypodium and rice. Although it implies that rice and Brachypodium are closer to each other than to wheat, DNA sequence would indicate rice and Brachypodium/Triticeae lineage split much earlier, with Brachypodium and the Triticeae splitting later as shown in [39]. The large proximal inversion of the segment 5 would have occurred after rice and Brachypodium diverged from a common ancestor of the Triticeae, leading to a collinear block between barley and Ae. tauschii (Figure 3).
The complexity of the rearrangements inside segment 5 between rice/Brachypodium and Ae. tauschii showed that the large inversion might actually be a series of inversions nested in each other. It might correspond to larger rearrangements between the adjacent bins 3S-0.57-0.78 and 3S-0.45-0.55 and rice chromosome 1 observed in the EST group 3 chromosome bin map [29]. Other inversion events have occurred in the distal region of Ph2 leading to different rearrangements of the rice/Brachypodium-barley-Ae. tauschii synteny. The disruption of co-linearity between wheat and barley showed that inversions would have occurred after the separation between Hordeum and Triticum/Aegilops species about 6-16 Mya [45]. However the inversions of segments 1 and 3 between barley and Ae. tauschii would need to be validated by increasing the resolution of the barley genetic map where the markers anchored on ctg 4 and 5, and ctg 6 and 7 co-segregated respectively at 8.2 cM and 12.5 cM.
Conclusions
The construction of physical maps for a 5 Gb size genome is a complex and laborious task. Although the map reported here is only for a portion of the wheat genome, it is nevertheless the first report using fingerprinting of a complete Triticeae genome. It provided significant data showing chromosomal rearrangements between wheat, Brachypodium and barley, and it will be a useful resource for gene cloning of chromosome 3D. Whether or not the composition of Triticeae genomes with 90% of repeat DNA sequence [47] precludes the assembly of BAC contigs from whole-genome BAC libraries, this study showed that global fingerprinting of the large Triticeae genomes is capable of generating such physical maps despite the complexity of and high repeated sequence content in these genomes. This strategy is now used by the International Barley Sequencing Consortium which will soon release a physical map of barley [48].
Methods
Probe design and BAC filter hybridization
Plant material (Triticum aestivum cv Chinese Spring and nullisomic-tetrasomic derivatives) and the DNA extraction were respectively described in [13] and [49]. The BAC libraries and assembly of Ae. tauschii AL8/78 are described in [10] and http://wheatdb.ucdavis.edu.
The primers were designed to amplify unique 150-400 bp fragments of unigenes defined in the DFCI wheat gene index v11 http://compbio.dfci.harvard.edu/tgi by using Primer 3 program (See Additional File 3). The PCR reaction was carried out in 20 μl final volume with 400 nM of both primers, 200 μM of each dNTP, 1.5 mM of MgCl2, 0.5 U Taq Platinum polymerase (Invitrogen) with its 1× buffer and 40 ng of DNA. The PCR amplification followed a touch-down procedure: 1 cycle 2 min at 94°C, 10 cycles (20 sec 95°C, 20 sec Tm minus 0.5°C each cycle, 2 min 72°C), 35 cycles (20 sec 95°C, 20 sec Tm minus 5°C, 2 min 72°C) and an additional extension step of 10 min at 72°C (See Additional File 3). Probes were purified by cleaning up the PCR product or by electrophoresis on agarose gel using the MinElute kit (Qiagen) and verified by Sanger's method sequencing prior hybridization. We used the ABI PRISM® BigDye™ Terminators V 3.1 for the sequencing reaction according to the manufacturer's instructions. The southern blot analyses onto membranes of restricted DNA from nullisomic-tetrasomic lines and onto Ae. tauschii BAC filters were performed according to [49] by pooling up to 8 probes. The positive BAC clones were cultivated in 96-well plates overnight in 200 μl of LB medium supplemented with 15 mg/l tetracycline for the BB and HB clones and with 15 mg/l chloramphenicol for HD, HI and RI clones. The cultures were centrifuged for 10 min at 5,000 rpm, and then the supernatant was replaced by MilliQ water. The anchoring of the markers on the positive BAC clones was validated by direct PCR with 1 μl of bacterial solution.
Re- assembly of the BAC from targeted region
To minimize false assembly caused by genome-wide network, a subset of BAC clones including all clones of original contigs which contain positive clone(s) and singletons were re-assembled into contigs by using the FPC program [50,51]. The contig number(s) used were results of the re-assembly.
