Abstract
The lack of reliable measures of alcohol intake is a major obstacle to the diagnosis, treatment, and research of alcohol abuse and alcoholism. Successful development of a biomarker that allows for accurate assessment of alcohol intake and drinking patterns would not only be a major advance in clinical care but also a valuable research tool. A number of advances have been made in testing the validity of proposed biomarkers as well as in identifying potential new biomarkers through systems biology approaches. This commentary will examine the definition of a biomarker of heavy drinking, the types of potential biomarkers, the steps in biomarker development, the current state of biomarker development, and critical obstacles for the field. The challenges in developing biomarkers for alcohol treatment and research are similar to those found in other fields. However, the alcohol research field must reach a competitive level of rigor and organization. We recommend that NIAAA consider taking a leadership role in organizing investigators in the field and providing a common set of clinical specimens for biomarker validation studies.
Introduction
The National Institute on Alcohol Abuse and Alcoholism (NIAAA) has been a leading sponsor of grants and contracts related to the development of biomarkers for alcohol consumption and alcohol-induced tissue injury for over two decades. Advancements in genomic, proteomic, metabolomic, and bioinformatic technologies have increased the potential for development of clinical biomarker diagnostics for these conditions. Prior to the 2008 annual meeting of the Research Society on Alcoholism in Washington DC., NIAAA sponsored two days of meetings focused on biomarkers of alcohol consumption. A one-day closed meeting of NIAAA grantees who had been awarded funding via biomarker-specific requests for applications (RFAs) preceded a one-day open meeting which was attended by NIAAA grantees, academicians with an interest in biomarker discovery and development, members of industry and representatives of regulatory agencies. The companion article by Hoek et al., summarizes the Workshop discussion panel’s perspective and the Commentary by Raye Litten provides further insight into the state of clinical alcohol consumption biomarkers today.
The research presented and discussed at the Workshop provided clear and encouraging evidence of advances in alcohol-related biomarker development, which included the characterization of a number of promising RNA, protein, and small molecule biomarker candidates. Also clear in these discussions were the challenges to translating these initial research findings into clinical diagnostic tests. This commentary will focus on the definition of biomarkers, the potential applications and development strategies for clinical biomarkers, current findings in the field, and challenges in biomarker development.
Clinical tests for alcohol consumption, typography, and alcohol-induced disorders are critically needed in alcoholism and alcohol abuse treatment and research. While detailed efforts have been made to construct interview formats that correctly quantify alcohol intake, such as AUDIT-C (Alcohol Use Disorders Identification Test Consumption) (Bradley et al., 2007; Bush et al., 1998), CAGE (Ewing, 1984), or including reports from collateral individuals (individuals [family and friends] who interact with the subject) (Connors and Maisto, 2003), these approaches have their limitations. This is especially true in cases where individuals are motivated to deny or minimize the magnitude of drinking behavior in order to mitigate personal, professional, or legal ramifications of alcohol abuse (Pernanen, 1974; Fuller et al., 1988). Also, self-reporting accuracy varies 1) between populations (Frank et al., 2008; Dhalla and Kopec, 2007) and 2) based on the manner and milieu of the interview (Steinweg and Worth, 1993). One difficulty in quantifying the validity of alcohol self-reports is that, unlike other abused substances, alcohol abuse lacks a clinical test correlate to verify self-report data (Brener et al., 2003). Self-reporting mechanisms will continue to have utility in clinical and research settings, but their use is constrained by limitations in time, resources, and training of personnel when using self-report mechanisms (Roche et al., 2006). These limitations of self-reports and have led to the search for a reliable biochemical biomarker of heavy drinking. A measureable and accurate clinical biomarker test that could provide an objective assessment of drinking behavior would alleviate the uncertainties of self-reporting. Additionally, clinical diagnostics that identify alcohol-induced tissue damage could improve clinical care and have applications in alcohol abuse treatment.
What is a biomarker?
NIH working group definition
The term ‘biomarker’ is often used indiscriminately to describe any gene or protein expression change – in this case, associated with drinking behavior. This broad usage blurs the statistical classification and clinical diagnostic power of a biomarker. As a correction and clarification, the NIH Biomarkers Definitions Working Group has defined a biomarker strictly as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or pharmacologic responses to a therapeutic intervention” (Biomarkers Definitions Working Group, 2001). The difference between the broad usage and the more precise definition is that the broad usage relates to a difference in the means between two populations, while the more specific definition from the NIH Biomarkers Working Group requires that a biomarker be informative for individual subjects. This difference is the heart of the utility of biomarkers – they allow highly confident classification of individuals.
Therefore, in general terms, a biomarker may be thought of as the analyte (or analytes) in a diagnostic test. With regard to alcohol consumption specifically, a biomarker would be an accurate indicator of an individual’s alcohol consumption. Similarly, a biomarker could be an indicator of pathogenic processes in alcohol-induced tissue damage and dysfunction. Note, however, that there is no such thing as a universal biomarker for all aspects of alcohol consumption and alcohol-induced tissue damage. Rather, a given biomarker (or biomarker panel) applies only to a specific physiological or behavioral state (e.g., heavy vs. light drinking or drinking vs. abstinent). For the purposes of this review, we will also not use the term biomarker of alcoholism or alcohol abuse because such a diagnosis necessitates certain psychological criteria (American Psychiatric Association, 2000). Rather, we will use the terms heavy drinking and/or alcohol-induced tissue damage.
