Summary
Deciphering the non-coding regulatory genome has proved a formidable challenge. Despite the wealth of available gene expression data, there currently exists no broadly applicable method for characterizing the regulatory elements that shape the rich underlying dynamics. We present a general framework for detecting such regulatory DNA and RNA motifs that relies on directly assessing the mutual information between sequence and gene expression measurements. Our approach makes minimal assumptions about the background sequence model and the mechanisms by which elements affect gene expression. This provides a versatile motif discovery framework, across all data types and genomes, with exceptional sensitivity and near-zero false-positive rates. Applications from yeast to human uncover putative and established transcription-factor binding and miRNA target sites, revealing rich diversity in their spatial configurations, pervasive co-occurrences of DNA and RNA motifs, context-dependent selection for motif avoidance, and the strong impact of post-transcriptional processes on eukaryotic transcriptomes.
Keywords: cis-regulatory element discovery, transcription factor binding sites, miRNA regulation, computational genomics, transcriptional regulation, post-transcriptional regulation, information-theory, mutual information, motif-discovery
Introduction
The emergence of whole-genome microarrays (Fodor et al., 1993; Schena et al., 1995) and high throughput in situ hybridization (Tomancak et al., 2002), has made it possible to probe the expression of all or most genes in an organism, as a function of space, time, genetic background, and environmental conditions. A major effort is now focused on decoding the transcriptional and post-transcriptional regulatory programs that mediate these expression dynamics. Transcription is regulated by proteins that bind specific short DNA sequences and then act to modulate the activity of the RNA polymerase. Transcript stability, localization, and translation are also regulated by proteins and RNAs (e.g., miRNAs), which also bind specific short RNA sequences, generally in 3′UTRs. A comprehensive characterization of these DNA and RNA regulatory elements is a formidable challenge, especially within complex metazoan genomes. Experimental (Gerber et al., 2004; Harbison et al., 2004) and computational approaches are emerging to meet these challenges. Several methods compare the intergenic regions of different genomes, aiming to detect sequence elements that are highly conserved across related species (Elemento and Tavazoie, 2005; Kellis et al., 2003; Xie et al., 2005). Other approaches perform a reverse engineering process that aims to infer the regulatory mechanisms underlying the observed expression dynamics (Beer and Tavazoie, 2004).
Various ab initio motif discovery methods have been developed and applied to gene expression data in recent years, e.g., (Bussemaker et al., 2001; Roth et al., 1998). These methods strive towards the same goal: finding a pattern in promoters that shows a statistically significant dependency with the observed expression levels, or variables associated with these expression levels (e.g., clusters of co-expressed genes). Typically, these methods rely on statistical assumptions. AlignACE (Hughes et al., 2000) looks for over-represented patterns in the promoters of pre-specified sets of genes with respect to a background model of the overall nucleotide statistics in the genome. REDUCE (Bussemaker et al., 2001; Foat et al., 2005) predicts motifs via linear regression, with the assumption that the number of occurrences of a putative motif in a given promoter is linearly correlated with the gene’s expression. Neither the extent to which such assumptions are valid nor the behavior of these methods upon violation of these assumptions has been widely explored.
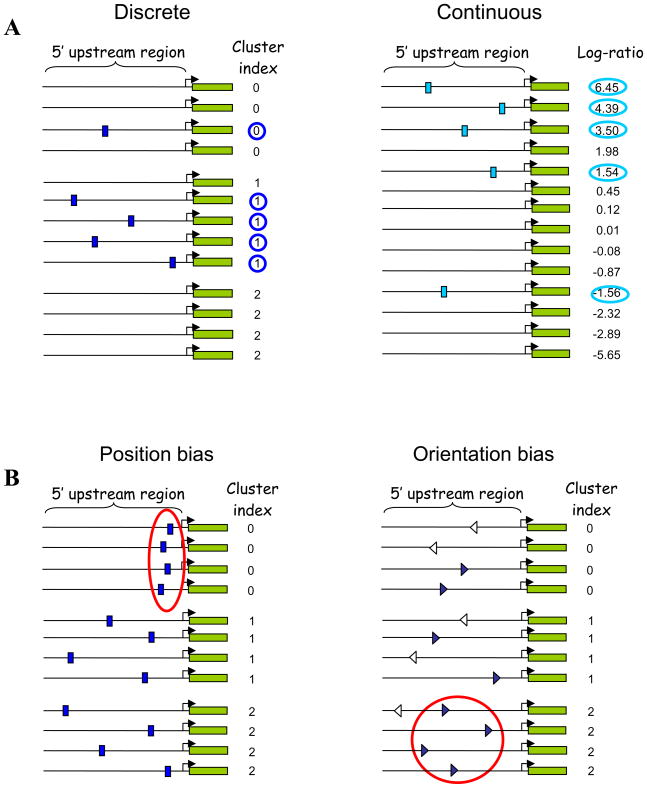
Here, we describe a novel approach for inferring motifs from gene expression data that aims at making as few a priori assumptions as possible. Our approach does not use any complex statistical models, but rather involves directly quantifying the dependency between the presence or absence of a given motif in a regulatory region and the expression of the corresponding gene. To capture this dependency in its most general form, we use the concept of mutual information (Cover and Thomas, 2006). Simply stated, we seek to discover motifs whose patterns of presence/absence across all considered regulatory regions are most informative about the expression of the corresponding genes (Figure 1A). Thus, knowing whether such a motif is present or absent within the regulatory region of a given gene provides significant information regarding the expression of that gene (e.g., the identity of the cluster to which the gene is assigned; see Figure 1A, left panel).
Figure 1. Examples of dependencies between motifs and expression.
(A) FIRE seeks motifs whose pattern of presence and absence across all promoters (or 3′UTRs) is highly informative about the expression profile for the same genes. The left panel presents a simple example for discrete expression data (e.g., a clustering partition). Here, knowing whether the motif is present or absent, provides significant information regarding the identity of the cluster to which the gene is assigned. The right panel presents a simple example for continuous data (e.g., expression log-ratios from a single microarray experiment). Again, knowing whether the motif is present or absent, provides significant information regarding the differential expression of the corresponding gene.