Genetic mapping and recombination rate
For map alignments, wheat unigenes and markers sequences were compared to the 8× B. distachyon genome sequence http://www.brachypodium.org and to the rice pseudomolecules of Rice Genome Annotation Project - MSU Rice Genome Annotation (Osa1) Release 6.0 using BLASTN and TBLASTX. The homologues were assigned based on the best match to the rice pseudomolecules with significant threshold for the expectation E value of e-10. The same wheat sequences were used to identify the barley homologous unigenes of HarvEST:Barley (v1.76) assembly #35 by using BLASTN. The best matches were selected with a cut-off E value of e-20. The Ae. tauschii genetic map derived from the F2 population AL8/78 × AS75 [11] with additional markers described in Table S3. The genetic map of barley derived from the Steptoe x Morex doubled haploid population [52] was described in [15] and [16]. Recombination rate was calculated as a coefficient of exchange (CE) in cM per Mb.
Authors' contributions
DF participated to the project design, designed the primers, assembled the BAC contigs based on EST anchoring, did the sequence alignment and the comparative maps, and drafted the manuscript. MCL designed the Ae. tauschii physical mapping research project and assembled the contigs based on fingerprinting. JD designed the Ae. tauschii physical mapping research project, analyzed the comparative maps and helped to draft the manuscript. LR analyzed the comparative maps and participated to the sequence alignment. BSG and ODA designed the Ae. tauschii physical mapping research project. FMY performed the fingerprint editing of the BAC libraries of Ae. tauschii and help to assemble contigs. ZS designed a subset of primers and performed the wet-lab work of BAC screening. KRD did the Ae. tauschii genetic mapping. PL designed and coordinated the research, analyzed the comparative maps and help to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Description of the markers mapped onto Ae. tauschii physical map of distal region of chromosome 3DS.
Primer sequences and Tm used for the touch-down PCR of the wheat markers.
Physical map of the Ph2 locus in Ae. tauschii compared to the orthologous genomic sequence of rice Os1 and B. distachyon Bd2 (8× release). The names to the left are rice genes as listed in MSU Rice Genome Annotation (Osa1) Release 6.0.
Contributor Information
Delphine Fleury, Email: delphine.fleury@acpfg.com.au.
Ming-Cheng Luo, Email: mcluo@plantsciences.ucdavis.edu.
Jan Dvorak, Email: jdvorak@ucdavis.edu.
Luke Ramsay, Email: luke.ramsay@scri.ac.uk.
Bikram S Gill, Email: bsg@ksu.edu.
Olin D Anderson, Email: Olin.Anderson@ARS.USDA.GOV.
Frank M You, Email: Frank.You@ARS.USDA.GOV.
Zahra Shoaei, Email: zahra.shoaei@acpfg.com.au.
Karin R Deal, Email: krdeal@ucdavis.edu.
Peter Langridge, Email: peter.langridge@acpfg.com.au.
Acknowledgements
This work is supported by the University of Adelaide, the Australian Research Council, the Grains Research and Development Corporation, the Government of South Australia, the International Science Linkages CG120174 and the US National Science Foundation by grant DBI-0077766. We thank Margaret Pallotta for supplying DNA of wheat nullisomic-tetrasomic lines and Southern blot membranes, Etienne Paux, Frédéric Choulet and Catherine Feuillet for the marker information of the chromosome 3B physical map, Alina Akhunova for providing the Ae. tauschii BAC clones, Ute Baumann and Julian Schwerdt for bioinformatics support.
References
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Gu Y, Ma Y, Huo N, Vogel J, Lazo G, Hill T, Coleman-Derr D, Hayden D, Dvorak J, Olin Anderson. et al. Construction of physical map for Brachypodium distachyon and its comparative analysis with rice. BMC Genomics. 2009;10:496–509. doi: 10.1186/1471-2164-10-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt M, Samuel D. In: 1st International Workshop on Hulled wheats. Padulosi S, Hammer K, Heller J, editor. Rome, Italy: International Plant Genetic Resources Institute; 1996. From staple crop to extinction? The archaeology and history of hulled wheats. Hulled Wheats. Promoting the conservation and use of underutilized and neglected crops; pp. 41–100. [Google Scholar]
- Kihara H. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric and Hort Tokyo. 1944;19:13–14. [Google Scholar]
- Dvorak J, Luo M, Yang Z, Zhang H. In: The Origins of Agriculture and Crop Domestication. Damania A, Valkoun J, Willcox G, Qualset C, editor. Aleppo, Syria: ICARDA; 1998. Genetic evidence on the origin of T. aestivum L; pp. 235–251. [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W. et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322:101–104. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- Luo MC, Ma Y, You FM, Anderson OD, Kopecky D, Simkova H, Safar J, Dolezel J, Gill BS, McGuire PE. et al. Feasibility of physical map construction from fingerprinted bacterial artificial chromosome libraries of polyploid plant species. BMC Genomics. 2010;11:122. doi: 10.1186/1471-2164-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Kubalakova M, Paux E, Bartos J, Feuillet C. Chromosome-based genomics in the cereals. Chromosome Res. 2007;15:51–66. doi: 10.1007/s10577-006-1106-x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Deal KR, Li W, Covaleda L, Chang Y-L, Dvorak J, Luo M-C, Gill BS, Anderson OD, Zhang HB. 10th Intern Plant and Animal Genome Conference: 2002. San Diego, U.S.A.: Scherago Internatl. Inc; 2002. Construction and characterization of five large-insert BAC and BIBAC libraries of Aegilops tauschii, the diploid donor of the wheat D genome; p. 101. [Google Scholar]