Not all biomolecule changes are biomarkers
As described above, the term biomarker is often used indiscriminately to describe any biomolecule with a difference in abundance between two conditions – a statistically significant difference in population means. However, a statistically significant difference between two populations can still contain many overlapping values for individual subjects. To comply with the NIH Biomarkers Working Group definition above, there must be a measure of how well a given biomolecule (or biomolecules) serves as a classifier of a certain state for individual subjects. It may be useful to apply new or more specific nomenclature for biomarkers in the process of validation such as ‘potential biomarker’ or ‘biomarker target’. The term ‘validated biomarkers’ may be appropriate for more fully developed indicators. The Food and Drug Administration (FDA) has already proposed rigorous standards for the term biomarker and criteria for validating a biomarker (US Food and Drug Administration, 2005). We will discuss this point in more detail below.
Measures of biomarker efficacy
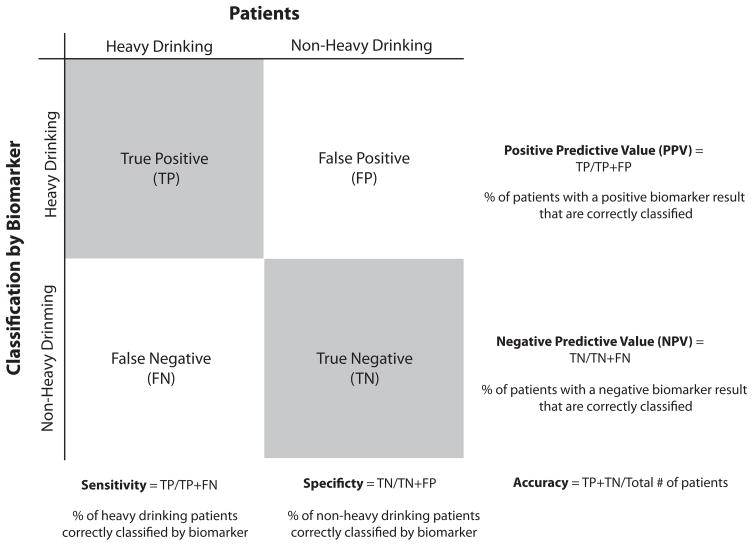
The statistical measures applied in determining the utility of biomarkers differ from those commonly used in basic research, and it is important to use these measures and terms precisely. Since the utility of a biomarker lies in its indicative or diagnostic power, the ability of a biomarker to correctly classify subjects is of primary importance. In alcohol abuse research as well as treatment settings, a biomarker could be used to categorize subjects as, for example, heavy drinkers vs. non-heavy drinkers (either light drinkers or abstainers, Fig. 1). In this example, a test can return four potential results: 1) a True Positive of a heavy drinker classified as such, 2) a True Negative of a non-heavy drinker correctly identified, 3) a False Negative of a heavy drinker classified as a non-heavy drinker, and 4) a False Positive where a non-heavy drinker is incorrectly identified as a heavy drinker. To quantify these outcomes, sensitivity, specificity, positive predictive value, negative predictive value, and accuracy measures are used (See Figure 1 for definitions). Sensitivity and specificity are the most commonly used terms. They indicate respectively, in this example, the percentage of heavy drinkers and non-heavy drinkers correctly identified by the test.
Figure 1. Metrics for assessing biomarker performance.
Unlike traditional statistics that examine differences between populations, diagnostics are intended to be informative of the individual patient. Sensitivity, specificity, positive predictive value, negative predictive value, and accuracy measures can be used to summarize a biomarker’s ability to correct classify subjects. In this example, the ability to classify heavy alcohol drinking and non-heavy alcohol drinking (abstainers or light drinkers) is portrayed. Sensitivity and specificity are the most commonly used measures, and, in this case, they respectively give the percentage of alcohol abusing and non-alcohol abusing subjects correctly identified.
Ideally, a test would have 100% sensitivity or specificity but this is often not possible due to individual differences in genetics, environment, co-morbidities, and drinking phenotype. Although achieving the highest possible sensitivity and specificity is most desirable, certain instances exist in which either high sensitivity or high specificity is more important. For example, if a test were to have legal ramifications, specificity would be of utmost importance to avoid a false positive outcome (i.e., incorrectly classifying a parolee as a heavy drinker, resulting in revocation of parole), even at the expense of failing to detect alcohol abuse in some parolees. Alternatively, if a test was intended to spur a discussion of individuals’ drinking behavior or referral to a treatment specialist, sensitivity at the expense of a degree of specificity would be acceptable (i.e., detecting all cases of heavy drinking, while incorrectly including some non-heavy drinkers).
Types of biomolecules used as biomarkers
The type of biomolecule that may serve as the best biomarker of alcohol consumption and alcohol-induced tissue injury is unknown. Nucleic acids, proteins, small molecules, or protein adducts could potentially be of use, or even non-biochemical tests such as in vivo imaging could serve as a biomarker. Here, we focus on biomarkers such as nucleic acids, proteins, or other small molecules that can be easily and non-invasively obtained from blood, plasma, urine, or hair. Previous reports provide examples of the technologies that can be used in analyses of nucleic acids (Biermann et al., 2007; Walker and Grant, 2006), proteins (Freeman et al., 2006; Nomura et al., 2007), or small molecules (Bradford et al., 2008; Stephanson et al., 2007; De et al., 2007) in alcohol abuse research. It is tempting to prejudge what type of sample and molecule will have the best diagnostic value, but researchers should remember that treatment professionals are pragmatic on this issue – whatever form of biomarker works best and can help effectively diagnose and treat patients is the best biomarker. Lastly, while it may be appealing to think of biomarkers as single molecules (e.g., a single mRNA or protein), a growing body of evidence indicates that panels of biomolecules in combination may function as the best biomarkers in terms of sensitivity and specificity as will be discussed below (Spira et al., 2007; Ray et al., 2007).