(B) FIRE also uses mutual information to determine whether a predicted motif has a position bias or an orientation bias. If the distance between the motif and the TSS is significantly informative about gene expression (left panel), a position bias is reported. If the occurrences of the motif on one strand (blue triangles in right panel) are significantly informative about gene expression while the occurrences on the other strand (white triangles) are randomly scattered, an orientation bias is reported.
Importantly, mutual information can capture any type of dependency, with no need to specify the nature of this dependency in advance. Thus, our approach is directly applicable to both discrete and continuous data (Figure 1A) whereas existing methods (Bussemaker et al., 2001; Hughes et al., 2000) are often designed to handle only one of these data types.
We also harness mutual information to examine various features of the predicted motifs. To detect motif position bias, we examine the information conveyed by the relative position of the motif in promoters over gene expression (Figure 1B). To highlight orientation preferences, we ask whether the occurrences of the motif on one strand are significantly informative about expression while the occurrences on the other strand are not (Figure 1B). Functional interactions between motifs are predicted by asking whether the presence of one motif in a promoter is informative about the presence of another motif within the same promoter (Figure S1, left panel). Spatial co-localization of motifs within the regulatory regions is explored via similar ideas (Figure S2). RNA motifs with a possible post-transcriptional role, located within 3′UTR sequences, are predicted using precisely the same approach, and functional correlations between DNA and RNA motifs are systematically explored.
Relying on mutual information allows us to capture different types of dependencies that have so far drawn little attention. For example, the presence of one motif in a promoter may be informative about the presence of another motif, but also about the absence of other motifs (Figure S1, right panel). Functional motifs are typically over-represented in coherent sets of genes (Tavazoie et al., 1999), but our results indicate that motifs can also be significantly under-represented. Also, while many motifs are preferentially located near the transcription start site (TSS), others tend to be preferentially located relatively far away from the TSS. We present examples of such dependencies and suggest possible biological interpretations.
To emphasize the versatility of our approach, we report results for various types of experimental data. These include single microarray experiments, gene clustering partitions, in situ expression data, and the “phase” associated with periodically expressed genes. These datasets are derived from several organisms, including yeast, worm, fly, mouse, and human, as well as P. falciparum, the malaria parasite. Many of the motifs we predict match known regulatory elements, but many more are novel predictions.
As a shorthand for our approach we use the acronym FIRE, standing for Finding Informative Regulatory Elements.
Results
Yeast clustering partition: Methodology
We first analyze a compendium of 173 microarray experiments assessing the transcriptional response of yeast (S. cerevisiae) to various stress conditions (Gasch et al., 2000). We clustered the ~6000 genes into 78 non-overlapping clusters using Iclust (Slonim et al., 2005). Each gene is then associated with an index, representing the cluster to which it belongs. Our goal is to discover motifs whose profile of presence and absence across the corresponding promoter sequences is highly informative about these cluster indices (Figure 1A, left panel). The same methodology is used to discover DNA motifs in 5′ upstream regions and RNA motifs in 3′UTRs.
FIRE starts by considering all possible 7-mers (e.g., CGATGAG). For each 7-mer, the mutual information between its presence/absence profile and the expression cluster indices is calculated. Next, all 7-mers are sorted by their information values and the most informative ones are retained as “seeds”. These seeds are then optimized into more general motif representations (using the degenerate code). The optimization procedure involves changing the set of allowed nucleotides at individual motif positions, and only retaining changes that lead to more informative motifs. This procedure is repeated until no further improvements can be made, i.e., the motif is maximally informative.
Importantly, seeds that provide little novel information over the gene expression (with respect to the information already provided by previously optimized seeds) are discarded so as to avoid redundant output. Using the same concept, we also constrain the optimization process in order to avoid optimizing different seeds into the same motif. Thus, the optimization process produces a concise set of motifs, each of which is highly informative about the expression data, but in a distinct way, as illustrated in Figure S3. Finally, each motif is subjected to extensive randomization tests, and only motifs with statistically significant and highly robust information are reported. The optimization procedure and statistical tests are described in detail in Supplementary Methods.
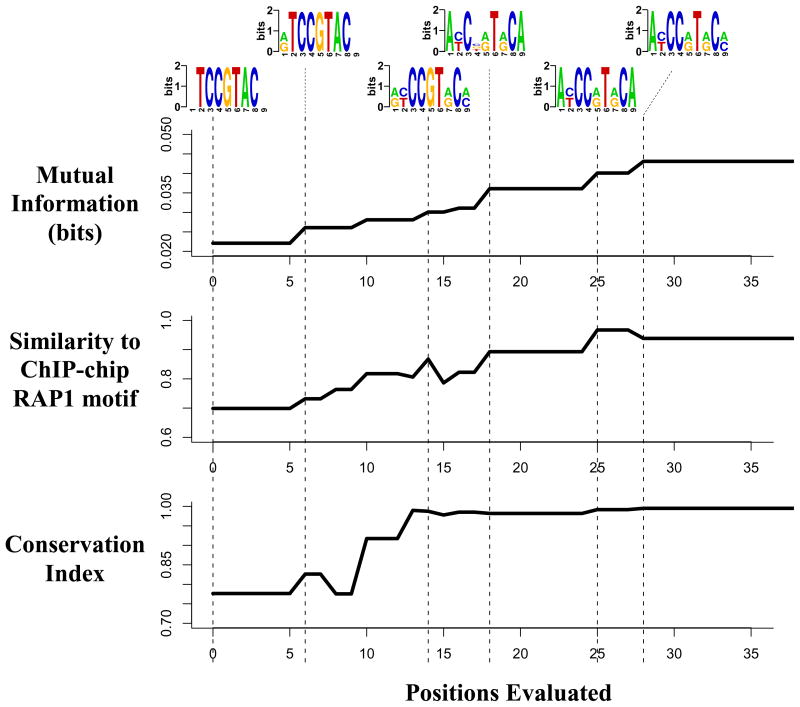
Figure 2 depicts the optimization process that converted TCCGTAC into the more informative A[CT]CC[AG]T[AG]C[AC], which matches the yeast Rap1 binding site obtained from ChIP-chip experiments (Harbison et al., 2004). The upper panel shows the intermediate motifs that progressively increase the mutual information. The middle panel shows the similarity between these intermediate motifs and the Rap1 motif obtained from a ChIP-Chip experiment (Harbison et al., 2004) using an independent motif-finding program (Hughes et al., 2000). Clearly, maximizing the information gradually leads to more accurate representation of the Rap1 motif. The lower panel illustrates the conservation level of these intermediate motifs with respect to the related yeast S. bayanus, using network-level conservation analysis (Elemento and Tavazoie, 2005). Remarkably, conservation increases as we move towards more informative motif definitions, although our information maximization algorithm does not use the S. bayanus genome.