- Luo MC, Thomas CS, Deal KR, You FM, Anderson OD, Gu Y-Q, Li W, Kuraparthy V, Gill BS, McGuire PE, Proceedings of the 10th International Wheat Genetic Symposium: 2003. Rome, Italy: Institute Sperimentale per la Cerealicoltura; 2003. Construction of contigs of Aegilops tauschii genomic DNA fragments cloned in BAC and BiBAC vectors; pp. 293–296. [Google Scholar]
- Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ. et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106:15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, Kota R, Varshney RK, Perovic D, Grosse I. et al. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor Appl Genet. 2007;114:823–839. doi: 10.1007/s00122-006-0480-2. [DOI] [PubMed] [Google Scholar]
- Sutton T, Whitford R, Baumann U, Dong C, Able JA, Langridge P. The Ph2 pairing homoeologous locus of wheat (Triticum aestivum): identification of candidate meiotic genes using a comparative genetics approach. Plant J. 2003;36:443–456. doi: 10.1046/j.1365-313X.2003.01891.x. [DOI] [PubMed] [Google Scholar]
- Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorak J, Linkiewicz AM, Ratnasiri A. et al. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics. 2004;168(2):701–712. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Close T, Bhat P, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson J. et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-582. 1186/1471-2164-1110-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potokina E, Druka A, Luo Z, Wise R, Waugh R, Kearsey M. eQTL analysis of 16,000 barley genes reveals a complex pattern of genome wide transcriptional regulation. Plant J. 2008;53:90–101. doi: 10.1111/j.1365-313X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Feuillet C, Langridge P, Waugh R. Cereal breeding takes a walk on the wild side. Trends Genet. 2008;24:24–32. doi: 10.1016/j.tig.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Tadesse W, Schmolke M, Hsam SLK, Mohler V, Wenzel G, Zeller FJ. Molecular mapping of resistance genes to tan spot (Pyrenophora tritici-repentis race 1) in synthetic wheat lines. Theor Appl Genet. 2007;114:855–862. doi: 10.1007/s00122-006-0484-y. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Fujii Y, Kato N, Ban T, Martinek P. Microsatellite mapping of the genes for brittle rachis on homoeologous group 3 chromosomes in tetraploid and hexaploid wheats. J Appl Genet. 2006;47(2):93–98. doi: 10.1007/BF03194606. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Liu MS, Li JR, Guan CM, Zhang XS. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol Biol. 2005;58:53–64. doi: 10.1007/s11103-005-4162-2. [DOI] [PubMed] [Google Scholar]
- Salina E, Borner A, Leonova I, Korzun V, Laikova L, Maystrenko O, Roder MS. Microsatellite mapping of the induced sphaerococcoid mutation genes in Triticum aestivum. Theor Appl Genet. 2000;100(5):686–689. doi: 10.1007/s001220051340. [DOI] [Google Scholar]
- McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, Cloutier S, McCallum BD. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × 'AC Domain'. Genome. 2005;48:870–883. doi: 10.1139/g05-055. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusic D, Waterman E, Weyen J. et al. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring X SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet. 2005;110:865–880. doi: 10.1007/s00122-004-1902-7. [DOI] [PubMed] [Google Scholar]
- Pestsova EG, Borner A, Roder MS. Development and QTL assessment of Triticum aestivum-Aegilops tauschii introgression lines. Theor Appl Genet. 2006;112:634–647. doi: 10.1007/s00122-005-0166-1. [DOI] [PubMed] [Google Scholar]
- Chu CG, Xu SS, Friesen TL, Faris JD. Whole genome mapping in a wheat doubled haploid population using SSRs and TRAPs and the identification of QTL for agronomic traits. Mol Breed. 2008;22:251–266. doi: 10.1007/s11032-008-9171-9. [DOI] [Google Scholar]
- Christopher MJ, Williamson PM, Michalowitz M, Jennings R, Lehmensiek A, Sheppard J, Banks P. Simple sequence repeat markers associated with three quantitative trait loci for black point resistance can be used to enrich selection populations in bread wheat. Aust J Agric Res. 2007;58:867–873. doi: 10.1071/AR05435. [DOI] [Google Scholar]
- Shen XR, Ittu M, Ohm HW. Quantitative trait loci conditioning resistance to Fusarium head blight in wheat line F201R. Crop Sci. 2003;43:850–857. [Google Scholar]