Clinical Biomarkers of Alcohol Use
There are a number of potential aspects of alcohol abuse and alcohol-induced tissue damage that biomarkers could be used to diagnose. These can be broadly defined as indicators of cumulative intake over a given time period, drinking patterns, and alcohol-induced organ damage. As discussed above, it is not possible for one biomarker to serve as an accurate indicator of all of these conditions, therefore biomarker development projects need to focus on the specific aspect of alcohol abuse that the biomarker is intended to diagnose. Without a clearly defined diagnostic goal, biomarker development projects are unlikely to be successful.
Alcohol Intake
There are a number of research programs seeking to develop a biomarker that quantifies total alcohol intake. The goal of these efforts is to create a biomarker test that returns either a categorization of the subject or a numeric estimation of their average daily alcohol intake. A categorical test would differentiate subjects into non-drinking, non-abusive drinking, and heavy drinking classes. This would correlate to 0, <2 and >2 drink equivalents per day, respectively. Alternatively, a biomarker could provide a dose-response relationship of alcohol intake and provide a numeric (drinks/day) estimate of daily intake over a period of time. Either of these approaches will allow subjects with ongoing, heavy drinking behaviors to be identified. Importantly, validation of biomarkers will require testing of how quickly a heavy drinking signature returns to normal levels with abstinence.
Drinking patterns
Of growing interest to researchers and treatment specialists are biomarkers of drinking patterns. This stems from a growing understanding of the dangers of binge drinking episodes (Dawson, 2000). That is, 2 drinks per day (14 drinks per week) are different from 7 drinks a day during the weekend (14 drinks per week) even though total alcohol intake is the same over the week. A clinical diagnostic for alcohol intake patterns that include periods of intense intoxication would be valuable for both treatment and harm reduction strategies.
Alcohol-induced organ damage
The remaining area of investigation for biomarker development is in diagnosis of alcohol-induced tissue damage. While cessation of excessive drinking is the primary goal in subjects who are developing alcohol-induced organ disease, diagnostics that identify early stages of disease development, such as liver damage, would enable earlier and more effective treatment. By revealing the onset and extent of organ damage, such diagnostics may also provide a needed “wake-up call” to heavy drinkers, and therefore increase the biomarkers’ treatment utility through altering patients’ drinking behavior.
Settings for use of alcohol biomarkers
The clinical settings that would benefit from new diagnostics of heavy alcohol use and alcohol-induced tissue damage are wide ranging. In standard clinical practice, diagnostics of excessive alcohol intake would aid in identifying individuals in need of treatment referral. For subjects in treatment, a diagnostic could also serve to monitor abstinence in a cessation regimen or a reduced level of drinking. Further, compliance testing of individuals under agreements or orders that prohibit alcohol consumption in the parole system, the military, and through professional licensing boards would also benefit from the development of such a diagnostic. National security protection and clinical trials of potential treatments are two additional settings in which implementation of alcohol intake diagnostics would be advantageous.
Current biomarkers
As stated before, the search for diagnostics of alcohol abuse is not new. For several decades, research and development programs have worked on developing diagnostics of alcohol intake with the notable success being in measuring acute alcohol intake. A number of accurate methods for determining blood alcohol content (BAC) through breath and blood now exist. This technology has developed to the point that small and inexpensive instruments are used by tens of thousands of law enforcement, medical, and security personnel.
What has remained more elusive are diagnostics that can retrospectively examine alcohol intake across days or weeks. A number of potential plasma, blood, urine, and hair biomarkers have been examined and this literature has been extensively reviewed elsewhere (Conigrave et al., 2002; Hannuksela et al., 2007b; Conigrave et al., 2003; Helander, 2003; Hannuksela et al., 2007a; Das et al., 2008; Litten, 2009). The most common laboratory tests for alcohol intake include: gamma-glutamyltransferase (GGT) (Taracha et al., 2001), mean corpuscular volume (MCV) (Hock et al., 2005), aspartate aminotransferase (AST) and alanine aminotransferase (ALT) (Niemela, 2007), sialylation of apoliprotein J (SIJ) (Ghosh et al., 2001), carbohydrate-deficient transferrin (CDT) (Golka and Wiese, 2004) (Koch et al., 2004), ethyl glucuronide (EtG) (Kissack et al., 2008), and 5-hydroxytryptophol (5HTOL) (Helander and Eriksson, 2002). While a detailed comparison of these proposed or potential markers is beyond the scope of this review, a common finding from the literature is that the sensitivity and specificity of tests varies greatly between study sites, and are frequently lower than required for diagnostic purposes. This may be due to several factors. 1) Most of these biomarkers have been proposed as unitary measures; however, combining multiple markers has been demonstrated to be more effective, suggesting that a panel of markers may result in better sensitivity and specificity (Anton et al., 2002; Korzec et al., 2005; Rinck et al., 2007). 2) Many biomarkers relate to hepatic function, which is well known to be altered with heavy alcohol consumption. Hepatic function is also impaired, however, in a number of other conditions which leads to false positives and reduced specificity. 3) Some biomarkers may occur only with extremely high intake or in conjunction with co-morbidities. This results in higher levels of false negatives and reduced sensitivity. While all of the biomarkers described above have some utility they have not been universally accepted or generally adopted in clinical practice, or have lower diagnostic accuracy than is desired.
How are biomarkers developed?