Figure 2. Optimization process for the motif matching the binding site for Rap1 in yeast.
Starting from the seed, FIRE gradually finds motif definitions that are more informative about gene expression (upper panel). These more informative motif definitions are more similar to the known binding site for Rap1 (middle panel), as measured using CompareACE (Hughes et al., 2000). They are also more conserved (with respect to S. bayanus, lower panel).
Yeast clustering partition: Results
Given the yeast clustering partition described above, the entire FIRE analysis with default parameters takes approximately 90 minutes on a standard desktop PC and leads to 17 predicted DNA motifs and 6 predicted RNA motifs. The full results are summarized in Figure 3, automatically generated as part of the default FIRE output. In this heat-map, rows correspond to predicted motifs and columns correspond to gene clusters. Yellow entries indicate over-representation of a given motif in a given cluster; significant over-representation (p<0.05 after Bonferroni correction) are highlighted using red frames. A similar, automatically generated figure with the actual fraction of promoters that contain the motif within each cluster is shown in Figure S4. In addition, Figure S5 depicts the information initially conveyed by each seed, and the information gained through optimization. Of the 23 predicted motifs, we found that 14 closely match a distinct known motif in yeast (see Supplementary Methods). When we shuffle the columns of these 23 motifs, we obtain, on average, less than 0.5 matches to known motifs.
Figure 3. All predicted DNA and RNA motifs for the yeast gene clustering partition.
Columns correspond to gene clusters and rows correspond to predicted motifs arranged into putative functional modules. For each cluster, the most significant GO enrichment is shown at the top. The yellow color-map indicates over-representation of a motif in a given cluster; significant over-representation (p<0.05 after Bonferroni correction) is highlighted using red frames. Similarly, the blue color-map and blue frames indicate under-representation. For each motif, we indicate 1) location, i.e., 5′ upstream region or 3′UTR, 2) mutual information (MI) value, 3) Z-score associated with the MI value, calculated using 10,000 randomization tests, 4) robustness score ranging from 0/10 to 10/10 obtained from 10 jack-knife trials of randomly removing 1/3 of the genes and reassessing the statistical significance of the resulting MI values, 5) position bias indicator (“Y” if a position bias is observed), 6) orientation bias indicator, 7) conservation index, 8) seed that gave rise to the motif, and 9) name of the closest known motif in our motif database (with CompareACE score > 0.8). For more details, see the Supplementary Methods section about FIRE p-value heat-maps.
The most informative motif matches the PAC element which was previously identified from sets of co-expressed genes using Gibbs-sampling (Tavazoie et al., 1999). It is present in ~11% of all yeast promoters, but in nearly 70% of promoters associated with cluster c8 (p<1e-37) and is also over-represented in four additional clusters. As expected, two of these clusters are enriched with genes whose product are localized to the nucleolus (p<1e-42) and genes involved in ribosomal biogenesis (p<1e-69). A more systematic analysis of the variance in the expression data that is explained by the 23 motifs is presented in Supplementary Results.
Importantly, a motif can be informative due to its over-representation in particular clusters, but also due to its under-representation in other clusters. Indeed, we observe many cases where motifs are significantly under-represented in specific clusters as indicated by the blue entries and blue frames in Figure 3. In many cases, the same motif is highly over-represented in one or several clusters, while also highly under-represented in others. For example, the 3′UTR motif in Figure 3 that matches the binding site for Puf3, an mRNA degradation factor in yeast (Olivas and Parker, 2000), is over-represented within two clusters enriched with mitochondrial ribosomal genes, consistent with (Gerber et al., 2004). However, it is also highly under-represented in cluster c9 that is enriched with genes coding for components of the cytosolic ribosome, suggesting a strong selection for regulatory de-coupling between cytoplasmic and mitochondrial ribosome expression.
Constraints on motif position
There is strong evidence that the functionality of some regulatory elements is affected by their distance to the TSS (Beer and Tavazoie, 2004). Thus, observing a significant position bias for a particular motif suggests that its functionality may be affected by its relative location. In FIRE, this is explored using the same concept of mutual information: we ask whether the distance (in bp or nt) between the motif and the TSS (or ATG when the TSS is not annotated; or between the stop codon and the motif, for RNA motifs) is significantly informative about gene expression (Figure 1B). For 9 out of the 17 predicted yeast DNA motifs, we find such significant information. Consistent with (Beer and Tavazoie, 2004), both PAC and RRPE display strong positional biases (Figures S6 and S7, respectively). More interestingly, FIRE detects a positional bias of a very different nature for the motif matching the Rap1 binding site, which seems to be preferentially located between 200 to 500 bp from ATG (Figure S8). We also find a position bias for 3 of the 6 RNA motifs. Closer inspection reveals that these motifs tend to be located close to the stop codon (Figure S9).