- Prasad M, Kumar N, Kulwal PL, Roder MS, Balyan HS, Dhaliwal HS, Gupta PK. QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor Appl Genet. 2003;106:659–667. doi: 10.1007/s00122-002-1114-y. [DOI] [PubMed] [Google Scholar]
- Munkvold JD, Greene RA, Bertmudez-Kandianis CE, La Rota CM, Edwards H, Sorrells SF, Dake T, Benscher D, Kantety R, Linkiewicz AM. et al. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics. 2004;168:639–650. doi: 10.1534/genetics.104.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Caldwell KS, Dvorak J, Lagudah ES, Akhunov E, Luo MC, Wolters P, Powell W. Sequence polymorphism in polyploid wheat and their D-genome diploid ancestor. Genetics. 2004;167:941–947. doi: 10.1534/genetics.103.016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin Gustafson JP, Lazo G, Chao SM. et al. The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 2003;13:753–763. doi: 10.1101/gr.808603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C, Falque M, Martin O, Paux E, Feuillet C, Sourdille P. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.) Genetics. 2009;181:393–403. doi: 10.1534/genetics.108.097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M, Namiki N, Mizuno H, Yamamoto K, Antonio BA, Baba T. et al. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A. et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Mezard C. Meiotic recombination hotspots in plants. Biochem Soc Trans. 2006;34:531–534. doi: 10.1042/BST0340531. [DOI] [PubMed] [Google Scholar]
- See DR, Brooks S, Nelson JC, Brown-Guedira G, Friebe B, Gill BS. Gene evolution at the ends of wheat chromosomes. Proc Natl Acad Sci USA. 2006;103:6775–6775. doi: 10.1073/pnas.0508942102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells ME, La Rota CM, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin Mahmoud A, Gustafson JP, Qi LL. et al. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 2003;13:1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Luo MC, Zhong GY, Bransteitter R, Desai A, Kilian A, Kleinhofs A, Dvorak J. Genetic map of diploid wheat, Triticum monococcum L, and its comparison with maps of Hordeum vulgare L. Genetics. 1996;143:983–999. doi: 10.1093/genetics/143.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Lazar MD. Comparison of homoeologous group-6 short arm physical maps of wheat and barley reveals a similar distribution of recombinogenic and gene-rich regions. Theor Appl Genet. 2002;104:1078–1085. doi: 10.1007/s00122-001-0804-1. [DOI] [PubMed] [Google Scholar]
- Dilbirligi M, Erayman M, Gill KS. Analysis of recombination and gene distribution in the 2L1.0 region of wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) Genomics. 2005;86:47–54. doi: 10.1016/j.ygeno.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Mochida K, Saisho D, Yoshida T, Sakurai T, Shinozaki K. TriMEDB: A database to integrate transcribed markers and facilitate genetic studies of the tribe Triticeae. BMC Plant Biol. 2008;8:72–80. doi: 10.1186/1471-2229-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P. Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA. 2008;105:9691–9696. doi: 10.1073/pnas.0803981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossolini E, Wicker T, Knobel PA, Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J. 2007;49:704–717. doi: 10.1111/j.1365-313X.2006.02991.x. [DOI] [PubMed] [Google Scholar]
- Dvorak J. In: Genetics and Genomics of the Triticeae. Feuillet C, Muehlbauer G, editor. New York: Springer; 2009. Triticeae genome structure and evolution; pp. 685–711. full_text. [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP. et al. The International Barley Sequencing Consortium - at the threshold of efficient access to the barley genome. Plant Physiol. 2009;149:142–147. doi: 10.1104/pp.108.128967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta M, Graham R, Langridge P, Sparrow D, Barker S. RFLP mapping of manganese efficiency in barley. Theor Appl Genet. 2000;101:1100–1108. doi: 10.1007/s001220051585. [DOI] [Google Scholar]
- Soderlund C, Humphrey S, Dunhum A, French L. Contigs built with fingerprints, markers and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.GR-1375R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Thomas C, You F, Hsiao J, Ouyang S, Buell C, Malandro M, McGuire P, Anderson O, Dvorak J. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics. 2003;82:378–389. doi: 10.1016/S0888-7543(03)00128-9. [DOI] [PubMed] [Google Scholar]
- Kleinhofs A, Kilian A, Saghai Maroof MA, Biyashev RM, Hayes P. Molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the markers mapped onto Ae. tauschii physical map of distal region of chromosome 3DS.
Primer sequences and Tm used for the touch-down PCR of the wheat markers.
Physical map of the Ph2 locus in Ae. tauschii compared to the orthologous genomic sequence of rice Os1 and B. distachyon Bd2 (8× release). The names to the left are rice genes as listed in MSU Rice Genome Annotation (Osa1) Release 6.0.