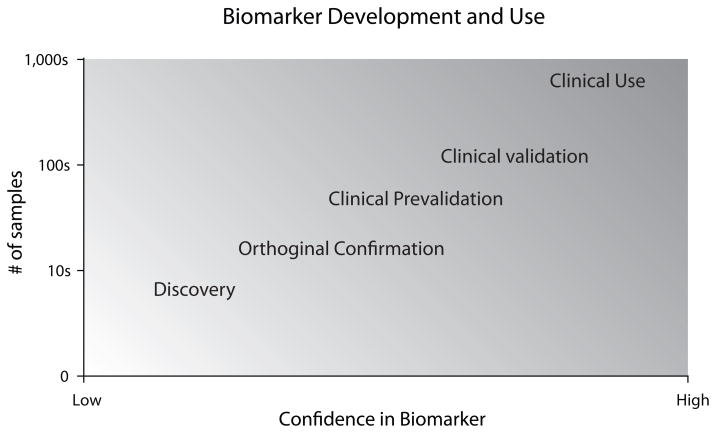
With the above definitions in mind, the issue in question is how to identify and conclusively demonstrate that a biomarker is an accurate indicator/diagnostic. Several excellent reports have examined the use and validation/qualification of clinical biomarkers in other fields and can serve as potential models for biomarker development in alcohol use and abuse (Lesko and Atkinson, 2001; Rolan et al., 2003; Williams et al., 2006). The process of validating or qualifying biomarkers requires explicit attention to translate initial findings into clinically applicable diagnostics. For diagnostics of heavy alcohol use and alcohol-induced injury, we propose five steps for development of a biomarker (Figure 2). As described in Figure 3, with each step of the development process more samples are examined and more confidence can be placed in a biomarker.
Figure 2. Steps in developing biomarkers of alcohol-related disorders.
As described in the text, development of a clinical biomarker requires multiple steps/stages of development. Successful completion of each of these steps results in a validated biomarker that is both sensitive and specific.
Figure 3. Biomarker development and use.
Through the process of biomarker development steadily increasing numbers of samples are examined. With successful completion of each stage the confidence, the likelihood that a biomarker will be clinically useful, increases.
Define the conditions to be classified
First, it is critical to define precisely what the biomarker is intended to diagnose or classify (the definition of a specific pathology). As noted above, it is unlikely that one biomarker will be able to accurately diagnose the many different aspects of alcohol use and abuse, and alcohol-induced tissue damage. A clear definition of what the biomarker is intended to reflect is therefore a first step that cannot be overlooked or bypassed. To use an example from our ongoing research, we have decided to focus on biomarkers of alcohol intake that will accurately segregate subjects into non-drinking (abstaining), light drinking, and heavy drinking categories. While we remain interested in what biomarkers tell us about drinking patterns or alcohol-induced tissue damage, the ultimate success or failure of our biomarker will be judged by patient diagnosis and segregation of consumption phenotypes.
Target discovery
Second, with the biomarker definition and goal clearly set, potential biomarkers for development must be identified (target discovery). Biomarker targets can come from pre-existing hypotheses or can come from systems biology discovery approaches (e.g., transcriptomics, proteomics, or metabolomics). The data presented at the Workshop on Alcohol Biomarkers detailed genomic, proteomic, and metabolic screening studies in preclinical model systems as well as in clinical samples to identify biomarkers for further development. As we discuss later, mRNA, protein, and small molecules all have potential strengths and weakness as biomarkers of alcoholism or alcohol abuse. Nonetheless, these initial screening studies are providing a wealth of new potential biomarkers for further development. Some disagreement exists in the scientific community on the ultimate utility of the results of ‘omic discovery approaches, in part because they have yielded a limited number of biomarkers (Baker, 2005; Marrer and Dieterle, 2007). It is important to note that many ‘omic screening technologies have been available in fully optimized forms for only a few years, so it is premature to expect them to have produced fully validated diagnostics in this short period. Prostate-specific antigen (PSA), one of the more broadly known biomarkers, took over 8 years to translate from initial discovery to widespread adoption as a clinical test (Rao et al., 2008; DeAntoni et al., 1993), and even now PSA possesses less diagnostic value than was originally hoped.
Validation by orthogonal methods
In the third step of biomarker development (orthogonal confirmation), it is necessary to confirm the findings of the initial screening studies in independent samples and by alternate techniques, if possible. With the wealth of information generated from genomic, proteomic, and metabolomic studies, there is always the potential for false positives arising from statistical Type I errors, sample specific factors, or artifacts of a particular analytical approach. Commonly used orthogonal technologies are qPCR for genomic studies and immunoblots, ELISAs, or multiple reaction monitoring (MRM) (Anderson and Hunter, 2006) for proteomic studies. Inevitably, some of the initial potential biomarkers identified in the screening studies will not be confirmed and can be set aside in favor of reproducible changes. This filtering approach is not dissimilar from the approach used in pharmacotherapy development, in which a large number of candidate compounds are reduced in number at each step of development. The goal in biomarker development is to have unsuitable potential biomarkers fail quickly - thus leaving further efforts to focus on the most promising biomarkers. These initial steps can be applied in either model systems or using clinical samples.
Determine classification ability
With biomolecules that have been demonstrated to have altered levels in multiple sample sets, the next hurdle is to determine if these potential biomarkers are accurate classifiers. This includes examining candidate biomarkers in other pathologies which may share some symptoms (pathobiological specificity). The determination of specificity is important, as has been demonstrated by previously proposed biomarkers of alcohol consumption related to hepatic function. Elevation of AST/ALT enzyme levels in the plasma, for instance, can indicate excessive alcohol intake, but can also be indicators of unrelated hepatic dysfunction (Giannini et al., 2005). As noted in Figure 1, in the present context, sensitivity is defined as the percentage of heavy drinkers correctly identified as ‘positive’, while specificity is the percentage of non-heavy drinkers (abstainers or light drinkers) that are correctly diagnosed as ‘negative’. A biomarker of heavy alcohol use based on hepatic function may lack pathophysiological specificity and produce false positives in patients with non-alcoholic liver disease.