Constraints on motif orientation
Certain transcription factors need be oriented in a particular direction relative to the gene in order to adequately fulfill their regulatory function (Beer and Tavazoie, 2004; Erives and Levine, 2004). Correspondingly, their binding sites are functional when located on one strand but not on the other. To systematically explore motif orientation biases, we compare the information conveyed over expression by the motif occurrences on the transcribed strand versus the information conveyed over expression by its occurrences on the non-transcribed strand. An orientation preference is reported when only one of these information values is significant. This reveals an orientation preference for 8 out of our 17 predicted DNA motifs, as shown in Figure S8 for the Rap1 motif. Specifically, in cluster c9, where this motif is over-represented, 83% of its occurrences are on the transcribed strand (p<1e-4), suggesting a strong functional orientation preference, as previously noted (Beer and Tavazoie, 2004). In addition, we observe a clear orientation bias for all 6 predicted RNA motifs, consistent with RNA regulatory elements being located on the transcribed strand.
Validating predicted motifs using independent genomic datasets
Functional regulatory elements tend to be conserved between closely related genomes (Chan et al., 2005; Elemento and Tavazoie, 2005; Kellis et al., 2003). As part of the default FIRE analysis, when a closely related genome is available, the conservation of predicted motifs is assessed by comparing their network-level conservation scores (Elemento and Tavazoie, 2005) to the conservation scores obtained for all possible 7-mers. A conservation index is defined as the fraction of 7-mers that are less conserved than the motif itself; i.e., a conservation index of 1.0 implies that the motif is more conserved than all 8,192 7-mers. Remarkably, all the motifs predicted by FIRE in our yeast example have a conservation index above 0.95 with respect to S. bayanus (Figure 3). Thus, motifs that are informative about expression and hence revealed by FIRE are highly conserved in S. bayanus. A comparison between our predicted motifs and the motifs discovered using an alignment-based approach (Kellis et al., 2003) further supports this conclusion (see Supplementary Results).
Next, FIRE automatically examines the functional coherence of the target genes of each predicted motif. The target genes of a given motif are defined as the genes whose promoters contain the motif and are members of a cluster where the motif is significantly over-represented. Specifically, we ask whether these target genes significantly overlap with any Gene Ontology (GO) category (Ashburner et al., 2000). For almost all motifs in Figure 3, we find GO functional enrichments that resonate well with previous studies (Table S1). For example, the target genes of the predicted Bas1 motif overlap significantly (p<1e-14) with genes involved in amino acid metabolism (Daignan-Fornier and Fink, 1992) while the target genes of [AU]UAUAUUC (an RNA motif) are associated with oxidative phosphorylation (Foat et al., 2005). This analysis also reveals a possible function for some of the motifs we predict. For example, the target genes of two RNA motifs, U[ACG][GU]AAUU[AGU] and UU[AU]AUUU[AU], are highly enriched in genes whose products are localized to the cytosolic ribosome (p<1e-54 and p<1e-48, respectively). The target genes of C[CGT]CCG[CG].[CGT], a DNA motif, are highly enriched with genes involved in oxidative phosphorylation (p<1e-17).
Predicting motif-motif interactions
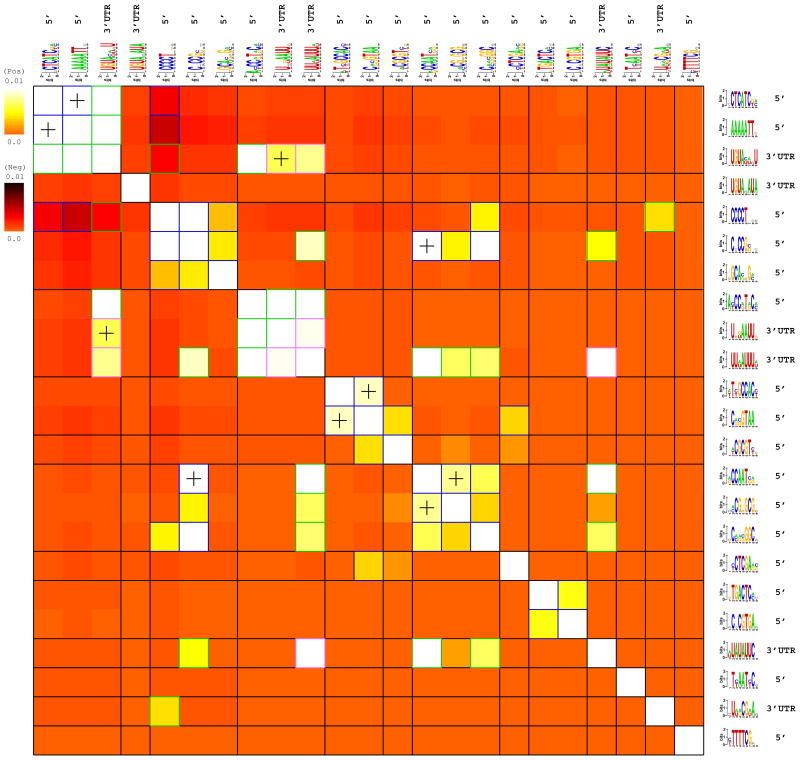
The temporal and spatial regulation of gene expression often involves combinatorial interactions between transcription factors (Beer and Tavazoie, 2004; Pilpel et al., 2001). It is also expected that post-transcriptional regulators (e.g., miRNAs) engage in combinatorial interactions although, to date, little data support this hypothesis. In FIRE, we reveal interactions between predicted motifs based on the mutual information shared between their presence/absence profiles (Figure S1). We further exploit these information values to partition the motifs into putative functional modules (see Supplementary Methods). Figure 4 shows all pairwise information values obtained for the 23 predicted motifs and the corresponding modules. Statistically significant information values between DNA motif pairs, or RNA motif pairs, are indicated in Figure 4 by blue and pink frames, respectively. Statistically significant information values between DNA motifs and RNA motifs are indicated by green frames and highlight possible cooperation between transcriptional and post-transcriptional processes. Importantly, high information values between two motifs may arise from a positive correlation, where the presence of one motif in a promoter implies the presence of its putative counterpart (lighter colors in Figure 4), but also due to a significant negative correlation where the presence of one motif implies the absence of another motif (darker colors). Finally, when two motifs are found to co-occur within the same promoters or 3′UTRs, we ask whether the distance between the two closest occurrences of each motif in this pair is significantly informative about the expression (Figure S2). Such constraints (‘+’ signs in Figure 4) may indicate close physical interactions between the bound factors.