As was discussed previously, with continued development of a biomarker, the sensitivity and specificity of the diagnostic can be determined. When determining the diagnostic utility of a biomarker it is critical to use a training set/test set design to avoid over fitting of the data. This happens when the same set of data is used both to determine (train) the biomarker level and to test the classification ability. As a result, the classification accuracy can be inflated. For example, if a single analyte is being used, a biomarker will have a cutoff value, under which is one classification and above which is the other. If the same set of data is tested with this cutoff value as was used to determine the cutoff level, the test may give much better results than if tested in an independent set of samples. If a biomarker panel is being used, a multi-variant classification algorithm (e.g., Support Vector Machines or Random Forest) can be used to produce the diagnosis. These algorithms require a set of samples with known classifications to train the algorithm. If the same set of samples is used to both train and test the algorithm (this is called cross-validation), the accuracy of the test is overestimated. To avoid these problems when determining the sensitivity and specificity of the test, independent samples should be used.
Clinical Trial
In the fifth step of biomarker development, potential biomarkers are validated in clinical samples. This need arises for several reasons. First, biomarker discovery frequently takes place in highly controlled animal models that are not completely faithful to the human condition. For instance, in our own work, discovery is being conducted in a non-human primate model of alcohol self-administration that has a high level of face-validity. However, these subjects have excellent dietary history and are not co-morbid for conditions common to the human alcohol abusing population. Therefore, there is a substantial risk that biomarkers initially developed in animal models will fall short, in terms of sensitivity and specificity, in the broader and more heterogeneous human population.
The utility of biomarkers in different geographic, ethnic, gender, and age groups is important to quantify as exemplified by differences in the sensitivity and specificity of biomarkers according to the type of alcohol consumed (Sakutata et al., 2008). While initial discovery can occur in clinical samples, it may be limited to a selected population. One example would be initial discovery from a single hospital or clinic. Such a population is unlikely to reflect the broader community in either demographics or behavior. Therefore, even if initial discovery occurs in clinical samples, validation in a well-characterized, broad collection of samples of mixed age, gender, ethnic background, and socioeconomic status is needed. Most importantly, this validation should be conducted in a blinded fashion to provide the maximal confidence in the validation process. The need for a cohort of well-annotated samples for translational study in the alcohol research field is the subject of discussion later in this commentary.
Challenges in biomarker development for EtOH
Moving from discovery to validation
Regardless of the particular form of the biomarker and the technology for its assessment, a recurring theme from the two days of discussion was the challenge in translating preclinical or discovery phase biomarkers into clinically useful biomarkers. The need for well curated clinical samples was particularly obvious. Initially promising findings in rodent or non-human primate models need to be demonstrated in human samples. Moreover, because of the genetic, environmental, and behavioral heterogeneity of the human population, promising biomarker candidates identified in human samples also need to be validated in broader independent samples. Generally, the initial discovery researchers do not have the requisite human samples and generation of new samples through a clinical trial would be both costly and time-consuming for each individual laboratory to undertake.
To address this issue, we propose the concept of clinical pre-validation. Animal model study results need to be confirmed in human subjects, but these are expensive and time-consuming projects. Due to the failure rate in clinical trials, it is also risky to undertake a full clinical trial based solely on animal model results. An intermediate phase of clinical testing would allow for data to be generated from human samples, and if successful, provide a rationale for larger clinical studies. We have termed this stage of development as pre-validation. A similar concept has been proposed to speed drug development through testing of pharmacodynamics in Phase 0 cancer trials (Kummar et al., 2007).
The creation of a high-quality pre-validation sample repository would be an important advance in alcohol use biomarker development. The basic vision is the establishment of a bank of archived and aliquoted samples for testing with new candidate biomarkers. Such a bank could include blood samples (plasma, serum, cells), blood RNAs, and urines. Moreover, such a group of samples could be characterized in terms of commonly accepted tests (MCV, AST/ALT, CDT, etc.) and include demographic and clinical data. It is important to note that for several reasons this set would not be used for discovery, but specifically for clinical pre-validation of biomarkers which have already undergone several steps of development. Discovery work generally requires large quantities of sample material, with milliliter volumes required for plasma proteomics and would quickly deplete any archive. Validation studies often use high-sensitivity assays that can be conducted on microliter volumes or equivalents. Therefore, a pre-validation bank could service the need for dozens of independent assays. Finally, the administration of a generally-available pre-validation bank would permit opportunities to conduct assays in masked fashion for optimal confidence in the findings.
Longitudinal samples and biomarkers
Longitudinal samples within-subjects over a time course of heavy alcohol consumption and abstinence from individual patients would be the ideal samples for a repository accessible to NIAAA funded investigators. These samples could serve two purposes. First, they would allow for a careful analysis of the kinetics of a biomarker (e.g., duration of drinking required for a positive test outcome and duration of abstinence required for a negative test outcome). Second, while the alcohol research field is focused on classification of subjects from a single sample, this may still prove to be too difficult in the heterogeneous human population. The possibility exists for within-subject biomarkers in which a percent change from a baseline measurement at a known drinking state may serve as the biomarker. Within-subject biomarkers would have utility not only as diagnostics in patient care, but also in clinical trials to measure either pharmacodynamics or pharmacoefficacy (Blasio-Smith et al., 2008; Bateman et al., 2006; Biomarkers Definitions Working Group, 2001).
Analytical Standardization
With the discovery and validation of biomarkers, there are additional analytical considerations with the implementation of these biomarkers as standard clinical tests. As with any diagnostic test, analytical standardization efforts will be needed to ensure the reliability of measurements, both in test-retest of individual samples and between testing sites. These efforts are currently underway for CDT testing (Jeppsson et al., 2007) and any newly validated biomarkers will need to undergo the same process.
National Alcohol Biomarkers Working Group
Another initiative of considerable importance is the need for central organization of national efforts, which could take the form of an NIAAA-sponsored working group of investigators. This group could share best practices, keep the entire group aware of progress (although not necessarily releasing confidential intellectual property information), and serve as a facilitator for technical and sample needs. The working group would also serve to bring investigators together (both at national meetings and via electronic means). Biomarker development is a considerable undertaking, quite different from traditional basic research. To reach the goal of sensitive and specific clinical biomarkers that improve clinical treatment and research studies of alcohol abuse, collaboration between research groups with different expertise and resources will be needed.
Summary
In this commentary, we have discussed methods and potential pitfalls in biomarker development for alcohol abuse and alcohol-induced tissue damage. While we have identified a number of challenges to successful development of diagnostic biomarkers, the future prospects for development of diagnostic tests are bright. The expertise of the investigators, the new technologies, and the more advanced analytical methods available to current research efforts are reasons for great optimism. These efforts will, of course, continue to require resources as more candidate biomarkers are identified and reach the clinical validation phase. The support and organization of investigators across the alcohol research field is needed. Biomarker development does not fit the standard basic science framework; rather it is translational research focused on one practical goal – accurate diagnosis of the patient to improve treatment. Together with the ongoing research efforts in alcohol abuse treatment regimens, new diagnostics offer the best opportunities for helping patients and their families.
Acknowledgments
This work is supported by NIH grant 5R01AA016613-03 (to KEV). The authors would like to thank Heather VanGuilder and Joseph Freeman for editorial assistance. The Workshop on Alcohol Biomarkers, sponsored by the National Institute on Alcohol Abuse and Alcoholism, took place in Rockville, Maryland, on June 26–27, 2008. The organizers were Jose Velazquez, Lisa Neuhold, Raye Litten, Q. Max Guo, Howard Moss and Katherine Jung.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders DSM-IV-TR. American Psychiatric Association; 2000. [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Anton RF, Lieber C, Tabakoff B. Carbohydrate-deficient transferrin and gamma-glutamyltransferase for the detection and monitoring of alcohol use: results from a multisite study. Alcohol Clin Exp Res. 2002;26:1215–1222. doi: 10.1097/01.ALC.0000023986.42254.F5. [DOI] [PubMed] [Google Scholar]
- Baker M. In biomarkers we trust? Nat Biotechnol. 2005;23:297–304. doi: 10.1038/nbt0305-297. [DOI] [PubMed] [Google Scholar]
- Bateman RJ, Munsell LY, Morris JC, Swarm R, Yarasheski KE, Holtzman DM. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12:856–861. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann T, Bonsch D, Reulbach U, Kornhuber J, Bleich S. Dopamine and N-methyl-D-aspartate receptor expression in peripheral blood of patients undergoing alcohol withdrawal. J Neural Transm. 2007;114:1081–1084. doi: 10.1007/s00702-007-0661-4. [DOI] [PubMed] [Google Scholar]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- Blasio-Smith EA, Arai M, Quinet EM, Evans MJ, Kornaga T, Basso MD, Chen L, Feingold I, Halpern AR, Liu QY, Nambi P, Savio D, Wang S, Mounts WM, Isler JA, Slager AM, Burczynski ME, Dorner AJ, LaVallie ER. Discovery and implementation of transcriptional biomarkers of synthetic LXR agonists in peripheral blood cells. J Transl Med. 2008;6:59. doi: 10.1186/1479-5876-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford BU, O’Connell TM, Han J, Kosyk O, Shymonyak S, Ross PK, Winnike J, Kono H, Rusyn I. Metabolomic profiling of a modified alcohol liquid diet model for liver injury in the mouse uncovers new markers of disease. Toxicol Appl Pharmacol. 2008;232:236–243. doi: 10.1016/j.taap.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KA, DeBenedetti AF, Volk RJ, Williams EC, Frank D, Kivlahan DR. AUDIT-C as a brief screen for alcohol misuse in primary care. Alcohol Clin Exp Res. 2007;31:1208–1217. doi: 10.1111/j.1530-0277.2007.00403.x. [DOI] [PubMed] [Google Scholar]
- Brener ND, Billy JO, Grady WR. Assessment of factors affecting the validity of self-reported health-risk behavior among adolescents: evidence from the scientific literature. J Adolesc Health. 2003;33:436–457. doi: 10.1016/s1054-139x(03)00052-1. [DOI] [PubMed] [Google Scholar]