Figure 4. Interactions among all predicted yeast motifs.
Each row/column corresponds to a predicted motif. The color-map indicates the level by which the presence of one motif implies the presence (light color-map) or the absence (dark color-map) of another motif within the same promoter, as quantified by their interaction information (see Supplementary Methods). Very light colors indicate strong positive co-occurrences between pairs of motifs that have further been used to construct putative functional modules, indicated on the figure (and in Figure 3). Very dark colors indicate that the two motifs tend to avoid being present within the same promoter. Statistically significant information values (p<1e-4) that involve homotypic motif pairs are highlighted using blue (DNA-DNA) and pink (RNA-RNA) frames, while those that involve heterotypic pairs (a DNA motif and a RNA motif) are highlighted using green frames. Significant spatial co-localization between pairs of motifs are denoted with ‘+’. For more details, see the Supplementary Methods section about FIRE interaction heat-maps.
FIRE correctly predicts the well-known co-occurrence between the PAC and RRPE motifs (Beer and Tavazoie, 2004). Likewise, FIRE predicts an association between Rpn4 and Reb1, also predicted independently in (Beer and Tavazoie, 2004). In both cases, FIRE finds that the distance between the two closest motifs on the DNA is significantly informative about the cluster indices. Closer inspection (figures available at our Web site) shows that in both cases, the motifs tend to be located very close to each other, suggesting a physical interaction between the factors binding these motifs, as postulated before for PAC and RRPE. Interestingly, the predicted PAC and RRPE motifs and the RNA motif bound by Puf4 tend to co-occur upstream and in the 3′UTRs, respectively, of the same genes, suggesting a functional interaction between these motifs. Finally, FIRE reveals a significant negative correlation between the predicted PAC (and RRPE) motifs and the predicted Msn2/4 motif (Figure S10). We hypothesize that the strong co-avoidance of these motifs is related to the almost opposite expression patterns observed for genes putatively regulated by PAC/RRPE versus genes regulated by Msn2/Msn4 (Gasch et al., 2000). In other words, we suggest that the observed co-avoidance reflects a strong selection against co-occurrence of motifs with opposing functions.
Minimizing false positive predictions - a comparison with AlignACE
To determine the rate of false positive motif predictions, we generated 100 random clustering partitions by shuffling the cluster indices analyzed above. Applying FIRE to these random partitions with the same default parameters yields an average of 0.07 “motifs”, as opposed to the 23 motifs for the original partition. We then applied AlignACE (Hughes et al., 2000) to the same dataset. Whereas FIRE is applied only once to a given clustering partition, AlignACE needs to be applied repeatedly and independently to each cluster, resulting in longer runtime and redundant output. For example, in Figure 3, many motifs are associated with more than one cluster; hence independently analyzing each cluster is expected to predict multiple variants of the same motif. Indeed, applying AlignACE to the original partition with default parameters and a MAP score cutoff of 10.0 (Hughes et al., 2000) returns 1,129 predicted DNA motifs (AlignACE does not support single-strand analyses, so it cannot be used on 3′UTRs), as opposed to the 23 motifs predicted by FIRE. Moreover, applying AlignACE over the 100 random partitions yields an average of 880 predicted “motifs”, in contrast to 0.07 “motifs” obtained by FIRE. These results further underscore the importance of the extensive statistical validation steps incorporated in FIRE.
Yeast single array analysis
As mentioned earlier, the mutual information can quantify the dependency between both discrete and continuous random variables (Cover and Thomas, 2006). Thus, FIRE can be used to find motifs that are informative about continuous expression data, e.g., single microarray experiments (Figure 1A, right panel). Here, each gene is associated with a continuous value, typically an expression log-ratio (e.g., resulting from a two-channel microarray). To demonstrate that, we re-examine the same compendium of yeast microarray experiments (Gasch et al., 2000). Applying FIRE independently to each of the 173 two-channel microarrays yields a total of 403 predicted DNA motifs and 54 RNA motifs (many motifs are found multiple times from distinct arrays), versus 3 DNA “motifs” and 4 RNA “motifs” when applied to the same arrays after expression values are randomly shuffled. In comparison, applying MatrixREDUCE (Foat et al., 2005) to the same data returns a total of 3,018 DNA motifs and 2,688 RNA motifs, or 2,070 DNA “motifs” and 2,324 RNA “motifs” when applied to the randomly shuffled arrays. Again, these results imply that the FIRE output contains very few false positives, at the potential cost of increasing the number of false negatives.
Figure 5 shows the four motifs found to be informative about the genome-wide response of a MSN2/MSN4 mutant strain when exposed to oxidative stress, at 0.3mM H2O2 (Gasch et al., 2000). One of these motifs (PAC) is associated with down-regulated genes, while the three others (Rpn4, Yap1 and Puf3) are associated with different populations of up-regulated genes. Yap1 is indeed known to induce the transcription of many genes in response to oxidative stress (Schnell et al., 1992). Notice that this motif was not found when we applied FIRE to the clustering partition above, highlighting the complementary role of array-by-array analyses. The full results for all 173 arrays are available at our Web site.
Figure 5. Single array analysis in yeast.
All informative motifs obtained from a single microarray experiment, analyzing the genome-wide response of a MSN2/MSN4 mutant strain when treated with 0.3mM H2O2. Rows correspond to motifs and columns to (equally sized) groups of genes with a similar expression value (log-ratio). One of these motifs (PAC) is associated with down-regulated genes, while the three others (Rpn4, Yap1, Puf3) are associated with different populations of up-regulated genes.
Plasmodium falciparum intra-erythrocytic developmental cycle
During the 48h intra-erythrocytic developmental cycle (IDC) of the malaria parasite, Plasmodium falciparum, ~2,700 of its genes exhibit a remarkably periodic expression pattern, characterized by an expression peak at a particular time point during the cycle (Bozdech et al., 2003). Each gene can therefore be associated with a “phase” between −π to p, indicating its peak of gene expression across the time-series (Bozdech et al., 2003). Understanding the regulatory mechanisms underlying this genome-wide periodic behavior may have important therapeutic implications. However, in contrast to S. cerevisiae, currently virtually nothing is known about the regulation of gene expression in Plasmodium. Furthermore, the intergenic regions of the Plasmodium falciparum genome are 90% A/T and are not well described by a simple model where individual bases are independent of their context. This has hampered traditional motif-finding approaches (e.g., AlignACE) that work well on more typical genome background compositions such as that of yeast. In addition, conventional motif-finding approaches are either designed to analyze continuous data that directly quantify expression levels (e.g., REDUCE), or categorical data that implicitly reflect co-regulation properties (e.g., AlignACE). However, the continuous phase data fits neither of these definitions. Nonetheless, applying FIRE to the IDC phase is straightforward; it also has a simple intuitive interpretation: searching for motifs whose presence/absence profile is highly informative about the associated phase values.
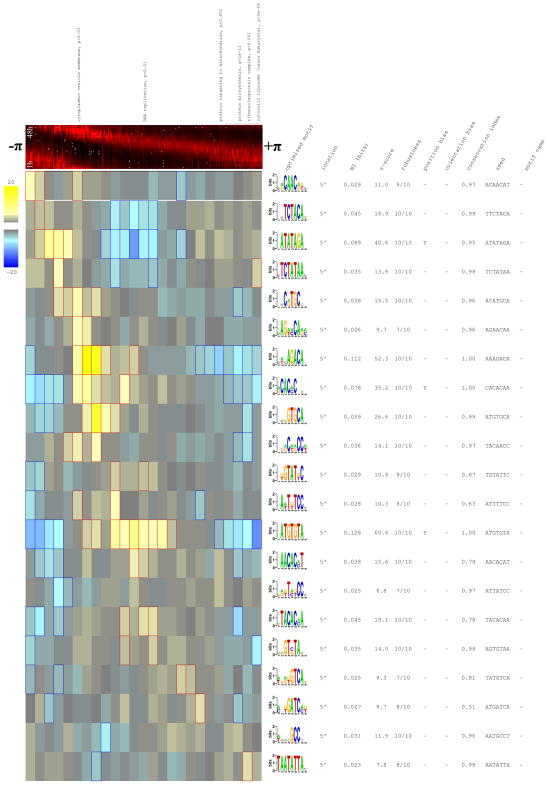
With default parameters, FIRE predicts 21 highly informative DNA motifs, versus an average of only 0.08 “motifs” when applied to 100 randomly shuffled phase profiles. The over/under-representation patterns of these predicted motifs, shown in Figure 6, reveals a remarkable periodic pattern embedded within the intergenic regions of the ~2,700 genes, where each motif is predicted to be involved in the expression of genes at a particular time window during the 48-hour cycle. Notably, the under-representation patterns approximately represent the mirror image of the over-representation patterns, suggesting strong selection against out-of-phase motif occurrences.
Figure 6. All predicted motifs for the P. falciparum phase dataset.
Motifs shown here are highly informative about the P. falciparum expression phase profile that indicates, for each of ~2,700 periodically expressed genes, the timing of its maximal expression during the parasite’s intra-erythrocytic developmental cycle (Bozdech et al., 2003). Rows correspond to motifs while columns to (equally sized) groups of genes with a similar phase value. A heat-map representing the expression profile of all periodically expressed genes (ordered by phase) is shown at the top. Motifs are sorted according to the phase range in which they are most over-represented.
Most (71%) of the 21 predicted motifs are highly conserved with respect to the related Plasmodium yoelli genome (conservation index ≥ 0.95), further supporting these predictions. GO analysis provides additional insights; the most informative predicted motif, ATGTGTA, is found upstream of many genes involved in DNA replication (p<1e-4) and the genes whose promoters contain the nearly palindromic TAATATTA[CGT] motif are enriched with ribosomal genes (p<1e-3). We also note that the predicted motifs contain only 57% A/T, compared to the overall 90% AT-content of the intergenic regions in which they reside. Finally, the FIRE analysis of the phase profile yields no RNA motifs, suggesting that sequence-specific post-transcriptional regulation does not play a significant role in shaping the IDC transcriptome.
Mammalian genomes: human and mouse tissue expression patterns
Motif discovery in mammalian genomes poses unique challenges. Large intergenic regions, distal regulatory elements, local and large-scale chromosomal composition trends (e.g., CpG islands and isochores), and a multitude of cell types, all contribute to the difficulty. On the other hand, a better understanding of these regulatory mechanisms is fundamental for understanding mammalian biology and human disease. Here, we show that applying FIRE to mouse and human expression data yields very promising results.
Given the expression levels of 17,390 human genes across 79 distinct human tissues (Su et al., 2004), we follow the same procedure as for yeast and cluster all genes using Iclust (Slonim et al., 2005). Applying FIRE to the obtained clustering partition yields 73 predicted DNA motifs and 42 predicted RNA motifs (Figure S11) versus an average of only 0.01 DNA and 0.03 RNA “motifs” when applied 100 times to the same clustering partition after randomly shuffling the cluster indices.
Among our predicted DNA motifs, 21 match 20 distinct known motifs in TRANSFAC or JASPAR. Also, 6 RNA motifs match different miRNA target sites (see Supplementary Methods). As in previous examples, independent conservation analysis supports many of our predictions; 46% of our predicted motifs are highly conserved (conservation index ≥ 0.9) with respect to the chicken genome, despite the high divergence between human and chicken. The predicted motifs are further partitioned into 71 modules (Figure S12), many of which consist of both DNA and RNA motifs, suggesting that cooperation between transcriptional and post-transcriptional regulatory mechanisms plays a strong role in shaping the human transcriptome.