- Bush K, Kivlahan DR, McDonell MB, Fihn SD, Bradley KA. The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP) Alcohol Use Disorders Identification Test. Arch Intern Med. 1998;158:1789–1795. doi: 10.1001/archinte.158.16.1789. [DOI] [PubMed] [Google Scholar]
- Conigrave KM, Davies P, Haber P, Whitfield JB. Traditional markers of excessive alcohol use. Addiction. 2003;98(Suppl 2):31–43. doi: 10.1046/j.1359-6357.2003.00581.x. [DOI] [PubMed] [Google Scholar]
- Conigrave KM, Degenhardt LJ, Whitfield JB, Saunders JB, Helander A, Tabakoff B. CDT, GGT, and AST as markers of alcohol use: the WHO/ISBRA collaborative project. Alcohol Clin Exp Res. 2002;26:332–339. [PubMed] [Google Scholar]
- Connors GJ, Maisto SA. Drinking reports from collateral individuals. Addiction. 2003;98(Suppl 2):21–29. doi: 10.1046/j.1359-6357.2003.00585.x. [DOI] [PubMed] [Google Scholar]
- Das SK, Dhanya L, Vasudevan DM. Biomarkers of alcoholism: an updated review. Scand J Clin Lab Invest. 2008;68:81–92. doi: 10.1080/00365510701532662. [DOI] [PubMed] [Google Scholar]
- Dawson DA. Drinking patterns among individuals with and without DSM-IV alcohol use disorders. J Stud Alcohol. 2000;61:111–120. doi: 10.15288/jsa.2000.61.111. [DOI] [PubMed] [Google Scholar]
- De GN, Donadio G, Chiarotti M. The reliability of fatty acid ethyl esters (FAEE) as biological markers for the diagnosis of alcohol abuse. J Anal Toxicol. 2007;31:93–97. doi: 10.1093/jat/31.2.93. [DOI] [PubMed] [Google Scholar]
- Dhalla S, Kopec JA. The CAGE questionnaire for alcohol misuse: a review of reliability and validity studies. Clin Invest Med. 2007;30:33–41. doi: 10.25011/cim.v30i1.447. [DOI] [PubMed] [Google Scholar]
- Ewing JA. Detecting alcoholism. The CAGE questionnaire. JAMA. 1984;252:1905–1907. doi: 10.1001/jama.252.14.1905. [DOI] [PubMed] [Google Scholar]
- Frank D, DeBenedetti AF, Volk RJ, Williams EC, Kivlahan DR, Bradley KA. Effectiveness of the AUDIT-C as a screening test for alcohol misuse in three race/ethnic groups. J Gen Intern Med. 2008;23:781–787. doi: 10.1007/s11606-008-0594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Gooch RS, Lull ME, Worst TJ, Walker SJ, Xu AS, Green H, Pierre PJ, Grant KA, Vrana KE. Apo-aii is an elevated biomarker of chronic non-human primate ethanol self-administration. Alcohol Alcohol. 2006;41:300–305. doi: 10.1093/alcalc/agl021. [DOI] [PubMed] [Google Scholar]
- Fuller RK, Lee KK, Gordis E. Validity of self-report in alcoholism research: results of a Veterans Administration Cooperative Study. Alcohol Clin Exp Res. 1988;12:201–205. doi: 10.1111/j.1530-0277.1988.tb00180.x. [DOI] [PubMed] [Google Scholar]
- Ghosh P, Hale EA, Lakshman MR. Plasma sialic-acid index of apolipoprotein J (SIJ): a new alcohol intake marker. Alcohol. 2001;25:173–179. doi: 10.1016/s0741-8329(01)00187-2. [DOI] [PubMed] [Google Scholar]
- Giannini EG, Testa R, Savarino V. Liver enzyme alteration: a guide for clinicians. CMAJ. 2005;172:367–379. doi: 10.1503/cmaj.1040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golka K, Wiese A. Carbohydrate-deficient transferrin (CDT)--a biomarker for long-term alcohol consumption. J Toxicol Environ Health B Crit Rev. 2004;7:319–337. doi: 10.1080/10937400490432400. [DOI] [PubMed] [Google Scholar]
- Hannuksela ML, Liisanantti MK, Nissinen AE, Savolainen MJ. Biochemical markers of alcoholism. Clin Chem Lab Med. 2007b;45:953–961. doi: 10.1515/CCLM.2007.190. [DOI] [PubMed] [Google Scholar]
- Hannuksela ML, Liisanantti MK, Nissinen AE, Savolainen MJ. Biochemical markers of alcoholism. Clin Chem Lab Med. 2007a;45:953–961. doi: 10.1515/CCLM.2007.190. [DOI] [PubMed] [Google Scholar]
- Helander A. Biological markers in alcoholism. J Neural Transm Suppl. 2003:15–32. doi: 10.1007/978-3-7091-0541-2_2. [DOI] [PubMed] [Google Scholar]
- Helander A, Eriksson CJ. Laboratory tests for acute alcohol consumption: results of the WHO/ISBRA Study on State and Trait Markers of Alcohol Use and Dependence. Alcohol Clin Exp Res. 2002;26:1070–1077. [PubMed] [Google Scholar]
- Hock B, Schwarz M, Domke I, Grunert VP, Wuertemberger M, Schiemann U, Horster S, Limmer C, Stecker G, Soyka M. Validity of carbohydrate-deficient transferrin (%CDT), gamma-glutamyltransferase (gamma-GT) and mean corpuscular erythrocyte volume (MCV) as biomarkers for chronic alcohol abuse: a study in patients with alcohol dependence and liver disorders of non-alcoholic and alcoholic origin. Addiction. 2005;100:1477–1486. doi: 10.1111/j.1360-0443.2005.01216.x. [DOI] [PubMed] [Google Scholar]
- Jeppsson JO, Arndt T, Schellenberg F, Wielders JP, Anton RF, Whitfield JB, Helander A. Toward standardization of carbohydrate-deficient transferrin (CDT) measurements: I. Analyte definition and proposal of a candidate reference method. Clin Chem Lab Med. 2007;45:558–562. doi: 10.1515/CCLM.2007.107. [DOI] [PubMed] [Google Scholar]
- Kissack JC, Bishop J, Roper AL. Ethylglucuronide as a biomarker for ethanol detection. Pharmacotherapy. 2008;28:769–781. doi: 10.1592/phco.28.6.769. [DOI] [PubMed] [Google Scholar]