Most of our expression clusters are enriched in core cellular processes such as protein biosynthesis, cell cycle, energy generation, etc. (Figure 7). Accordingly, many of the motifs revealed by FIRE are associated with these processes. The most informative motif, CCGGAA[AG], is highly associated with genes whose products are localized to the ribosome (p<1e-34), spliceosome (p<1e-10), and proteasome (p<1e-4). It is over-represented in many clusters, but also under-represented in many others (Figure 7). It best matches Elk4, a member of the ETS family of transcription factors, known to be involved in cell proliferation, differentiation, and to operate downstream of several important signal transduction pathways (e.g., MAP kinases, PI3 kinases, Erk1 and 2, p38 and JNK, etc.) (Yordy and Muise-Helmericks, 2000).
Figure 7. Predicted DNA and RNA motifs for the human gene clustering partition.
FIRE was applied to a clustering partition of 17,390 human genes, clustered based on tissue expression data (Su et al., 2004). The format of this figure is identical to the format of Figure 3. Due to space limitations, only a selection of the predicted motifs is presented. The complete figure is given in Figure S11. Motif names are reported based on the closest known motif in JASPAR or TRANSFAC, with CompareACE score > 0.8. The miRNAs whose 5′ extremity matches 3′UTR elements with high specificity are also reported (see Supplementary Methods).
The second module includes several C-rich DNA and RNA motifs, including a motif that matches the binding site for the Sp1 regulator (Jackson et al., 1990), and an RNA motif that matches the target sites of several known miRNAs. The third module contains a motif that matches the binding site for NF-Y, a known cell cycle regulator (Bolognese et al., 1999). This motif is over-represented in cluster c114, a cluster that is indeed highly enriched with genes involved in mitosis (p<1e-40). Our automated analysis shows that this motif has a strong positional preference towards the TSS (Figure S13), as previously noted (Bucher, 1990). Two other predicted motifs match binding sites of known cell cycle regulators, E2F and E2F1, but belong to separate modules, implying that each of these cell cycle related motifs is involved in regulating somewhat different sets of genes. One of these modules also contains an E-box-like motif, CGTGA[CGT][AC][AGT], that best matches the Pax2 binding site. The motif is over-represented in cluster c56, a cluster that is highly enriched for genes involved in energy generation (Figure 7). Indeed, this motif is often found upstream of genes whose products have electron carrier activity (p<1e-7).
Other gene clusters capture certain tissue-specific patterns. For example, genes in c0 are highly expressed in adult and fetal liver (Figure S14); two predicted motifs that are over-represented in this cluster match the binding sites for the CHOP-C/EBP-alpha and TCF11/MafG heterodimers. Genes within cluster c112 are highly expressed in heart, skeletal muscle, and tongue tissues (Figure S14), and correspondingly this cluster is found to be enriched in genes coding for proteins that make up contractile fibers (p<1e-25) and other muscle-related processes. Thus, the motifs that are highly over-represented in cluster c112, (e.g., ATG[AG][GT]CT), may play a role in the regulation of gene expression in muscle tissues.
Finally, some gene clusters are associated with olfactory receptor activity (Figure 7) and FIRE detects several motifs that are over-represented among genes in these clusters. One module consists of four predicted DNA motifs (two are shown in Figure 7) and one predicted RNA motif, all over-represented in clusters highly enriched with olfactory receptor genes like c3 and c92 (p<1e-43 and p<1e-45, respectively). We expect these results to advance our understanding of the transcriptional regulatory program of the mammalian olfactory system (Serizawa et al., 2003).
We also applied FIRE to whole-genome expression data reported for mouse tissues (Su et al., 2004). Following the same procedure, we obtained 80 predicted DNA motifs and 10 predicted RNA motifs, many of which are highly conserved with respect to the chicken genome, and/or match known regulatory elements. A substantial fraction of the motifs we detect for human tissue data match the motifs obtained independently while analyzing mouse tissue data, despite the fact that the two sets of tissues studied only partially overlap (Su et al., 2004). Specifically, 31 (42%) of the 73 predicted human DNA motifs have a highly similar counterpart among the 80 predicted mouse DNA motifs (CompareACE score ≥ 0.75), far more than what would be expected by chance; when we repeat this analysis 10 times while the positions of each predicted mouse DNA motif are randomly shuffled, we obtain a similar counterpart to an average of only 8.1 (11%) predicted human motifs.
Additional analyses
We have applied FIRE to several other datasets, in other species. These include a large compendium of gene expression profiles in C. elegans, and pre-specified groups of genes in D. melanogaster, including groups of genes with similar spatio-temporal patterns of gene expression as revealed by in situ hybridization. The results complement the ones described here and are elaborated on in detail in Supplementary Results.
Discussion
Implied assumptions and possible limitations
The approach presented here may have its own limitations. Our analysis may overlook certain highly degenerate motifs, as it currently starts by considering non-degenerate motif representations (k-mers). Nonetheless, we note that many of our predicted motifs are quite degenerate, e.g., the predicted Rap1 motif in yeast (Figure 2), demonstrating that even degenerate motifs often have highly informative non-degenerate seeds, possibly corresponding to different affinity classes. Another potential limitation comes from the use of relatively coarse motif representations via the discrete degenerate code, as opposed to continuous representations like weight matrices (Stormo, 2000). However, the degenerate code representation can be seen as an approximation of weight matrices, and it is used in FIRE merely because it allows for a very efficient search of motif space; there is no conceptual difficulty in using weight matrices instead.
To emphasize the generality of our approach, all datasets presented here were analyzed using the same default parameter values. Some of these parameters define highly stringent significance tests, leading to low false positive rates. Indeed, the default FIRE settings were chosen so as to favor specificity over sensitivity. Thus, FIRE typically returns a manageable number of high-confidence motifs, highly suitable for experimental testing. However, in its default configuration, FIRE may miss weaker, less informative motifs. Less stringent statistical tests can be used to increase sensitivity (see Supplementary Results and Methods).