- Koch H, Meerkerk GJ, Zaat JO, Ham MF, Scholten RJ, Assendelft WJ. Accuracy of carbohydrate-deficient transferrin in the detection of excessive alcohol consumption: a systematic review. Alcohol Alcohol. 2004;39:75–85. doi: 10.1093/alcalc/agh031. [DOI] [PubMed] [Google Scholar]
- Korzec A, de BC, van LM. The Bayesian Alcoholism Test had better diagnostic properties for confirming diagnosis of hazardous and harmful alcohol use. J Clin Epidemiol. 2005;58:1024–1032. doi: 10.1016/j.jclinepi.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Kummar S, Kinders R, Rubinstein L, Parchment RE, Murgo AJ, Collins J, Pickeral O, Low J, Steinberg SM, Gutierrez M, Yang S, Helman L, Wiltrout R, Tomaszewski JE, Doroshow JH. Compressing drug development timelines in oncology using phase ‘0’ trials. Nat Rev Cancer. 2007;7:131–139. doi: 10.1038/nrc2066. [DOI] [PubMed] [Google Scholar]
- Lesko LJ, Atkinson AJ. Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annu Rev Pharmacol Toxicol. 2001;41:347–366. doi: 10.1146/annurev.pharmtox.41.1.347. [DOI] [PubMed] [Google Scholar]
- Litten RZ. Alcohol biomarkers in applied settings: recent advances and future research opportunities. Alcoholism: Clinical and Experimental Research. 2009 doi: 10.1111/j.1530-0277.2010.01170.x. Submitted. [DOI] [PubMed] [Google Scholar]
- Marrer E, Dieterle F. Promises of biomarkers in drug development--a reality check. Chem Biol Drug Des. 2007;69:381–394. doi: 10.1111/j.1747-0285.2007.00522.x. [DOI] [PubMed] [Google Scholar]
- Niemela O. Biomarkers in alcoholism. Clin Chim Acta. 2007;377:39–49. doi: 10.1016/j.cca.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Nomura F, Tomonaga T, Sogawa K, Wu D, Ohashi T. Application of proteomic technologies to discover and identify biomarkers for excessive alcohol consumption: a review. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855:35–41. doi: 10.1016/j.jchromb.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Pernanen K. In: Validity of survey data on alcohol use. Gibbons, et al., editors. Research Advances in Alcohol and Drug Problems; New York: 1974. pp. 355–374. [Google Scholar]
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A, Kaye JA, Leszek J, Miller BL, Minthon L, Quinn JF, Rabinovici GD, Robinson WH, Sabbagh MN, So YT, Sparks DL, Tabaton M, Tinklenberg J, Yesavage JA, Tibshirani R, Wyss-Coray T. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- Rinck D, Frieling H, Freitag A, Hillemacher T, Bayerlein K, Kornhuber J, Bleich S. Combinations of carbohydrate-deficient transferrin, mean corpuscular erythrocyte volume, gamma-glutamyltransferase, homocysteine and folate increase the significance of biological markers in alcohol dependent patients. Drug Alcohol Depend. 2007;89:60–65. doi: 10.1016/j.drugalcdep.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Roche AM, Freeman T, Skinner N. From data to evidence, to action: findings from a systematic review of hospital screening studies for high risk alcohol consumption. Drug Alcohol Depend. 2006;83:1–14. doi: 10.1016/j.drugalcdep.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Rolan P, Atkinson AJ, Lesko LJ. Use of biomarkers from drug discovery through clinical practice: report of the Ninth European Federation of Pharmaceutical Sciences Conference on Optimizing Drug Development. Clin Pharmacol Ther. 2003;73:284–291. doi: 10.1016/s0009-9236(02)17625-9. [DOI] [PubMed] [Google Scholar]
- Sakutata H, Suzuki T, Yasuda H, Ito T. Beverage-specific effects of ethanol consumption on its biological markers. Clin Chem Lab Med. 2008;46:699–702. doi: 10.1515/cclm.2008.124. [DOI] [PubMed] [Google Scholar]
- Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, Sridhar S, Beamis J, Lamb C, Anderson T, Gerry N, Keane J, Lenburg ME, Brody JS. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- Steinweg DL, Worth H. Alcoholism: the keys to the CAGE. Am J Med. 1993;94:520–523. doi: 10.1016/0002-9343(93)90088-7. [DOI] [PubMed] [Google Scholar]
- Stephanson N, Helander A, Beck O. Alcohol biomarker analysis: simultaneous determination of 5-hydroxytryptophol glucuronide and 5-hydroxyindoleacetic acid by direct injection of urine using ultra-performance liquid chromatography-tandem mass spectrometry. J Mass Spectrom. 2007;42:940–949. doi: 10.1002/jms.1231. [DOI] [PubMed] [Google Scholar]
- Taracha E, Habrat B, Wozniak P, Walkowiak J, Szukalski B. The activity of beta-hexosaminidase (uHex) and gamma-glutamyl-transferase (uGGT) in urine as non-invasive markers of chronic alcohol abuse: I. Alcohol-dependent subjects. World J Biol Psychiatry. 2001;2:184–189. doi: 10.3109/15622970109026807. [DOI] [PubMed] [Google Scholar]
- US Food and Drug Administration. Guidance for Industry: Pharmacogenomic Data Submissions. 2005. [Google Scholar]
- Walker SJ, Grant KA. Peripheral blood alpha-synuclein mRNA levels are elevated in cynomolgus monkeys that chronically self-administer ethanol. Alcohol. 2006;38:1–4. doi: 10.1016/j.alcohol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Williams SA, Slavin DE, Wagner JA, Webster CJ. A cost-effectiveness approach to the qualification and acceptance of biomarkers. Nat Rev Drug Discov. 2006;5:897–902. doi: 10.1038/nrd2174. [DOI] [PubMed] [Google Scholar]