Other parameters include the length of the predicted motifs. For example, our candidate seeds consist of the entire set of 7-mers and for computational efficiency, optimized motifs are set to be 9 nt-long. We note, however, that using 6-mers as seeds leads to very similar predictions (results available for yeast at our Web site), and that given enough computational resources, predicting longer motifs is fully supported.
Finally, it seems that somewhat arbitrary decisions regarding certain aspects of the input data are inevitable. An important issue is the length of the upstream promoters and 3′UTR sequences to be analyzed. Here, we have attempted to use similar lengths to those used by previous reports. Applying FIRE to longer intergenic regions represents an important direction for future work.
A model independent analysis
The model independence properties of mutual information as a general statistical dependency measure are particularly appealing in our context. Indeed, we use mutual information not only to predict motifs but also to characterize their functional constraints and relationships, where in all cases the underlying principle is the same: looking for sequence patterns that show maximal dependency with the expression data.
The fact that there is no need to specify in advance the nature of the dependency, nor to postulate any a priori statistical model, yields a versatile machinery that can be applied to any type of experimental data related to gene expression. We predict motifs in yeast, Plasmodium, fly, worm, mouse, and human, by using precisely the same methodology and algorithms, and without tuning any parameters. The different statistical properties of each genome seem to have no impact on FIRE performance. For example, the intergenic regions of the Plasmodium genome are 90% A/T and are not well described by a simple model where individual bases are independent of their context. While this may have hampered conventional motif discovery methods, it seems to cause no difficulties in our case.
Model-independency also allows revealing diverse types of biologically relevant dependencies, not readily captured by other methods. For example, our results suggest that regulatory elements are often under-represented in specific coherent sets of genes. In particular, our results for Plasmodium (Figure 6) and to some extent for other species, argue that there sometimes exists a strong selection against occurrence of regulatory elements within an inappropriate regulatory context. In addition, our results suggest that selecting against the joint presence of certain motifs in the same intergenic region (Figure S10) plays an important role in shaping the regulatory genome. Furthermore, some dependencies discovered by our approach involve specific motifs being preferentially located neither close nor far, but at an intermediate distance from the TSS or start codon (Figure S8). In addition, elucidating functional relations between DNA motifs and RNA motifs is naturally embedded within our analysis, and our results indeed suggest that such heterotypic correlations are quite common across eukaryotic genomes.
Importance of post-transcriptional regulation
For most species considered here, roughly one-third of the detected informative motifs are RNA elements; this holds for unicellular eukaryotes (S. cerevisiae), invertebrates (C. elegans), as well as mammals (human). Moreover, these RNA motifs are quantitatively indistinguishable from the predicted DNA motifs in terms of the amount of information they carry over gene expression (e.g., Figures S5 and S17). This suggests that post-transcriptional regulation based on specific binding to 3′UTR motifs is widespread in eukaryotes, including metazoans, with significant and measurable consequences for mRNA levels.
While many of the RNA motifs predicted by FIRE in metazoans match the 5′ extremity of known miRNAs, many more do not. Many of these motifs may be bound by RNA-binding proteins (Keene and Tenenbaum, 2002) (e.g., PUF proteins, for which we found binding sites in our yeast and worm analyses). However, many of these RNA motifs may represent targets for miRNAs that have not yet been identified.
Concluding remarks
We have introduced an information-theoretic framework for motif discovery and characterization within large genomic datasets. The model-independent nature of our approach allows the discovery of DNA and RNA motifs that show generic dependency with diverse aspects of gene expression, including differential expression observed in single microarray experiments, cluster indices associated with co-expressed genes, expression phases in a periodic time-series, spatial patterns of gene expression from in situ hybridization experiments, and even classification of enhancers based on genetic/biochemical data (see Supplementary Results).
In all analyses reported here, many of the predicted motifs are supported by inter-species conservation and functional enrichments. Moreover, upon randomly shuffling the expression profiles, FIRE typically returns zero predictions. Thus, we expect that the majority of our predictions, encompassing hundreds of DNA and RNA motifs, correspond to novel transcription factor binding sites, RNA-binding protein sites, or miRNA targets. The follow-up experimental validation of such high-yield predictions represents an important challenge to experimental biologists.
Finally, we have put much emphasis on the practical aspects of our approach. FIRE can be downloaded and used on standard workstations or via a Web interface. For commonly studied model organisms, a simple command line executes the entire analysis. The results are presented via automatically generated figures that summarize the most important biological predictions in a concise visual manner. In particular, almost all figures presented here were automatically generated by FIRE.
Supplementary Material
Acknowledgments
We thank W. Bialek for many helpful comments, and Chang Chan, David Gresham, Alison Hottes, Zia Khan, Manuel Llinás, and Ilias Tagkopoulos, for their comments on earlier versions of this article. We are also grateful to Bambi Tsui for making FIRE available through a Web interface. ST is supported by grants from NIH and NSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer M, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Bolognese F, Wasner M, Dohna CL, Gurtner A, Ronchi A, Muller H, Manni I, Mossner J, Piaggio G, Mantovani R, Engeland K. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene. 1999;18:1845–1853. doi: 10.1038/sj.onc.1202494. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- Chan CS, Elemento O, Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover T, Thomas J. Elements of Information Theory. 2. Hoboken, NJ: Wiley-Interscience; 2006. [Google Scholar]
- Daignan-Fornier B, Fink GR. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci U S A. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives A, Levine M. Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci U S A. 2004;101:3851–3856. doi: 10.1073/pnas.0400611101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci U S A. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Herschlag D, Brown P. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Jackson SP, MacDonald JJ, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schnell N, Krems B, Entian KD. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Slonim N, Atwal GS, Tkacik G, Bialek W. Information-based clustering. Proc Natl Acad Sci U S A. 2005;102:18297–18302. doi: 10.1073/pnas.0507432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes J, Campbell M, Cho R, Church G. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:R88. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas E, Golub T, Mootha V, Lindblad-Toh K, Lander E, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.